Submitted:
27 May 2024
Posted:
27 May 2024
You are already at the latest version
Abstract
Keywords:
1. Challenges and Limitations with Traditional Acquisition of Structural and Binding Affinity Data
2. Concept of In Silico Generation of Structural and Intermolecular Kd Data
- a real GIBAC needs to take genetic variations into account; and
- a real GIBAC needs to work even without structural information; and
- for a real GIBAC, a variety of factors need to be taken into account, such as temperature, pH [22,23], site-specific protonation states (e.g., side chain pKa of protein) [24,25], post-translational modifications (PTMs) [26,27,28], post-expression modifications (PEMs) [29,30], buffer conditions [31], et cetera; and
- a real GIBAC requires a general forcefield for all types of molecules [3]; and
- a real GIBAC is able to be used the other way around, i.e., to be used as a search engine for therapeutic candidate(s). With such a GIBAC-based search engine, a list of therapeutic candidates can be retrieved and ranked according to drug-target Kd value(s), with input parameters including drug target(s) and a desired drug-target Kd value or a range of it.
3. Rationale Behind In Silico Generation of Structural and Intermolecular Kd Data
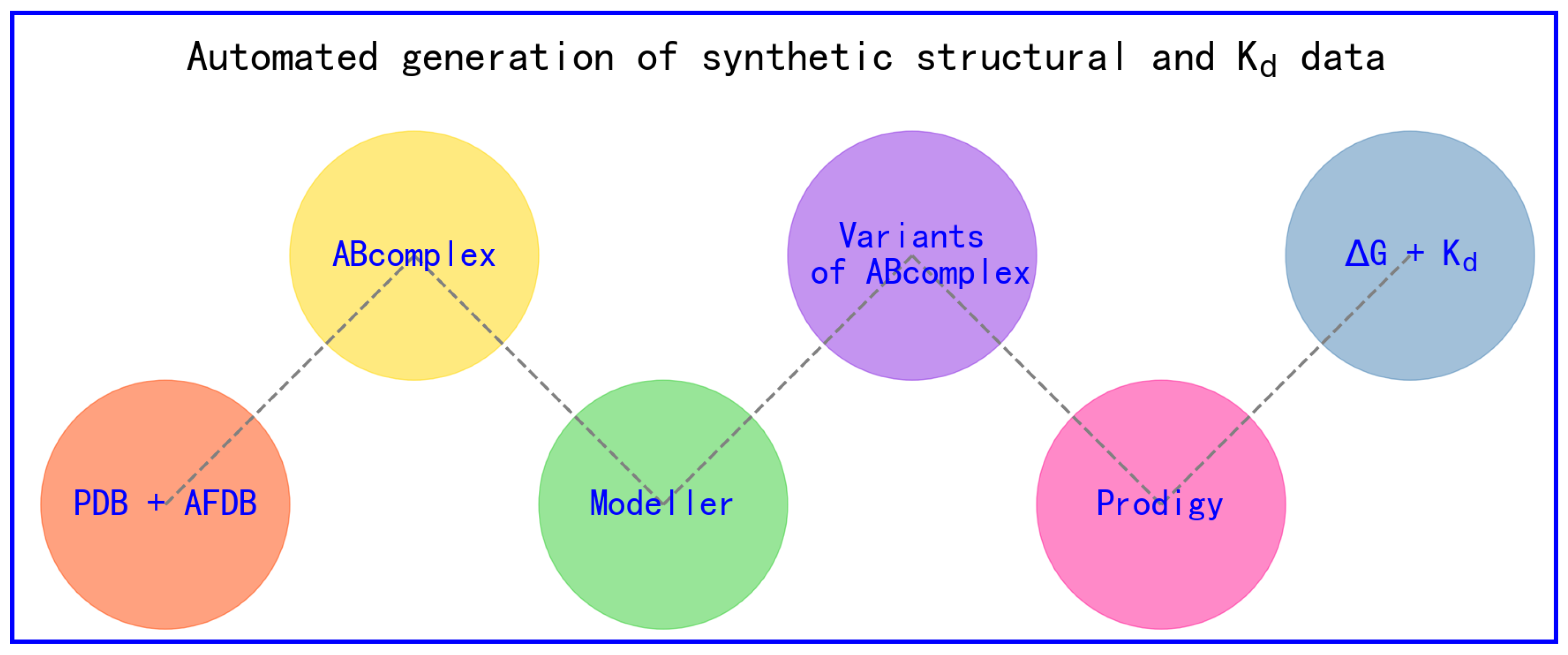
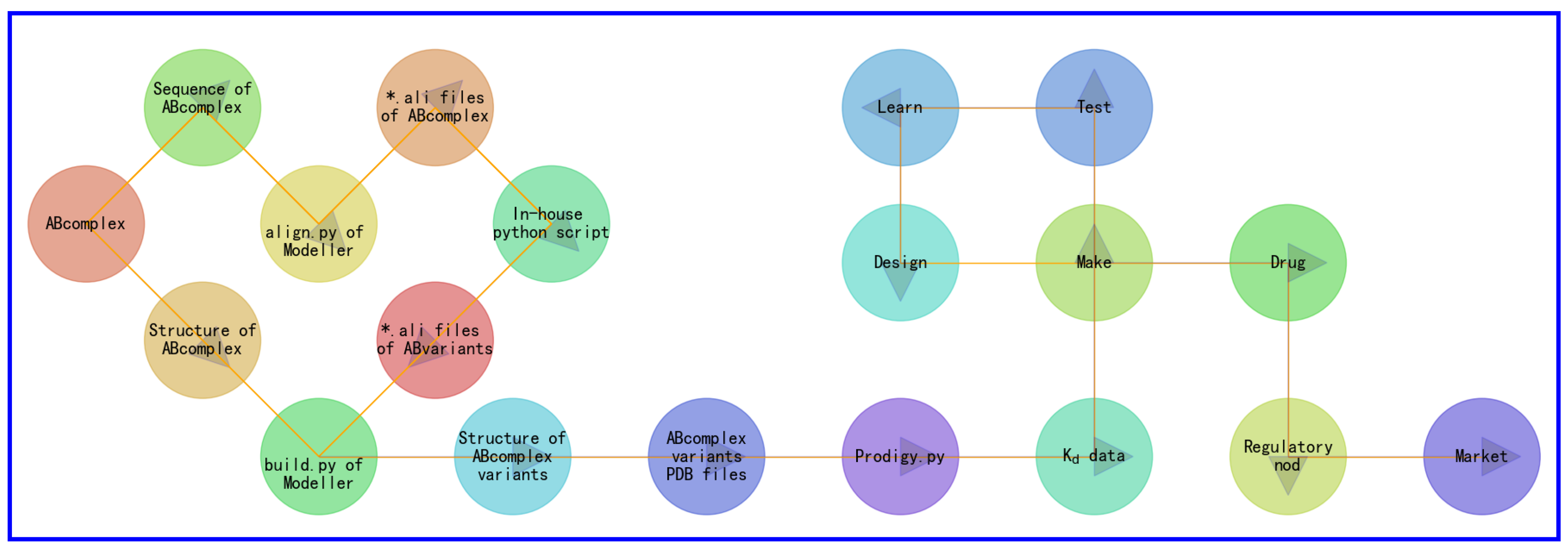
4. Workflow Steps Involved in Generating Synthetic Structural Data and Intermolecular Kd Data
- the experimental complex structure of molecule A and molecule B (ABcomplex) is retrived from PDB.
- amino acid sequences of molecules A and B are retrieved from the PDB file of ABcomplex.
- amino acid sequences of molecules A and B are plugged into align.py of Modeller [37] to generate alignment files of ABcomplex (*.ali files) for each set of site-specific mutations for the amino acid sequences of molecules A and B. Here, for each set of site-specific mutations for the amino acid sequences of molecules A and B, the number of missense mutations are restricted to ensure that the overall accuracy of the subsequent structural modeling and Kd calculations.
- alignment files of ABcomplex (*.ali files) are plugged into a set of in-house python scripts to produce a set of *ali files for variants of molecules A and B.
- the structure of ABcomplex and the set of *ali files for variants of molecules A and B are plugged into build.py of Modeller [37] to generate a set of homology complex structural models for variants of molecules A and B for each set of site-specific mutations for the amino acid sequences of molecules A and B.
5. Synthetic Structural Data and Intermolecular Kd Data: Validation and Benchmarking
- cross-validation is a widely used technique to assess the robustness of predictive models and evaluate their generalization performance. In the context of in silico data generation, cross-validation involves partitioning the dataset into training and validation sets and iteratively training the model on different subsets of the data. This process allows researchers to assess the model’s performance on unseen data and identify any potential biases or overfitting issues [45].
- external validation involves testing the predictive model on an independent dataset that was not used during the model training phase. By evaluating the model’s performance on unseen data from different sources or experimental conditions, researchers can assess its ability to generalize and make accurate predictions in real-world scenarios. External validation provides a more rigorous assessment of the model’s performance and its applicability to diverse biological systems [46].
- various performance metrics can be used to quantify the agreement between in silico predictions and experimental data. For structural data, metrics such as root-mean-square deviation (RMSD) or pairwise atomic distance distributions can assess the similarity between predicted and experimental structures. For Kd data, metrics such as Pearson correlation coefficient or mean absolute error can evaluate the accuracy of predicted binding affinities compared to experimental measurements [47,48].
- benchmarking involves comparing the performance of the in silico workflow with other computational methods or experimental techniques. By benchmarking against established methods or gold standard datasets, researchers can assess the relative strengths and weaknesses of the workflow and identify areas for improvement. Benchmarking provides valuable insights into the performance of the workflow in comparison to existing approaches and helps establish its credibility and reliability in drug discovery applications [49].
6. Synthetic Structural Data and Intermolecular Kd Data: Applications and Implications
6.1. Data Augmentation and Training of Machine Learning Models
6.2. Broader Implications of Synthetic Structural Data and Intermolecular Kd Data

7. In Silico Generation of Structural and Intermolecular Kd Data: Future Directions
7.1. Integration of Multi-Scale Modeling
7.2. Incorporation of Structural Dynamics
7.3. Integration of Experimental and Computational Approaches
8. Conclusion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xiong, P.; Zhang, C.; Zheng, W.; Zhang, Y. BindProfX: Assessing Mutation-Induced Binding Affinity Change by Protein Interface Profiles with Pseudo-Counts. Journal of Molecular Biology 2017, 429, 426–434. [CrossRef] [PubMed]
- Liu, T.; Lin, Y.; Wen, X.; Jorissen, R.N.; Gilson, M.K. BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Research 2007, 35, D198–D201. [CrossRef] [PubMed]
- Hofmann, D.W.M.; Kuleshova, L.N. A general force field by machine learning on experimental crystal structures. Calculations of intermolecular Gibbs energy with iFlexCryst. Acta Crystallographica Section A Foundations and Advances 2023, 79, 132–144. [CrossRef]
- Vranken, W.F.; Boucher, W.; Stevens, T.J.; Fogh, R.H.; Pajon, A.; Llinas, M.; Ulrich, E.L.; Markley, J.L.; Ionides, J.; Laue, E.D. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins: Struct., Funct., Bioinf. 2005, 59, 687–696. [CrossRef] [PubMed]
- Bezencon, J.; Wittwer, M.B.; Cutting, B.; Smiesko, M.; Wagner, B.; Kansy, M.; Ernst, B. pKa determination by (1)H NMR spectroscopy-an old methodology revisited. J. Pharm. Biomed. Anal. 2014, 1, 147–155. [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology. 2011, 7, 539–543. [CrossRef] [PubMed]
- Lipinski, C.; Hopkins, A. Navigating chemical space for biology and medicine. Nature 2004, 432, 855–861. [CrossRef] [PubMed]
- Li, W. Towards a General Intermolecular Binding Affinity Calculator 2022.
- Li, W.; Vottevor, G. Towards a Truly General Intermolecular Binding Affinity Calculator for Drug Discovery & Design 2023.
- Wang, T.; He, X.; Li, M.; Shao, B.; Liu, T.Y. AIMD-Chig: Exploring the conformational space of a 166-atom protein Chignolin with ab initio molecular dynamics. Scientific Data 2023, 10. [CrossRef]
- Li, W. Characterising the interaction between caenopore-5 and model membranes by NMR spectroscopy and molecular dynamics simulations. PhD thesis, University of Auckland, 2016.
- Liu, W.; Chen, L.; Yin, D.; Yang, Z.; Feng, J.; Sun, Q.; Lai, L.; Guo, X. Visualizing single-molecule conformational transition and binding dynamics of intrinsically disordered proteins. Nature Communications 2023, 14. [CrossRef]
- Kawade, R.; Kuroda, D.; Tsumoto, K. How the protonation state of a phosphorylated amino acid governs molecular recognition: insights from classical molecular dynamics simulations. FEBS Letters 2019, 594, 903–912. [CrossRef]
- Tu, C.L.; Wang, Y.L.; Hu, T.M.; Hsu, L.F. Analysis of Pharmacokinetic and Pharmacodynamic Parameters in EU- Versus US-Licensed Reference Biological Products: Are In Vivo Bridging Studies Justified for Biosimilar Development? BioDrugs 2019, 33, 437–446. [CrossRef] [PubMed]
- Hirata, F.; Sugita, M.; Yoshida, M.; Akasaka, K. Perspective: Structural fluctuation of protein and Anfinsen’s thermodynamic hypothesis. The Journal of Chemical Physics 2018, 148, 020901. [CrossRef] [PubMed]
- Lee, J.; Pilch, P.F. The insulin receptor: structure, function, and signaling. American Journal of Physiology-Cell Physiology 1994, 266, C319–C334. [CrossRef]
- Rahuel-Clermont, S.; French, C.A.; Kaarsholm, N.C.; Dunn, M.F. Mechanisms of Stabilization of the Insulin Hexamer through Allosteric Ligand Interactions. Biochemistry 1997, 36, 5837–5845. [CrossRef] [PubMed]
- Nishimura, E.; Pridal, L.; Glendorf, T.; Hansen, B.F.; Hubálek, F.; Kjeldsen, T.; Kristensen, N.R.; Lützen, A.; Lyby, K.; Madsen, P.; Pedersen, T.Å.; Ribel-Madsen, R.; Stidsen, C.E.; Haahr, H. Molecular and pharmacological characterization of insulin icodec: a new basal insulin analog designed for once-weekly dosing. BMJ Open Diabetes Research & Care 2021, 9, e002301.
- Li, W. Designing Insulin Analogues with Lower Binding Affinity to Insulin Receptor than That of Insulin Icodec 2024. [CrossRef]
- Li, W. How Structural Modifications of Insulin Icodec Contributes to Its Prolonged Duration of Action: A Structural and Biophysical Perspective 2023. [CrossRef]
- Wong, C.H.; Siah, K.W.; Lo, A.W. Estimation of clinical trial success rates and related parameters. Biostatistics 2018, 20, 273–286. [CrossRef]
- Yang, A.S.; Honig, B. On the pH Dependence of Protein Stability. Journal of Molecular Biology 1993, 231, 459–474. [CrossRef]
- Harris, T.K.; Turner, G.J. Structural Basis of Perturbed pKa Values of Catalytic Groups in Enzyme Active Sites. IUBMB Life (International Union of Biochemistry and Molecular Biology: Life) 2002, 53, 85–98. [CrossRef]
- Li, W. Gravity-driven pH adjustment for site-specific protein pKa measurement by solution-state NMR. Measurement Science and Technology 2017, 28, 127002. [CrossRef]
- Hansen, A.L.; Kay, L.E. Measurement of histidine pKa values and tautomer populations in invisible protein states. Proceedings of the National Academy of Sciences 2014, 111, E1705–E1712. [CrossRef]
- Herget, S.; Ranzinger, R.; Maass, K.; Lieth, C.W. GlycoCT—a unifying sequence format for carbohydrates. Carbohydrate Research 2008, 343, 2162–2171. [CrossRef] [PubMed]
- Foster, J.M.; Moreno, P.; Fabregat, A.; Hermjakob, H.; Steinbeck, C.; Apweiler, R.; Wakelam, M.J.O.; Vizcaíno, J.A. LipidHome: A Database of Theoretical Lipids Optimized for High Throughput Mass Spectrometry Lipidomics. PLoS ONE 2013, 8, e61951. [CrossRef] [PubMed]
- Sud, M.; Fahy, E.; Subramaniam, S. Template-based combinatorial enumeration of virtual compound libraries for lipids. Journal of Cheminformatics 2012, 4. [CrossRef] [PubMed]
- Li, W. Strengthening Semaglutide-GLP-1R Binding Affinity via a Val27-Arg28 Exchange in the Peptide Backbone of Semaglutide: A Computational Structural Approach. Journal of Computational Biophysics and Chemistry 2021, 20, 495–499. [CrossRef]
- Weiss, M. Design of ultra-stable insulin analogues for the developing world. Journal of Health Specialties 2013, 1, 59. [CrossRef]
- Olsson, M.H.M.; Søndergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. Journal of Chemical Theory and Computation 2011, 7, 525–537. [CrossRef] [PubMed]
- Müller, C.E.; Hansen, F.K.; Gütschow, M.; Lindsley, C.W.; Liotta, D. New Drug Modalities in Medicinal Chemistry, Pharmacology, and Translational Science. ACS Pharmacology & Translational Science 2021, 4, 1712–1713.
- Yuan, Y.; Cao, D.; Zhang, Y.; Ma, J.; Qi, J.; Wang, Q.; Lu, G.; Wu, Y.; Yan, J.; Shi, Y.; Zhang, X.; Gao, G.F. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nature Communications 2017, 8. [CrossRef] [PubMed]
- Vangone, A.; Bonvin, A.M. Contacts-based prediction of binding affinity in protein-protein complexes. eLife 2015, 4. [CrossRef]
- Li, W.; Shi, G. How CaV1.2-bound verapamil blocks Ca2+ influx into cardiomyocyte: Atomic level views. Pharmacological Research 2019, 139, 153–157. [CrossRef]
- Li, W. How do SMA-linked mutations of SMN1 lead to structural/functional deficiency of the SMA protein? PLOS ONE 2017, 12, e0178519. [CrossRef] [PubMed]
- Webb, B.; Sali, A. Protein Structure Modeling with MODELLER. In Methods in Molecular Biology; Springer US, 2020; pp. 239–255.
- Berman, H.; Henrick, K.; Nakamura, H. Announcing the worldwide Protein Data Bank. Nature Structural & Molecular Biology 2003, 10, 980–980. [CrossRef]
- Li, W. Half-a-century Burial of ρ, θ and φ in PDB 2021.
- Li, W. Visualising the Experimentally Uncharted Territories of Membrane Protein Structures inside Protein Data Bank 2020.
- Li, W. A Local Spherical Coordinate System Approach to Protein 3D Structure Description 2020.
- Xue, L.C.; Rodrigues, J.P.; Kastritis, P.L.; Bonvin, A.M.; Vangone, A. PRODIGY: a web server for predicting the binding affinity of protein-protein complexes. Bioinformatics 2016, p. btw514.
- Hartshorn, M.J.; Verdonk, M.L.; Chessari, G.; Brewerton, S.C.; Mooij, W.T.M.; Mortenson, P.N.; Murray, C.W. Diverse, High-Quality Test Set for the Validation of Protein-Ligand Docking Performance. Journal of Medicinal Chemistry 2007, 50, 726–741. [CrossRef] [PubMed]
- Rohrer, S.G.; Baumann, K. Maximum Unbiased Validation (MUV) Data Sets for Virtual Screening Based on PubChem Bioactivity Data. Journal of Chemical Information and Modeling 2009, 49, 169–184. [CrossRef] [PubMed]
- Song, Q.C.; Tang, C.; Wee, S. Making Sense of Model Generalizability: A Tutorial on Cross-Validation in R and Shiny. Advances in Methods and Practices in Psychological Science 2021, 4, 251524592094706. [CrossRef]
- Zeng, Y.; Chen, X.; Luo, Y.; Li, X.; Peng, D. Deep drug-target binding affinity prediction with multiple attention blocks. Briefings in Bioinformatics 2021, 22. [CrossRef] [PubMed]
- Antunes, D.A.; Abella, J.R.; Devaurs, D.; Rigo, M.M.; Kavraki, L.E. Structure-based Methods for Binding Mode and Binding Affinity Prediction for Peptide-MHC Complexes. Current Topics in Medicinal Chemistry 2019, 18, 2239–2255. [CrossRef] [PubMed]
- Kastritis, P.L.; Rodrigues, J.P.; Folkers, G.E.; Boelens, R.; Bonvin, A.M. Proteins Feel More Than They See: Fine-Tuning of Binding Affinity by Properties of the Non-Interacting Surface. Journal of Molecular Biology 2014, 426, 2632–2652. [CrossRef] [PubMed]
- Agrawal, P.; Singh, H.; Srivastava, H.K.; Singh, S.; Kishore, G.; Raghava, G.P.S. Benchmarking of different molecular docking methods for protein-peptide docking. BMC Bioinformatics 2019, 19. [CrossRef]
- Zhang, C.; Han, Q.; Chen, R.; Zhao, X.; Tang, P.; Song, H. SSDRec: Self-Augmented Sequence Denoising for Sequential Recommendation, 2024. [CrossRef]
- Carracedo-Reboredo, P.; Liñares-Blanco, J.; Rodríguez-Fernández, N.; Cedrón, F.; Novoa, F.J.; Carballal, A.; Maojo, V.; Pazos, A.; Fernandez-Lozano, C. A review on machine learning approaches and trends in drug discovery. Computational and Structural Biotechnology Journal 2021, 19, 4538–4558. [CrossRef]
- Stefano, M.D.; Galati, S.; Piazza, L.; Granchi, C.; Mancini, S.; Fratini, F.; Macchia, M.; Poli, G.; Tuccinardi, T. VenomPred 2.0: A Novel In Silico Platform for an Extended and Human Interpretable Toxicological Profiling of Small Molecules. Journal of Chemical Information and Modeling 2023. [CrossRef] [PubMed]
- Cheng, J.; Novati, G.; Pan, J.; Bycroft, C.; Žemgulytė, A.; Applebaum, T.; Pritzel, A.; Wong, L.H.; Zielinski, M.; Sargeant, T.; Schneider, R.G.; Senior, A.W.; Jumper, J.; Hassabis, D.; Kohli, P.; Avsec, Ž. Accurate proteome-wide missense variant effect prediction with AlphaMissense. Science 2023. [CrossRef] [PubMed]
- Sadybekov, A.A.; Sadybekov, A.V.; Liu, Y.; Iliopoulos-Tsoutsouvas, C.; Huang, X.P.; Pickett, J.; Houser, B.; Patel, N.; Tran, N.K.; Tong, F.; Zvonok, N.; Jain, M.K.; Savych, O.; Radchenko, D.S.; Nikas, S.P.; Petasis, N.A.; Moroz, Y.S.; Roth, B.L.; Makriyannis, A.; Katritch, V. Synthon-based ligand discovery in virtual libraries of over 11 billion compounds. Nature 2021, 601, 452–459. [CrossRef] [PubMed]
- Kinghorn, A.; Fraser, L.; Liang, S.; Shiu, S.; Tanner, J. Aptamer Bioinformatics. International Journal of Molecular Sciences 2017, 18, 2516. [CrossRef]
- Lobato, M.; Rabbitts, T. Intracellular Antibodies as Specific Reagents for Functional Ablation: Future Therapeutic Molecules. Current Molecular Medicine 2004, 4, 519–528. [CrossRef] [PubMed]
- Weng, G.; Shen, C.; Cao, D.; Gao, J.; Dong, X.; He, Q.; Yang, B.; Li, D.; Wu, J.; Hou, T. PROTAC-DB: an online database of PROTACs. Nucleic Acids Research 2020, 49, D1381–D1387. [CrossRef] [PubMed]
- Weng, G.; Cai, X.; Cao, D.; Du, H.; Shen, C.; Deng, Y.; He, Q.; Yang, B.; Li, D.; Hou, T. PROTAC-DB 2.0: an updated database of PROTACs. Nucleic Acids Research 2022, 51, D1367–D1372. [CrossRef]
- Zhang, W.; Chen, Y.; Liu, F.; Luo, F.; Tian, G.; Li, X. Predicting potential drug-drug interactions by integrating chemical, biological, phenotypic and network data. BMC Bioinformatics 2017, 18. [CrossRef]
- Rahnama, R.; Kizerwetter, M.; Yang, H.; Christodoulou, I.; Fearnow, A.; Vorri, S.; Spangler, J.; Bonifant, C. Poster: AML-496 Study of Affinity Tuning in AML-Targeted CAR-NK Cells. Clinical Lymphoma Myeloma and Leukemia 2022, 22, S136. [CrossRef]
- Fakhrai-Rad, H. Insulin-degrading enzyme identified as a candidate diabetes susceptibility gene in GK rats. Human Molecular Genetics 2000, 9, 2149–2158. [CrossRef]
- González-Casimiro, C.M.; Merino, B.; Casanueva-Álvarez, E.; Postigo-Casado, T.; Cámara-Torres, P.; Fernández-Díaz, C.M.; Leissring, M.A.; Cózar-Castellano, I.; Perdomo, G. Modulation of Insulin Sensitivity by Insulin-Degrading Enzyme. Biomedicines 2021, 9, 86. [CrossRef] [PubMed]
- Ferreira, L.G.; Santos, R.N.; Andricopulo, A.D. Computational methods for calculation of protein-ligand binding affinities. Frontiers in chemistry 2019, 7, 559.
- Petukh, M.; Stefl, S.; Alexov, E. The Role of Protonation States in Ligand-Receptor Recognition and Binding. Current Pharmaceutical Design 2013, 19, 4182–4190. [CrossRef]
- Jubb, H.C.; Pandurangan, A.P.; Turner, M.A.; Ochoa-Montaño, B.; Blundell, T.L.; Ascher, D.B. Mutations at protein-protein interfaces: Small changes over big surfaces have large impacts on human health. Progress in Biophysics and Molecular Biology 2017, 128, 3–13. [CrossRef]
- Li, W. Calcium Channel Trafficking Blocker Gabapentin Bound to the α-2-δ-1 Subunit of Voltage-Gated Calcium Channel: A Computational Structural Investigation 2020.
- Elliott, W.J.; Ram, C.V.S. Calcium Channel Blockers. The Journal of Clinical Hypertension 2011, 13, 687–689. [CrossRef]
- Wu, D.; Piszczek, G. Rapid Determination of Antibody-Antigen Affinity by Mass Photometry. Journal of Visualized Experiments 2021. [CrossRef]
- Mason, D.M.; Friedensohn, S.; Weber, C.R.; Jordi, C.; Wagner, B.; Meng, S.M.; Ehling, R.A.; Bonati, L.; Dahinden, J.; Gainza, P.; Correia, B.E.; Reddy, S.T. Optimization of therapeutic antibodies by predicting antigen specificity from antibody sequence via deep learning. Nature Biomedical Engineering 2021, 5, 600–612. [CrossRef] [PubMed]
- Li, W. Extracting the Interfacial Electrostatic Features from Experimentally Determined Antigen and/or Antibody-Related Structures inside Protein Data Bank for Machine Learning-Based Antibody Design 2020.
- Lane, D.P. p53, guardian of the genome. Nature 1992, 358, 15–16. [CrossRef]
- Sola, I.; Mateos-Gomez, P.A.; Almazan, F.; Zuñiga, S.; Enjuanes, L. RNA-RNA and RNA-protein interactions in coronavirus replication and transcription. RNA Biology 2011, 8, 237–248. [CrossRef]
- Iida, K.; Itoh, E.; Kim, D.S.; Rincon, J.P.D.; Coschigano, K.T.; Kopchick, J.J.; Thorner, M.O. Muscle mechano growth factor is preferentially induced by growth hormone in growth hormone-deficientlit/litmice. The Journal of Physiology 2004, 560, 341–349. [CrossRef]
- Zhang, S.; Karthikeyan, R.; Fernando, S.D. Evaluating apoenzyme-coenzyme-substrate interactions of methane monooxygenase with an engineered active site for electron harvesting: a computational study. Journal of Molecular Modeling 2018, 24. [CrossRef] [PubMed]
- Ham, S.W.; Jeon, H.Y.; Kim, H. Verapamil augments carmustine- and irradiation-induced senescence in glioma cells by reducing intracellular reactive oxygen species and calcium ion levels. Tumor Biology 2017, 39, 101042831769224. [CrossRef]
- Rhee, S.G. Regulation of Phosphoinositide-Specific Phospholipase C. Annual Review of Biochemistry 2001, 70, 281–312. [CrossRef] [PubMed]
- Robert, C.; Schachter, J.; Long, G.V.; Arance, A.; Grob, J.J.; Mortier, L.; Daud, A.; Carlino, M.S.; McNeil, C.; Lotem, M.; Larkin, J.; Lorigan, P.; Neyns, B.; Blank, C.U.; Hamid, O.; Mateus, C.; Shapira-Frommer, R.; Kosh, M.; Zhou, H.; Ibrahim, N.; Ebbinghaus, S.; Ribas, A. Pembrolizumab versus Ipilimumab in Advanced Melanoma. New England Journal of Medicine 2015, 372, 2521–2532. [CrossRef] [PubMed]
- Mroczko, B.; Groblewska, M.; Litman-Zawadzka, A.; Kornhuber, J.; Lewczuk, P. Amyloid β oligomers (AβOs) in Alzheimer’s disease. Journal of Neural Transmission 2017, 125, 177–191. [CrossRef] [PubMed]
- Pinheiro, F.; Santos, J.; Ventura, S. AlphaFold and the amyloid landscape. Journal of Molecular Biology 2021, p. 167059. [CrossRef]
- Lin, Z.; Akin, H.; Rao, R.; Hie, B.; Zhu, Z.; Lu, W.; dos Santos Costa, A.; Fazel-Zarandi, M.; Sercu, T.; Candido, S.; Rives, A. Language models of protein sequences at the scale of evolution enable accurate structure prediction. bioRxiv 2022.
- Lounkine, E.; Keiser, M.J.; Whitebread, S.; Mikhailov, D.; Hamon, J.; Jenkins, J.L.; Lavan, P.; Weber, E.; Doak, A.K.; Côté, S.; Shoichet, B.K.; Urban, L. Large-scale prediction and testing of drug activity on side-effect targets. Nature 2012, 486, 361–367. [CrossRef] [PubMed]
- Di Russo, N.V.; Estrin, D.A.; Martí, M.A.; Roitberg, A.E. pH-Dependent conformational changes in proteins and their effect on experimental pKas: the case of nitrophorin 4. PLoS Comput. Biol. 2012, 8, e1002761. [CrossRef] [PubMed]
- Shulman, R.; Wüthrich, K.; Yamane, T.; Patel, D.J.; Blumberg, W. Nuclear magnetic resonance determination of ligand-induced conformational changes in myoglobin. Journal of Molecular Biology 1970, 53, 143–157. [CrossRef]
- Masureel, M.; Martens, C.; Stein, R.A.; Mishra, S.; Ruysschaert, J.M.; Mchaourab, H.S.; Govaerts, C. Protonation drives the conformational switch in the multidrug transporter LmrP. Nature Chemical Biology 2013, 10, 149–155. [CrossRef]
- Peirce, S.M.; Gabhann, F.M.; Bautch, V.L. Integration of experimental and computational approaches to sprouting angiogenesis. Current Opinion in Hematology 2012, 19, 184–191. [CrossRef] [PubMed]
- Cramer, P. AlphaFold2 and the future of structural biology. Nature Structural & Molecular Biology 2021, 28, 704–705. [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; Bodenstein, S.W.; Evans, D.A.; Hung, C.C.; O’Neill, M.; Reiman, D.; Tunyasuvunakool, K.; Wu, Z.; Žemgulytė, A.; Arvaniti, E.; Beattie, C.; Bertolli, O.; Bridgland, A.; Cherepanov, A.; Congreve, M.; Cowen-Rivers, A.I.; Cowie, A.; Figurnov, M.; Fuchs, F.B.; Gladman, H.; Jain, R.; Khan, Y.A.; Low, C.M.R.; Perlin, K.; Potapenko, A.; Savy, P.; Singh, S.; Stecula, A.; Thillaisundaram, A.; Tong, C.; Yakneen, S.; Zhong, E.D.; Zielinski, M.; Žídek, A.; Bapst, V.; Kohli, P.; Jaderberg, M.; Hassabis, D.; Jumper, J.M. Accurate structure prediction of biomolecular interactions with AlphaFold3. Nature 2024. [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




