Submitted:
04 June 2024
Posted:
05 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Methods
2.1. Human Subjects, Informed Consent Statement and Institutional Review Board Statement
2.2. Cells
2.3. Proviral DNA Constructs
2.4. Virus Production and Infection
2.5. Antibodies and Flow Cytometry
2.6. Transfection of Primary CD4+ T-Cells
2.7. RNA Extraction, Reverse-Transcription, and Real-Time qPCR Analyses
2.8. CD59 3′UTR Validation Assay
2.9. Generation of the CEM-CCR5 CD59 Knock-Out Cell Line
2.10. Purification of Antibodies
2.11. Antibody-Dependent Complement-Mediated Lysis Assay
2.12. Virion Capture Assay
2.13. RNA-Seq of Sorted HIV-1-Infected CD4+ T-cells for mRNA/miRNA Profiling and Analyses of Their Expression
2.14. Quantification and Statistical Analyses
3. Results
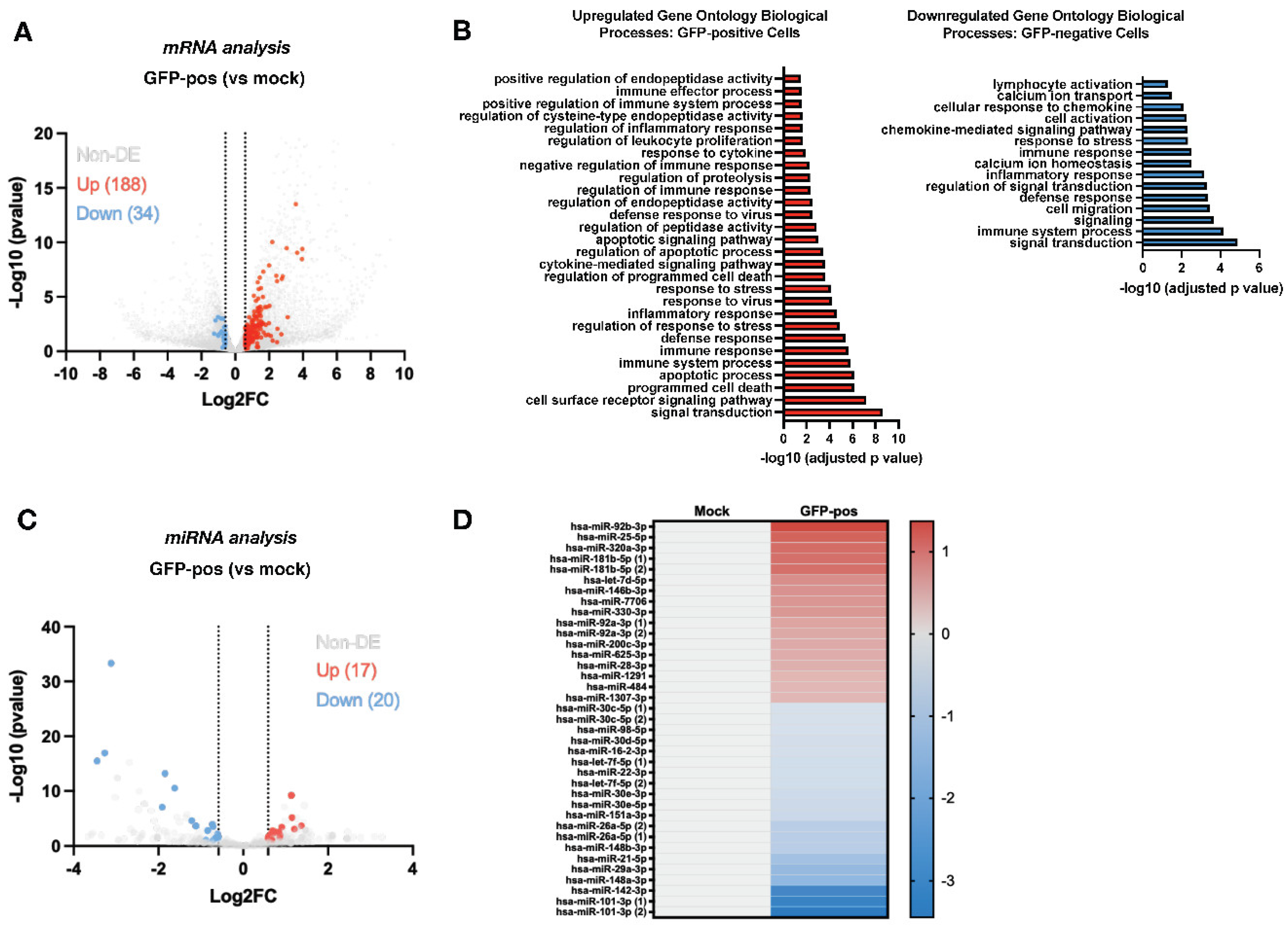
3.1. HIV-1 Alters the miRNA and mRNA Expression Profiles during Infection of CD4+ T-Cells
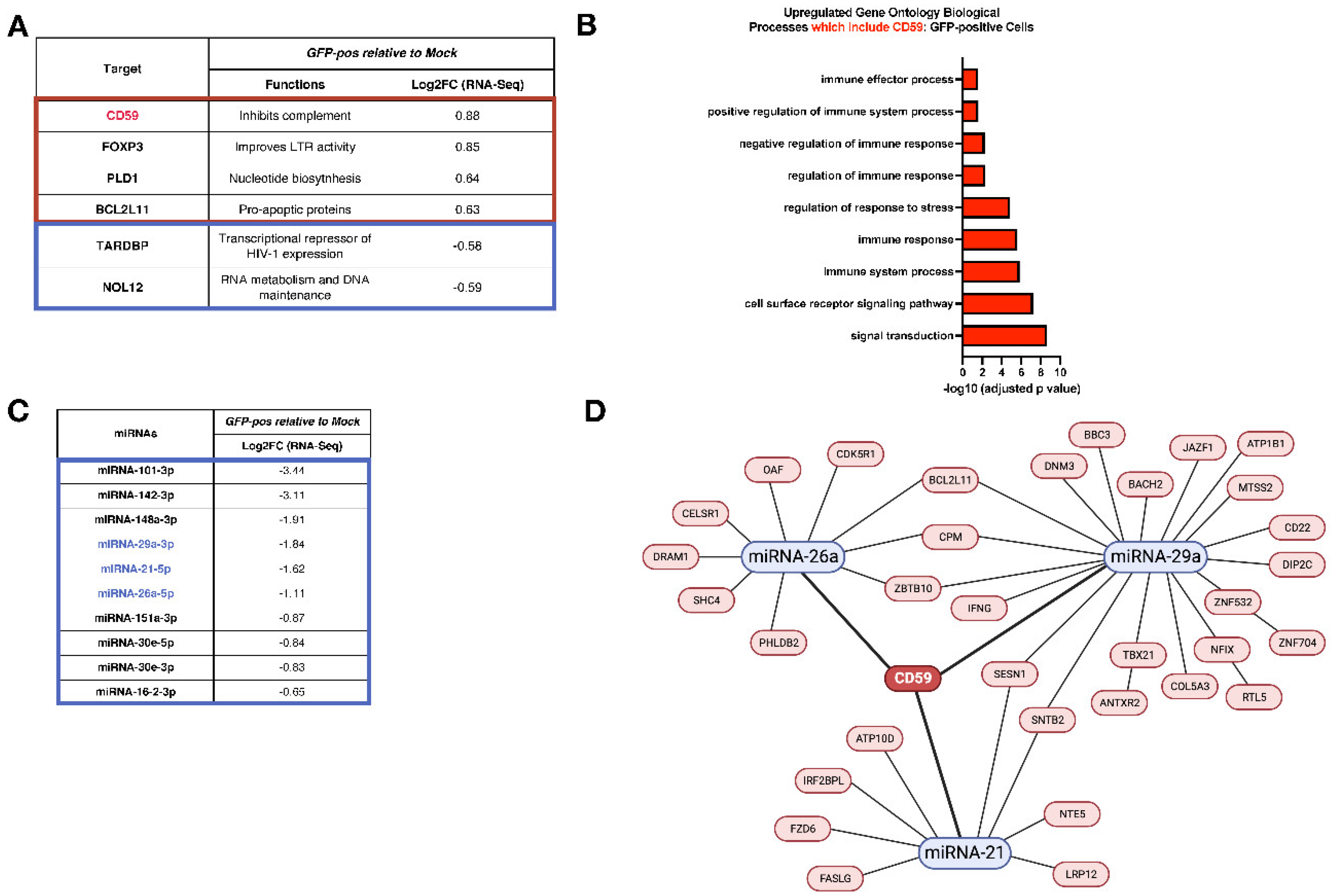
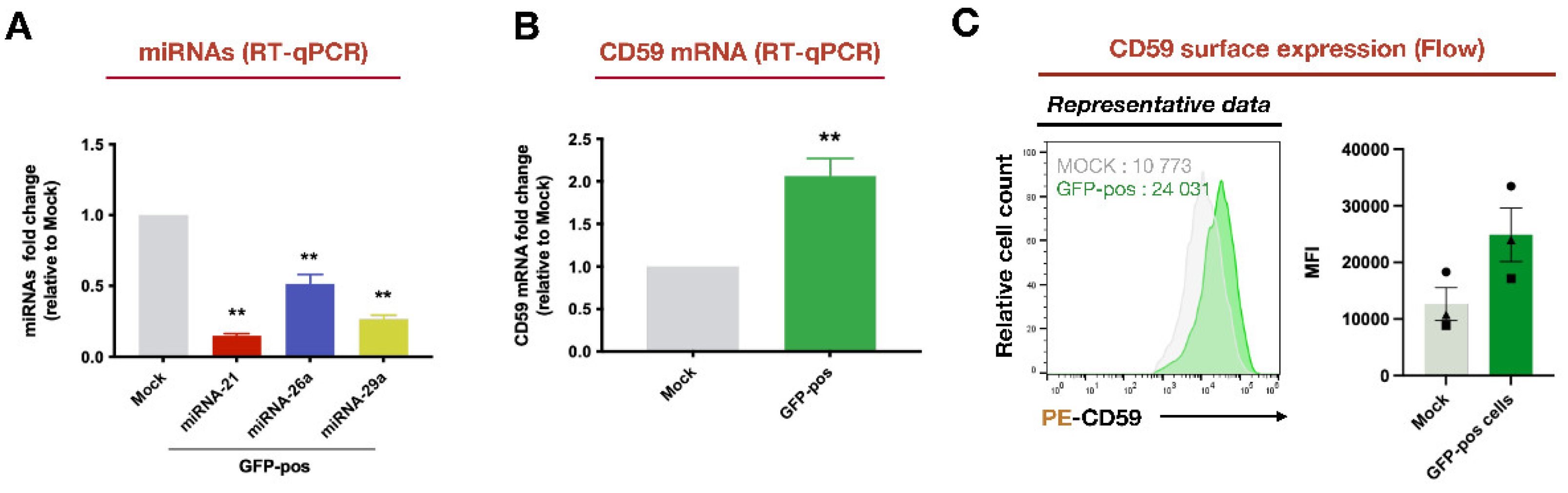
3.2. Analyses of mRNA and miRNA Profiles in Infected CD4+ T-Cells Link CD59 Upregulation to a Decrease of Putative Targeting miRNAs
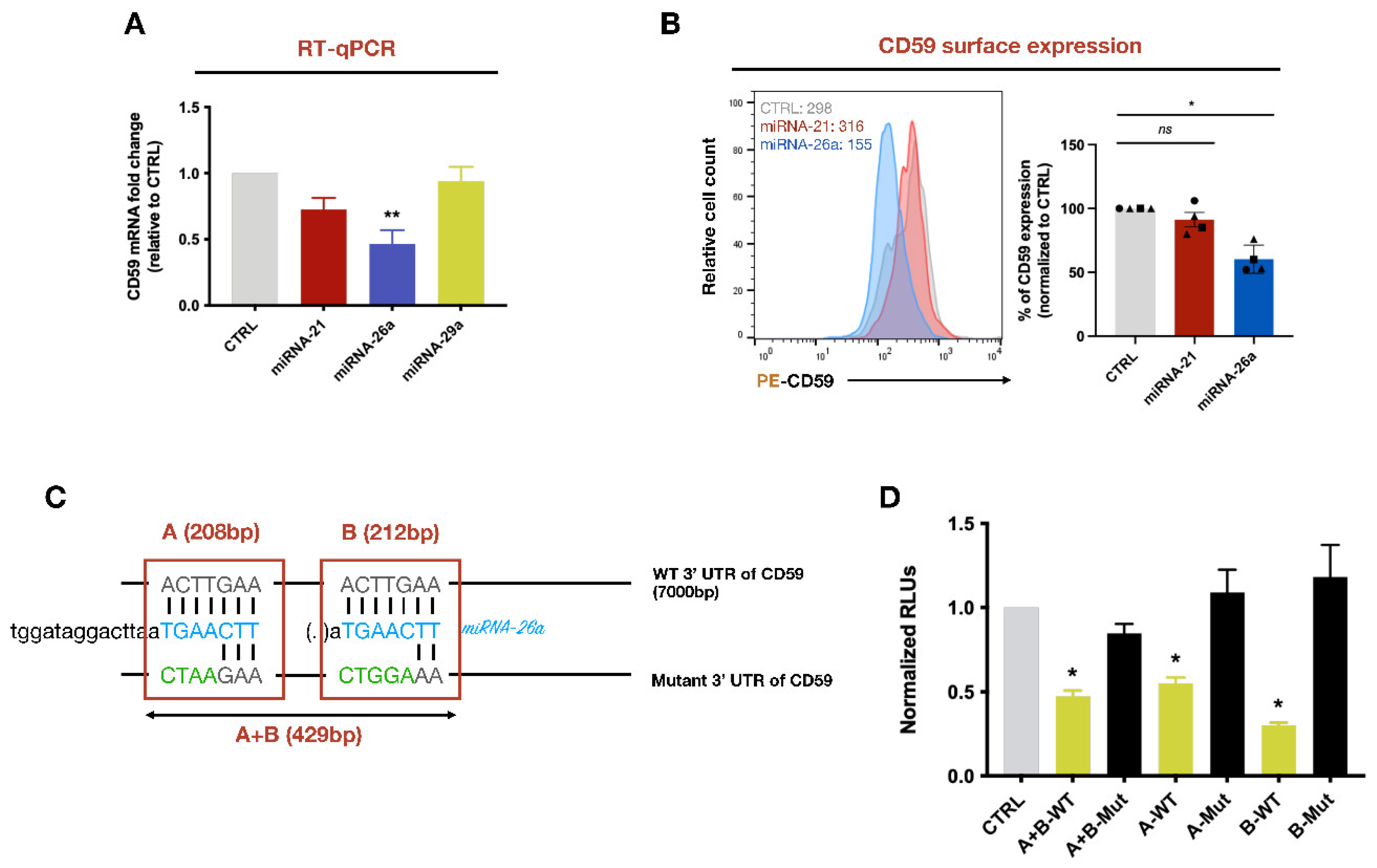
3.3. CD59 Is a Target of miRNA-26a
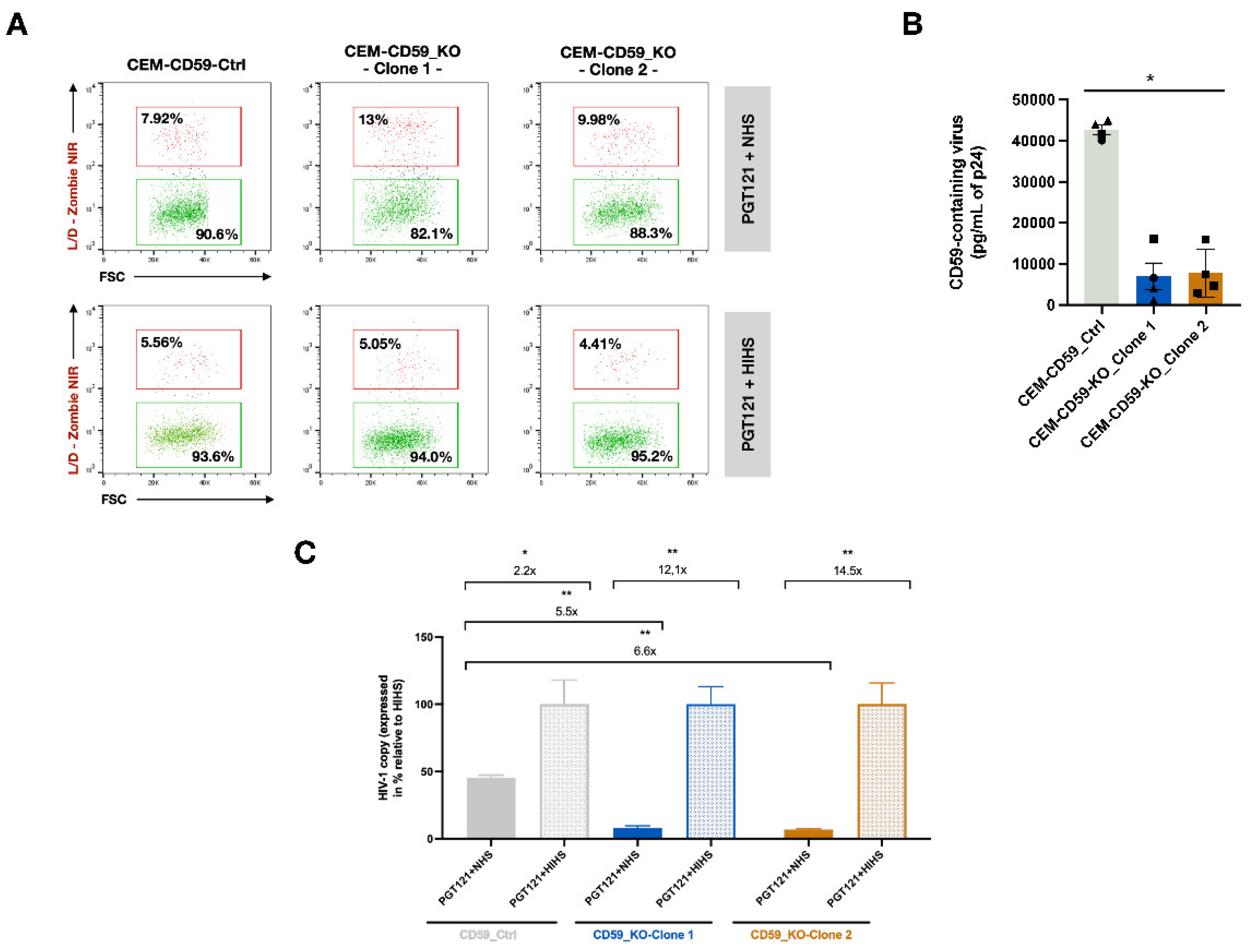
3.4. CD59 affects the Susceptibility of HIV-1 to Antibody-Dependent Complement-Mediated Lysis
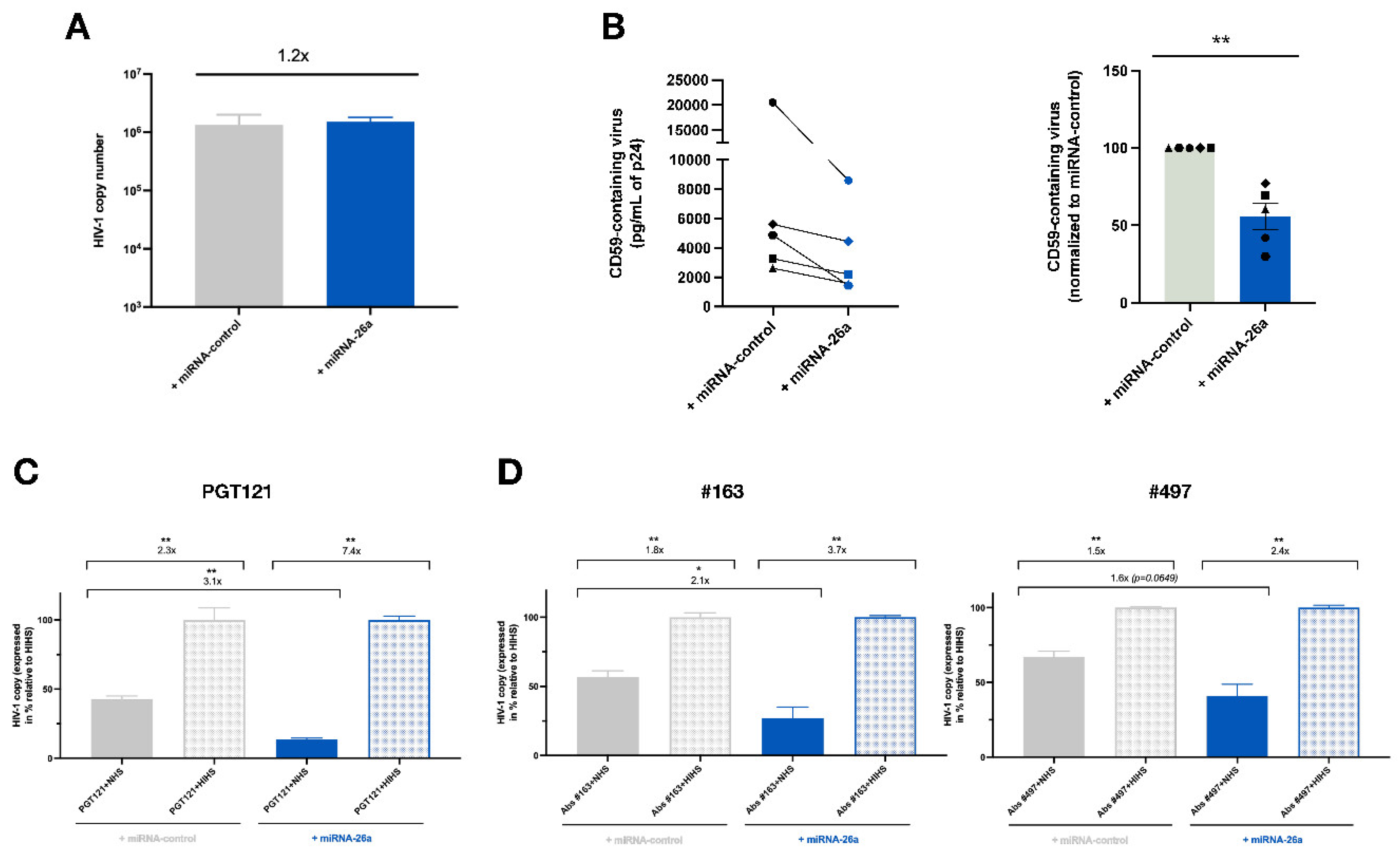
3.5. Enhanced Levels of miRNA-26a Promote ADCML of HIV-1 by Reducing CD59 Packaging into Released Virions
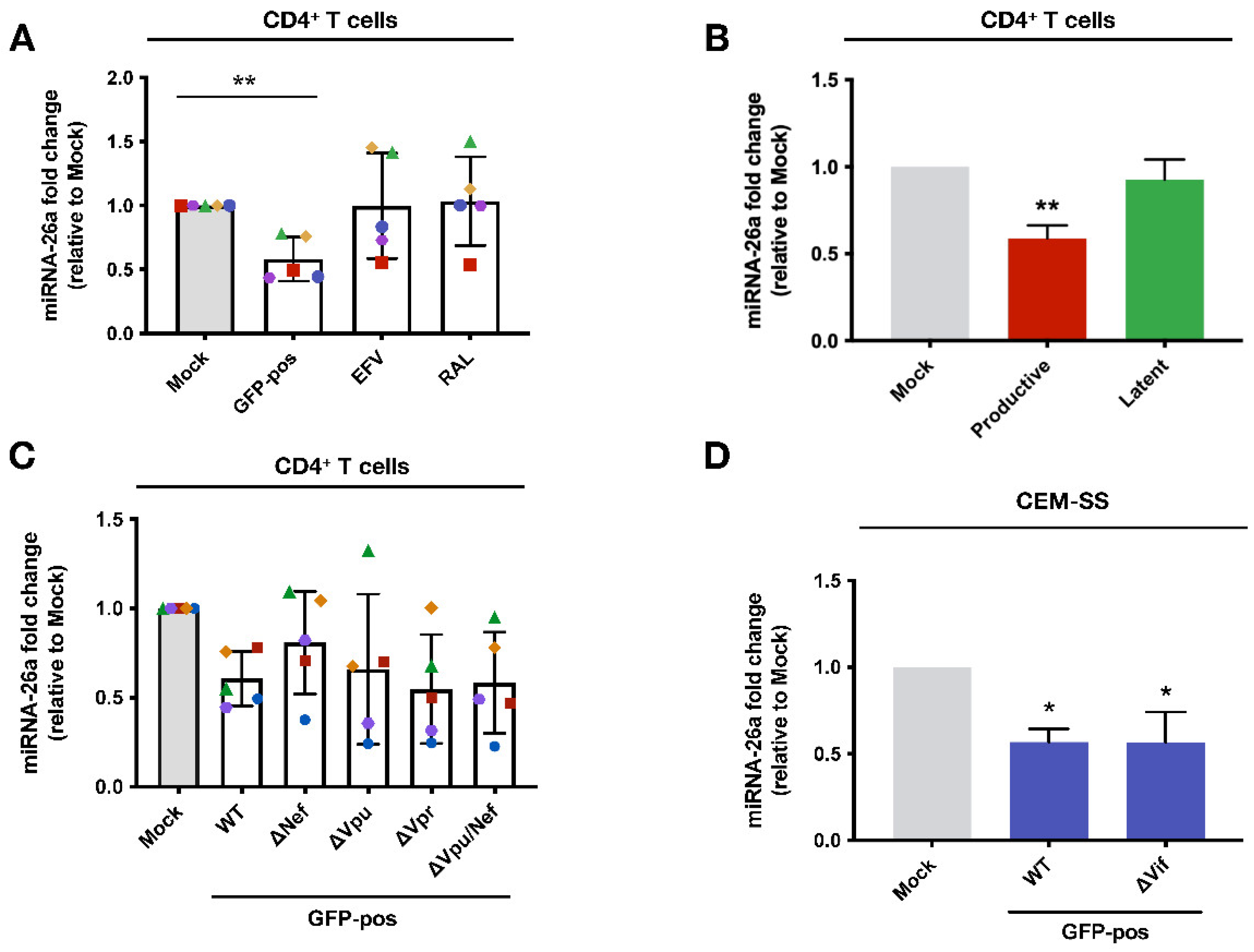
3.6. The Downregulation of miRNA-26a during HIV-1 Infection of CD4+ T-lymphocytes Is Contingent on viral DNA Integration and Expression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alter, G.; Moody, M.A. The humoral response to HIV-1: new insights, renewed focus. The Journal of Infectious Diseases 2010, 202 Suppl 2, S315-322. [CrossRef]
- Tomaras, G.D.; Yates, N.L.; Liu, P.; Qin, L.; Fouda, G.G.; Chavez, L.L.; Decamp, A.C.; Parks, R.J.; Ashley, V.C.; Lucas, J.T.; et al. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. Journal of Virology 2008, 82, 12449-12463. [CrossRef]
- Poignard, P.; Moulard, M.; Golez, E.; Vivona, V.; Franti, M.; Venturini, S.; Wang, M.; Parren, P.W.H.I.; Burton, D.R. Heterogeneity of envelope molecules expressed on primary human immunodeficiency virus type 1 particles as probed by the binding of neutralizing and nonneutralizing antibodies. Journal of Virology 2003, 77, 353-365. [CrossRef]
- Bunnik, E.M.; Pisas, L.; van Nuenen, A.C.; Schuitemaker, H. Autologous neutralizing humoral immunity and evolution of the viral envelope in the course of subtype B human immunodeficiency virus type 1 infection. Journal of Virology 2008, 82, 7932-7941. [CrossRef]
- Gray, E.S.; Moore, P.L.; Choge, I.A.; Decker, J.M.; Bibollet-Ruche, F.; Li, H.; Leseka, N.; Treurnicht, F.; Mlisana, K.; Shaw, G.M.; et al. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. Journal of Virology 2007, 81, 6187-6196. [CrossRef]
- Haynes, B.F.; Shaw, G.M.; Korber, B.; Kelsoe, G.; Sodroski, J.; Hahn, B.H.; Borrow, P.; McMichael, A.J. HIV-Host Interactions: Implications for Vaccine Design. Cell Host & Microbe 2016, 19, 292-303. [CrossRef]
- Liu, Y.; Cao, W.; Sun, M.; Li, T. Broadly neutralizing antibodies for HIV-1: efficacies, challenges and opportunities. Emerging Microbes & Infections 2020, 9, 194-206. [CrossRef]
- Ebenbichler, C.F.; Thielens, N.M.; Vornhagen, R.; Marschang, P.; Arlaud, G.J.; Dierich, M.P. Human immunodeficiency virus type 1 activates the classical pathway of complement by direct C1 binding through specific sites in the transmembrane glycoprotein gp41. The Journal of Experimental Medicine 1991, 174, 1417-1424. [CrossRef]
- Walport, M.J. Complement. First of two parts. The New England Journal of Medicine 2001, 344, 1058-1066. [CrossRef]
- Dunkelberger, J.R.; Song, W.-C. Complement and its role in innate and adaptive immune responses. Cell Research 2010, 20, 34-50. [CrossRef]
- Couves, E.C.; Gardner, S.; Voisin, T.B.; Bickel, J.K.; Stansfeld, P.J.; Tate, E.W.; Bubeck, D. Structural basis for membrane attack complex inhibition by CD59. Nature Communications 2023, 14, 890. [CrossRef]
- Zipfel, P.F.; Skerka, C. Complement regulators and inhibitory proteins. Nature Reviews. Immunology 2009, 9, 729-740. [CrossRef]
- Saifuddin, M.; Hedayati, T.; Atkinson, J.P.; Holguin, M.H.; Parker, C.J.; Spear, G.T. Human immunodeficiency virus type 1 incorporates both glycosyl phosphatidylinositol-anchored CD55 and CD59 and integral membrane CD46 at levels that protect from complement-mediated destruction. The Journal of General Virology 1997, 78 ( Pt 8), 1907-1911. [CrossRef]
- Marschang, P.; Sodroski, J.; Wurzner, R.; Dierich, M.P. Decay-accelerating factor (CD55) protects human immunodeficiency virus type 1 from inactivation by human complement. Eur J Immunol 1995, 25, 285-290. [CrossRef]
- Huber, M.; Fischer, M.; Misselwitz, B.; Manrique, A.; Kuster, H.; Niederöst, B.; Weber, R.; von Wyl, V.; Günthard, H.F.; Trkola, A. Complement lysis activity in autologous plasma is associated with lower viral loads during the acute phase of HIV-1 infection. PLoS medicine 2006, 3, e441. [CrossRef]
- Aasa-Chapman, M.M.I.; Holuigue, S.; Aubin, K.; Wong, M.; Jones, N.A.; Cornforth, D.; Pellegrino, P.; Newton, P.; Williams, I.; Borrow, P.; et al. Detection of antibody-dependent complement-mediated inactivation of both autologous and heterologous virus in primary human immunodeficiency virus type 1 infection. Journal of Virology 2005, 79, 2823-2830. [CrossRef]
- Hammond, S.M. An overview of microRNAs. Advanced Drug Delivery Reviews 2015, 87, 3-14. [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Frontiers in Endocrinology 2018, 9, 402. [CrossRef]
- Balasubramaniam, M.; Pandhare, J.; Dash, C. Are microRNAs Important Players in HIV-1 Infection? An Update. Viruses 2018, 10, 110. [CrossRef]
- Orecchini, E.; Doria, M.; Michienzi, A.; Giuliani, E.; Vassena, L.; Ciafrè, S.A.; Farace, M.G.; Galardi, S. The HIV-1 Tat protein modulates CD4 expression in human T cells through the induction of miR-222. RNA biology 2014, 11, 334-338. [CrossRef]
- Lodge, R.; Ferreira Barbosa, J.A.; Lombard-Vadnais, F.; Gilmore, J.C.; Deshiere, A.; Gosselin, A.; Wiche Salinas, T.R.; Bego, M.G.; Power, C.; Routy, J.-P.; et al. Host MicroRNAs-221 and -222 Inhibit HIV-1 Entry in Macrophages by Targeting the CD4 Viral Receptor. Cell Reports 2017, 21, 141-153. [CrossRef]
- Lodge, R.; Bellini, N.; Laporte, M.; Salahuddin, S.; Routy, J.-P.; Ancuta, P.; Costiniuk, C.T.; Jenabian, M.-A.; Cohen, É.A. Interleukin-1β Triggers p53-Mediated Downmodulation of CCR5 and HIV-1 Entry in Macrophages through MicroRNAs 103 and 107. mBio 2020, 11, e02314-02320. [CrossRef]
- Bellini, N.; Lodge, R.; Pham, T.N.Q.; Jain, J.; Murooka, T.T.; Herschhorn, A.; Bernard, N.F.; Routy, J.-P.; Tremblay, C.L.; Cohen, É.A. MiRNA-103 downmodulates CCR5 expression reducing human immunodeficiency virus type-1 entry and impacting latency establishment in CD4+ T cells. iScience 2022, 25, 105234. [CrossRef]
- Riess, M.; Fuchs, N.V.; Idica, A.; Hamdorf, M.; Flory, E.; Pedersen, I.M.; König, R. Interferons Induce Expression of SAMHD1 in Monocytes through Down-regulation of miR-181a and miR-30a. The Journal of Biological Chemistry 2017, 292, 264-277. [CrossRef]
- Ruelas, D.S.; Chan, J.K.; Oh, E.; Heidersbach, A.J.; Hebbeler, A.M.; Chavez, L.; Verdin, E.; Rape, M.; Greene, W.C. MicroRNA-155 Reinforces HIV Latency. The Journal of Biological Chemistry 2015, 290, 13736-13748. [CrossRef]
- Xu, Z.; Lodge, R.; Power, C.; Cohen, E.A.; Hobman, T.C. The HIV-1 Accessory Protein Vpu Downregulates Peroxisome Biogenesis. mBio 2020, 11, e03395-03319. [CrossRef]
- Lodge, R.; Xu, Z.; Eklund, M.; Stürzel, C.; Kirchhoff, F.; Tremblay, M.J.; Hobman, T.C.; Cohen, É.A. MicroRNA-25/93 induction by Vpu as a mechanism for counteracting MARCH1-restriction on HIV-1 infectivity in macrophages. mBio 2023, 14, e0195023. [CrossRef]
- Shen, C.-J.; Jia, Y.-H.; Tian, R.-R.; Ding, M.; Zhang, C.; Wang, J.-H. Translation of Pur-α is targeted by cellular miRNAs to modulate the differentiation-dependent susceptibility of monocytes to HIV-1 infection. FASEB journal: official publication of the Federation of American Societies for Experimental Biology 2012, 26, 4755-4764. [CrossRef]
- Ou, S.H.; Wu, F.; Harrich, D.; Garcia-Martinez, L.F.; Gaynor, R.B. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J Virol 1995, 69, 3584-3596. [CrossRef]
- Cabrera-Rodríguez, R.; Pérez-Yanes, S.; Montelongo, R.; Lorenzo-Salazar, J.M.; Estévez-Herrera, J.; García-Luis, J.; Íñigo-Campos, A.; Rubio-Rodríguez, L.A.; Muñoz-Barrera, A.; Trujillo-González, R.; et al. Transactive Response DNA-Binding Protein (TARDBP/TDP-43) Regulates Cell Permissivity to HIV-1 Infection by Acting on HDAC6. International Journal of Molecular Sciences 2022, 23, 6180. [CrossRef]
- Scott, D.D.; Trahan, C.; Zindy, P.J.; Aguilar, L.C.; Delubac, M.Y.; Van Nostrand, E.L.; Adivarahan, S.; Wei, K.E.; Yeo, G.W.; Zenklusen, D.; et al. Nol12 is a multifunctional RNA binding protein at the nexus of RNA and DNA metabolism. Nucleic Acids Research 2017, 45, 12509-12528. [CrossRef]
- Chen, D.; Wang, M.; Zhou, S.; Zhou, Q. HIV-1 Tat targets microtubules to induce apoptosis, a process promoted by the pro-apoptotic Bcl-2 relative Bim. The EMBO journal 2002, 21, 6801-6810. [CrossRef]
- Taylor, H.E.; Simmons, G.E.; Mathews, T.P.; Khatua, A.K.; Popik, W.; Lindsley, C.W.; D’Aquila, R.T.; Brown, H.A. Phospholipase D1 Couples CD4+ T Cell Activation to c-Myc-Dependent Deoxyribonucleotide Pool Expansion and HIV-1 Replication. PLoS pathogens 2015, 11, e1004864. [CrossRef]
- Jiang, Q.; Zhang, L.; Wang, R.; Jeffrey, J.; Washburn, M.L.; Brouwer, D.; Barbour, S.; Kovalev, G.I.; Unutmaz, D.; Su, L. FoxP3+CD4+ regulatory T cells play an important role in acute HIV-1 infection in humanized Rag2-/-gammaC-/- mice in vivo. Blood 2008, 112, 2858-2868. [CrossRef]
- Trkola, A.; Matthews, J.; Gordon, C.; Ketas, T.; Moore, J.P. A cell line-based neutralization assay for primary human immunodeficiency virus type 1 isolates that use either the CCR5 or the CXCR4 coreceptor. Journal of Virology 1999, 73, 8966-8974. [CrossRef]
- Walker, L.M.; Huber, M.; Doores, K.J.; Falkowska, E.; Pejchal, R.; Julien, J.-P.; Wang, S.-K.; Ramos, A.; Chan-Hui, P.-Y.; Moyle, M.; et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 2011, 477, 466-470. [CrossRef]
- Ratnapriya, S.; Harris, M.; Chov, A.; Herbert, Z.T.; Vrbanac, V.; Deruaz, M.; Achuthan, V.; Engelman, A.N.; Sodroski, J.; Herschhorn, A. Intra- and extra-cellular environments contribute to the fate of HIV-1 infection. Cell Reports 2021, 36, 109622. [CrossRef]
- Dave, V.P.; Hajjar, F.; Dieng, M.M.; Haddad, É.; Cohen, É.A. Efficient BST2 antagonism by Vpu is critical for early HIV-1 dissemination in humanized mice. Retrovirology 2013, 10, 128. [CrossRef]
- Richard, J.; Sindhu, S.; Pham, T.N.Q.; Belzile, J.-P.; Cohen, E.A. HIV-1 Vpr up-regulates expression of ligands for the activating NKG2D receptor and promotes NK cell-mediated killing. Blood 2010, 115, 1354-1363. [CrossRef]
- Pham, T.N.Q.; Lukhele, S.; Hajjar, F.; Routy, J.-P.; Cohen, É.A. HIV Nef and Vpu protect HIV-infected CD4+ T cells from antibody-mediated cell lysis through down-modulation of CD4 and BST2. Retrovirology 2014, 11, 15. [CrossRef]
- Stopak, K.; de Noronha, C.; Yonemoto, W.; Greene, W.C. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Molecular Cell 2003, 12, 591-601. [CrossRef]
- Han, Y.; Wang, X.; Dang, Y.; Zheng, Y.-H. APOBEC3G and APOBEC3F require an endogenous cofactor to block HIV-1 replication. PLoS pathogens 2008, 4, e1000095. [CrossRef]
- Dey, B.K.; Gagan, J.; Yan, Z.; Dutta, A. miR-26a is required for skeletal muscle differentiation and regeneration in mice. Genes & Development 2012, 26, 2180-2191. [CrossRef]
- Fu, X.; Dong, B.; Tian, Y.; Lefebvre, P.; Meng, Z.; Wang, X.; Pattou, F.; Han, W.; Wang, X.; Lou, F.; et al. MicroRNA-26a regulates insulin sensitivity and metabolism of glucose and lipids. The Journal of Clinical Investigation 2015, 125, 2497-2509. [CrossRef]
- Hutter, K.; Lindner, S.E.; Kurschat, C.; Rülicke, T.; Villunger, A.; Herzog, S. The miR-26 family regulates early B cell development and transformation. Life Science Alliance 2022, 5, e202101303. [CrossRef]
- Spear, G.T.; Lurain, N.S.; Parker, C.J.; Ghassemi, M.; Payne, G.H.; Saifuddin, M. Host cell-derived complement control proteins CD55 and CD59 are incorporated into the virions of two unrelated enveloped viruses. Human T cell leukemia/lymphoma virus type I (HTLV-I) and human cytomegalovirus (HCMV). Journal of Immunology (Baltimore, Md.: 1950) 1995, 155, 4376-4381.
- Amet, T.; Ghabril, M.; Chalasani, N.; Byrd, D.; Hu, N.; Grantham, A.; Liu, Z.; Qin, X.; He, J.J.; Yu, Q. CD59 incorporation protects hepatitis C virus against complement-mediated destruction. Hepatology (Baltimore, Md.) 2012, 55, 354-363. [CrossRef]
- Zhuang, C.; Wang, P.; Huang, D.; Xu, L.; Wang, X.; Wang, L.; Hu, L. A double-negative feedback loop between EZH2 and miR-26a regulates tumor cell growth in hepatocellular carcinoma. International Journal of Oncology 2016, 48, 1195-1204. [CrossRef]
- Liu, J.; Li, X.; Wang, M.; Xiao, G.; Yang, G.; Wang, H.; Li, Y.; Sun, X.; Qin, S.; Du, N.; et al. A miR-26a/E2F7 feedback loop contributes to tamoxifen resistance in ER-positive breast cancer. International Journal of Oncology 2018, 53, 1601-1612. [CrossRef]
- Lukacik, P.; Roversi, P.; White, J.; Esser, D.; Smith, G.P.; Billington, J.; Williams, P.A.; Rudd, P.M.; Wormald, M.R.; Harvey, D.J.; et al. Complement regulation at the molecular level: the structure of decay-accelerating factor. Proceedings of the National Academy of Sciences of the United States of America 2004, 101, 1279-1284. [CrossRef]
- Riley-Vargas, R.C.; Gill, D.B.; Kemper, C.; Liszewski, M.K.; Atkinson, J.P. CD46: expanding beyond complement regulation. Trends in Immunology 2004, 25, 496-503. [CrossRef]
- Lan, J.; Yang, K.; Byrd, D.; Hu, N.; Amet, T.; Shepherd, N.; Desai, M.; Gao, J.; Gupta, S.; Sun, Y.; et al. Provirus activation plus CD59 blockage triggers antibody-dependent complement-mediated lysis of latently HIV-1-infected cells. Journal of Immunology (Baltimore, Md.: 1950) 2014, 193, 3577-3589. [CrossRef]
- Yang, K.; Lan, J.; Shepherd, N.; Hu, N.; Xing, Y.; Byrd, D.; Amet, T.; Jewell, C.; Gupta, S.; Kounga, C.; et al. Blockage of CD59 Function Restores Activities of Neutralizing and Nonneutralizing Antibodies in Triggering Antibody-Dependent Complement-Mediated Lysis of HIV-1 Virions and Provirus-Activated Latently Infected Cells. Journal of Virology 2015, 89, 9393-9406. [CrossRef]
- Amet, T.; Lan, J.; Shepherd, N.; Yang, K.; Byrd, D.; Xing, Y.; Yu, Q. Glycosylphosphatidylinositol Anchor Deficiency Attenuates the Production of Infectious HIV-1 and Renders Virions Sensitive to Complement Attack. AIDS research and human retroviruses 2016, 32, 1100-1112. [CrossRef]
- Dufloo, J.; Guivel-Benhassine, F.; Buchrieser, J.; Lorin, V.; Grzelak, L.; Dupouy, E.; Mestrallet, G.; Bourdic, K.; Lambotte, O.; Mouquet, H.; et al. Anti-HIV-1 antibodies trigger non-lytic complement deposition on infected cells. EMBO reports 2020, 21, e49351. [CrossRef]
- Sather, D.N.; Armann, J.; Ching, L.K.; Mavrantoni, A.; Sellhorn, G.; Caldwell, Z.; Yu, X.; Wood, B.; Self, S.; Kalams, S.; et al. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J Virol 2009, 83, 757-769. [CrossRef]
- Wang, G.; de Jong, R.N.; van den Bremer, E.T.J.; Beurskens, F.J.; Labrijn, A.F.; Ugurlar, D.; Gros, P.; Schuurman, J.; Parren, P.W.H.I.; Heck, A.J.R. Molecular Basis of Assembly and Activation of Complement Component C1 in Complex with Immunoglobulin G1 and Antigen. Molecular Cell 2016, 63, 135-145. [CrossRef]
- Zhou, T.; Georgiev, I.; Wu, X.; Yang, Z.-Y.; Dai, K.; Finzi, A.; Kwon, Y.D.; Scheid, J.F.; Shi, W.; Xu, L.; et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science (New York, N.Y.) 2010, 329, 811-817. [CrossRef]
- Norman, J.M.; Mashiba, M.; McNamara, L.A.; Onafuwa-Nuga, A.; Chiari-Fort, E.; Shen, W.; Collins, K.L. The antiviral factor APOBEC3G enhances the recognition of HIV-infected primary T cells by natural killer cells. Nature Immunology 2011, 12, 975-983. [CrossRef]
- Murooka, T.T.; Deruaz, M.; Marangoni, F.; Vrbanac, V.D.; Seung, E.; von Andrian, U.H.; Tager, A.M.; Luster, A.D.; Mempel, T.R. HIV-infected T cells are migratory vehicles for viral dissemination. Nature 2012, 490, 283-287. [CrossRef]
- Kim, J.H.; Lee, S.-R.; Li, L.-H.; Park, H.-J.; Park, J.-H.; Lee, K.Y.; Kim, M.-K.; Shin, B.A.; Choi, S.-Y. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PloS One 2011, 6, e18556. [CrossRef]
- Lodge, R.; Lalonde, J.P.; Lemay, G.; Cohen, E.A. The membrane-proximal intracytoplasmic tyrosine residue of HIV-1 envelope glycoprotein is critical for basolateral targeting of viral budding in MDCK cells. The EMBO journal 1997, 16, 695-705. [CrossRef]
- Androvic, P.; Valihrach, L.; Elling, J.; Sjoback, R.; Kubista, M. Two-tailed RT-qPCR: a novel method for highly accurate miRNA quantification. Nucleic Acids Research 2017, 45, e144. [CrossRef]
- Bego, M.G.; Mercier, J.; Cohen, E.A. Virus-activated interferon regulatory factor 7 upregulates expression of the interferon-regulated BST2 gene independently of interferon signaling. Journal of Virology 2012, 86, 3513-3527. [CrossRef]
- Thielen, A.J.F.; van Baarsen, I.M.; Jongsma, M.L.; Zeerleder, S.; Spaapen, R.M.; Wouters, D. CRISPR/Cas9 generated human CD46, CD55 and CD59 knockout cell lines as a tool for complement research. Journal of Immunological Methods 2018, 456, 15-22. [CrossRef]
- Sanjana, N.E.; Shalem, O.; Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nature Methods 2014, 11, 783-784. [CrossRef]
- Vandergeeten, C.; Fromentin, R.; Merlini, E.; Lawani, M.B.; DaFonseca, S.; Bakeman, W.; McNulty, A.; Ramgopal, M.; Michael, N.; Kim, J.H.; et al. Cross-clade ultrasensitive PCR-based assays to measure HIV persistence in large-cohort studies. Journal of Virology 2014, 88, 12385-12396. [CrossRef]
- Burnie, J.; Tang, V.A.; Welsh, J.A.; Persaud, A.T.; Thaya, L.; Jones, J.C.; Guzzo, C. Flow Virometry Quantification of Host Proteins on the Surface of HIV-1 Pseudovirus Particles. Viruses 2020, 12, 1296. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




