1. Introduction
Cellular metabolism includes various biochemical processes such as glucose metabolism, fatty acid metabolism, amino acid metabolism, and mitophagy, which are closely related to signal transduction [
1]. Cellular metabolism not only provides energy for cell growth and proliferation, but also regulates cell fate through transcriptional and epigenetic regulatory mechanisms, thereby affecting cell survival, proliferation, and stem cell function [
2]. Stem cells can maintain an undifferentiated state, possess self-renewal potential and pluripotency [
3]. Research has found that energy metabolism is crucial for maintaining stem cell homeostasis and regulating biological activities, and stem cells can also maintain metabolic homeostasis through various mechanisms [
4].
Glucose metabolism is considered to be the core process of energy metabolism in stem cells, which has a significant impact on the survival, proliferation and self-renewal of stem cells [
5]. Unlike other types of cells, most stem cells rely mainly on glycolysis to obtain the required energy [
6]. The self-renewal of stem cells are highly dependent on amino acid metabolism, including threonine metabolism, carbon metabolism, methionine metabolism, glutamine metabolism, and L-proline metabolism [
7]. Mitophagy is an important regulatory factor for metabolic homeostasis and a necessary condition for alleviating stem cell stress [
8]. In addition, fatty acid metabolism is also crucial for the pluripotency of stem cells [
9,
10].
In this article, we summarize the latest research progress on the correlation between glucose metabolism, amino acid metabolism, fatty acid metabolism, and mitophagy with stem cell function, and discuss in detail how nutritional sensors promote the transformation of stem cell fate. The regulatory mechanism of stem cells is very complex. We rely on the dynamic changes in metabolism to study the signaling regulatory pathways of stem cells, which will help regulate the self-renewal and lineage specific differentiation of stem cells through energy metabolism, providing new theoretical and empirical support for the development of regenerative medicine.
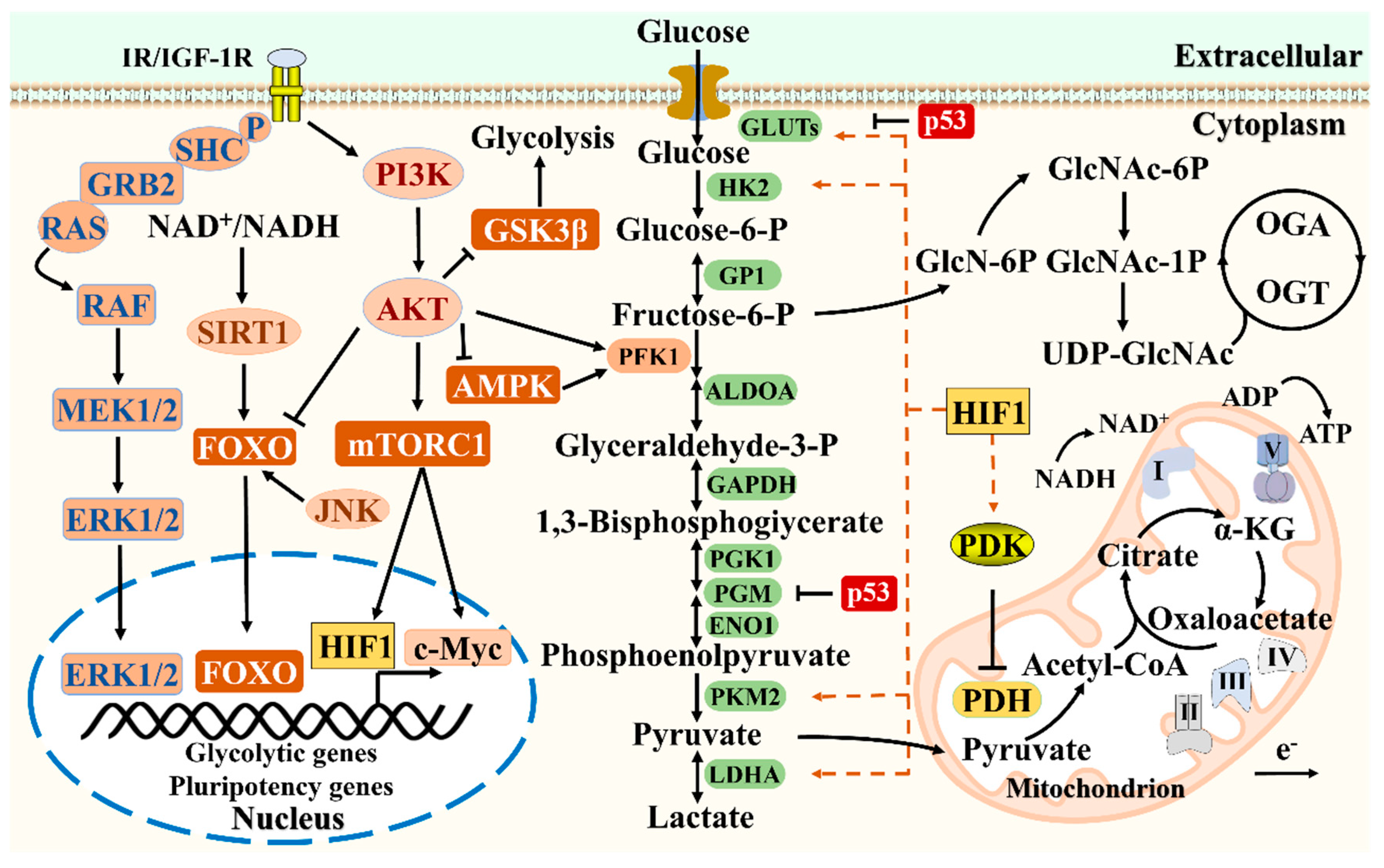
2. Glucose Metabolism Determines the Fate and Function of Stem Cells
Research has shown that glucose metabolism has a significant impact on the survival, proliferation, self-renewal, and differentiation potential of stem cells [
11]. Glucose metabolism mainly involves oxidative phosphorylation (OXPHOS), glycolysis, pentose phosphate pathway (PPP), etc. OXPHOS and Glycolysis are the main sources of ATP in stem cells [
12]. PPP and Glycolysis produce precursors for nucleotide and amino acid biosynthesis, thereby promoting rapid stem cell proliferation [
13]. We will discuss the relationship between glucose metabolism and stem cell function, describe the metabolic transition mechanism from somatic cell reprogramming to induced pluripotent stem cells (iPSCs), and the mechanism of action of hypoxia inducible factors (HIF) in stem cells (
Figure 1).
2.1. Oxidative Phosphorylation
Research has found that mitochondria in pluripotent stem cells (PSCs) are in a hyperactive state, and OXPHOS is the main production mode of PSCs. OXPHOS coupled with the hexosamine biosynthesis pathway (HBP) regulates self-renewal of PSCs through aminoglycosylation of pluripotent factors [
14]. OXPHOS maintains bioenergy homeostasis by connecting to the tricarboxylic acid (TCA) cycle pathway, which is a metabolite of the TCA cycle, such as acetyl CoA, α-Ketoglutarate (α-KG), nicotinamide adenine dinucleotide (NAD), and S-adenosylmethionine (SAM) can diffuse through nuclear pores and play important roles in regulating epigenetics and transcription [
15]. Phosphate serine aminotransferase 1 (Psat1) is a target protein associated with the pluripotent factor Oct4/Sox2/Nanog. Psat1 can maintain intracellular α-KG levels and is crucial for the self-renewal and pluripotency of embryonic stem cells (ESCs) [
16]. TCA cycle enzymes such as pyruvate dehydrogenase (PDH), pyruvate carboxylase (PCB), aconitase (ACO), and isocitrate dehydrogenase 3 (IDH3A) play a crucial role in pluripotency regulation [
17]. Uncoupling protein 2 (UCP2) is a mitochondrial inner membrane transport protein, and the expression of UCP2 in PSCs maintains low levels of reactive oxygen species (ROS) produced by OXPHOS [
18]. In addition, the reprogramming process is accompanied by a significant decrease in mitochondrial quantity, mtDNA copy number, and mitochondrial density. Mitochondria degenerate from a slender tubular morphology rich in cristae to a spherical structure lacking cristae [
19]. Due to the fact that mitochondrial cristae are the main site of OXPHOS, the reduction of cristae facilitates the transition of OXPHOS to Glycolysis metabolism [
20]. At present, our understanding of OXPHOS regulating the fate and function of stem cells is limited, but it is meaningful for OXPHOS to serve as a target for stem cell therapy in the future.
2.2. Glycolysis
Although glycolysis has a lower efficiency in producing ATP compared to OXPHOS, stem cells rely more on glycolysis. Glycolysis can quickly provide a large amount of ATP to meet the needs of stem cell life activities [
21]. Glycolysis can produce a large number of metabolic precursors, preparing for the subsequent biogenesis of nucleic acids, amino acids, and fatty acids [
19]. Compared with ESCs, iPSCs exhibit a high metabolic flux similar to cancer cells, known as the Warburg effect, which can be established by reducing the activity and expression of the key subunit β-F1-ATPase of mitochondrial ATP synthase [
22,
23]. The high metabolic flux of Glycolysis is a common feature of many stem cells, and ESCs achieve high metabolic flux of glycolysis by regulating key glycolytic enzymes such as hexokinase 2 (HK2) and pyruvate kinase 2 (PKM2) [
24]. The high metabolic flux of Glycolysis can enhance the levels of Acetyl-CoA and lactate, thereby enhancing H3K27Ac acetylation and H3K18la lactate at pluripotent gene loci and promoting somatic reprogramming [
25].
The core pluripotent factors Oct4, Sox2, and Nanog coordinate the regulation of Glycolysis and participate in somatic reprogramming [
26]. The core pluripotent factor Oct4 can directly regulate the promoter activity of HK2 and PKM2 in ESCs, thereby regulating Glycolysis to maintain stem cell characteristics [
24]. In the early stages of reprogramming, upregulation of Glycolysis leads to epigenetic changes in iPSCs [
4]. The metabolic transition of iPSCs from OXPHOS to glycolysis occurs prior to the expression of pluripotent markers. The expression of glycolysis related genes GLUT, HK2, PKM2, and lactate dehydrogenase A (LDHA) significantly increases during the first week of reprogramming, while the expression of pluripotent genes Nanog, Oct4, and Sox2 remains at low levels [
27,
28]. The transcription factor Glis1 directly binds and opens the chromatin of the Glycolysis gene, upregulating Glycolysis to promote pluripotency induction [
29]. PDK1 is a key factor in the association between Glycolysis and the TCA cycle. It has been reported that using PDK1 to inhibit PDH activity can increase Glycolysis levels, thereby improving the reprogramming efficiency of iPSCs [
19]. PDK1 activator PS48 can prevent pyruvate from entering the TCA cycle, induce metabolic transition, and improve reprogramming efficiency [
30]. In addition, miR-31 is a type of non-coding small RNA that participates in metabolic regulation during reprogramming by inhibiting succinate dehydrogenase complex subunit A (SDHA) [
31]. Therefore, Glycolysis is a key factor in somatic reprogramming, and glucose metabolism is the switch that determines stem cell self-renewal.
2.3. Metabolic Changes during Somatic Reprogramming
The key events in the process of somatic reprogramming mainly include the inhibition of somatic genes, activation of pluripotent genes, mesenchymal epithelial transition (MET), and metabolic transition from OXPHOS to Glycolysis [
32]. After the introduction of reprogramming factors in the early stages of reprogramming, estrogen related nuclear receptors (ERRα/ERRγ) andperoxisome proliferator-activated receptor-gamma coactivator-1α (PGC-1α) were instantaneously upregulated, inducing transient high-energy metabolism, mainly including transient bursts of OXPHOS and glycolysis, which are necessary for reprogramming somatic cells into iPSCs [
33]. The transcription factor NRF2 protects cells from oxidative stress during reprogramming, and overexpression or knockdown of NRF2 reduces reprogramming efficiency [
34]. This outbreak in the early stages of reprogramming resulted in a surge in ROS production, mediating an increase in NRF2 activity, which can promote metabolic transition by activating hypoxia inducible factor 1 (HIF1) [
35].
The transcription regulatory factor HIF1 can participate in gene transcription of stem cell metabolism, and the hypoxic environment provided by HIF1 is necessary for stem cell survival, maintenance of redox homeostasis, and pluripotency (
Figure 2) [
36]. Under hypoxic conditions, HIF1 upregulates Glycolysis by encoding glucose transporter protein (GLUT), phosphofructose kinase (PFK), aldolase (ALDOA), phosphoglycerate kinase-1 (PGK1), enolase (ENO), pyruvate dehydrogenase kinase 1 (PDK1), and LDHA [
37]. The upregulation of HIF1 leads to an increase in PDK1 expression, which in turn inactivates the PDH complex, maintaining the high glycolytic metabolism of ESCs [
38]. During the reprogramming process, HIF1 controls the transcription of many target genes to initiate metabolic changes. HIF1 upregulates the transcriptional regulatory enzymes of Glycolysis genes such as HK2, PKM2, and LDHA, shifting the metabolic mode from OXPHOS to Glycolysis and improving reprogramming efficiency [
39]. In addition, the transcription regulatory factor c-Myc can significantly upregulate the expression levels of Glycolysis and OXPHOS related enzymes, and c-Myc and HIF1 cooperate to induce metabolic transition during iPSCs reprogramming [
40]. The PI3K/AKT signaling pathway regulates glucose metabolism by stabilizing HIF1, but further research is needed to determine to what extent the PI3K/AKT signaling pathway is involved in HIF1 dependent hypoxia induced glucose metabolism [
41]. Considering that HIF1 maintains high glycolytic flux in the later stages of reprogramming, early induction of HIF1 stabilizers and related glycolytic modulators may help establish pluripotency and have the potential for efficient production of iPSCs [
42]. In the early stage of reprogramming, the transcription regulatory factor HIF2α promotes the expression of pluripotent markers Oct4, Sox2, and Nanog, regulates Wnt/β-catenin signaling, and improves reprogramming efficiency by inhibiting p53 [
43]. HIF2α upregulates the expression of tumor necrosis factor related apoptosis inducing ligand (TRAIL) in the late stage of reprogramming, and TRAIL inhibits the generation of iPSCs by inhibiting the activity of apoptotic caspase3 [
44].
Regarding metabolic changes during reprogramming, T cell leukemia/lymphoma protein 1 (TCL1) is a downstream effector of the pluripotent gene Klf4. Klf4 enhances glycolysis by upregulating TCL1 expression, while TCL1 inhibits mitochondrial biogenesis and OXPHOS by inhibiting mitochondrial polynucleoside phosphorylase (PnPase) (
Figure 1) [
45]. TCL1 promotes the transition from OXPHOS to Glycolysis in an AKT dependent manner, thereby improving reprogramming efficiency [
46]. The circadian rhythm gene 1 (CRY1) is crucial for regulating biological rhythms. The metabolic transformation of iPSCs leads to the accumulation of CRY1, which can maintain the metabolic characteristics, self-renewal ability, and improve reprogramming efficiency of iPSCs [
47]. Signal Transduction and Transcription Activating Factor 3 (STAT3), as a signaling transcription protein, can regulate cell proliferation and survival. The metabolic transition of iPSCs can be activated by STAT3, which plays a crucial role in the maintenance of pluripotency and somatic reprogramming of ESCs [
48]. RNA binding protein LIN28 is an important regulatory factor in the reprogramming process, which can promote the metabolic transition from immature state to initiating pluripotent state, maintain low mitochondrial function, and regulate One-C metabolism, nucleotide metabolism, etc. [
49]. Considering the role of LIN28 and STAT3 in regulating energy metabolism, it is meaningful to study their interaction in the process of reprogramming metabolic transition.
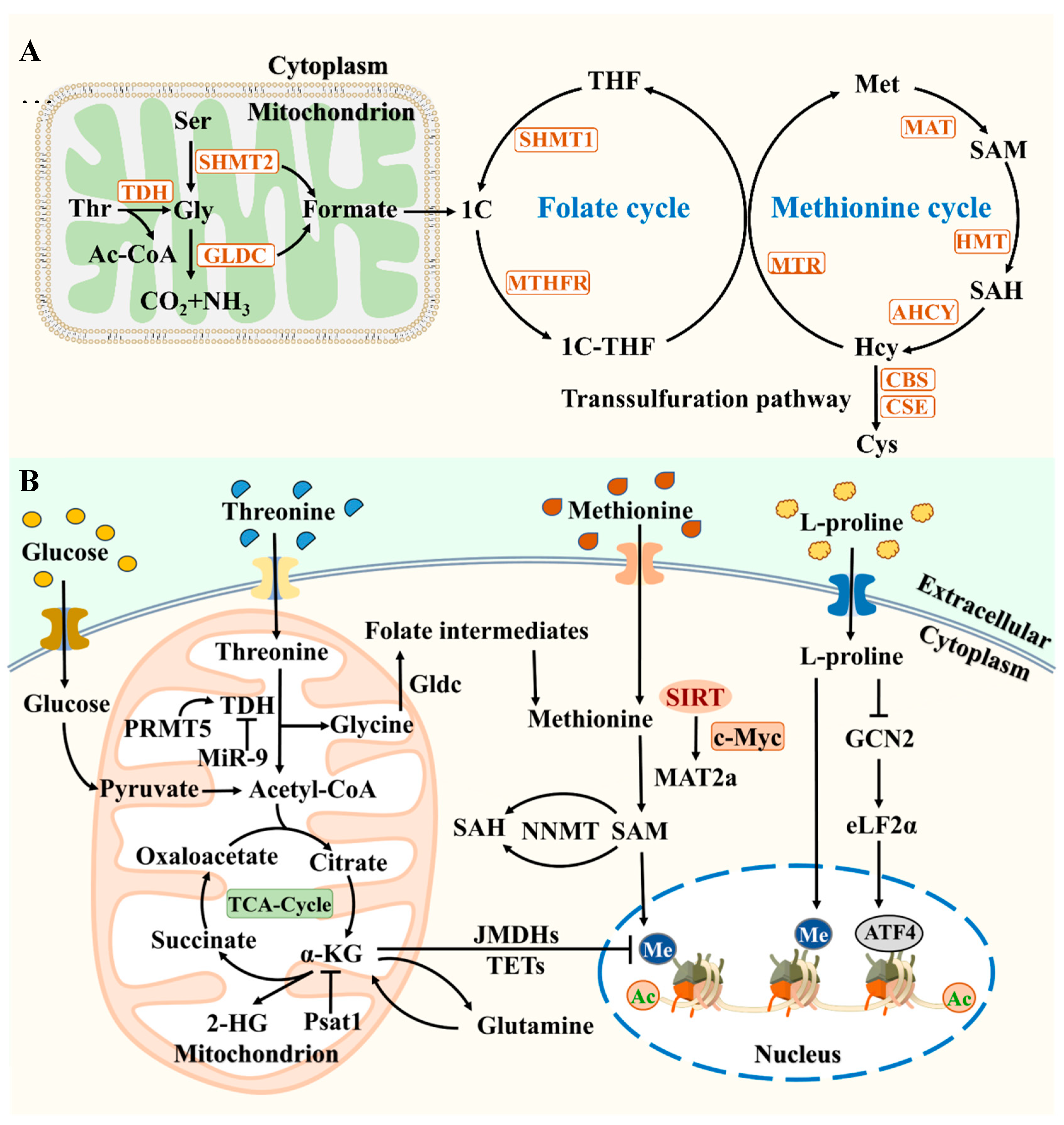
3. Amino Acid (AA) Metabolism Regulates Stem Cell Function
Amino acid metabolism is closely related to epigenetics, organelle remodeling, and cellular signal transduction pathways, and can participate in regulating the pluripotency of stem cells [
50]. The self-renewal of stem cells is highly dependent on AA metabolism, including threonine metabolism, One-C metabolism, methionine cycle, glutamine metabolism, and L-Pro metabolism, indicating that AA metabolism is essential for the growth of stem cells (
Figure 2) [
51,
52]. It is worth noting that mESCs mainly rely on the breakdown metabolism of threonine, while hESCs achieve self-renewal through the breakdown metabolism of methionine [
53,
54]. We summarize the regulation of stem cell pluripotency, self-renewal, and somatic reprogramming by AA metabolism, and these research findings reveal the close relationship between AA metabolism and stem cells.
3.1. Threonine Metabolism
Current research suggests that thyroid metabolism plays an important role in mESCs pluripotency and self-renewal [
52]. Wang et al. used a method of removing 20 amino acids one by one and found that after removing threonine for 1 to 4 days, the expression level of pluripotent marker genes decreased, and the high-throughput metabolic state of mESCs was highly dependent on threonine metabolism [
55]. Threonine mainly regulates the proliferation of mESCs through signaling pathways such as PI3K/AKT, mTOR, MAPK, p70S6K, and 6E-BP1 [
56]. Threonine metabolism mediated by threonine dehydrogenase (TDH) is a specific metabolic trait of mESCs. As a novel positive regulatory factor in somatic reprogramming, inducing TDH expression can improve reprogramming efficiency [
57]. Non-coding small RNA miR-9 negatively regulates the expression of TDH after transcription, inhibiting reprogramming efficiency. Protein arginine methyltransferase (PRMT5) plays an important role in regulating DNA repair, cell cycle, and transcriptional regulation processes, improving reprogramming efficiency by positively regulating TDH expression [
57]. SATB homeobox 1 (SATB1) is a tissue-specific nuclear matrix binding protein that promotes trophoblast stem cell renewal by regulating the expression of TDH in trophoblast stem cells [
53]. The driving protein binding protein (KBP) plays an important role in neural development. TDH hydrolyzes threonine into acetyl-CoA and glycine, resulting in acetyl-CoA acetylation of KBP. The KBP protein further hydrolyzes, limiting the accumulation of mitochondria in mESCs to maintain their optimal adaptability [
58]. Glycine generates folate intermediates through the catalysis of glycine decarboxylase (Gldc), enhancing One-C metabolism and promoting purine biosynthesis [
59].
In addition, thyroid metabolism maintains the pluripotency of mESCs by regulating SAM levels and histone H3 lysine 4 trimethylation (H3K4me3) [
52]. The consumption of threonine or TDH leads to a decrease in SAM consumption and H3K4me3 levels, resulting in slower stem cell growth. Combined supplementation of glycine and pyruvate restores the supply of Acetyl CoA, rescuing H3K4me3 levels [
54]. Unlike the case of mESCs, the absence of threonine in hESCs does not significantly affect pluripotency, as the TDH gene is a non-functional pseudogene in humans [
60]. These research results indicate that thyroid metabolism is crucial for the function and reprogramming process of mESCs, which helps us to gain a deeper understanding of signaling pathways related to stem cell survival and pluripotency.
3.2. One-C Metabolism and Methionine Cycle
One-C metabolism is crucial for various biological processes, including methylation, nucleic acid and amino acid synthesis, and can directly control the levels of methionine, serine, and glycine [
51]. One-C metabolism is involved in regulating cell proliferation and embryonic development, and is a universal substrate for all protein methylation reactions in cells. It mainly involves the folate cycle, methionine cycle, and trans sulfuration cycle [
12]. The folate cycle is a major contributor to amino acid homeostasis, promoting nucleotide synthesis and demethylation of homocysteine (Hcy) to form methionine and SAM, thereby maintaining the methylation potential of cells [
59]. In addition, folate cycle mediated NADPH production is crucial for mitochondrial redox homeostasis and cell proliferation [
61]. Folic acid maintains the immature state of ESCs under the cultivation conditions of GSK-3 inhibitor CHIR99021 for a long time, supporting pluripotency and reprogramming by regulating the LIF/STAT3 and MAPK/ERK signaling pathways [
62]. Transient deficiency of folic acid promotes demethylation of Oct4 and Nanog promoters in MEF, thereby improving reprogramming efficiency [
63].
The metabolism cycle is closely related to One-C metabolism, which is crucial for redox homeostasis, epigenetics, and self-renewal of stem cells. Methionine is catalyzed by methionine adenosine transferase 2a (MAT2a) to form SAM, which is independent of threonine metabolism [
64]. The dependence of stem cells on methionine is related to multiple mechanisms. SIRT1 is a conserved mammalian NAD dependent protein deacetylase, and it coordinates with the transcription factor c-Myc to regulate the expression of MAT2a, combining methionine metabolism with cellular energy status, which is crucial in mESCs function and embryonic development [
65]. PGC-1α is the main regulator of lipid metabolism and fatty acid oxidation (FAO), and PPAR-α triggers the expression of pluripotent reprogramming genes. Depriving cells of methionine reduces SIRT1 levels, leading to energy metabolism disorders in PGC-1α/PPAR-α [
66]. As a sensor for methionine metabolism, SAM consumption leads to a rapid decrease in SAM, activating p53/p38 signaling and reducing Nanog expression. SAM is closely related to the maintenance of PSC pluripotency and cell survival in methionine metabolism [
60]. Methionine metabolism maintains a multi energy network with histone markers, and transient consumption of methionine triggers rapid metabolic changes [
67]. High levels of DNA methylation play a barrier role in somatic reprogramming, and reports suggest that SAM exists at high levels in iPSCs. Methionine metabolism regulates the pluripotency of PSCs through zinc mobilization [
7]. SAM releases methyl groups and produces S-adenosine homocysteine (SAH). Nicotinamide N-methyltransferase (NNMT) controls the conversion of SAM-SAH. The epigenetic changes caused by NNMT levels inhibit the Wnt pathway and electron transport chain activity, activate the HIF pathway and lipid synthesis, and are crucial for the metabolic transition of hESCs [
68]. Polycomb inhibition complex 2 (PRC2) is an important epigenetic modifying enzyme composed of Suz12, EED, and EZH2 [
69]. Although it is known that NNMT regulates the substrate level of PRC2, the factors regulating the position control of this methylation and its function in pluripotency have not yet been determined. Although these studies emphasize the crucial role of One-C metabolism and metabolic cycle in regulating stem cell fate determination, further research is needed to investigate the direct relationship between them and epigenetic changes.
3.3. Glutamine Metabolism and L-Pro Metabolism
Glutamine is predicted to be a positive regulator of stem cells and the main energy source of OXPHOS in hPSCs, which is essential for maintaining the dryness of hPSCs [
70]. Glutamine is essential for maintaining intracellular levels of glutathione and low levels of ROS, and high levels of glutamine metabolism are crucial for preventing the degradation of the pluripotent transcription factor Oct4 [
71]. Glutamine is different from glucose and lipids. The main function of glutamine metabolism is to produce various metabolic intermediates involved in energy supply [
72]. Supplementing with glutamine and glucose can provide fuel for de novo synthesis of nucleotides [
5]. Glutamine is converted to glutamic acid through glutaminase and further converted to α-KG. The transport of glutamic acid into the cytoplasm facilitates the synthesis of glutathione and amino acids [
73]. α-KG is a substrate for histone demethylases (JHDMs) and DNA demethylation translocatases (TETs), which affect the embryonic development of mESCs by regulating metabolism and epigenetics [
74]. Juvenile mESCs utilize glucose and glutamine catabolism to maintain high levels of α-KG, which helps maintain low levels of H3K27me3 in cells and increases TET dependent DNA demethylation, regulating the expression of pluripotency related genes [
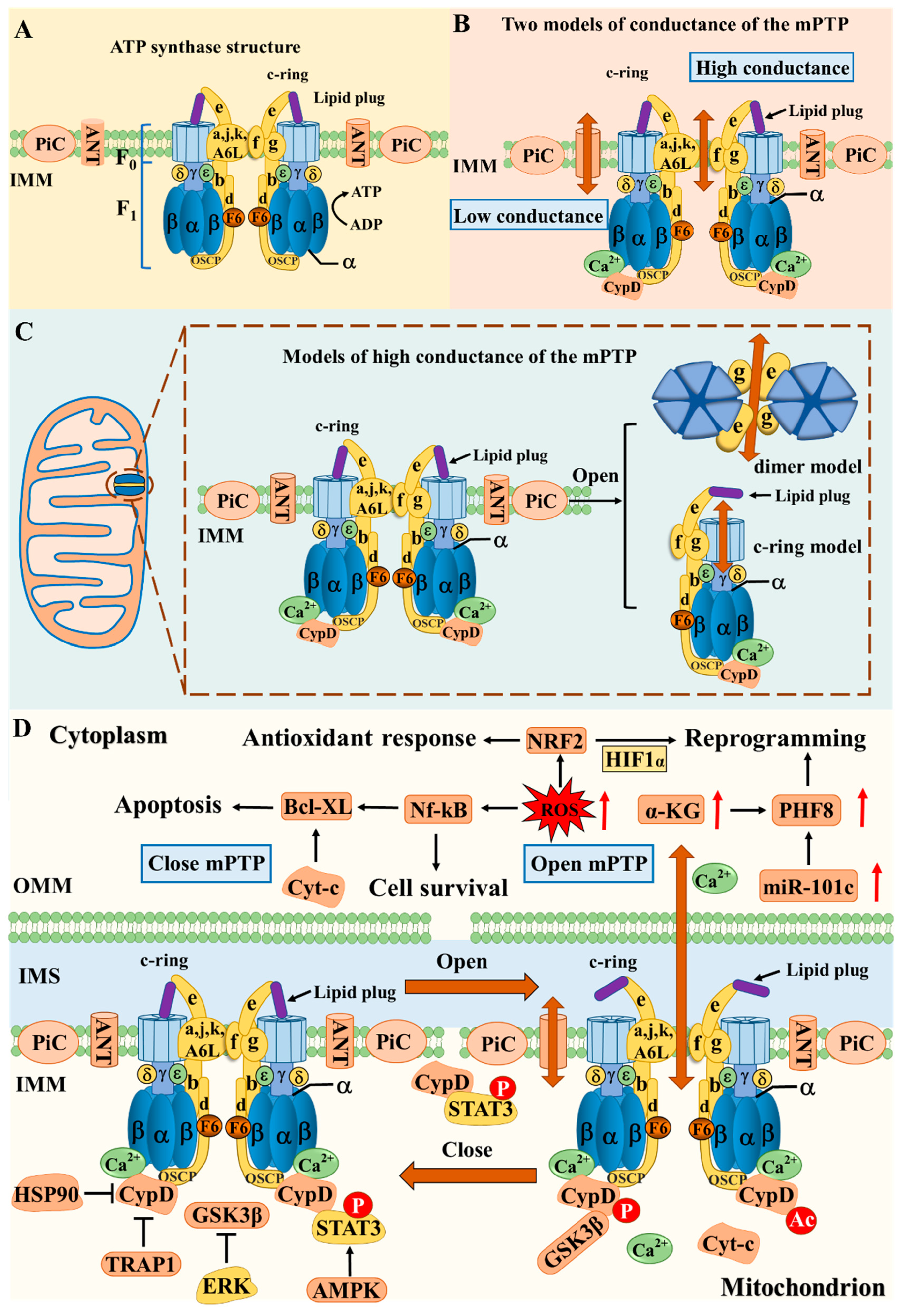
75]. In the early stages of somatic reprogramming, mitochondrial permeability transition pore (mPTP) regulates the fate of stem cells through mitochondrial metabolism. The transient opening of mPTP promotes the expression of structurally homologous plant domain finger protein (PHF8) through the ROS/miR-101c pathway, which is mediated by increased PHF8 activity driven by elevated α-KG [
76]. Mitochondrial ATP synthase is a negative regulator of mPTP [
77]. Understanding the general structure and protein components of mPTP is necessary for regulating stem cell life activities (
Figure 3).
L-Proline, as a signaling molecule, induces embryonic stem cell to mesenchymal like transition (esMT) in ESCs [
78]. After discontinuation of L-proline or addition of ascorbic acid (Vit C), L-proline induced esMT is completely reversible, inducing a decrease in H3K9 and H3K36 methylation levels and promoting mesenchymal stem cell to ESCs transformation (MesT) [
79]. The MAPK and PI3K/mTOR signaling pathways stimulated by L-proline can drive the transition of mESCs to primary exoderm like (EPL) cells [
80]. The process of somatic reprogramming involves mesenchymal to epithelial transition (MET), and VitC can promote the production of iPSCs by regulating histone demethylases [
81]. L-proline antagonizes VitC in DNA methylation and chromatin structure, but has opposite regulatory effects on pluripotency [
82]. In summary, these research findings demonstrate the important role of amino acid metabolism in regulating stem cell fate. Future work needs to understand and utilize the relationship between amino acid metabolism and stem cell fate, providing insights into the mechanisms by which metabolites control stem cell fate.
4. Fatty Acid Metabolism Regulates Stem Cell Function
At present, the regulatory effects of glucose and amino acid metabolism on stem cells have been determined, but there are few reports on the role of fatty acid metabolism in regulating stem cell function [
83]. Fatty acid metabolism is crucial for the pluripotency and proliferation of stem cells. FAO and de novo fat synthesis (DNL) are two important processes in fatty acid metabolism, which can maintain appropriate levels of fatty acids and promote pluripotency regulation [
9,
84].
4.1. Fatty Acid Oxidation
In the early stages of somatic reprogramming, a highly expressed C-terminal domain binding protein Wdr82 in oocytes can phosphorylate RNA polymerase II, transferring carbon energy from FAO to glycolysis to promote the generation of iPSCs [
85]. The premature myeloid leukemia peroxisome proliferator activated receptor δ (PML-PPAR-δ) pathway of FAO is used to maintain stem cell characteristics [
86]. Protein kinase C (PKC) is a family of protein kinases involved in many signaling cascades, which can regulate the self-renewal of hPSCs [
87]. Inhibiting PKC signal transduction helps maintain the pluripotency of ESCs [
88]. Carnitine palmitoyltransferase (Cpt1) is a key regulatory factor mediating FAO regulation of reprogramming. A significant increase in Cpt1 during early reprogramming is necessary for FAO upregulation. FAO promotes reprogramming by enhancing OXPHOS and inhibiting PKC [
89].
4.2. De Novo Lipogenesis
Through a comprehensive analysis of the metabolic flux of hPSCs, it was found that Essential 8 (E8) medium containing lipid supplements can maintain pluripotency and enhance mitochondrial metabolism [
90]. Exogenous lipids have availability in regulating human pluripotency, and hPSCs cultured in E8 medium capture an intermediate state of pluripotency from infancy to initiation [
91]. De novo lipogenesis (DNL) is one of the main pathways for intracellular synthesis of fatty acids, and stem cell proliferation is always accompanied by the enhancement of DNL and the accumulation of various lipids [
92]. Recent studies have shown that enhanced DNL promotes somatic reprogramming and maintains pluripotency of ESCs by regulating mitochondrial fission, providing previously overlooked connections between DNL, mitochondrial fission, and cellular pluripotency [
93]. In summary, the activation of fatty acid synthesis metabolism and catabolism support the production of iPSCs through different mechanisms. How is fatty acid metabolism and other metabolism balanced in stem cells? Exploring this issue will provide new insights into the relationship between fatty acid metabolism and stem cells.
5. Mitophagy Regulates Stem Cell Function
Mitophagy is an important regulatory factor for metabolic homeostasis and a necessary condition for alleviating cellular stress [
94]. Mitophagy can regulate the integrity, dynamics, and function of mitochondria in stem cells [
95]. During the process of somatic reprogramming, mitophagy reduces the number of mitochondria to facilitate the metabolic transition from OXPHOS to Glycolysis [
96]. We review the relationship between mitophagy and stem cell self-renewal, discuss how mitophagy regulates stem cell fate, and focus on the role of mitophagy in somatic cell reprogramming.
5.1. The Role of Mitophagy in Maintaining Stem Cell Characteristics
Mitophagy is the process by which cells selectively clear damaged or unwanted mitochondria in order to survive by initiating mitophagy mechanisms, maintaining stem cell characteristics and regenerative potential by controlling oxidative metabolism [
97,
98]. The initial stage of mitophagy is inhibited by mTORC1, which integrates different upstream nutrient and stress signals and promotes biosynthesis [
94]. The AKT and MAPK signaling pathways inhibit mitophagy by activating mTORC1, while the AMPK and p53 signaling pathways promote mitophagy by negatively regulating mTORC1 [
8]. AMPK and mTORC1 kinases catalyze the phosphorylation of ULK1. The absence of AMPK or ULK1 leads to abnormal accumulation of p62 and impaired mitochondrial mitophagy. The coordinated phosphorylation of ULK1 by mTORC1 and AMPK provides mechanistic insights for signal integration [
99]. Mitophagy is crucial for the pluripotency acquisition and maintenance of mitochondrial homeostasis in mESCs [
100]. MESCs exhibit high autophagic flux and are maintained by coordinating the expression of autophagic genes of the forkhead family transcription factor FOXO1 [
101]. Mitochondrial mitophagy dependent removal of p53 can express Nanog and promote stem cell survival [
102]. Mitophagy plays a role in avoiding aging. Adipose derived mesenchymal stem cells (ADSCs) accelerate mitochondrial mitophagy, eliminate intracellular ROS, improve mitochondrial quality, and regulate cellular metabolic homeostasis to delay the aging process [
103,
104]. In addition to maintaining stemness, mitophagy also helps establish stemness in oocytes after fertilization [
105]. In summary, mitophagy and mTOR signaling constitute a complex mechanism that is necessary for inducing and maintaining stem cell pluripotency.
5.2. The Role of Mitophagy in Somatic Reprogramming
In the process of somatic reprogramming, the deficiency of major autophagic proteins Atg3, Atg5, and Atg7 can eliminate the formation of iPSCs colonies [
106]. On the 2nd to 3rd day of reprogramming, mitophagy reached its peak. The transcription factor Sox2 binds to the repressor region in the mTOR promoter and recruits NuRD complexes, promoting somatic reprogramming by inhibiting mTOR transcription induced mitophagy [
107]. Transcription factors Klf4 and c-Myc induce mitophagy related genes, while transcription factors Oct4 and Sox2 inhibit mitophagy related genes. The four synergistically inhibit mTORC1 during somatic reprogramming, and mitophagy promotes reprogramming by degrading p62 [
108]. The activity of mTORC1 and mTORC2 will both decrease during reprogramming, but only downregulation of mTORC1 is essential [
109]. Inhibiting mTORC1 not only promotes mitophagy, but also helps overcome cellular reprogramming barriers and prevent cellular aging [
110]. In addition, mTORC1/PGC1 axis regulates mitochondrial mass during reprogramming [
10]. Treatment with mTOR inhibitors such as rapamycin or PP242, or mitophagy inducer spermidine, can significantly increase the rate of iPSCs generation [
111]. Studies have shown that stimulating mTOR independent ULK1 kinase mediated mitophagy can enhance the lifespan of iPSCs derived endothelium [
112]. However, little is currently known about whether ULK complexes contribute to other aspects of somatic reprogramming, and further research is needed.
Mitophagy driven metabolic switches help guide stem cell bioenergy conversion [
113]. The decrease in mitochondrial count during somatic reprogramming is associated with mitophagy. iPSCs produce new immature mitochondria during induction, which are cleared by Atg5 independent mitophagy and promote the metabolic transition from OXPHOS to glycolysis [
114]. The PINK1 dependent mitophagy pathway determines the efficiency and quality of somatic reprogramming [
96]. The HIF1 target gene BNIP3L mediated mitochondrial mitophagy is crucial for mitochondrial remodeling and maintenance of pluripotency, and knocking down BNIP3L significantly reduces reprogramming efficiency [
115]. BNIP3L/NIX dependent mitochondrial mitophagy specifically regulates mitochondrial clearance during reprogramming, thereby achieving mitochondrial remodeling [
116,
117]. These pieces of evidence suggest that the mechanism relationship between reprogramming factors, mTOR pathways, and mitochondrial mitophagy may be more complex and requires further research.
Mitochondrial dynamics are crucial for the pluripotency and embryonic development of stem cells [
118,
119]. The fusion and fission processes can regulate mitochondrial morphology and dynamics. Changes in mitochondrial dynamics lead to an increase in mitochondrial fission, promoting mitochondrial mitophagy [
120,
121]. It has been confirmed that mitochondrial fission is induced during reprogramming, and the fatty acid synthesis pathway promotes the formation of iPSCs by regulating mitochondrial fission [
93]. The cell cycle regulatory factor CDK1 and protein kinase ERK phosphorylate mitochondrial motility related protein 1 (DRP1) during early reprogramming, and DRP1 phosphorylation is associated with downregulation of MAP kinase phosphatase Dusp6 [
122]. DRP1 impairs the production of iPSCs colonies by downregulating the pluripotency related genes Nanog and Oct4 [
123]. Mitochondrial fission controlled by the ERK-DRP1 axis is a necessary step in the early stages of reprogramming, while SIRT2 regulates mitochondrial dynamics and reprogramming through the MEK1-ERK-DRP1 and AKT1-DRP1 axes [
124]. Unlike mitochondrial fission, mitochondrial fusion is believed to inhibit mitochondrial mitophagy [
125]. The consumption of mitochondrial fusion protein (MFN) leads to the activation of Ras Raf and HIF1α, as well as inhibition of p53/p21. Inhibition of mitochondrial fusion can promote metabolic transformation and maintenance of pluripotency in the early stages of reprogramming [
120]. Mitochondrial dynamics is an upstream regulatory factor that controls stem cell self-renewal, and changes in mitochondrial dynamics guide stem cell fate by modifying ROS signals [
95]. Recent reports have found that the Sirtuins protein family affects cell reprogramming efficiency by regulating mitochondrial dynamics, including the regulation of fission proteins, mitochondrial mitophagy, mTOR signaling, and control of ROS production [
126,
127]. In summary, mitochondrial mitophagy is related to the efficiency of somatic reprogramming, and studying the relationship between the two will provide new insights into the regulation of stem cell fate by mitophagy.
6. Molecular Mechanisms Regulating Stem Cell Metabolism
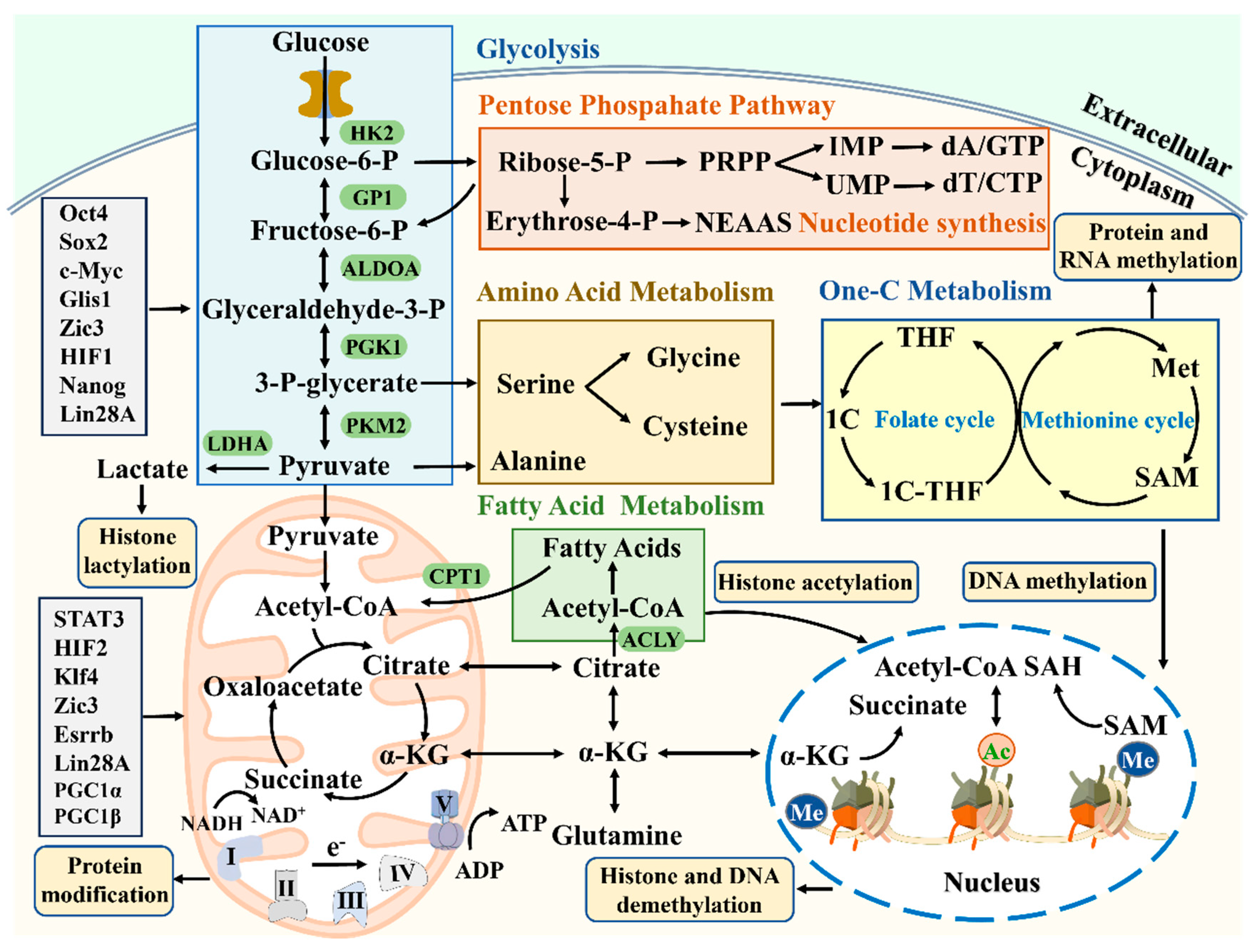
The molecular mechanisms of stem cell metabolism and energy regulation will provide new opportunities for regulating stem cell fate, contributing to the development of regenerative medicine (
Figure 4) [
4]. O-linked β-N-acetylglucosaminylation (O-GlcNAcylation) modification is the covalent connection of N-acetylglucosamine (GlcNAc) to the serine or threonine hydroxyl groups of proteins through β-glycosidic bonds. This post-translational modification regulates stem cell self-renewal and pluripotency through epigenetic mechanisms [
128]. The phosphoinositol 3 kinase/protein kinase B/mammalian rapamycin target (PI3K/AKT/mTOR) signaling pathway plays an important role in regulating cellular metabolism and is closely related to the functional homeostasis of various stem cells [
129]. Therefore, it is very important to study the molecular mechanisms that regulate the metabolic homeostasis of stem cells.
6.1. O-GlcNACylation Modification
O-GlcNAcylation modification is only regulated by two enzymes, namely O-GlcNAc transfer (OGT) and O-GlcNAcase (OGA) with residue removal [
130]. The hexosamine biosynthesis pathway (HBP) is located at a crossroads of major metabolic pathways, including the synthesis of carbohydrates, amino acids, nucleotides, and fatty acids [
131]. O-GlcNAcylation modification is controlled by HBP, and under hypoxic conditions, stem cells increase glucose and glutamine uptake, thereby activating HBP and OGT [
132]. In addition, OGT activity is highly sensitive to glucose concentration [
133], regulating the transcription of genes involved in embryonic development by regulating the abundance of methylcytosine oxidative modification epigenomic markers mediated by TET enzyme in primordial germ cells (PGCs) [
134]. The TET-OGT interaction promotes O-GlcNAcylation of host cytokine 1 (HCF1), which helps to recruit SET1/COMPASS complexes and H3K4Me3 in mESCs, highlighting a novel approach for TET enzyme induced transcriptional activation [
135]. The OGT gene is crucial for mESCs, and knocking out OGT is lethal for mESCs, as its chemical inhibition reduces overall O-GlcNAcylation levels [
136].
Transcription factors Oct4, Sox2, Klf4, and Nanog are core factors that control the self-renewal and pluripotency network of mESCs. O-GlcNAcylation controls pluripotency by directly regulating the transcriptional activity of these core factors, and blocking O-GlcNAcylation inhibits the self-renewal of mESCs and the generation efficiency of iPSCs [
137,
138]. The pluripotent transcription factors Oct4 and Esrrb are O-GlcNAcylated at the threonine 228 and serine 25 sites, respectively. The O-GlcNAcylation of these residues enhances the self-renewal and pluripotency of mESCs [
139]. In mESCs, the interaction between transcription factor Sox2 and poly ADP ribopolymerase 1 (PARP1) inhibits the binding of Sox2/Oct4 to enhancers, and this mechanism of fine-tuning Sox2 activity can maintain the pluripotency of mESCs [
140]. The transcription factor Sox2 regulates the self-renewal and early cell fate of mESCs through O-GlcNACylation at the 258 site of threonine [
141]. O-GlcNAcylation regulates the methionine cycle and promotes pluripotency of stem cells [
128]. Adenosine homocysteine enzyme (AHCY) is an important enzyme in the methionine cycle. AHCY undergoes the O-GlcNAcycle of threonine 136, which is beneficial for maintaining the trimethylation of histone H3 lysine 4 (H3K4me3) and the pluripotency of mESCs [
142]. The O-GlcNAcylation of pluripotency related transcription factors is complex, and O-GlcNAcylation affects the fate of ESCs through different mechanisms. These findings will expand our understanding of reprogramming processes and pluripotency at the molecular level [
143]. O-GlcNAcylation constitutes a molecular mechanism associated with stem cell signaling pathways, and further research is needed to fully demonstrate the interaction between O-GlcNAcylation and stem cell fate determination.
6.2. PI3K/AKT/mTOR Signaling Pathway
The PI3K/AKT/mTOR signaling pathway is closely related to cell growth, proliferation, transcription, translation, mitophagy, and metabolism, and is crucial for maintaining the pluripotency of mESCs [
144]. The activation of the PI3K/AKT signaling pathway phosphorylates downstream targets such as the homologous gene 2 (MDM2), the protein complex subunit 2 (TSC2) of nodular sclerosis, glycogen synthase kinase 3β (GSK3β), the forkhead transcription family O subfamily regulatory factor (FOXO1), and mTOR, thereby regulating the life activities of stem cells [
145]. In the early stages of reprogramming, the PI3K/AKT signaling pathway enhances glycolytic metabolism by inhibiting GSK3β and FOXO1, thereby improving the production efficiency of iPSCs [
146]. The PI3K/AKT/mTOR signaling pathway is the main regulatory factor of aerobic glycolysis. AKT increases glucose uptake by upregulating GLUT expression and activates Glycolysis by stimulating key glycolytic genes such as HK2 and PFK [
147]. In addition, AKT participates in TET mediated DNA demethylation during somatic reprogramming, maintaining mitochondrial integrity and preventing cytochrome C release, thereby inhibiting cell apoptosis [
129]. AKT enhances the conversion of citrate to Acetyl-CoA by activating citrate lyase (ACLY), leading to histone acetylation and chromatin activation, thereby increasing the availability of Acetyl-CoA precursors for lipid synthesis [
148]. The PI3K/AKT/Sox2 axis is a stem cell specific branch of the PI3K/AKT signaling pathway, which improves reprogramming efficiency and addresses safety issues associated with inducing iPSCs [
149]. MTOR is a central regulatory factor for cell growth and metabolism, existing in the form of Mechanical target of rapamycin complex 1 (mTORC1) and complex 2 (mTORC2), with a more complex role in metabolic regulation [
150]. MTORC1 upregulates the expression of HIF1, the main regulatory factor of Glycolysis metabolism [
151]. MTORC1 controls mitochondrial activity and biogenesis by selectively promoting the translation of mitochondrial related mRNA encoded by the nucleus [
152].
Nutrients and amino acids are effective activators of the mTORC1 pathway, while leucine, glutamine, and arginine further increase nutrient absorption through the RAG GTP enzyme mechanism to promote synthetic metabolic reactions [
153]. The mTORC1 pathway phosphorylates downstream targets of the eukaryotic translation initiation factor 4E binding protein (4EBP1) and ribosomal protein S6 kinase 1 (S6K1) to initiate mRNA translation, glycolysis, and biosynthesis [
154]. ULK1/2 directly phosphorylates key glycolytic enzymes such as PFK and enolase 1 (ENO1), as well as gluconeogenic enzyme fructose-1,6-diphosphatase (FBP1), initiating mitophagy to maintain glucose metabolism flux, cellular energy, and redox homeostasis [
155]. In addition, mTOR signaling is an important pathway involved in the survival of hESCs. Undifferentiated hESCs maintain pluripotency by expressing mTOR, while inhibiting mTOR significantly reduces the expression of pluripotency markers Oct4, Sox2, and Nanog, disrupting the dense morphology of hESCs colonies [
156]. The PI3K/AKT/mTOR signaling pathway plays different roles in somatic reprogramming. mTOR is initially suppressed to clear mitochondria, and subsequent activation requires the establishment of new gene expression profiles and metabolic activity [
157]. In summary, the PI3K/AKT/mTOR signaling pathway is essential for regulating self-renewal and pluripotency in ESCs. However, this signaling pathway is not isolated and further research is needed to investigate how it interacts with other signaling pathways or transcription factors.
7. Energy Metabolism Balance of Stem Cells
Stem cell metabolomics plays an important role in regulating proteomics, epigenetics, and transcriptomics (
Figure 5) [
158]. In the past few years, many studies have described the regulatory role of epigenetics in stem cell metabolism, where metabolic changes regulate chromatin modification levels and specific genome expression [
159]. The regulation of gene expression is achieved through various epigenetic events, including histone modifications, DNA methylation, and chromatin remodeling [
160]. It is worth noting that cellular metabolism provides metabolites for epigenetic modifications of histones and DNA, such as NAD, SAM α- Metabolic intermediates such as KG, Acetyl CoA, and O-GlcNAc can regulate pluripotency through nuclear pore diffusion, thereby determining the fate of PSCs [
161]. The histone demethylase JMJD3 plays a dual role in the process of somatic reprogramming. On the one hand, it can inhibit reprogramming by upregulating the expression of tumor suppressor genes INK4a/Arf through ubiquitination of PHF20, and on the other hand, it synergistically promotes reprogramming with Klf4 [
162]. Therefore, stabilizing the unique epigenetic characteristics in stem cells is crucial for maintaining their pluripotency. However, how pluripotent transcription factors in turn affect cellular metabolism remains a mystery [
163]. The core pluripotent transcription factor Oct4 plays a role in transcriptional regulation of multiple metabolic genes, directly encoding glycolytic rate limiting enzymes HK2 and PKM2 [
24]. Transcription factors Oct4, Sox2, and Nanog induce GLUT1 expression by directly activating the enhancer of GLUT1, promoting glucose uptake and glycolysis in hESCs [
164].
The key genes HK2, PFKP, and LDHA promoters of glycolysis contain binding sites for these transcription factors [
165]. The LIF/STAT3 pathway is connected to the pluripotent transcription factors Oct4, Sox2, and Nanog, maintaining the pluripotency of mESCs [
166]. The transcription factor Tfcp2l1 is a downstream target of STAT3 and participates in the metabolic regulation of mESCs. Among the metabolic genes regulated by Tfcp2l1, the Cpt1a promoter directly binds to Tfcp2l1 to regulate FAO [
167]. The transcription factor Glis1 mediates epigenetic and metabolic remodeling of stem cells, reprogramming aging cells into pluripotent states and improving genomic stability [
29]. The transcription factors Zic3 and Esrrb synergistically activate glycolysis to improve reprogramming efficiency, but Zic3 inhibits OXPHOS, while Esrrb activates OXPHOS, which is antagonistic to OXPHOS [
168]. With the rapid development of omics technologies such as proteomics, epigenetics, transcriptomics, and metabolomics, our understanding of pluripotent metabolic regulation has gradually deepened, greatly promoting the development of regenerative medicine.
8. Conclusion
Given the mixed relationship between metabolism, signal transduction, and epigenetic modifications, in this review, we extensively discuss how key pathways of energy metabolism, such as glucose metabolism, amino acid metabolism, fatty acid metabolism, and mitochondrial mitophagy, regulate stem cells. In addition, we also described the molecular mechanisms that regulate the energy metabolism balance of stem cells and the metabolic changes that occur during the reprogramming process. Nowadays, people are increasingly aware of the role of metabolism in regulating the fate of stem cells, and elucidating these molecular mechanisms is crucial for the application of stem cells in regenerative medicine and tissue engineering. At the same time, analyzing the genome, transcriptome, metabolome, and proteome to connect multiple levels of information is of great value in determining the regulatory network that controls stem cell function and fate determination. This will strengthen the understanding of development, aging, tumor occurrence, and disease. It is hoped that this review will bring promising improvements to the fields of stem cell technology and regenerative medicine.
Author Contributions
Conceptualization, M.W., X.Y. and H.C.; writing—original draft preparation, M.W.; writing—review and editing, M.W., X.Y. and H.C.; visualization, M.W.; funding acquisition, H.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (32272875) and Anhui Natural Science Foundation (2208085MC75).
Informed Consent Statement
Not applicable.
Acknowledgments
We sincerely appreciate the support of the Anhui Province Key Laboratory of Local 582 Livestock and Poultry Genetic Resource Conservation and Bio-breeding, College of Animal Science 583 and Technology, Anhui Agricultural University.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviation
| Acetyl-CoA |
Acetyl-Coenzyme A |
| ADP |
Adenosine diphosphate |
| AHCY |
Adenosylhomocysteinase |
| AMPK |
Adenosine monophosphate activated protein kinase |
| ANT |
Adenine nucleotide translocator |
| ATP |
Adenosine triphosphate |
| α-KG |
α-Ketoglurate |
| CBS |
Cystathionine-β-synthase |
| CSE |
Cystathionine-γ-lyase |
| CypD |
Cyclophilin D |
| Cys |
Cysteine |
| DRP1 |
Dynamin-related 1 |
| DUSP6 |
Dual specificity protein phosphatase 6 |
| ERK |
Extracellular signal-related kinase |
| ERR |
Estrogen-related nuclear receptor |
| ESCs |
Embryonic stem cells |
| ETC |
Electron transport chain |
| GLDC |
Glycine Decarboxylase |
| GSK3β |
Glycogen synthase kinase-3β |
| Hcy |
Homocysteine |
| HIF1α |
Hypoxia-inducible factor 1-alpha |
| HIF2α |
Hypoxia-inducible factor 2-alpha |
| HK2 |
Hexokinase 2 |
| HMT |
Histone melthyltransferases |
| iPSCs |
induced pluripotent stem cells |
| LDHA |
Lactate dehydrogenase |
| MAC |
Mitochondrial apoptosis-induced channel |
| MAPK |
Mitogen-activated protein kinase |
| MAT2a |
Methionine Adenosyltransferase 2A |
| Met |
Methionine |
| MFN |
Mitochondrial fusion protein |
| mPTP |
Mitochondrial permeability transition pore |
| MTHFR |
5,10-methylenetetrahydrofolate reductase |
| mTOR |
mammalian target of rapamycin |
| NAD |
Nicotinamide adenine dinucleotide |
| NADPH |
Nicotinamide adenine dinucleotide phosphate |
| Nf-kB |
Nuclear factor kappa B |
| NRF2 |
Nuclear factor (erythroid-derived 2)-like-2 |
| OXPHOS |
Oxidative phosphorylation |
| PDH |
Pyruvate dehydrogenase |
| PDK1 |
3-Phosphoinositide-dependent protein kinase-1 |
| PGC-1α |
Peroxisome proliferator-activated receptor-1α |
| PI3K-AKT |
Phosphatidyl inositol 3-kinase-protein kinase B |
| PKM2 |
Pyruvate kinase M2 |
| PnPase |
Polynucleotide phosphorylase |
| PSCs |
Pluripotent stem cells |
| ROS |
Reactive oxygen species |
| SAH |
S-adenosylmethionine |
| SHMT2 |
Serine Hydroxymethyltransferase 2 |
| TCA |
Tricarboxylic acid |
| TDH |
Threonine dehydratase |
| UCP2 |
Uncoupling protein 2 |
References
- Liu, W.; Chen, G. Regulation of energy metabolism in human pluripotent stem cells. Cellular and molecular life sciences: CMLS 2021, 78, 8097–8108. [Google Scholar] [CrossRef] [PubMed]
- Tyurin-Kuzmin, P.A.; Molchanov, A.Y.; Chechekhin, V.I.; Ivanova, A.M.; Kulebyakin, K.Y. Metabolic Regulation of Mammalian Stem Cell Differentiation. Biochemistry. Biokhimiia 2020, 85, 264–278. [Google Scholar] [CrossRef] [PubMed]
- Meacham, C.E.; DeVilbiss, A.W.; Morrison, S.J. Metabolic regulation of somatic stem cells in vivo. Nature reviews. Molecular cell biology 2022, 23, 428–443. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Jiang, O.; Wang, S. Molecular mechanisms of cellular metabolic homeostasis in stem cells. International journal of oral science 2023, 15, 52. [Google Scholar] [CrossRef]
- Oburoglu, L.; Tardito, S.; Fritz, V.; de Barros, S.C.; Merida, P.; Craveiro, M.; Mamede, J.; Cretenet, G.; Mongellaz, C.; An, X.; et al. Glucose and glutamine metabolism regulate human hematopoietic stem cell lineage specification. Cell stem cell 2014, 15, 169–184. [Google Scholar] [CrossRef] [PubMed]
- Arthur, T.D.; Nguyen, J.P.; D’Antonio-Chronowska, A.; Matsui, H.; Silva, N.S.; Joshua, I.N.; Luchessi, A.D.; Greenwald, W.W.Y.; D’Antonio, M.; Pera, M.F.; et al. Complex regulatory networks influence pluripotent cell state transitions in human iPSCs. Nature communications 2024, 15, 1664. [Google Scholar] [CrossRef] [PubMed]
- Sim, E.Z.; Enomoto, T.; Shiraki, N.; Furuta, N.; Kashio, S.; Kambe, T.; Tsuyama, T.; Arakawa, A.; Ozawa, H.; Yokoyama, M.; et al. Methionine metabolism regulates pluripotent stem cell pluripotency and differentiation through zinc mobilization. Cell reports 2022, 40, 111120. [Google Scholar] [CrossRef] [PubMed]
- García-Prat, L.; Martínez-Vicente, M.; Muñoz-Cánoves, P. Autophagy: a decisive process for stemness. Oncotarget 2016, 7, 12286–12288. [Google Scholar] [CrossRef] [PubMed]
- Visweswaran, M.; Arfuso, F.; Warrier, S.; Dharmarajan, A. Aberrant lipid metabolism as an emerging therapeutic strategy to target cancer stem cells. Stem cells (Dayton, Ohio) 2020, 38, 6–14. [Google Scholar] [CrossRef]
- Wang, L.; Xu, X.; Jiang, C.; Ma, G.; Huang, Y.; Zhang, H.; Lai, Y.; Wang, M.; Ahmed, T.; Lin, R.; et al. mTORC1-PGC1 axis regulates mitochondrial remodeling during reprogramming. The FEBS journal 2020, 287, 108–121. [Google Scholar] [CrossRef]
- Jasra, I.T.; Cuesta-Gomez, N.; Verhoeff, K.; Marfil-Garza, B.A.; Dadheech, N.; Shapiro, A.M.J. Mitochondrial regulation in human pluripotent stem cells during reprogramming and β cell differentiation. Frontiers in endocrinology 2023, 14, 1236472. [Google Scholar] [CrossRef] [PubMed]
- Ryall, J.G.; Cliff, T.; Dalton, S.; Sartorelli, V. Metabolic Reprogramming of Stem Cell Epigenetics. Cell stem cell 2015, 17, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Vaupel, P.; Schmidberger, H.; Mayer, A. The Warburg effect: essential part of metabolic reprogramming and central contributor to cancer progression. International journal of radiation biology 2019, 95, 912–919. [Google Scholar] [CrossRef]
- Cao, J.; Li, M.; Liu, K.; Shi, X.; Sui, N.; Yao, Y.; Wang, X.; Li, S.; Tian, Y.; Tan, S.; et al. Oxidative phosphorylation safeguards pluripotency via UDP-N-acetylglucosamine. Protein & cell 2023, 14, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Teslaa, T.; Teitell, M.A. Pluripotent stem cell energy metabolism: an update. The EMBO journal 2015, 34, 138–153. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.Y.; Kwak, S.; Lee, S.; Kim, H.; Lee, S.E.; Kim, J.H.; Kim, Y.A.; Jeon, Y.K.; Chung, D.H.; Jin, X.; et al. Psat1-Dependent Fluctuations in α-Ketoglutarate Affect the Timing of ESC Differentiation. Cell metabolism 2016, 24, 494–501. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Long, Q.; Wu, H.; Zhou, Y.; Duan, L.; Yuan, H.; Ding, Y.; Huang, Y.; Wu, Y.; Huang, J.; et al. Nuclear localization of mitochondrial TCA cycle enzymes modulates pluripotency via histone acetylation. Nature communications 2022, 13, 7414. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Khvorostov, I.; Hong, J.S.; Oktay, Y.; Vergnes, L.; Nuebel, E.; Wahjudi, P.N.; Setoguchi, K.; Wang, G.; Do, A.; et al. UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells. The EMBO journal 2016, 35, 899. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, K.; Fukuda, A.; Hisatake, K. Mechanisms of the Metabolic Shift during Somatic Cell Reprogramming. International journal of molecular sciences 2019, 20. [Google Scholar] [CrossRef]
- Cogliati, S.; Enriquez, J.A.; Scorrano, L. Mitochondrial Cristae: Where Beauty Meets Functionality. Trends Biochem Sci 2016, 41, 261–273. [Google Scholar] [CrossRef]
- Chandel, N.S. Glycolysis. Cold Spring Harbor perspectives in biology 2021, 13. [Google Scholar] [CrossRef]
- Kocianova, E.; Piatrikova, V.; Golias, T. Revisiting the Warburg Effect with Focus on Lactate. Cancers 2022, 14. [Google Scholar] [CrossRef]
- Cuezva, J.M.; Domínguez-Zorita, S. The ATPase Inhibitory Factor 1 (IF1) Contributes to the Warburg Effect and Is Regulated by Its Phosphorylation in S39 by a Protein Kinase A-like Activity. Cancers 2024, 16. [Google Scholar] [CrossRef]
- Kim, H.; Jang, H.; Kim, T.W.; Kang, B.H.; Lee, S.E.; Jeon, Y.K.; Chung, D.H.; Choi, J.; Shin, J.; Cho, E.J.; et al. Core Pluripotency Factors Directly Regulate Metabolism in Embryonic Stem Cell to Maintain Pluripotency. Stem cells (Dayton, Ohio) 2015, 33, 2699–2711. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Chen, K.; Wang, T.; Wu, Y.; Xing, G.; Chen, M.; Hao, Z.; Zhang, C.; Zhang, J.; Ma, B.; et al. Author Correction: Glis1 facilitates induction of pluripotency via an epigenome-metabolome-epigenome signalling cascade. Nature metabolism 2020, 2, 1179. [Google Scholar] [CrossRef]
- Ding, Y.; Yuan, X.; Zou, Y.; Gao, J.; Xu, X.; Sun, H.; Zuo, Q.; Zhang, Y.; Li, B. OCT4, SOX2 and NANOG co-regulate glycolysis and participate in somatic induced reprogramming. Cytotechnology 2022, 74, 371–383. [Google Scholar] [CrossRef] [PubMed]
- Folmes, C.D.; Terzic, A. Energy metabolism in the acquisition and maintenance of stemness. Seminars in cell & developmental biology 2016, 52, 68–75. [Google Scholar] [CrossRef]
- Folmes, C.D.; Nelson, T.J.; Martinez-Fernandez, A.; Arrell, D.K.; Lindor, J.Z.; Dzeja, P.P.; Ikeda, Y.; Perez-Terzic, C.; Terzic, A. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell metabolism 2011, 14, 264–271. [Google Scholar] [CrossRef]
- Li, L.; Chen, K.; Wang, T.; Wu, Y.; Xing, G.; Chen, M.; Hao, Z.; Zhang, C.; Zhang, J.; Ma, B.; et al. Glis1 facilitates induction of pluripotency via an epigenome-metabolome-epigenome signalling cascade. Nature metabolism 2020, 2, 882–892. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Li, W.; Zhou, H.; Wei, W.; Ambasudhan, R.; Lin, T.; Kim, J.; Zhang, K.; Ding, S. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell stem cell 2010, 7, 651–655. [Google Scholar] [CrossRef]
- Lee, M.R.; Mantel, C.; Lee, S.A.; Moon, S.H.; Broxmeyer, H.E. MiR-31/SDHA Axis Regulates Reprogramming Efficiency through Mitochondrial Metabolism. Stem cell reports 2016, 7, 1–10. [Google Scholar] [CrossRef]
- Hu, X.; Huang, X.; Yang, Y.; Sun, Y.; Zhao, Y.; Zhang, Z.; Qiu, D.; Wu, Y.; Wu, G.; Lei, L. Dux activates metabolism-lactylation-MET network during early iPSC reprogramming with Brg1 as the histone lactylation reader. Nucleic acids research 2024. [Google Scholar] [CrossRef]
- Kida, Y.S.; Kawamura, T.; Wei, Z.; Sogo, T.; Jacinto, S.; Shigeno, A.; Kushige, H.; Yoshihara, E.; Liddle, C.; Ecker, J.R.; et al. ERRs Mediate a Metabolic Switch Required for Somatic Cell Reprogramming to Pluripotency. Cell stem cell 2015, 16, 547–555. [Google Scholar] [CrossRef]
- Jang, J.; Wang, Y.; Kim, H.S.; Lalli, M.A.; Kosik, K.S. Nrf2, a regulator of the proteasome, controls self-renewal and pluripotency in human embryonic stem cells. Stem cells (Dayton, Ohio) 2014, 32, 2616–2625. [Google Scholar] [CrossRef]
- Hawkins, K.E.; Joy, S.; Delhove, J.M.; Kotiadis, V.N.; Fernandez, E.; Fitzpatrick, L.M.; Whiteford, J.R.; King, P.J.; Bolanos, J.P.; Duchen, M.R.; et al. NRF2 Orchestrates the Metabolic Shift during Induced Pluripotent Stem Cell Reprogramming. Cell reports 2016, 14, 1883–1891. [Google Scholar] [CrossRef]
- Ishida, T.; Nakao, S.; Ueyama, T.; Harada, Y.; Kawamura, T. Metabolic remodeling during somatic cell reprogramming to induced pluripotent stem cells: involvement of hypoxia-inducible factor 1. Inflammation and regeneration 2020, 40, 8. [Google Scholar] [CrossRef]
- Kierans, S.J.; Taylor, C.T. Regulation of glycolysis by the hypoxia-inducible factor (HIF): implications for cellular physiology. The Journal of physiology 2021, 599, 23–37. [Google Scholar] [CrossRef]
- Varum, S.; Rodrigues, A.S.; Moura, M.B.; Momcilovic, O.; Easley, C.A.t.; Ramalho-Santos, J.; Van Houten, B.; Schatten, G. Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PloS one 2011, 6, e20914. [Google Scholar] [CrossRef]
- Prigione, A.; Rohwer, N.; Hoffmann, S.; Mlody, B.; Drews, K.; Bukowiecki, R.; Blümlein, K.; Wanker, E.E.; Ralser, M.; Cramer, T.; et al. HIF1α modulates cell fate reprogramming through early glycolytic shift and upregulation of PDK1-3 and PKM2. Stem cells (Dayton, Ohio) 2014, 32, 364–376. [Google Scholar] [CrossRef]
- Prieto, J.; Seo, A.Y.; León, M.; Santacatterina, F.; Torresano, L.; Palomino-Schätzlein, M.; Giménez, K.; Vallet-Sánchez, A.; Ponsoda, X.; Pineda-Lucena, A.; et al. MYC Induces a Hybrid Energetics Program Early in Cell Reprogramming. Stem cell reports 2018, 11, 1479–1492. [Google Scholar] [CrossRef]
- Hossini, A.M.; Quast, A.S.; Plötz, M.; Grauel, K.; Exner, T.; Küchler, J.; Stachelscheid, H.; Eberle, J.; Rabien, A.; Makrantonaki, E.; et al. PI3K/AKT Signaling Pathway Is Essential for Survival of Induced Pluripotent Stem Cells. PloS one 2016, 11, e0154770. [Google Scholar] [CrossRef]
- Kao, T.W.; Bai, G.H.; Wang, T.L.; Shih, I.M.; Chuang, C.M.; Lo, C.L.; Tsai, M.C.; Chiu, L.Y.; Lin, C.C.; Shen, Y.A. Novel cancer treatment paradigm targeting hypoxia-induced factor in conjunction with current therapies to overcome resistance. Journal of experimental & clinical cancer research: CR 2023, 42, 171. [Google Scholar] [CrossRef]
- Arthur, S.A.; Blaydes, J.P.; Houghton, F.D. Glycolysis Regulates Human Embryonic Stem Cell Self-Renewal under Hypoxia through HIF-2α and the Glycolytic Sensors CTBPs. Stem cell reports 2019, 12, 728–742. [Google Scholar] [CrossRef]
- Mathieu, J.; Zhou, W.; Xing, Y.; Sperber, H.; Ferreccio, A.; Agoston, Z.; Kuppusamy, K.T.; Moon, R.T.; Ruohola-Baker, H. Hypoxia-inducible factors have distinct and stage-specific roles during reprogramming of human cells to pluripotency. Cell stem cell 2014, 14, 592–605. [Google Scholar] [CrossRef]
- Nishimura, K.; Aizawa, S.; Nugroho, F.L.; Shiomitsu, E.; Tran, Y.T.H.; Bui, P.L.; Borisova, E.; Sakuragi, Y.; Takada, H.; Kurisaki, A.; et al. A Role for KLF4 in Promoting the Metabolic Shift via TCL1 during Induced Pluripotent Stem Cell Generation. Stem cell reports 2017, 8, 787–801. [Google Scholar] [CrossRef]
- Fiorenza, M.T.; Rava, A. The TCL1 function revisited focusing on metabolic requirements of stemness. Cell cycle (Georgetown, Tex.) 2019, 18, 3055–3063. [Google Scholar] [CrossRef]
- Sato, S.; Hishida, T.; Kinouchi, K.; Hatanaka, F.; Li, Y.; Nguyen, Q.; Chen, Y.; Wang, P.H.; Kessenbrock, K.; Li, W.; et al. The circadian clock CRY1 regulates pluripotent stem cell identity and somatic cell reprogramming. Cell reports 2023, 42, 112590. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, D.; Xu, J.; Wang, Y.; Ma, F.; Li, Z.; Liu, N. Stat3 activation is critical for pluripotency maintenance. Journal of cellular physiology 2019, 234, 1044–1051. [Google Scholar] [CrossRef]
- Zhang, J.; Ratanasirintrawoot, S.; Chandrasekaran, S.; Wu, Z.; Ficarro, S.B.; Yu, C.; Ross, C.A.; Cacchiarelli, D.; Xia, Q.; Seligson, M.; et al. LIN28 Regulates Stem Cell Metabolism and Conversion to Primed Pluripotency. Cell stem cell 2016, 19, 66–80. [Google Scholar] [CrossRef]
- Kilberg, M.S.; Terada, N.; Shan, J. Influence of Amino Acid Metabolism on Embryonic Stem Cell Function and Differentiation. Advances in nutrition (Bethesda, Md.) 2016, 7, 780s–789s. [Google Scholar] [CrossRef]
- Ducker, G.S.; Rabinowitz, J.D. One-Carbon Metabolism in Health and Disease. Cell metabolism 2017, 25, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wang, J. A regulatory circuitry locking pluripotent stemness to embryonic stem cell: Interaction between threonine catabolism and histone methylation. Seminars in cancer biology 2019, 57, 72–78. [Google Scholar] [CrossRef]
- Kubota, K.; Iqbal, K.; Soares, M.J. SATB1 promotion of trophoblast stem cell renewal through regulation of threonine dehydrogenase. Biochim Biophys Acta Gen Subj 2021, 1865, 129757. [Google Scholar] [CrossRef] [PubMed]
- Shyh-Chang, N.; Locasale, J.W.; Lyssiotis, C.A.; Zheng, Y.; Teo, R.Y.; Ratanasirintrawoot, S.; Zhang, J.; Onder, T.; Unternaehrer, J.J.; Zhu, H.; et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science (New York, N.Y.) 2013, 339, 222–226. [Google Scholar] [CrossRef]
- Wang, J.; Alexander, P.; Wu, L.; Hammer, R.; Cleaver, O.; McKnight, S.L. Dependence of mouse embryonic stem cells on threonine catabolism. Science 2009, 325, 435–439. [Google Scholar] [CrossRef]
- Ryu, J.M.; Han, H.J. L-threonine regulates G1/S phase transition of mouse embryonic stem cells via PI3K/Akt, MAPKs, and mTORC pathways. J Biol Chem 2011, 286, 23667–23678. [Google Scholar] [CrossRef]
- Han, C.; Gu, H.; Wang, J.; Lu, W.; Mei, Y.; Wu, M. Regulation of L-threonine dehydrogenase in somatic cell reprogramming. Stem cells (Dayton, Ohio) 2013, 31, 953–965. [Google Scholar] [CrossRef]
- Donato, V.; Bonora, M.; Simoneschi, D.; Sartini, D.; Kudo, Y.; Saraf, A.; Florens, L.; Washburn, M.P.; Stadtfeld, M.; Pinton, P.; et al. The TDH-GCN5L1-Fbxo15-KBP axis limits mitochondrial biogenesis in mouse embryonic stem cells. Nature cell biology 2017, 19, 341–351. [Google Scholar] [CrossRef]
- Lan, X.; Field, M.S.; Stover, P.J. Cell cycle regulation of folate-mediated one-carbon metabolism. Wiley interdisciplinary reviews. Systems biology and medicine 2018, 10, e1426. [Google Scholar] [CrossRef] [PubMed]
- Shiraki, N.; Shiraki, Y.; Tsuyama, T.; Obata, F.; Miura, M.; Nagae, G.; Aburatani, H.; Kume, K.; Endo, F.; Kume, S. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell metabolism 2014, 19, 780–794. [Google Scholar] [CrossRef]
- Fan, J.; Ye, J.; Kamphorst, J.J.; Shlomi, T.; Thompson, C.B.; Rabinowitz, J.D. Quantitative flux analysis reveals folate-dependent NADPH production. Nature 2014, 510, 298–302. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Jia, W.; Qian, Z.; Zhao, L.; Yu, Y.; Li, L.; Wang, C.; Zhang, W.; Liu, Q.; Yang, D.; et al. Folic Acid Supports Pluripotency and Reprogramming by Regulating LIF/STAT3 and MAPK/ERK Signaling. Stem cells and development 2017, 26, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Xu, J.; Hu, W.; Li, Z.; Wu, J.; Zhang, S. Transient folate deprivation facilitates the generation of mouse-induced pluripotent stem cells. Cell biology international 2014, 38, 571–576. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.; Shima, H.; Matsuo, Y.; Long, N.C.; Matsumoto, M.; Ishii, Y.; Sato, N.; Sugiyama, T.; Nobuta, R.; Hashimoto, S.; et al. mTORC1-independent translation control in mammalian cells by methionine adenosyltransferase 2A and S-adenosylmethionine. The Journal of biological chemistry 2022, 298, 102084. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Fang, Y.; Huang, G.; Xu, X.; Padilla-Banks, E.; Fan, W.; Xu, Q.; Sanderson, S.M.; Foley, J.F.; Dowdy, S.; et al. Methionine metabolism is essential for SIRT1-regulated mouse embryonic stem cell maintenance and embryonic development. The EMBO journal 2017, 36, 3175–3193. [Google Scholar] [CrossRef] [PubMed]
- Siblini, Y.; Namour, F.; Oussalah, A.; Guéant, J.L.; Chéry, C. Stemness of Normal and Cancer Cells: The Influence of Methionine Needs and SIRT1/PGC-1α/PPAR-α Players. Cells 2022, 11. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, H.; Kambe, A.; Hibi, K.; Murakami, S.; Oikawa, A.; Handa, T.; Fujiki, K.; Nakato, R.; Shirahige, K.; Kimura, H.; et al. Transient Methionine Deprivation Triggers Histone Modification and Potentiates Differentiation of Induced Pluripotent Stem Cells. Stem cells (Dayton, Ohio) 2023, 41, 271–286. [Google Scholar] [CrossRef]
- Sperber, H.; Mathieu, J.; Wang, Y.; Ferreccio, A.; Hesson, J.; Xu, Z.; Fischer, K.A.; Devi, A.; Detraux, D.; Gu, H.; et al. The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nat Cell Biol 2015, 17, 1523–1535. [Google Scholar] [CrossRef] [PubMed]
- Glancy, E.; Ciferri, C.; Bracken, A.P. Structural basis for PRC2 engagement with chromatin. Current opinion in structural biology 2021, 67, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Tohyama, S.; Fujita, J.; Hishiki, T.; Matsuura, T.; Hattori, F.; Ohno, R.; Kanazawa, H.; Seki, T.; Nakajima, K.; Kishino, Y.; et al. Glutamine Oxidation Is Indispensable for Survival of Human Pluripotent Stem Cells. Cell metabolism 2016, 23, 663–674. [Google Scholar] [CrossRef]
- Marsboom, G.; Zhang, G.F.; Pohl-Avila, N.; Zhang, Y.; Yuan, Y.; Kang, H.; Hao, B.; Brunengraber, H.; Malik, A.B.; Rehman, J. Glutamine Metabolism Regulates the Pluripotency Transcription Factor OCT4. Cell reports 2016, 16, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Nesci, S. Glucose and glutamine in the mitochondrial oxidative metabolism of stem cells. Mitochondrion 2017, 35, 11–12. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.C.; Yu, Y.C.; Sung, Y.; Han, J.M. Glutamine reliance in cell metabolism. Exp Mol Med 2020, 52, 1496–1516. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; He, C.; Zhang, L.; Zhu, T.; Lv, D.; Li, G.; Song, Y.; Wang, J.; Wu, H.; Ji, P.; et al. Alpha-ketoglutarate affects murine embryo development through metabolic and epigenetic modulations. Reproduction 2019, 158, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Carey, B.W.; Finley, L.W.; Cross, J.R.; Allis, C.D.; Thompson, C.B. Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 2015, 518, 413–416. [Google Scholar] [CrossRef] [PubMed]
- Ying, Z.; Xiang, G.; Zheng, L.; Tang, H.; Duan, L.; Lin, X.; Zhao, Q.; Chen, K.; Wu, Y.; Xing, G.; et al. Short-Term Mitochondrial Permeability Transition Pore Opening Modulates Histone Lysine Methylation at the Early Phase of Somatic Cell Reprogramming. Cell metabolism 2019, 29, 502. [Google Scholar] [CrossRef] [PubMed]
- Pekson, R.; Liang, F.G.; Axelrod, J.L.; Lee, J.; Qin, D.; Wittig, A.J.H.; Paulino, V.M.; Zheng, M.; Peixoto, P.M.; Kitsis, R.N. The mitochondrial ATP synthase is a negative regulator of the mitochondrial permeability transition pore. Proceedings of the National Academy of Sciences of the United States of America 2023, 120, e2303713120. [Google Scholar] [CrossRef] [PubMed]
- Minchiotti, G.; D’Aniello, C.; Fico, A.; De Cesare, D.; Patriarca, E.J. Capturing Transitional Pluripotency through Proline Metabolism. Cells 2022, 11. [Google Scholar] [CrossRef] [PubMed]
- Comes, S.; Gagliardi, M.; Laprano, N.; Fico, A.; Cimmino, A.; Palamidessi, A.; De Cesare, D.; De Falco, S.; Angelini, C.; Scita, G.; et al. L-Proline induces a mesenchymal-like invasive program in embryonic stem cells by remodeling H3K9 and H3K36 methylation. Stem cell reports 2013, 1, 307–321. [Google Scholar] [CrossRef]
- Glover, H.J.; Holliday, H.; Shparberg, R.A.; Winkler, D.; Day, M.; Morris, M.B. Signalling pathway crosstalk stimulated by L-proline drives mouse embryonic stem cells to primitive-ectoderm-like cells. Development (Cambridge, England) 2023, 150. [Google Scholar] [CrossRef]
- Chen, J.; Liu, H.; Liu, J.; Qi, J.; Wei, B.; Yang, J.; Liang, H.; Chen, Y.; Chen, J.; Wu, Y.; et al. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat Genet 2013, 45, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Glover, H.J.; Shparberg, R.A.; Morris, M.B. L-Proline Supplementation Drives Self-Renewing Mouse Embryonic Stem Cells to a Partially Primed Pluripotent State: The Early Primitive Ectoderm-Like Cell. Methods Mol Biol 2022, 2490, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Shahbodi, M.; Emami, S.A.; Javadi, B.; Tayarani-Najaran, Z. Effects of Thymoquinone on Adipocyte Differentiation in Human Adipose-Derived Stem Cells. Cell biochemistry and biophysics 2022, 80, 771–779. [Google Scholar] [CrossRef] [PubMed]
- Mihaylova, M.M.; Cheng, C.W.; Cao, A.Q.; Tripathi, S.; Mana, M.D.; Bauer-Rowe, K.E.; Abu-Remaileh, M.; Clavain, L.; Erdemir, A.; Lewis, C.A.; et al. Fasting Activates Fatty Acid Oxidation to Enhance Intestinal Stem Cell Function during Homeostasis and Aging. Cell stem cell 2018, 22, 769–778.e764. [Google Scholar] [CrossRef] [PubMed]
- Cui, G.; Zhou, J.; Sun, J.; Kou, X.; Su, Z.; Xu, Y.; Liu, T.; Sun, L.; Li, W.; Wu, X.; et al. WD repeat domain 82 (Wdr82) facilitates mouse iPSCs generation by interfering mitochondrial oxidative phosphorylation and glycolysis. Cellular and molecular life sciences: CMLS 2023, 80, 218. [Google Scholar] [CrossRef] [PubMed]
- Chakravarti, B.; Akhtar Siddiqui, J.; Anthony Sinha, R.; Raza, S. Targeting autophagy and lipid metabolism in cancer stem cells. Biochem Pharmacol 2023, 212, 115550. [Google Scholar] [CrossRef]
- Kinehara, M.; Kawamura, S.; Tateyama, D.; Suga, M.; Matsumura, H.; Mimura, S.; Hirayama, N.; Hirata, M.; Uchio-Yamada, K.; Kohara, A.; et al. Protein kinase C regulates human pluripotent stem cell self-renewal. PloS one 2013, 8, e54122. [Google Scholar] [CrossRef] [PubMed]
- Baral, I.; Shirude, M.B.; Jothi, D.L.; Mukherjee, A.; Dutta, D. Characterization of a Distinct State in the Continuum of Pluripotency Facilitated by Inhibition of PKCζ in Mouse Embryonic Stem Cells. Stem Cell Rev Rep 2023, 19, 1098–1115. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Liu, F.; Shi, P.; Song, A.; Huang, Z.; Zou, D.; Chen, Q.; Li, J.; Gao, X. Fatty acid oxidation promotes reprogramming by enhancing oxidative phosphorylation and inhibiting protein kinase C. Stem cell research & therapy 2018, 9, 47. [Google Scholar] [CrossRef]
- Zhang, H.; Badur, M.G.; Divakaruni, A.S.; Parker, S.J.; Jäger, C.; Hiller, K.; Murphy, A.N.; Metallo, C.M. Distinct Metabolic States Can Support Self-Renewal and Lipogenesis in Human Pluripotent Stem Cells under Different Culture Conditions. Cell reports 2016, 16, 1536–1547. [Google Scholar] [CrossRef]
- Cornacchia, D.; Zhang, C.; Zimmer, B.; Chung, S.Y.; Fan, Y.; Soliman, M.A.; Tchieu, J.; Chambers, S.M.; Shah, H.; Paull, D.; et al. Lipid Deprivation Induces a Stable, Naive-to-Primed Intermediate State of Pluripotency in Human PSCs. Cell stem cell 2019, 25, 120–136.e110. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Lu, C.W.; Diem, E.C.; Li, W.; Guderian, M.; Lindenberg, M.; Kruse, F.; Buettner, M.; Floess, S.; Winny, M.R.; et al. Acetyl-CoA-Carboxylase 1-mediated de novo fatty acid synthesis sustains Lgr5(+) intestinal stem cell function. Nat Commun 2022, 13, 3998. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, T.; Wang, L.; Cai, Y.; Zhong, X.; He, X.; Hu, L.; Tian, S.; Wu, M.; Hui, L.; et al. Fatty acid synthesis is critical for stem cell pluripotency via promoting mitochondrial fission. The EMBO journal 2017, 36, 1330–1347. [Google Scholar] [CrossRef] [PubMed]
- Al-Bari, M.A.A.; Xu, P. Molecular regulation of autophagy machinery by mTOR-dependent and -independent pathways. Ann N Y Acad Sci 2020, 1467, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Khacho, M.; Clark, A.; Svoboda, D.S.; Azzi, J.; MacLaurin, J.G.; Meghaizel, C.; Sesaki, H.; Lagace, D.C.; Germain, M.; Harper, M.E.; et al. Mitochondrial Dynamics Impacts Stem Cell Identity and Fate Decisions by Regulating a Nuclear Transcriptional Program. Cell stem cell 2016, 19, 232–247. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Martin, A.; Van den Haute, C.; Cufí, S.; Corominas-Faja, B.; Cuyàs, E.; Lopez-Bonet, E.; Rodriguez-Gallego, E.; Fernández-Arroyo, S.; Joven, J.; Baekelandt, V.; et al. Mitophagy-driven mitochondrial rejuvenation regulates stem cell fate. Aging 2016, 8, 1330–1352. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Long, H.; Hou, L.; Feng, B.; Ma, Z.; Wu, Y.; Zeng, Y.; Cai, J.; Zhang, D.W.; Zhao, G. The mitophagy pathway and its implications in human diseases. Signal Transduct Target Ther 2023, 8, 304. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Chen, Z.; Wang, Y.; Tan, Z.; Zhu, C.; Li, Y.; Han, Z.; Chen, L.; Gao, R.; Liu, L.; et al. Mitophagy receptor FUNDC1 regulates mitochondrial dynamics and mitophagy. Autophagy 2016, 12, 689–702. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.; Jeong, S.H.; Yi, K.; Chung, K.M.; Hong, C.J.; Kim, S.W.; Kim, E.K.; Yu, S.W. Phosphorylation of p62 by AMP-activated protein kinase mediates autophagic cell death in adult hippocampal neural stem cells. J Biol Chem 2017, 292, 13795–13808. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, K.; Cao, J.; Wang, L.; Zhao, Q.; Li, Z.; Zhang, H.; Chen, Q.; Zhao, T. PINK1-mediated mitophagy maintains pluripotency through optineurin. Cell proliferation 2021, 54, e13034. [Google Scholar] [CrossRef]
- Liu, P.; Liu, K.; Gu, H.; Wang, W.; Gong, J.; Zhu, Y.; Zhao, Q.; Cao, J.; Han, C.; Gao, F.; et al. High autophagic flux guards ESC identity through coordinating autophagy machinery gene program by FOXO1. Cell Death Differ 2017, 24, 1672–1680. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Wang, Q.; Kao, Y.R.; Diaz, A.; Tasset, I.; Kaushik, S.; Thiruthuvanathan, V.; Zintiridou, A.; Nieves, E.; Dzieciatkowska, M.; et al. Chaperone-mediated autophagy sustains haematopoietic stem-cell function. Nature 2021, 591, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, X.Y.; Liu, Y.J.; Feng, J.; Wu, Y.; Shen, H.M.; Lu, G.D. Full-coverage regulations of autophagy by ROS: from induction to maturation. Autophagy 2022, 18, 1240–1255. [Google Scholar] [CrossRef]
- Lv, M.; Zhang, S.; Jiang, B.; Cao, S.; Dong, Y.; Cao, L.; Guo, S. Adipose-derived stem cells regulate metabolic homeostasis and delay aging by promoting mitophagy. FASEB journal: official publication of the Federation of American Societies for Experimental Biology 2021, 35, e21709. [Google Scholar] [CrossRef] [PubMed]
- Vijayakumar, K.; Cho, G.W. Autophagy: An evolutionarily conserved process in the maintenance of stem cells and aging. Cell Biochem Funct 2019, 37, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Zhao, Q.; Liu, P.; Cao, J.; Gong, J.; Wang, C.; Wang, W.; Li, X.; Sun, H.; Zhang, C.; et al. ATG3-dependent autophagy mediates mitochondrial homeostasis in pluripotency acquirement and maintenance. Autophagy 2016, 12, 2000–2008. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Xia, P.; Ye, B.; Huang, G.; Liu, J.; Fan, Z. Transient activation of autophagy via Sox2-mediated suppression of mTOR is an important early step in reprogramming to pluripotency. Cell stem cell 2013, 13, 617–625. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, Y.; Zhang, H.; Huang, Y.; Zhao, P.; Tang, Y.; Qiu, X.; Ying, Y.; Li, W.; Ni, S.; et al. Autophagy and mTORC1 regulate the stochastic phase of somatic cell reprogramming. Nature cell biology 2015, 17, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Xia, P.; Rehm, M.; Fan, Z. Autophagy and cell reprogramming. Cellular and molecular life sciences: CMLS 2015, 72, 1699–1713. [Google Scholar] [CrossRef] [PubMed]
- Noda, T. Regulation of Autophagy through TORC1 and mTORC1. Biomolecules 2017, 7. [Google Scholar] [CrossRef]
- Chen, T.; Shen, L.; Yu, J.; Wan, H.; Guo, A.; Chen, J.; Long, Y.; Zhao, J.; Pei, G. Rapamycin and other longevity-promoting compounds enhance the generation of mouse induced pluripotent stem cells. Aging Cell 2011, 10, 908–911. [Google Scholar] [CrossRef] [PubMed]
- Hekman, K.E.; Koss, K.M.; Ivancic, D.Z.; He, C.; Wertheim, J.A. Autophagy Enhances Longevity of Induced Pluripotent Stem Cell-Derived Endothelium via mTOR-Independent ULK1 Kinase. Stem cells translational medicine 2022, 11, 1151–1164. [Google Scholar] [CrossRef] [PubMed]
- Naik, P.P.; Birbrair, A.; Bhutia, S.K. Mitophagy-driven metabolic switch reprograms stem cell fate. Cellular and molecular life sciences: CMLS 2019, 76, 27–43. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Li, J.; Xu, Y.; Yu, C.; Xu, T.; Wang, H.; Liu, K.; Cao, N.; Nie, B.M.; Zhu, S.Y.; et al. Atg5-independent autophagy regulates mitochondrial clearance and is essential for iPSC reprogramming. Nature cell biology 2015, 17, 1379–1387. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Zhao, Q.; Sun, H.; Liu, L.; Wang, C.; Li, Z.; Xu, Y.; Wang, L.; Zhang, L.; Zhang, H.; et al. BNIP3 (BCL2 interacting protein 3) regulates pluripotency by modulating mitochondrial homeostasis via mitophagy. Cell death & disease 2022, 13, 334. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Yang, X. Autophagy and pluripotency: self-eating your way to eternal youth. Trends Cell Biol 2022, 32, 868–882. [Google Scholar] [CrossRef] [PubMed]
- Xiang, G.; Yang, L.; Long, Q.; Chen, K.; Tang, H.; Wu, Y.; Liu, Z.; Zhou, Y.; Qi, J.; Zheng, L.; et al. BNIP3L-dependent mitophagy accounts for mitochondrial clearance during 3 factors-induced somatic cell reprogramming. Autophagy 2017, 13, 1543–1555. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Cui, P.; Cai, Y.; Wang, L.; He, X.; Long, P.; Lu, K.; Yan, R.; Zhang, Y.; Pan, X.; et al. Mitochondrial Dynamics Is Critical for the Full Pluripotency and Embryonic Developmental Potential of Pluripotent Stem Cells. Cell metabolism 2019, 29, 979–992.e974. [Google Scholar] [CrossRef] [PubMed]
- Son, M.Y.; Choi, H.; Han, Y.M.; Cho, Y.S. Unveiling the critical role of REX1 in the regulation of human stem cell pluripotency. Stem cells (Dayton, Ohio) 2013, 31, 2374–2387. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Mihara, K.; Chen, Y.; Scorrano, L.; Dorn, G.W., 2nd. Mitochondrial fission and fusion factors reciprocally orchestrate mitophagic culling in mouse hearts and cultured fibroblasts. Cell metabolism 2015, 21, 273–286. [Google Scholar] [CrossRef]
- Chang, C.R.; Manlandro, C.M.; Arnoult, D.; Stadler, J.; Posey, A.E.; Hill, R.B.; Blackstone, C. A lethal de novo mutation in the middle domain of the dynamin-related GTPase Drp1 impairs higher order assembly and mitochondrial division. J Biol Chem 2010, 285, 32494–32503. [Google Scholar] [CrossRef] [PubMed]
- Prieto, J.; León, M.; Ponsoda, X.; Sendra, R.; Bort, R.; Ferrer-Lorente, R.; Raya, A.; López-García, C.; Torres, J. Early ERK1/2 activation promotes DRP1-dependent mitochondrial fission necessary for cell reprogramming. Nature communications 2016, 7, 11124. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ye, X.; Zhao, Q.; Zhou, Z.; Dan, J.; Zhu, Y.; Chen, Q.; Liu, L. Drp1 is dispensable for mitochondria biogenesis in induction to pluripotency but required for differentiation of embryonic stem cells. Stem cells and development 2014, 23, 2422–2434. [Google Scholar] [CrossRef] [PubMed]
- Cha, Y.; Kim, T.; Jeon, J.; Jang, Y.; Kim, P.B.; Lopes, C.; Leblanc, P.; Cohen, B.M.; Kim, K.S. SIRT2 regulates mitochondrial dynamics and reprogramming via MEK1-ERK-DRP1 and AKT1-DRP1 axes. Cell reports 2021, 37, 110155. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Meng, Y.; Han, J. Emerging roles of mitochondrial functions and epigenetic changes in the modulation of stem cell fate. Cellular and molecular life sciences: CMLS 2024, 81, 26. [Google Scholar] [CrossRef]
- Kim, A.Y.; Lee, E.M.; Lee, E.J.; Kim, J.H.; Suk, K.; Lee, E.; Hur, K.; Hong, Y.J.; Do, J.T.; Park, S.; et al. SIRT2 is required for efficient reprogramming of mouse embryonic fibroblasts toward pluripotency. Cell death & disease 2018, 9, 893. [Google Scholar] [CrossRef] [PubMed]
- Samant, S.A.; Zhang, H.J.; Hong, Z.; Pillai, V.B.; Sundaresan, N.R.; Wolfgeher, D.; Archer, S.L.; Chan, D.C.; Gupta, M.P. SIRT3 deacetylates and activates OPA1 to regulate mitochondrial dynamics during stress. Molecular and cellular biology 2014, 34, 807–819. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, M.A.; Emerald, B.S.; Ansari, S.A. Stem cell fate determination through protein O-GlcNAcylation. The Journal of biological chemistry 2021, 296, 100035. [Google Scholar] [CrossRef] [PubMed]
- Sekita, Y.; Sugiura, Y.; Matsumoto, A.; Kawasaki, Y.; Akasaka, K.; Konno, R.; Shimizu, M.; Ito, T.; Sugiyama, E.; Yamazaki, T.; et al. AKT signaling is associated with epigenetic reprogramming via the upregulation of TET and its cofactor, alpha-ketoglutarate during iPSC generation. Stem cell research & therapy 2021, 12, 510. [Google Scholar] [CrossRef]
- Czajewski, I.; van Aalten, D.M.F. The role of O-GlcNAcylation in development. Development (Cambridge, England) 2023, 150. [Google Scholar] [CrossRef]
- Paneque, A.; Fortus, H.; Zheng, J.; Werlen, G.; Jacinto, E. The Hexosamine Biosynthesis Pathway: Regulation and Function. Genes (Basel) 2023, 14. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.S.; Saluja, A.K.; Banerjee, S. “Nutrient-sensing” and self-renewal: O-GlcNAc in a new role. J Bioenerg Biomembr 2018, 50, 205–211. [Google Scholar] [CrossRef]
- Baldini, S.F.; Steenackers, A.; Olivier-Van Stichelen, S.; Mir, A.M.; Mortuaire, M.; Lefebvre, T.; Guinez, C. Glucokinase expression is regulated by glucose through O-GlcNAc glycosylation. Biochem Biophys Res Commun 2016, 478, 942–948. [Google Scholar] [CrossRef] [PubMed]
- Pastor, W.A.; Aravind, L.; Rao, A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nature reviews. Molecular cell biology 2013, 14, 341–356. [Google Scholar] [CrossRef] [PubMed]
- Deplus, R.; Delatte, B.; Schwinn, M.K.; Defrance, M.; Méndez, J.; Murphy, N.; Dawson, M.A.; Volkmar, M.; Putmans, P.; Calonne, E.; et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. The EMBO journal 2013, 32, 645–655. [Google Scholar] [CrossRef] [PubMed]
- Speakman, C.M.; Domke, T.C.; Wongpaiboonwattana, W.; Sanders, K.; Mudaliar, M.; van Aalten, D.M.; Barton, G.J.; Stavridis, M.P. Elevated O-GlcNAc levels activate epigenetically repressed genes and delay mouse ESC differentiation without affecting naïve to primed cell transition. Stem cells (Dayton, Ohio) 2014, 32, 2605–2615. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Li, X.; Qin, K.; Shi, Y.; He, Y.; Zhang, C.; Cheng, B.; Zhang, X.; Hu, G.; Liang, S.; et al. Chemoproteomic and Transcriptomic Analysis Reveals that O-GlcNAc Regulates Mouse Embryonic Stem Cell Fate through the Pluripotency Network. Angew Chem Int Ed Engl 2023, 62, e202300500. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.; Kim, T.W.; Yoon, S.; Choi, S.Y.; Kang, T.W.; Kim, S.Y.; Kwon, Y.W.; Cho, E.J.; Youn, H.D. O-GlcNAc regulates pluripotency and reprogramming by directly acting on core components of the pluripotency network. Cell stem cell 2012, 11, 62–74. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Fan, X.; Shi, Y.; Zhang, C.; Sun, D.E.; Qin, K.; Qin, W.; Zhou, W.; Chen, X. Next-generation unnatural monosaccharides reveal that ESRRB O-GlcNAcylation regulates pluripotency of mouse embryonic stem cells. Nat Commun 2019, 10, 4065. [Google Scholar] [CrossRef]
- Lai, Y.S.; Chang, C.W.; Pawlik, K.M.; Zhou, D.; Renfrow, M.B.; Townes, T.M. SRY (sex determining region Y)-box2 (Sox2)/poly ADP-ribose polymerase 1 (Parp1) complexes regulate pluripotency. Proc Natl Acad Sci U S A 2012, 109, 3772–3777. [Google Scholar] [CrossRef]
- Kim, D.K.; Lee, J.S.; Lee, E.Y.; Jang, H.; Han, S.; Kim, H.Y.; Hwang, I.Y.; Choi, J.W.; Shin, H.M.; You, H.J.; et al. O-GlcNAcylation of Sox2 at threonine 258 regulates the self-renewal and early cell fate of embryonic stem cells. Exp Mol Med 2021, 53, 1759–1768. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Cheng, X.; Cheng, Y.; Chen, J.; Xu, H.; Gao, Y.; Duan, X.; Ji, J.; Li, X.; Yi, W. O-GlcNAcylation regulates the methionine cycle to promote pluripotency of stem cells. Proc Natl Acad Sci U S A 2020, 117, 7755–7763. [Google Scholar] [CrossRef]
- Hart, G.W. Nutrient regulation of signaling and transcription. J Biol Chem 2019, 294, 2211–2231. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.S.; Cui, W. Proliferation, survival and metabolism: the role of PI3K/AKT/mTOR signalling in pluripotency and cell fate determination. Development (Cambridge, England) 2016, 143, 3050–3060. [Google Scholar] [CrossRef] [PubMed]
- Romorini, L.; Garate, X.; Neiman, G.; Luzzani, C.; Furmento, V.A.; Guberman, A.S.; Sevlever, G.E.; Scassa, M.E.; Miriuka, S.G. AKT/GSK3β signaling pathway is critically involved in human pluripotent stem cell survival. Scientific reports 2016, 6, 35660. [Google Scholar] [CrossRef]
- Yu, Y.; Liang, D.; Tian, Q.; Chen, X.; Jiang, B.; Chou, B.K.; Hu, P.; Cheng, L.; Gao, P.; Li, J.; et al. Stimulation of somatic cell reprogramming by ERas-Akt-FoxO1 signaling axis. Stem cells (Dayton, Ohio) 2014, 32, 349–363. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.H.; Su, C.C.; Deng, W.; Lock, L.F.; Donovan, P.J.; Kayala, M.A.; Baldi, P.; Lee, H.C.; Chen, Y.; Wang, P.H. Mitochondrial Akt Signaling Modulated Reprogramming of Somatic Cells. Scientific reports 2019, 9, 9919. [Google Scholar] [CrossRef] [PubMed]
- Ward, P.S.; Thompson, C.B. Signaling in control of cell growth and metabolism. Cold Spring Harbor perspectives in biology 2012, 4, a006783. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, T.; Steiner, R.; Lengerke, C. SOX2 and p53 Expression Control Converges in PI3K/AKT Signaling with Versatile Implications for Stemness and Cancer. International journal of molecular sciences 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- Szwed, A.; Kim, E.; Jacinto, E. Regulation and metabolic functions of mTORC1 and mTORC2. Physiological reviews 2021, 101, 1371–1426. [Google Scholar] [CrossRef]
- Dodd, K.M.; Yang, J.; Shen, M.H.; Sampson, J.R.; Tee, A.R. mTORC1 drives HIF-1α and VEGF-A signalling via multiple mechanisms involving 4E-BP1, S6K1 and STAT3. Oncogene 2015, 34, 2239–2250. [Google Scholar] [CrossRef] [PubMed]
- Morita, M.; Gravel, S.P.; Chénard, V.; Sikström, K.; Zheng, L.; Alain, T.; Gandin, V.; Avizonis, D.; Arguello, M.; Zakaria, C.; et al. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell metabolism 2013, 18, 698–711. [Google Scholar] [CrossRef] [PubMed]
- Carroll, B.; Maetzel, D.; Maddocks, O.D.; Otten, G.; Ratcliff, M.; Smith, G.R.; Dunlop, E.A.; Passos, J.F.; Davies, O.R.; Jaenisch, R.; et al. Correction: Control of TSC2-Rheb signaling axis by arginine regulates mTORC1 activity. eLife 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Lu, Y.; Piao, W.; Jin, H. The Translational Regulation in mTOR Pathway. Biomolecules 2022, 12. [Google Scholar] [CrossRef] [PubMed]
- Li, T.Y.; Sun, Y.; Liang, Y.; Liu, Q.; Shi, Y.; Zhang, C.S.; Zhang, C.; Song, L.; Zhang, P.; Zhang, X.; et al. ULK1/2 Constitute a Bifurcate Node Controlling Glucose Metabolic Fluxes in Addition to Autophagy. Molecular cell 2016, 62, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 169, 361–371. [Google Scholar] [CrossRef]
- Zunder, E.R.; Lujan, E.; Goltsev, Y.; Wernig, M.; Nolan, G.P. A continuous molecular roadmap to iPSC reprogramming through progression analysis of single-cell mass cytometry. Cell stem cell 2015, 16, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Ly, C.H.; Lynch, G.S.; Ryall, J.G. A Metabolic Roadmap for Somatic Stem Cell Fate. Cell metabolism 2020, 31, 1052–1067. [Google Scholar] [CrossRef] [PubMed]
- Godini, R.; Lafta, H.Y.; Fallahi, H. Epigenetic modifications in the embryonic and induced pluripotent stem cells. Gene expression patterns: GEP 2018, 29, 1–9. [Google Scholar] [CrossRef]
- Acharjee, S.; Chauhan, S.; Pal, R.; Tomar, R.S. Mechanisms of DNA methylation and histone modifications. Progress in molecular biology and translational science 2023, 197, 51–92. [Google Scholar] [CrossRef]
- Ding, Y.; Yao, Y.; Gong, X.; Zhuo, Q.; Chen, J.; Tian, M.; Farzaneh, M. JMJD3: a critical epigenetic regulator in stem cell fate. Cell communication and signaling: CCS 2021, 19, 72. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zhang, H.; Wang, L.; Tang, C.; Qin, X.; Wu, X.; Pan, M.; Tang, Y.; Yang, Z.; Babarinde, I.A.; et al. JMJD3 acts in tandem with KLF4 to facilitate reprogramming to pluripotency. Nature communications 2020, 11, 5061. [Google Scholar] [CrossRef]
- Rad, S.M.; Mohammadi-Sangcheshmeh, A.; Bamdad, T.; Langroudi, L.; Atashi, A.; Lotfinia, M.; Arefian, E.; Gastal, E.L.; Soleimani, M. Pluripotency Crossroads: Junction of Transcription Factors, Epigenetic Mechanisms, MicroRNAs, and Long Non-coding RNAs. Current stem cell research & therapy 2017, 12, 300–311. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Ji, K.Y.; Zhang, J.; Xu, Y.; Ying, Y.; Mai, T.; Xu, S.; Zhang, Q.B.; Yao, K.T.; Xu, Y. Core pluripotency factors promote glycolysis of human embryonic stem cells by activating GLUT1 enhancer. Protein & cell 2019, 10, 668–680. [Google Scholar] [CrossRef]
- Yuan, X.; Zhang, C.; Zhao, R.; Jiang, J.; Shi, X.; Zhang, M.; Sun, H.; Zuo, Q.; Zhang, Y.; Song, J.; et al. Glycolysis Combined with Core Pluripotency Factors to Promote the Formation of Chicken Induced Pluripotent Stem Cells. Animals: an open access journal from MDPI 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Stirparo, G.G.; Kurowski, A.; Yanagida, A.; Bates, L.E.; Strawbridge, S.E.; Hladkou, S.; Stuart, H.T.; Boroviak, T.E.; Silva, J.C.R.; Nichols, J. OCT4 induces embryonic pluripotency via STAT3 signaling and metabolic mechanisms. Proceedings of the National Academy of Sciences of the United States of America 2021, 118. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Malik, N.; Kim, Y.I.; He, Y.; Li, M.; Dubois, W.; Liu, H.; Peat, T.J.; Nguyen, J.T.; Tseng, Y.C.; et al. Fatty acid oxidation is required for embryonic stem cell survival during metabolic stress. EMBO reports 2021, 22, e52122. [Google Scholar] [CrossRef] [PubMed]
- Sone, M.; Morone, N.; Nakamura, T.; Tanaka, A.; Okita, K.; Woltjen, K.; Nakagawa, M.; Heuser, J.E.; Yamada, Y.; Yamanaka, S.; et al. Hybrid Cellular Metabolism Coordinated by Zic3 and Esrrb Synergistically Enhances Induction of Naive Pluripotency. Cell metabolism 2017, 25, 1103–1117.e1106. [Google Scholar] [CrossRef]
Figure 1.
Glucose metabolism transformation and its molecular correlation during reprogramming process. (A) Hypoxia plays an important role in the production of iPSCs, and the stable increase of HIF2α and HIF3α in the expression of transcription factors. Knock down HIF-2 α or HIF-3α, resulting in a decrease in the expression of Oct4, Nanog, and Sox2. (B) The TCL1 induced by Klf4 adopts a dual pathway mechanism. Klf4 upregulates the expression of TCL1 and activates the AKT pathway to enhance glycolysis during reprogramming. TCL1 inhibits mitochondrial biogenesis and OXPHOS by inhibiting mitochondrial polynucleoside phosphorylase (PnPase). (C) In the early stages of reprogramming, the OXPHOS outbreak caused a sharp increase in ROS production, and the increase in ROS activity was induced by NRF2 to increase HIF1 levels. NRF2 activates HIF1α through TRX1, and HIF2α upregulates the expression of TNF related apoptosis inducing ligand (TRAIL) in the late stage of reprogramming, inhibiting reprogramming.
Figure 1.
Glucose metabolism transformation and its molecular correlation during reprogramming process. (A) Hypoxia plays an important role in the production of iPSCs, and the stable increase of HIF2α and HIF3α in the expression of transcription factors. Knock down HIF-2 α or HIF-3α, resulting in a decrease in the expression of Oct4, Nanog, and Sox2. (B) The TCL1 induced by Klf4 adopts a dual pathway mechanism. Klf4 upregulates the expression of TCL1 and activates the AKT pathway to enhance glycolysis during reprogramming. TCL1 inhibits mitochondrial biogenesis and OXPHOS by inhibiting mitochondrial polynucleoside phosphorylase (PnPase). (C) In the early stages of reprogramming, the OXPHOS outbreak caused a sharp increase in ROS production, and the increase in ROS activity was induced by NRF2 to increase HIF1 levels. NRF2 activates HIF1α through TRX1, and HIF2α upregulates the expression of TNF related apoptosis inducing ligand (TRAIL) in the late stage of reprogramming, inhibiting reprogramming.

Figure 2.
Amino acid metabolism regulates stem cell homeostasis. (A)The folate cycle and methionine cycle play important roles in stem cell homeostasis. (B) Amino acid metabolism involves thyroid metabolism, One-C metabolism, metabolic cycle, glutamine metabolism, and L-Pro metabolism, playing important roles in various processes of stem cells.
Figure 2.
Amino acid metabolism regulates stem cell homeostasis. (A)The folate cycle and methionine cycle play important roles in stem cell homeostasis. (B) Amino acid metabolism involves thyroid metabolism, One-C metabolism, metabolic cycle, glutamine metabolism, and L-Pro metabolism, playing important roles in various processes of stem cells.
Figure 3.
The mPTP model and mitochondrial metabolism. (A) A schematic diagram of ATP synthase structure. (B) Models of high conductance and low conductance pore formation by ATP synthase and ANT. (C) A model of high conductivity channel formation mediated by ATP synthase. The binding of Ca2+and cyclophilic protein D (CypD) to ATP synthase leads to the instability of the dimer structure. (D) Schematic diagram of the open mechanism for regulating mPTP. ERK inhibits GSK3β activity, which promotes CyPD phosphorylation and induces mPTP opening. Phosphorylated STAT3 inhibits mPTP opening by binding to CyPD, while CyPD acetylation induces mPTP opening. MPTP is short-term open in the early stages of reprogramming, and is enhanced by PHF8 mediated reprogramming. NRF2 enhances cellular antioxidant stress capacity and enhances reprogramming by regulating HIF1α. The activation of Nf kB is beneficial for cell survival.
Figure 3.
The mPTP model and mitochondrial metabolism. (A) A schematic diagram of ATP synthase structure. (B) Models of high conductance and low conductance pore formation by ATP synthase and ANT. (C) A model of high conductivity channel formation mediated by ATP synthase. The binding of Ca2+and cyclophilic protein D (CypD) to ATP synthase leads to the instability of the dimer structure. (D) Schematic diagram of the open mechanism for regulating mPTP. ERK inhibits GSK3β activity, which promotes CyPD phosphorylation and induces mPTP opening. Phosphorylated STAT3 inhibits mPTP opening by binding to CyPD, while CyPD acetylation induces mPTP opening. MPTP is short-term open in the early stages of reprogramming, and is enhanced by PHF8 mediated reprogramming. NRF2 enhances cellular antioxidant stress capacity and enhances reprogramming by regulating HIF1α. The activation of Nf kB is beneficial for cell survival.
Figure 4.
Molecular Mechanisms of Stem Cell Metabolic Regulation. The HBP pathway integrates glucose, amino acids, lipids, and nucleotide metabolism, ultimately generating the substrate UDP-GlcNAc for the O-GlcNAcylation reaction. O-GlcNAcylation modification, PI3K/AKT/mTORC1, AMPK, and NAD/SIRT1 pathways regulate stem cell metabolism levels.
Figure 4.
Molecular Mechanisms of Stem Cell Metabolic Regulation. The HBP pathway integrates glucose, amino acids, lipids, and nucleotide metabolism, ultimately generating the substrate UDP-GlcNAc for the O-GlcNAcylation reaction. O-GlcNAcylation modification, PI3K/AKT/mTORC1, AMPK, and NAD/SIRT1 pathways regulate stem cell metabolism levels.
Figure 5.
Epigenetic regulation and major metabolic pathways of stem cells. Mitochondrial TCA cycle intermediate metabolites, such as Acetyl-CoA, citric acid, and α-KG, are transported to the cytoplasm and nucleus for the biosynthesis of amino acids and lipids or participate in gene expression through epigenetic regulation. The mitochondrial single carbon (One-C) metabolism, combined with the cytoplasmic folate and methionine cycle, produces SAM for histones, DNA methyltransferases, and other methylation reactions. Metabolites, epigenetic regulation, and interactions between nuclei directly affect chromatin structure and gene expression, controlling self-renewal of stem cells.
Figure 5.
Epigenetic regulation and major metabolic pathways of stem cells. Mitochondrial TCA cycle intermediate metabolites, such as Acetyl-CoA, citric acid, and α-KG, are transported to the cytoplasm and nucleus for the biosynthesis of amino acids and lipids or participate in gene expression through epigenetic regulation. The mitochondrial single carbon (One-C) metabolism, combined with the cytoplasmic folate and methionine cycle, produces SAM for histones, DNA methyltransferases, and other methylation reactions. Metabolites, epigenetic regulation, and interactions between nuclei directly affect chromatin structure and gene expression, controlling self-renewal of stem cells.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).