Submitted:
27 May 2024
Posted:
27 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Compound Synthesis
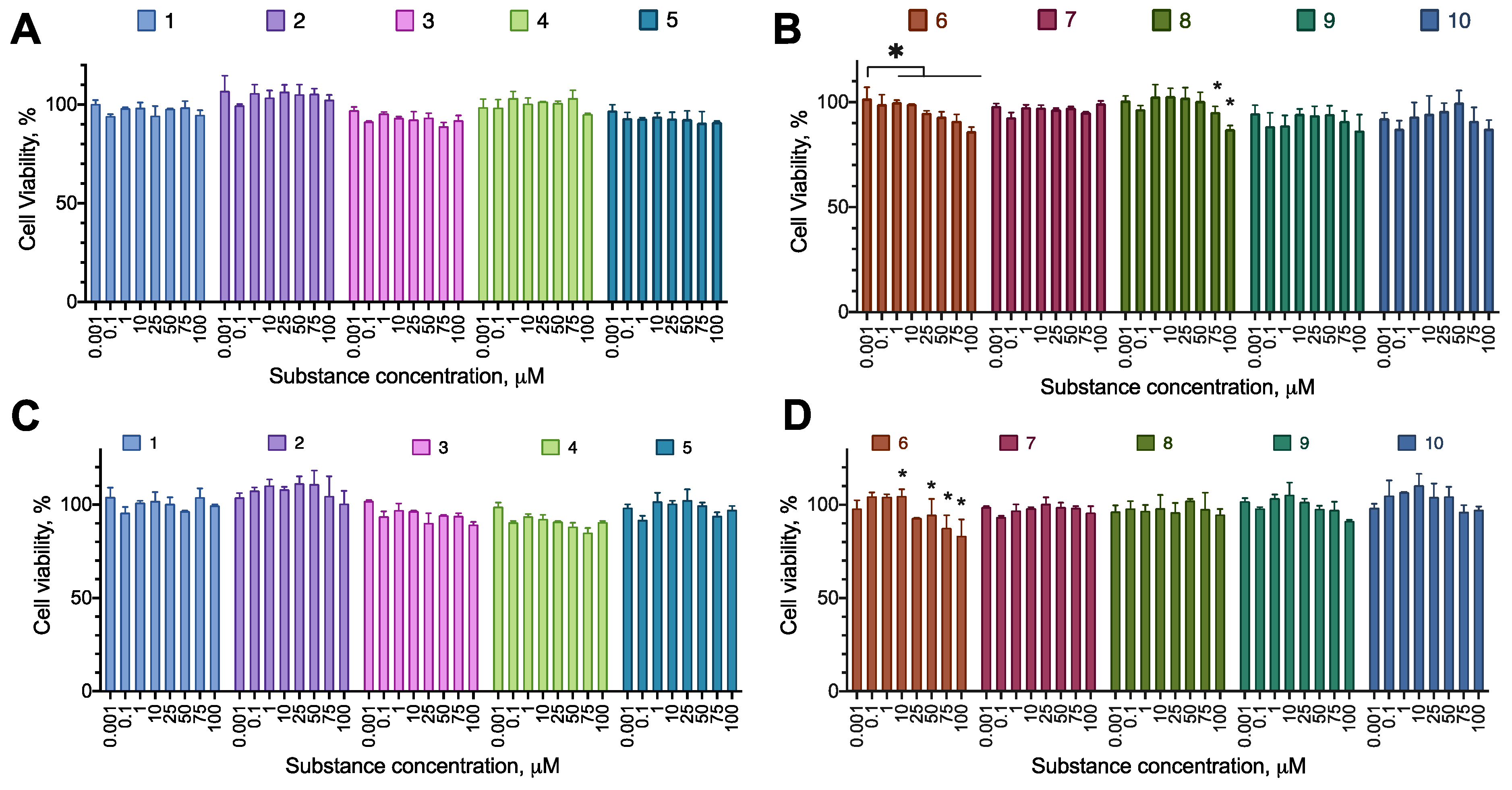
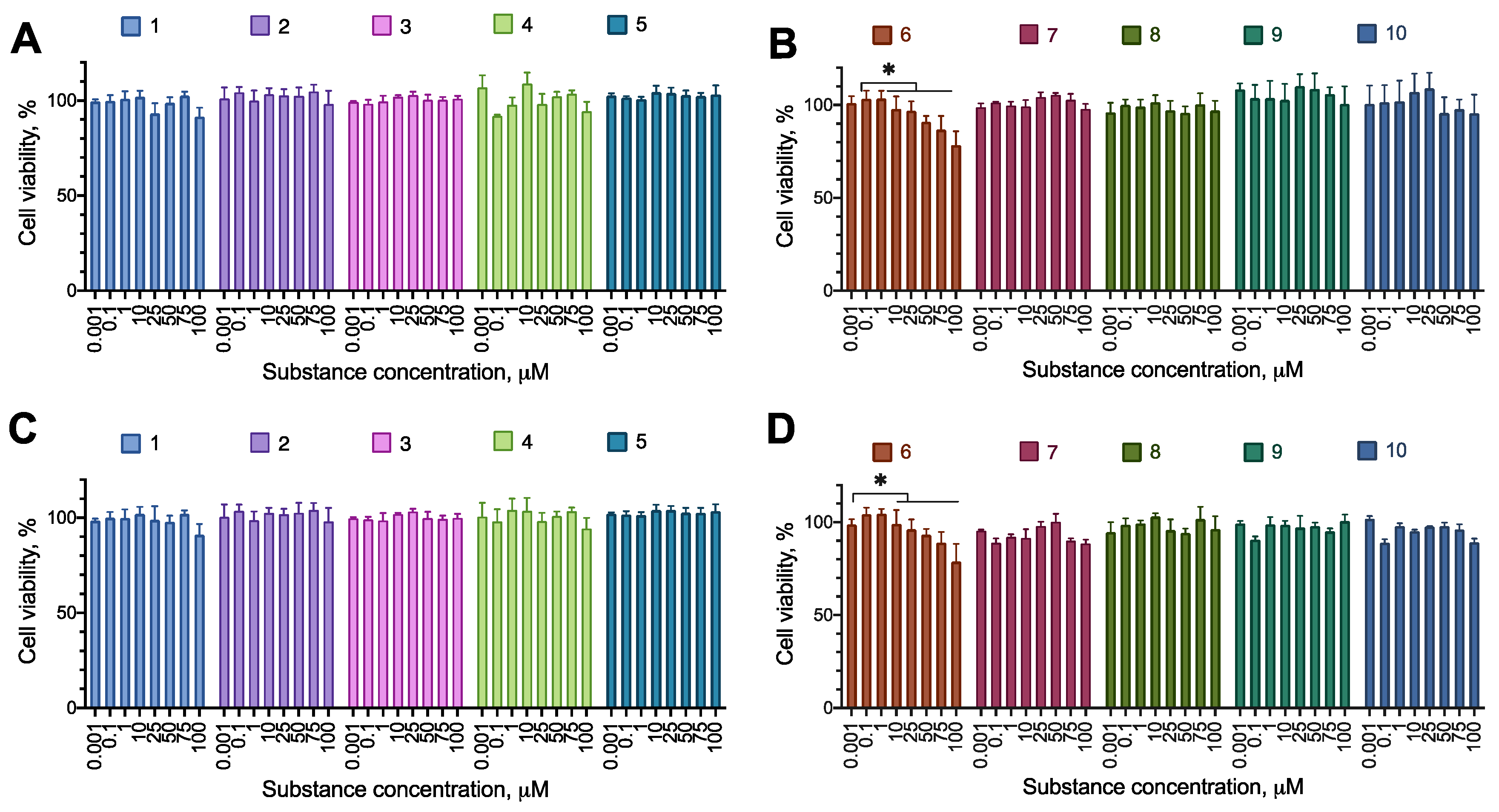
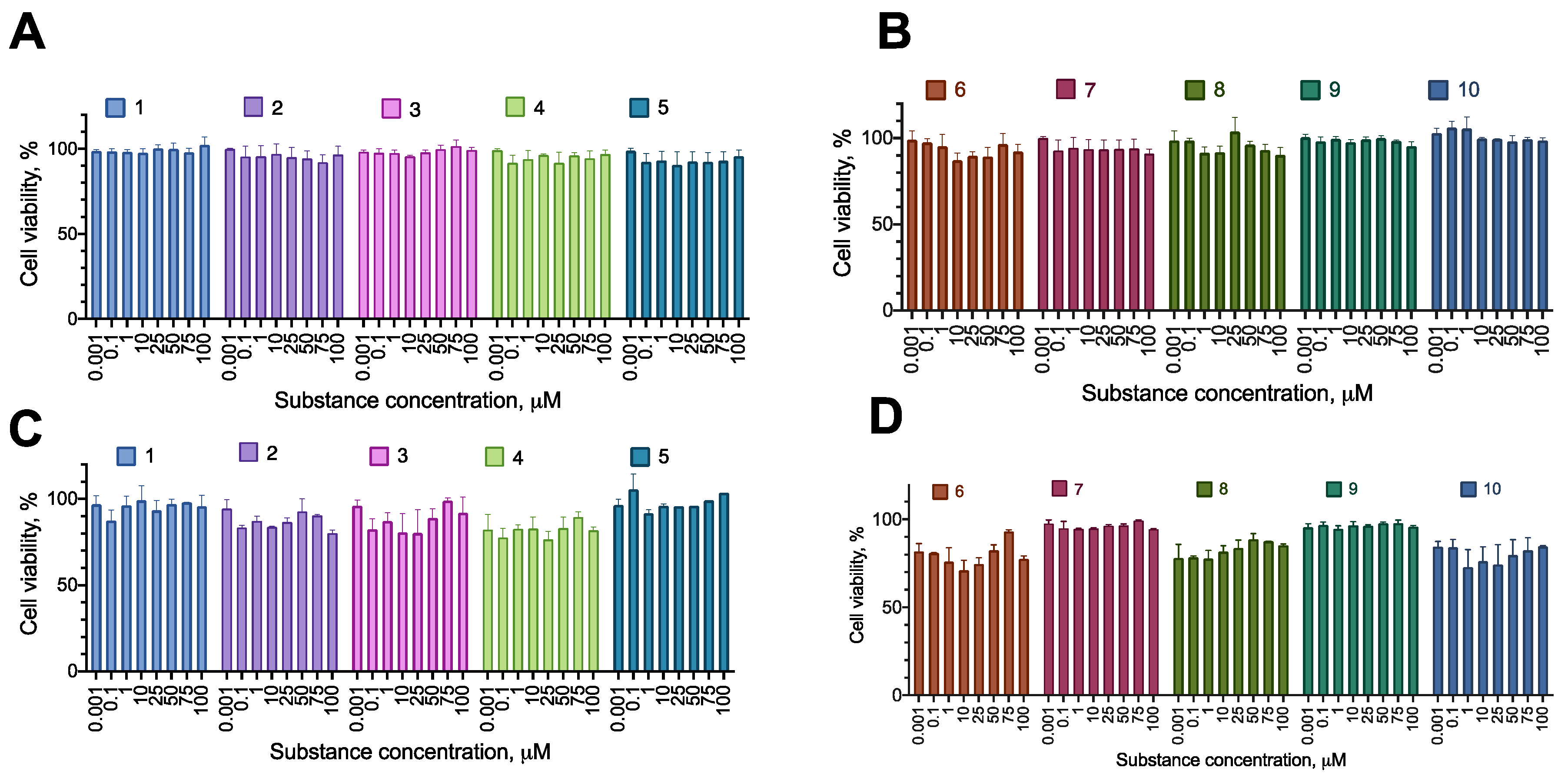
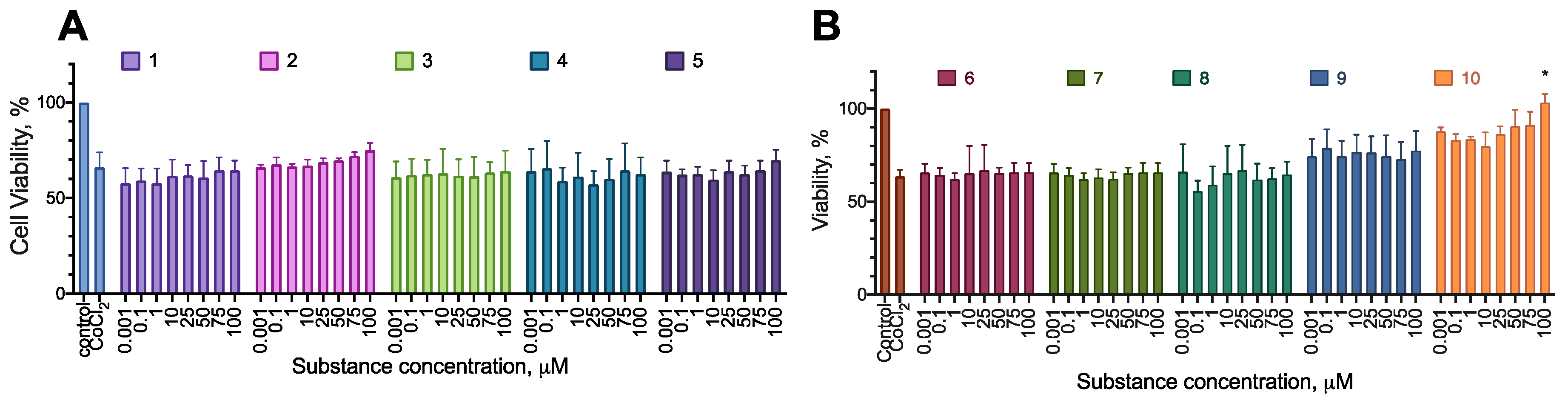
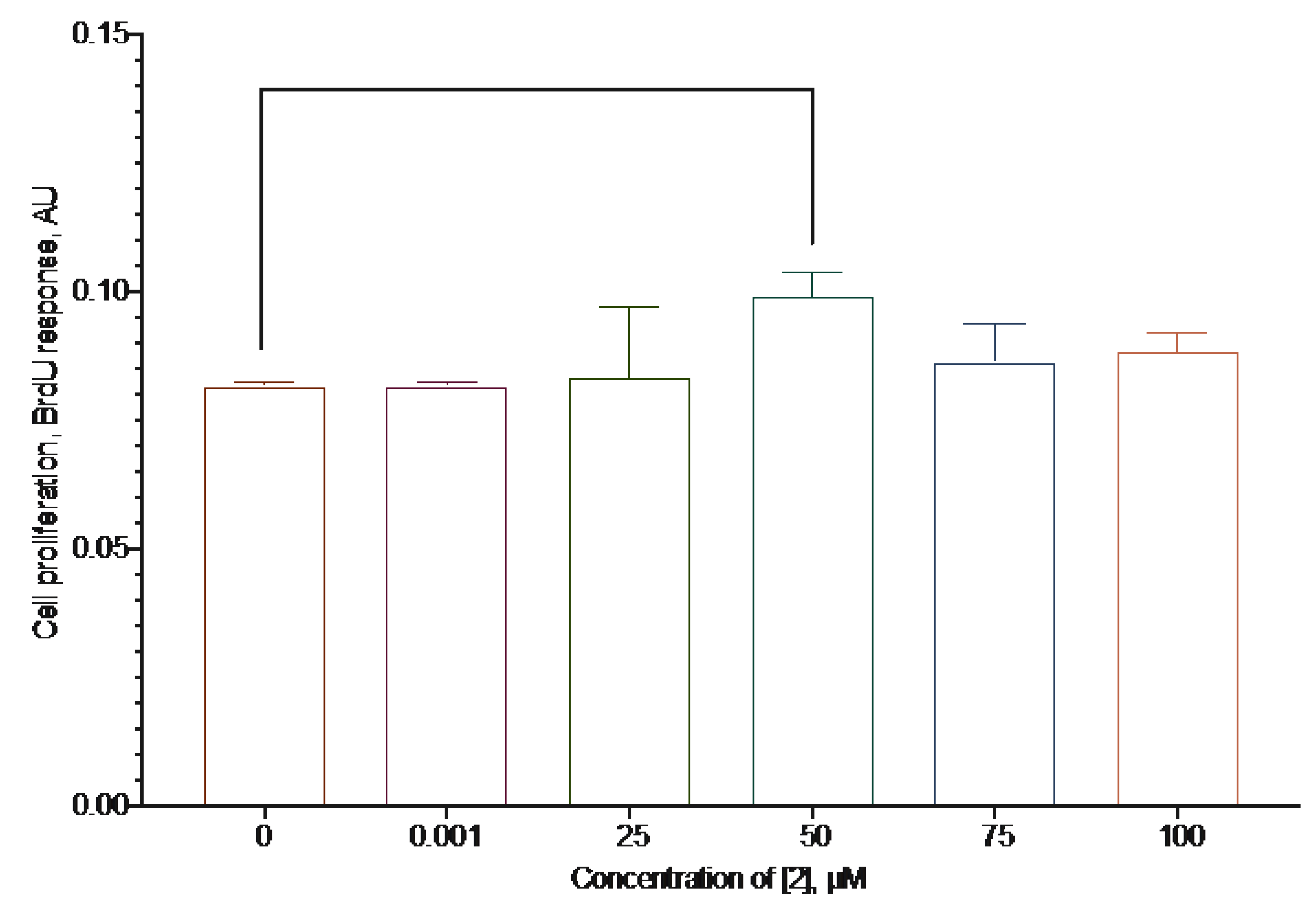
2.2. Anti-Proliferative Activity of the Synthesized Compounds
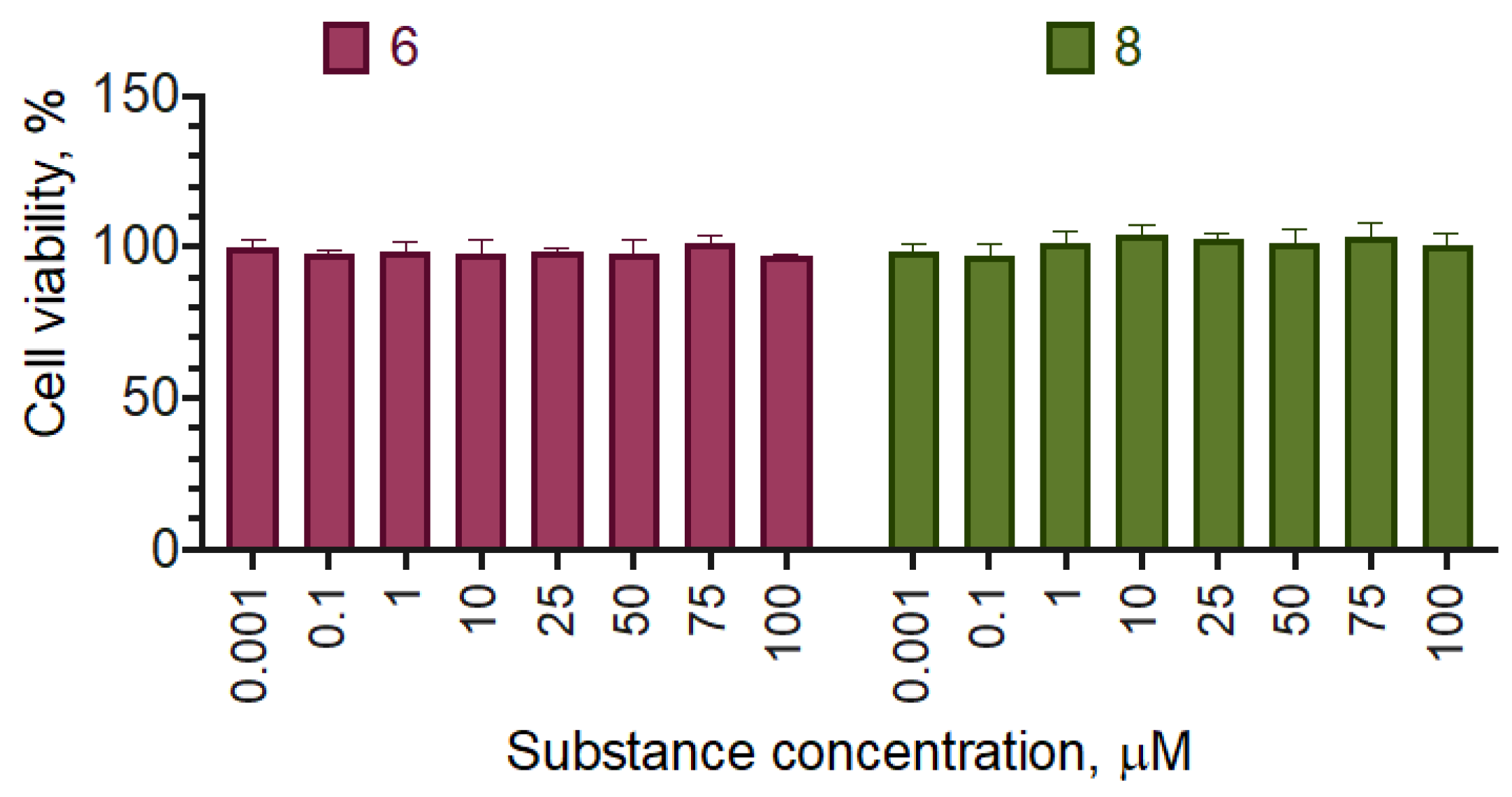
2.3. Selectivity of the Active Compounds
2.4. Mechanism of Action of Compound 6
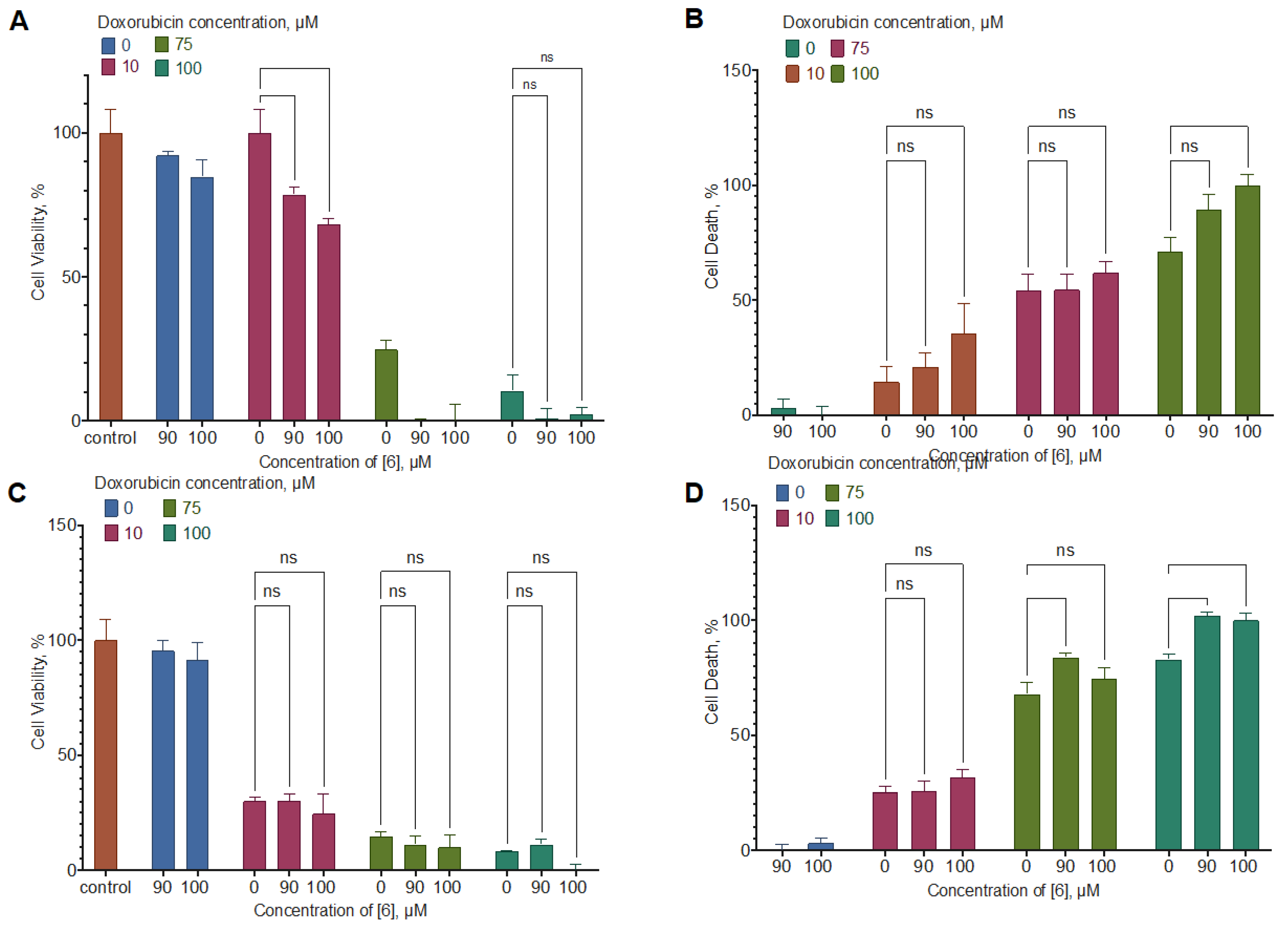
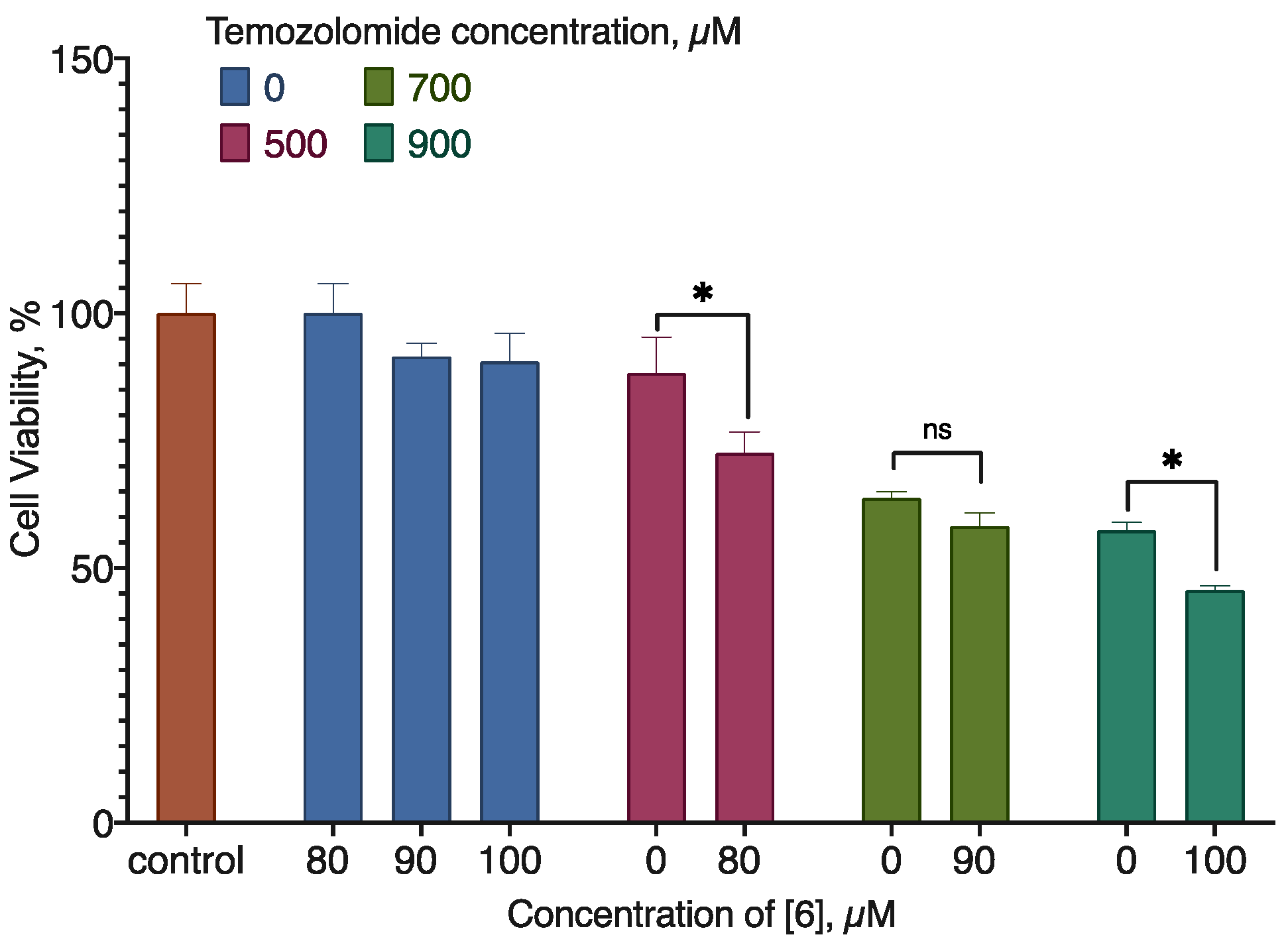
2.5. Combined Activity of Compound 6 with Doxorubicin and Temozolomide
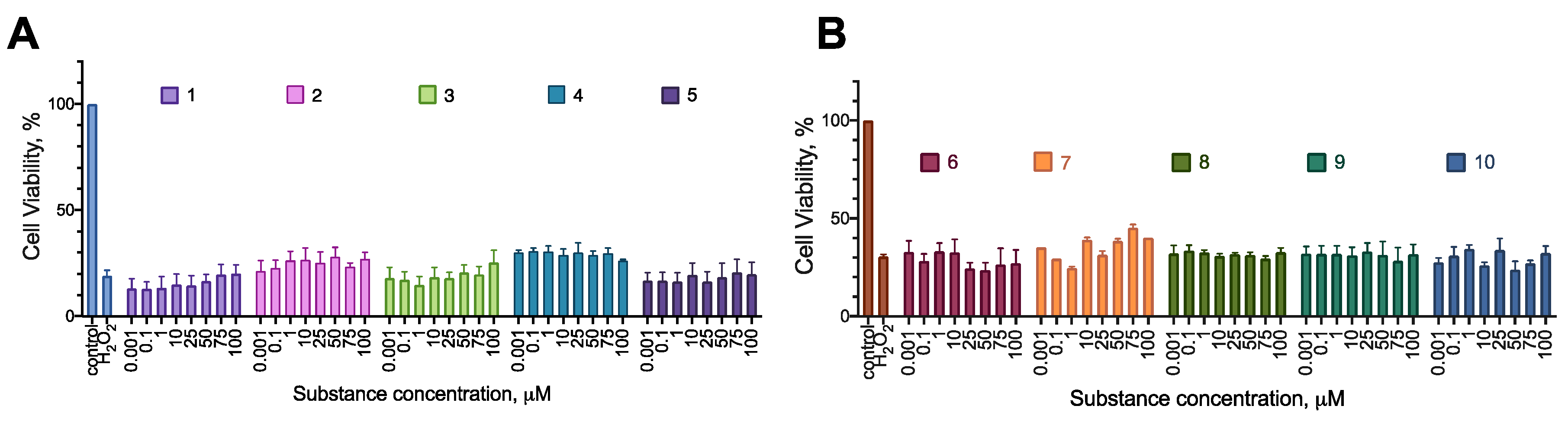
2.5. Cytoprotection
2.6. Molecular Docking
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Synthesized Compounds Characterization
4.3. Chemical Synthesis
4.4. Cell Culture
4.5. Oxidative Stress Induction
4.6. Cytotoxicity and Proliferation Stimulation
4.7. Cell Viability Assay
4.8. BrDU Cell Proliferation Assay
4.9. Compound Interaction Analysis
4.10. Molecular Docking
4.11. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Ramakrishna, W.; Kumari, A.; Rahman, N.; Mandave, P. Anticancer Activities of Plant Secondary Metabolites: Rice Callus Suspension Culture as a New Paradigm. Rice Sci. 2021, 28, 13–30. [Google Scholar] [CrossRef]
- Tuli, H.S.; Garg, V.K.; Bhushan, S.; Uttam, V.; Sharma, U.; Jain, A.; Sak, K.; Yadav, V.; Lorenzo, J.M.; Dhama, K.; et al. Natural Flavonoids Exhibit Potent Anticancer Activity by Targeting MicroRNAs in Cancer: A Signature Step Hinting towards Clinical Perfection. Transl. Oncol. 2023, 27, 101596. [Google Scholar] [CrossRef] [PubMed]
- de Luna, F.C.F.; Ferreira, W.A.S.; Casseb, S.M.M.; de Oliveira, E.H.C. Anticancer Potential of Flavonoids: An Overview with an Emphasis on Tangeretin. Pharmaceuticals (Basel) 2023, 16. [Google Scholar] [CrossRef] [PubMed]
- Kopustinskiene, D.M.; Jakstas, V.; Savickas, A.; Bernatoniene, J. Flavonoids as Anticancer Agents. Nutrients 2020, 12, 457. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Kwon, Y.-J.; Jin, H.; Liu, H.; Kang, W.; Chun, Y.-J.; Bae, J.; Choi, H.-K. Anticancer Activity and Metabolic Profile Alterations by Ortho-Topolin Riboside in in Vitro and in Vivo Models of Non-Small Cell Lung Cancer. FASEB J. 2022, 36, e22127. [Google Scholar] [CrossRef] [PubMed]
- Voller, J.; Béres, T.; Zatloukal, M.; Džubák, P.; Hajdúch, M.; Doležal, K.; Schmülling, T.; Miroslav, S. Anti-Cancer Activities of Cytokinin Ribosides. Phytochem. Rev. 2019, 18, 1101–1113. [Google Scholar] [CrossRef]
- Oshchepkov, M.S.; Kalistratova, A.V.; Savelieva, E.M.; Romanov, G.A.; Bystrova, N.A.; Kochetkov, K.A. Natural and Synthetic Cytokinins and Their Applications in Biotechnology, Agrochemistry and Medicine. Russ. Chem. Rev. 2020, 89, 787–810. [Google Scholar] [CrossRef]
- Gill, A.; Patranabis, S. Phytohormones as Potential Anticancer Agents. Int. j. res. appl. sci. biotechnol. 2021, 8. [Google Scholar] [CrossRef]
- Gonzalez, G.; Grúz, J.; D’Acunto, C.W.; Kaňovský, P.; Strnad, M. Cytokinin Plant Hormones Have Neuroprotective Activity in in Vitro Models of Parkinson’s Disease. Molecules 2021, 26, 361. [Google Scholar] [CrossRef]
- Atallah-Yunes, S.A.; Robertson, M.J. Cytokine Based Immunotherapy for Cancer and Lymphoma: Biology, Challenges and Future Perspectives. Front. Immunol. 2022, 13, 872010. [Google Scholar] [CrossRef]
- Oshchepkov, M.; Kovalenko, L.; Kalistratova, A.; Ivanova, M.; Sherstyanykh, G.; Dudina, P.; Antonov, A.; Cherkasova, A.; Akimov, M. Anti-Proliferative and Cytoprotective Activity of Aryl Carbamate and Aryl Urea Derivatives with Alkyl Groups and Chlorine as Substituents. Molecules 2022, 27, 3616. [Google Scholar] [CrossRef] [PubMed]
- Berraondo, P.; Sanmamed, M.F.; Ochoa, M.C.; Etxeberria, I.; Aznar, M.A.; Pérez-Gracia, J.L.; Rodríguez-Ruiz, M.E.; Ponz-Sarvise, M.; Castañón, E.; Melero, I. Cytokines in Clinical Cancer Immunotherapy. Br. J. Cancer 2019, 120, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Castiglioni, S.; Casati, S.; Ottria, R.; Ciuffreda, P.; Maier, J.A.M. N6-Isopentenyladenosine and Its Analogue N6-Benzyladenosine Induce Cell Cycle Arrest and Apoptosis in Bladder Carcinoma T24 Cells. Anticancer Agents Med. Chem. 2013, 13, 672–678. [Google Scholar] [CrossRef] [PubMed]
- Voller, J.; Zatloukal, M.; Lenobel, R.; Dolezal, K.; Béres, T.; Krystof, V.; Spíchal, L.; Niemann, P.; Dzubák, P.; Hajdúch, M.; et al. Anticancer Activity of Natural Cytokinins: A Structure-Activity Relationship Study. Phytochemistry 2010, 71, 1350–1359. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yu, D.L.; Zhang, H.W.; He, L.Y.; Wu, L. Ortho-Topolin Riboside Induces Apoptosis in Acute Myeloid Leukemia HL-60 Cells. Mol. Cell. Toxicol. 2016, 12, 159–166. [Google Scholar] [CrossRef]
- Yonova, P. ChemInform Abstract: Design, Synthesis and Properties of Synthetic Cytokinins. Recent Advances on Their Application. ChemInform 2012, 43, no. [Google Scholar] [CrossRef]
- Kim, K.K.; Singh, R.K.; Khazan, N.; Kodza, A.; Singh, N.A.; Jones, A.; Sivagnanalingam, U.; Towner, M.; Itamochi, H.; Turner, R.; et al. Development of Potent Forchlorfenuron Analogs and Their Cytotoxic Effect in Cancer Cell Lines. Sci. Rep. 2020, 10, 3241. [Google Scholar] [CrossRef] [PubMed]
- Blum, W.; Henzi, T.; Pecze, L.; Diep, K.-L.; Bochet, C.G.; Schwaller, B. The Phytohormone Forchlorfenuron Decreases Viability and Proliferation of Malignant Mesothelioma Cells in Vitro and in Vivo. Oncotarget 2019, 10, 6944–6956. [Google Scholar] [CrossRef]
- Weidner, C.; Rousseau, M.; Micikas, R.J.; Fischer, C.; Plauth, A.; Wowro, S.J.; Siems, K.; Hetterling, G.; Kliem, M.; Schroeder, F.C.; et al. Amorfrutin C Induces Apoptosis and Inhibits Proliferation in Colon Cancer Cells through Targeting Mitochondria. J. Nat. Prod. 2016, 79, 2–12. [Google Scholar] [CrossRef]
- Kalistratova, A.V.; Kovalenko, L.V.; Oshchepkov, M.S.; Gamisoniya, A.M.; Gerasimova, T.S.; Demidov, Y.A.; Akimov, M.G. Synthesis of New Compounds in the Series of Aryl-Substituted Ureas with Cytotoxic and Antioxidant Activity. Mendeleev Commun. 2020, 30, 153–155. [Google Scholar] [CrossRef]
- Chou, T.-C. Theoretical Basis, Experimental Design, and Computerized Simulation of Synergism and Antagonism in Drug Combination Studies. Pharmacol. Rev. 2006, 58, 621–681. [Google Scholar] [CrossRef] [PubMed]
- Cheng, R.K.Y.; Segala, E.; Robertson, N.; Deflorian, F.; Doré, A.S.; Errey, J.C.; Fiez-Vandal, C.; Marshall, F.H.; Cooke, R.M. Structures of Human A 1 and A 2A Adenosine Receptors with Xanthines Reveal Determinants of Selectivity. Structure 2017, 25, 1275–1285.e4. [Google Scholar] [CrossRef] [PubMed]
- Lebon, G.; Warne, T.; Edwards, P.C.; Bennett, K.; Langmead, C.J.; Leslie, A.G.W.; Tate, C.G. Agonist-Bound Adenosine A2A Receptor Structures Reveal Common Features of GPCR Activation. Nature 2011, 474, 521–525. [Google Scholar] [CrossRef] [PubMed]
- Ozeir, M.; Huyet, J.; Burgevin, M.-C.; Pinson, B.; Chesney, F.; Remy, J.-M.; Siddiqi, A.R.; Lupoli, R.; Pinon, G.; Saint-Marc, C.; et al. Structural Basis for Substrate Selectivity and Nucleophilic Substitution Mechanisms in Human Adenine Phosphoribosyltransferase Catalyzed Reaction. J. Biol. Chem. 2019, 294, 11980–11991. [Google Scholar] [CrossRef] [PubMed]
- Ludlow, R.F.; Verdonk, M.L.; Saini, H.K.; Tickle, I.J.; Jhoti, H. Detection of Secondary Binding Sites in Proteins Using Fragment Screening. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 15910–15915. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.R.; Lowe, E.D.; Petri, E.; Skamnaki, V.; Antrobus, R.; Johnson, L. Cyclin B and Cyclin A Confer Different Substrate Recognition Properties on CDK2. Cell Cycle 2007, 6, 1350–1359. [Google Scholar] [CrossRef] [PubMed]
- Wesolowski, J.R.; Rajdev, P.; Mukherji, S.K. Temozolomide (Temodar). AJNR Am. J. Neuroradiol. 2010, 31, 1383–1384. [Google Scholar] [CrossRef] [PubMed]
- Thorn, C.F.; Oshiro, C.; Marsh, S.; Hernandez-Boussard, T.; McLeod, H.; Klein, T.E.; Altman, R.B. Doxorubicin Pathways. Pharmacogenet. Genomics 2011, 21, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Ye, B.; Wang, P.; Yao, F.; Zhang, C.; Yu, G. Overview of MicroRNA-199a Regulation in Cancer. Cancer Manag. Res. 2019, 11, 10327–10335. [Google Scholar] [CrossRef]
- Naseem, M.; Othman, E.M.; Fathy, M.; Iqbal, J.; Howari, F.M.; AlRemeithi, F.A.; Kodandaraman, G.; Stopper, H.; Bencurova, E.; Vlachakis, D.; et al. Integrated Structural and Functional Analysis of the Protective Effects of Kinetin against Oxidative Stress in Mammalian Cellular Systems. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef]
- Spíchal, L.; Kryŝtof, V.; Paprskářová, M.; Lenobel, R.; Stýskala, J.; Binarová, P.; Cenklová, V.; De Veylder, L.; Inzé; , D. ; Kontopidis, G.; et al. Classical Anticytokinins Do Not Interact with Cytokinin Receptors but Inhibit Cyclin-Dependent Kinases. J. Biol. Chem. 2007, 282, 14356–14363. [Google Scholar] [CrossRef] [PubMed]
- Young, G.-H.; Lin, J.-T.; Cheng, Y.-F.; Ho, C.-F.; Kuok, Q.-Y.; Hsu, R.-C.; Liao, W.-R.; Chen, C.-C.; Chen, H.-M. Modulation of Adenine Phosphoribosyltransferase-mediated Salvage Pathway to Accelerate Diabetic Wound Healing. FASEB J. 2021, 35. [Google Scholar] [CrossRef] [PubMed]
- Gessi, S.; Bencivenni, S.; Battistello, E.; Vincenzi, F.; Colotta, V.; Catarzi, D.; Varano, F.; Merighi, S.; Borea, P.A.; Varani, K. Inhibition of A2A Adenosine Receptor Signaling in Cancer Cells Proliferation by the Novel Antagonist TP455. Front. Pharmacol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Merighi, S.; Mirandola, P.; Milani, D.; Varani, K.; Gessi, S.; Klotz, K.-N.; Leung, E.; Baraldi, P.G.; Borea, P.A. Adenosine Receptors as Mediators of Both Cell Proliferation and Cell Death of Cultured Human Melanoma Cells. J. Invest. Dermatol. 2002, 119, 923–933. [Google Scholar] [CrossRef] [PubMed]
- Spencer, S.L.; Cappell, S.D.; Tsai, F.-C.; Overton, K.W.; Wang, C.L.; Meyer, T. The Proliferation-Quiescence Decision Is Controlled by a Bifurcation in CDK2 Activity at Mitotic Exit. Cell 2013, 155, 369–383. [Google Scholar] [CrossRef] [PubMed]
- White, E.Z.; Pennant, N.M.; Carter, J.R.; Hawsawi, O.; Odero-Marah, V.; Hinton, C.V. Serum Deprivation Initiates Adaptation and Survival to Oxidative Stress in Prostate Cancer Cells. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef]
- Gong, X.; Liang, Z.; Yang, Y.; Liu, H.; Ji, J.; Fan, Y. A Resazurin-Based, Nondestructive Assay for Monitoring Cell Proliferation during a Scaffold-Based 3D Culture Process. Regen. Biomater. 2020, 7, 271–281. [Google Scholar] [CrossRef]
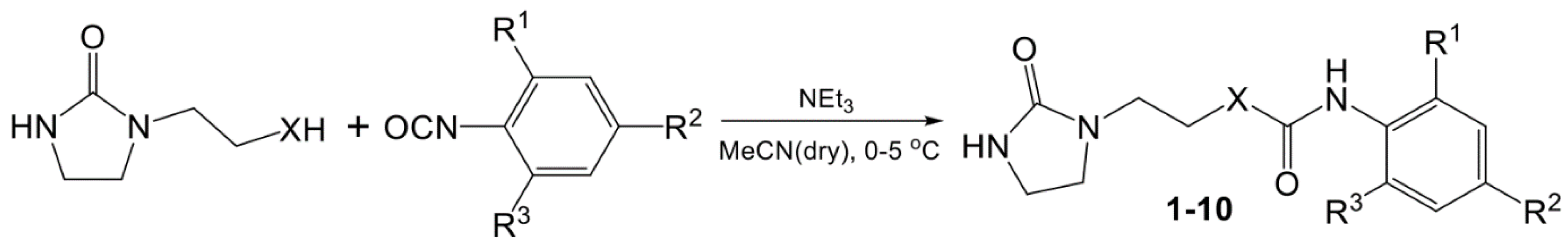


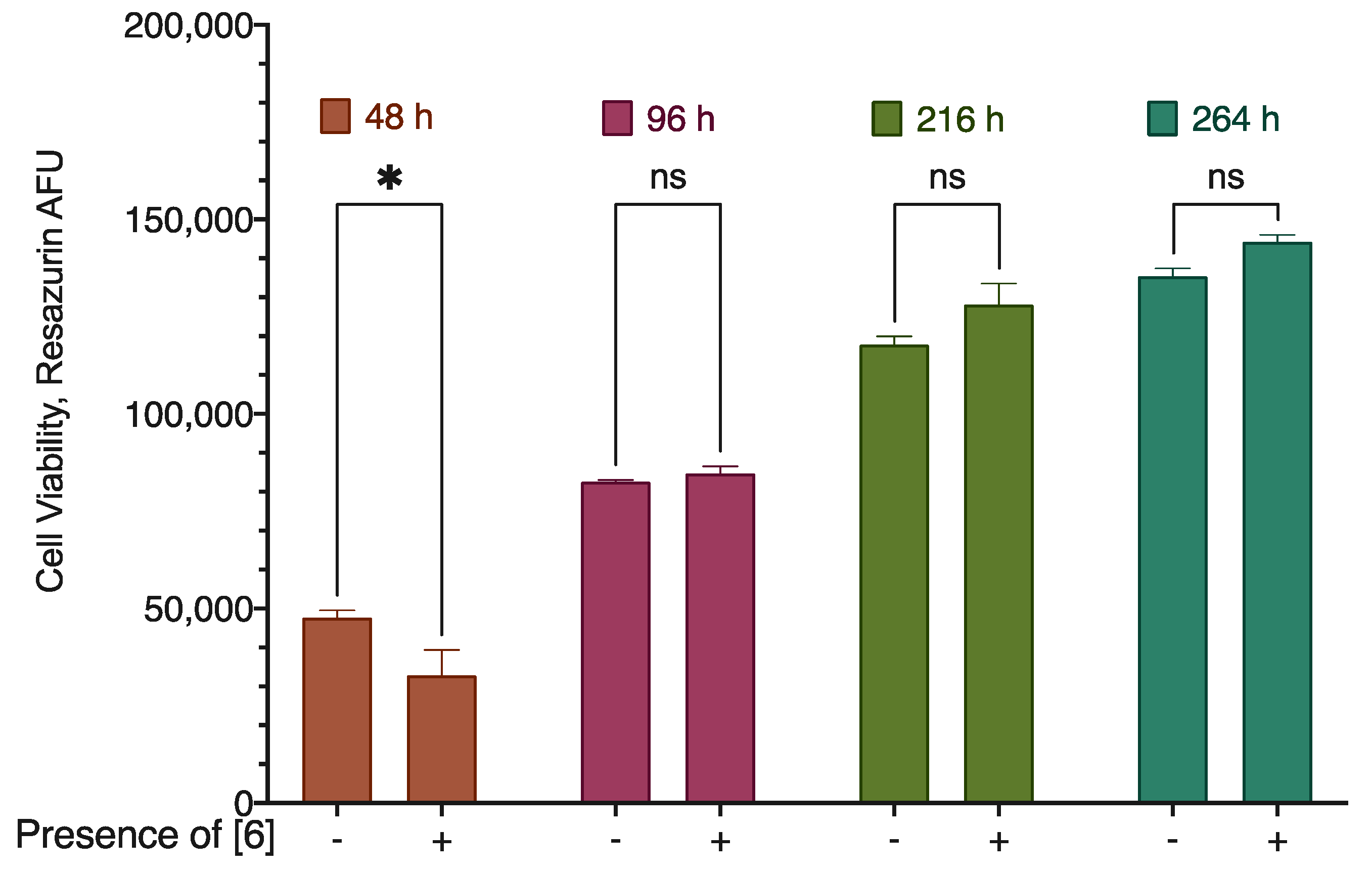
| Compound | R1 | R2 | R3 | X | Yield, % |
|---|---|---|---|---|---|
| 1 | CH3 | - | CH3 | NH | 59 |
| 2 | CH3 | - | CH3 | O | 30 |
| 3 | - | CH3 | - | NH | 31 |
| 4 | - | CH3 | - | O | 36 |
| 5 | C2H5 | - | - | NH | 62 |
| 6 | C2H5 | - | - | O | 50 |
| 7 | CH3 | CH3 | - | NH | 58 |
| 8 | - | OCH3 | - | NH | 35 |
| 9 | - | OCH3 | - | O | 18 |
| 10 | - | COOCH3 | - | O | 30 |
| Bj-5ta | MDA-MB-231 | U-87 MG | A-375 | SH-SY5Y | |
|---|---|---|---|---|---|
| Cell viability, % (95% C.I.) | |||||
| 6 | 96.8 (91.39-102.2) | 86.26 (62.59-109.94) | 102.83 (83.22-122.43) | 78.46 (46.77-110.16) | 92.2 (39.62-144.79) |
| 8 | 100.85 (55.45-146.25) | 87.15 (65.36-108.94) | 92.77 (-23.93-209.49) | 97.07 (74.48-119.67) | 90.27 (35.67-144.87) |
| MDA-MB-231 | U-87 MG | SH-SY5Y | |
|---|---|---|---|
| Cell viability, % (95% C.I.) | |||
| 6 | 83.25 (78.1-96.6) | 85.87 (80.19-99.93) | 77.44 (55.52-99.36) |
| 8 | 94.71 (56.93-132.48) | 107.81 (91.59-124.04) | 85.13 (74.19-96.06) |
| Doxorubicin, µm | (6), µM | CI | ||
|---|---|---|---|---|
| MDA-MB-231, Viability | MDA-MB-231,Cell death | U-87 MG, Viability | ||
| 10.0 | 90.0 | 1.50 | 1.81 | 1.09285 |
| 10.0 | 100.0 | 1.17 | 1.68 | 1.09440 |
| 75.0 | 90.0 | 1.35 | 1.40 | 0.32642 |
| 75.0 | 100.0 | 1.22 | 1.90 | 0.53187 |
| 100.0 | 90.0 | 1.65 | 0.79 | 0.57710 |
| 100.0 | 100.0 | 0.17 | 0.88 | 0.60758 |
| Temozolomide, µm | (6), µM | CI |
|---|---|---|
| 500.0 | 80.0 | 1.51722 |
| 700.0 | 90.0 | 1.68461 |
| 900.0 | 100.0 | 1.82996 |
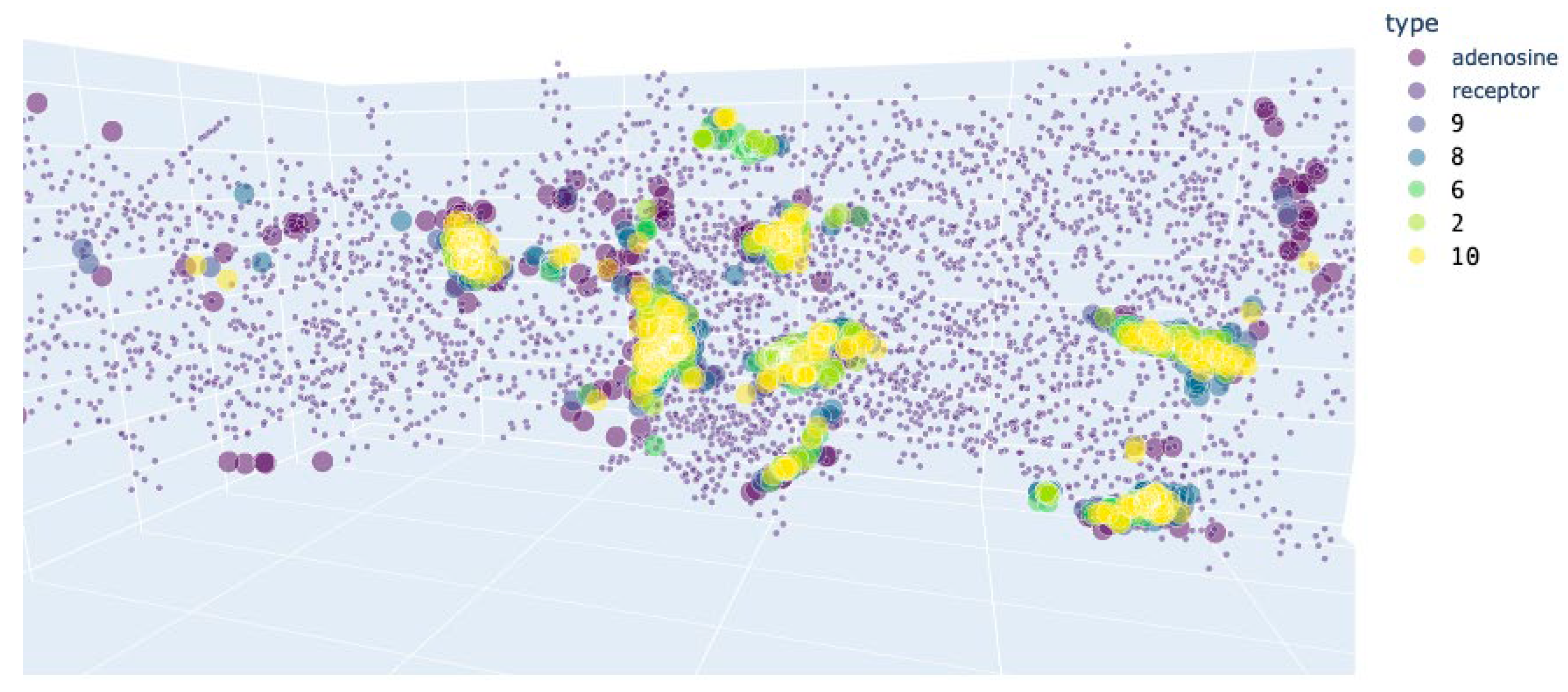
| A2AR | A2AR (active) + Adenosine (activator) | A2AR (inactivated) + Caffeine (blocker) | ||||
|---|---|---|---|---|---|---|
| Mean±SEM | p | Mean±SEM | p | Mean±SEM | p | |
| Adenosine | 5.32±0.05 | - | 3.06±0.15 | - | 5.17±0.06 | - |
| 9 | 5.11±0.04 | 0.340 | 3.87±0.11* | 0.000 | 5.09±0.04 | 1.000 |
| 8 | 5.53±0.04 | 0.310 | 4.67±0.09* | 0.000 | 5.26±0.04 | 1.000 |
| 7 | 5.79±0.03* | 0.000 | 4.66±0.19* | 0.000 | 5.75±0.04* | 0.000 |
| 1 | 5.8±0.04* | 0.000 | 4.96±0.09* | 0.000 | 5.71±0.04* | 0.000 |
| 5 | 5.67±0.03* | 0.000 | 4.8±0.14* | 0.000 | 5.61±0.03* | 0.000 |
| 6 | 5.42±0.03 | 0.999 | 3.54±0.2 | 0.072 | 5.38±0.04 | 0.166 |
| 2 | 5.56±0.04 | 0.078 | 3.66±0.18* | 0.002 | 5.59±0.03* | 0.000 |
| 10 | 5.28±0.04 | 1.000 | 4.37±0.11* | 0.000 | 5.2±0.03 | 1.000 |
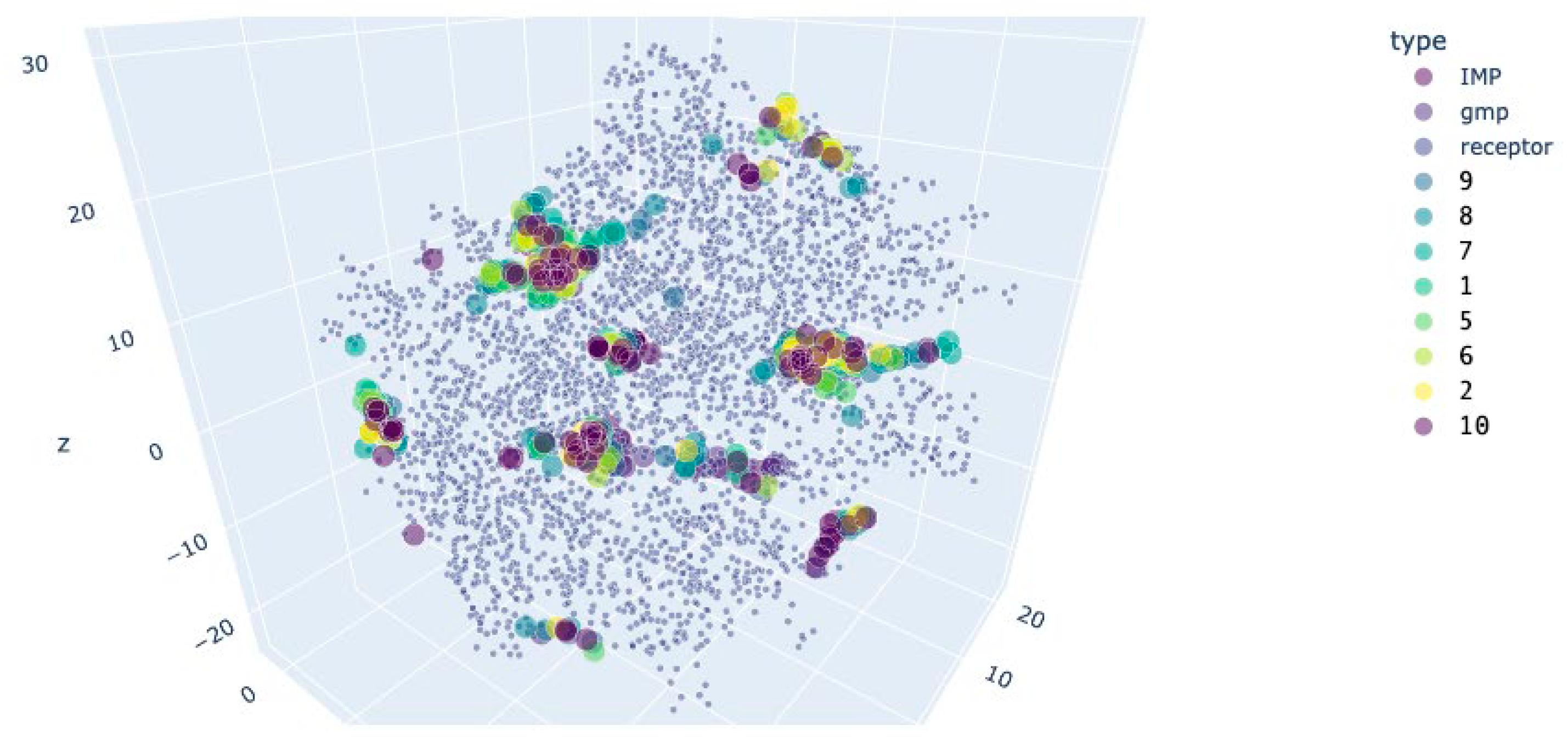
| APRT+GMP (active conformation) | APRT+IMP (inactivated conformation) | |||
|---|---|---|---|---|
| Mean±SEM | P | Mean±SEM | p | |
| IMP (blocker) | 7.32±0.08 | - | 7.69±0.07 | - |
| GMP (activator) | 6.23±0.06 | - | 6.24±0.07 | - |
| 9 | 5.33±0.06* | 0.000 | 5.55±0.06* | 0.000 |
| 8 | 5.63±0.05* | 0.000 | 5.7±0.04* | 0.000 |
| 7 | 6.22±0.06 | 1.000 | 6.27±0.06 | 1.000 |
| 1 | 6.1±0.07 | 0.918 | 6.4±0.06 | 0.720 |
| 5 | 5.95±0.05* | 0.044 | 6.0±0.05 | 0.088 |
| 6 | 5.93±0.05* | 0.020 | 5.65±0.07* | 0.000 |
| 2 | 6.29±0.06 | 0.999 | 6.21±0.06 | 1.000 |
| 10 | 5.4±0.05* | 0.000 | 5.57±0.05* | 0.000 |
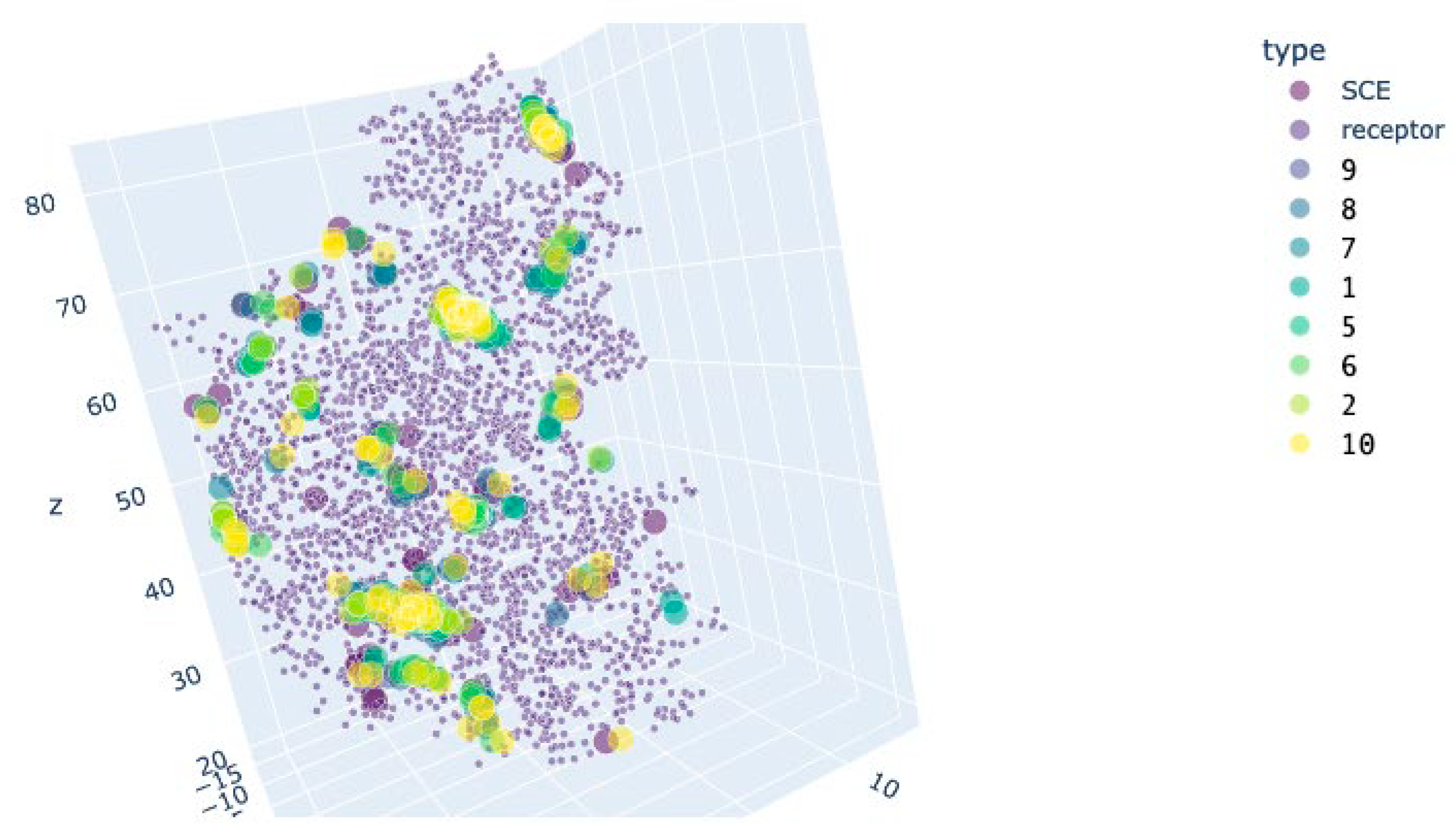
| CDK2 | CDK2+Cyclin D (active conformation) | |||
|---|---|---|---|---|
| Mean±SEM | p | Mean±SEM | p | |
| SCE (blocker) | 5.03±0.05 | - | 5.44±0.04 | 0.963 |
| 9 | 5.18±0.06 | 0.849 | 5.3±0.04* | 0.004 |
| 8 | 5.71±0.06* | 0.000 | 5.9±0.03* | 0.000 |
| 7 | 6.12±0.09* | 0.000 | 6.32±0.05* | 0.000 |
| 1 | 5.91±0.07* | 0.000 | 6.1±0.05* | 0.000 |
| 5 | 6.36±0.09* | 0.000 | 6.22±0.04* | 0.000 |
| 6 | 5.36±0.07* | 0.025 | 5.78±0.05 | 0.133 |
| 2 | 5.99±0.08* | 0.000 | 5.95±0.04* | 0.000 |
| 10 | 5.24±0.07 | 0.495 | 5.56±0.04 | 1.000 |
| Protein | Configuration Variant | x | y | z |
|---|---|---|---|---|
| A2AR 5mzj | 1 | −17.629 | −30.760 | 18.168 |
| 2 | −4.629 | −50.760 | 18.168 | |
| 3 | −17.629 | 6.760 | 18.168 | |
| A2AR 2ydo | 1 | −23.602 | 10.545 | −25.256 |
| 2 | −23.602 | 20.545 | −25.256 | |
| 3 | −4.629 | −50.760 | 18.168 | |
| A2AR 5mzp | 1 | −16.417 | −40.474 | 18.316 |
| 2 | −16.417 | 5.474 | 18.316 | |
| 3 | −1.417 | −50.474 | 18.316 | |
| APRT 6hgs | 1 | 22.572 | −3.082 | 4.313 |
| APRT 6hgr | 1 | 23.667 | −7.067 | 5.057 |
| APRT 6hgp | 1 | −24.642 | 0.247 | 1.919 |
| CDK2 5fp5 | 1 | 29.547 | 4.964 | 49.678 |
| CDK2 2jgz | 1 | 55.623 | 20.504 | −10.503 |
| 2 | 38.623 | 20.504 | 5.503 | |
| 3 | 38.623 | 20.504 | −30.503 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





