Submitted:
11 May 2024
Posted:
14 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction: Fluorescent Nucleoside Analogs – An Outlook
2. Purine Nucleoside Phosphorylase as a Synthetic Tool
3. Chemo-Enzymatic Synthesis of Fluorescent Ribosides of Purines and 8-Azapurines

3.1. 8-Azaadenosine, 8-Azainosine, 8-Azaxanthosine and Derivatives
3.2. 8-Azaguanosine and Analogs
3.3. Other 8-Azapurine Analogs and Their Enzymatic Ribosylation
3.4. 8-Aza-7-Deazapurine and 8-aza-9-Deazapurine Ribosides
3.5. Other Bicyclic Heteroaromatics
4. Chemo-Enzymatic Synthesis and Properties of the Tri-Cyclic Purine Analogs and Their Ribosides
4.1. Adenosine Analogs
4.2. Guanosine Analogs
4.3. 2-Aminopurine Riboside and Isoguanosine Analogs
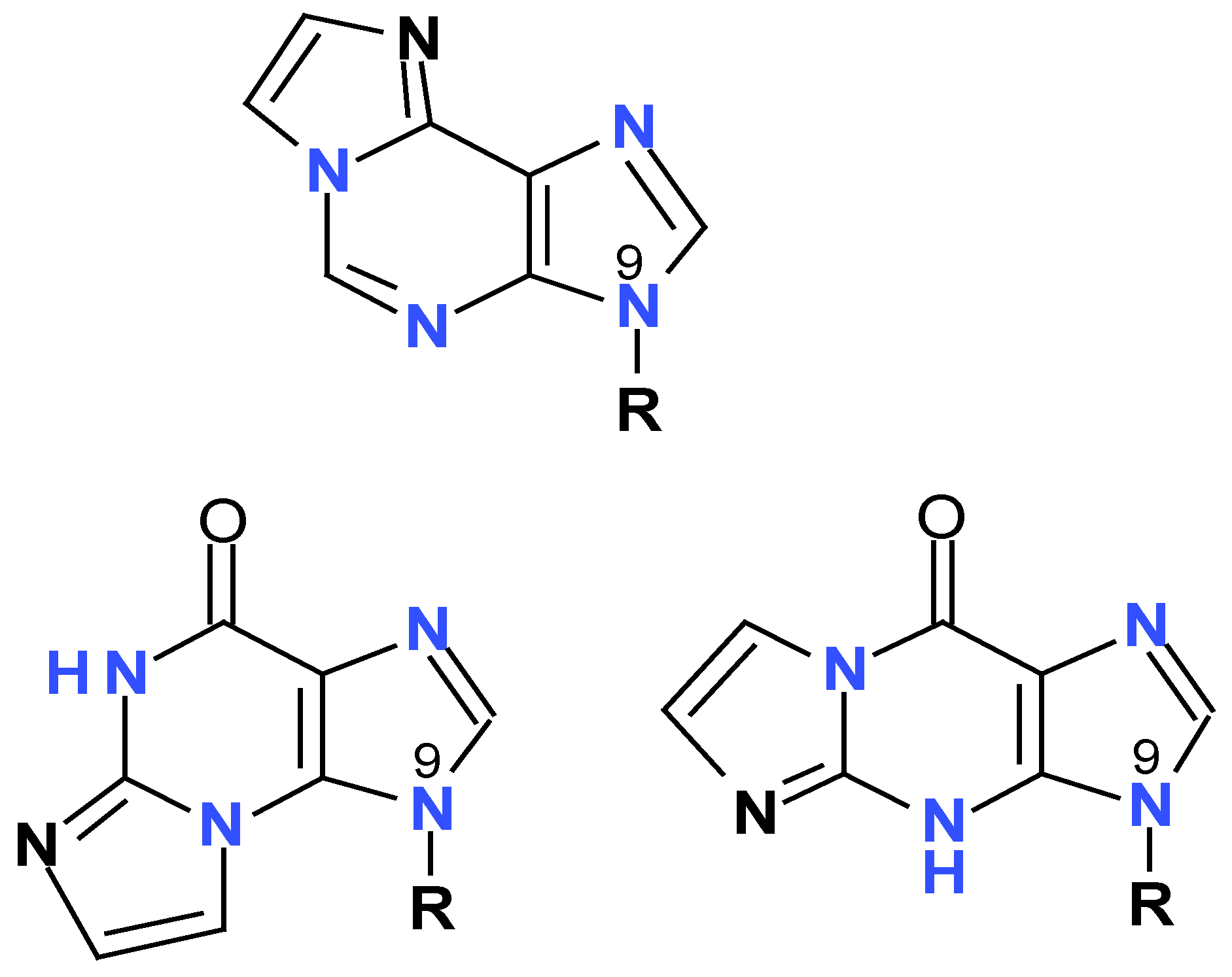
4.4. Tri-Cyclic aza-Purine Analogs
| Compound | excitation: λmax [nm] |
emission: λmax [nm] |
φ | τ [ns] |
|---|---|---|---|---|
| 1,N6-etheno-adenosine | 305 | 410 | 0.56 | 21 |
| 1,N6-etheno-6-β-D-ribosyl-adenine | 310 | 380 | 0.10 | nd |
| 1,N2-ethenoguanosine (anion) | 400 | <0.01 | nd | |
| N2,3-ethenoguanosine | 261 | 400 | 0.02 | nd |
| N2,3-etheno-O6-methylguanosine | 272 | 405 | 0.11 | nd |
| 1,N2-etheno-2-aminopurine | 248 | 473 | 0.18 | 6.9;10.3 |
| 1,N2-etheno-2-aminopurine-9-β-D- riboside |
295 | 463 | 0.14 | 3.8;8.5 |
| 1,N2-etheno-2-aminopurine-2-β-D- riboside |
338 | 406 | 0.73 | nd |
| N2,3-etheno-2-aminopurine | 315 | 357 | 0.29 | 2.15 |
| N2,3-etheno-2-aminopurine-2-β-D- riboside |
315 | 365 | 0.29 | nd |
| 1,N6-etheno-2-oxo-adenine | 295 | 415 | 0.17 | nd |
| 1,N6-etheno-2-oxo-adenosine | 295 | 415 | 0.34 | 6.1 |
| 1,N6-etheno-2-oxo-adenine-7-β-D- riboside |
294 | 360 | 0.036 | 0.8;5.2 |
| 1,N6-etheno-2-oxo-adenine-6-β-D- riboside |
303 | 425 | 0.66 | nd |
5. Conclusion and Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lakowicz, J. Principles of fluorescence spectroscopy, 3rd Edition. Springer, New York, 2006.
- Zalejski, J.; Sun, J.; Sharma, A. Unravelling the Mystery inside Cells by Using Single-Molecule Fluorescence Imaging. J. of Imaging 2023, 9, art#192. [Google Scholar] [CrossRef]
- Adhikari, S; Smit, R and Orrit, M. Future Paths in Cryogenic Single-Molecule Fluorescence Spectroscopy. J. Physical Chem. C, 2023, 128, 3-18. DOI:10.1021/acs.jpcc.3c06564; b) Wang, D.; Shalamberidze, A.; Arguello, A. E.; Purse, B. W.; Kleiner, R. E. Live-Cell RNA Imaging with Metabolically Incorporated Fluorescent Nucleosides. J. Am. Chem. Soc. 2022, 144, 14647–14656, DOI: 10.1021/jacs.2c04142.
- Wilhelmsson M, Tor Y. Fluorescent Analogues of Biomolecular Building Blocks: Design and Applications. John Wiley & Sons; New York, USA, 2016.
- Xu, W.; Ke, M.C.; Kool, E.T. Fluorescent nucleobases as tools for studying DNA and RNA. Nature Chem. 2017, 9, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Tanpure, A.A.; Pawar, G.; Srivatsan, S.G. Fluorescent Nucleoside Analogs: Probes for Investigating Nucleic Acid Structure and Function. Israel Journal of Chemistry 2013, 53, 366–378. [Google Scholar] [CrossRef]
- Hocek, M. Enzymatic Synthesis of Base-Functionalized Nucleic Acids for Sensing, Cross-linking, and Modulation of Protein–DNA Binding and Transcription. Accounts of Chemical Research 2019, 52, 1730–1737. [Google Scholar] [CrossRef]
- Michel, B.Y.; Dziuba, D.; Benhida, R.; Demchenko, A.P.; Burger, A. Probing of Nucleic Acid Structures, Dynamics, and Interactions With Environment-Sensitive Fluorescent Labels. Front. Chem. 2020, 8, 112. [Google Scholar] [CrossRef]
- a) Xu, Wang; Ke, Min Chan; Kool, E.T. Fluorescent nucleobases as tools for studying DNA and RNA. Nature Chem. 2017, 9, 1043–1055. https://doi.org/10.1038/nchem.2859 b) Wang, D.; Shalamberidze, A.; Arguello, A. E.; Purse, B. W.; Kleiner, R. E. Live-Cell RNA Imaging with Metabolically Incorporated Fluorescent Nucleosides. J. Am. Chem. Soc. 2022, 144, 14647– 14656, DOI: 10.1021/jacs.2c04142.
- Saito, Y.; Hudson, R.H.E. Base-modified fluorescent purine nucleosides and nucleotides for use in oligonucleotide probes. J. Photochem. Photobiol. C: Photochemistry Reviews 2018, 36, 48–73. [Google Scholar] [CrossRef]
- Singh, H.; K Tiwari, R. Tiwari, S.K. Pramanik, A. Das. Small molecules as fluorescent probes for monitoring intracellular enzymatic transformations. Chemical Revs. 2019, 119, 11718-11760. [CrossRef]
- Gilbault, G.G. in: Practical Fluorescence, editor: G. G. Gilbault, Marcel Dekker, New York, 1990, Chapter 12.
- Callis, P. R. Electronic States and Luminescence of Nucleic Acids Systems. Annu. Rev. Phys. Chem. 1983, 34, 329–357. [Google Scholar] [CrossRef]
- Leng, M.; Pochon, F.; Michelson, M. Photochemistry of polynucleotides.2. Mononucleotide and dinucleotide fluorescence at ordinary temperatures. Biochim Biophys Acta 1968, 169, 338–349. [Google Scholar] [CrossRef]
- Ward, D. C.; Reich, E.; Stryer, L. Fluorescence Studies of Nucleotides and Polynucleotides. 1. Formycin, 2-aminopurine riboside, 2,6-diaminopurine riboside and their derivatives. J. Biol. Chem. 1969, 244, 1228–1237. [Google Scholar] [CrossRef]
- Jones, A. C.; Neely, R. K. 2-aminopurine as a fluorescent probe of DNA conformation and the DNA–enzyme interface. Q. Rev. Biophys. 2015, 48, 244–279. [Google Scholar] [CrossRef]
- Kirk, S. R.; Luedtke, N. W.; Tor, Y. 2-Aminopurine as a Real-Time Probe of Enzymatic Cleavage and Inhibition of Hammerhead Ribozymes. Bioorg. Med. Chem. 2001, 9, 2295–2301. [Google Scholar] [CrossRef]
- P. Herdewijn, Ed. Modified nucleosides in biochemistry, biotechnology and medicine. Wiley-VCH, New York, 2008.
- Sinkeldam, R. W.; Greco, N. J.; Tor, Y. Fluorescent Analogs of Biomolecular Building Blocks: Design, Properties and Applications. Chem. Rev. 2010, 110, 2579–2619. [Google Scholar] [CrossRef]
- Okamoto, A.; Saito, Y.; Saito I. Design of base-discriminating fluorescent nucleosides. J. Photochem. Photobiol., C 2005, 6, 108–122. [CrossRef]
- Tor, Y. Isomorphic Fluorescent Nucleosides. Acc. Chem. Res. 2024, 57(9), 1325–1335. [Google Scholar] [CrossRef]
- Ludford, P.; Yang, Shengua; Bucardo, M.S.; Tor, Y. A New Variant of Emissive RNA Alphabets. Chemistry Europ. J. 2022, 28, e202104472. [CrossRef]
- Dziuba, D.; Didier, P.; Ciaco, S.; Barth, A.; Seidel, C.A.M.; Mély, Y. Fundamental photophysics of isomorphic and expanded fluorescent nucleoside analogues. Chemical Society Reviews 2021, 50, 7062–7107. [Google Scholar] [CrossRef] [PubMed]
- Mikhailopulo, A.I.; Miroshnikov, A.I. Biologically important nucleosides: Modern trends in biotechnology and application. Mendeleev Comm. 2011, 21, 57–68. [Google Scholar] [CrossRef]
- Cosgrove, S.C.; Miller, G.J. Advances in biocatalytic and chemoenzymatic synthesis of nucleoside analogues. Expert Opinion on Drug Discovery 2022, 17, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, P. A.; Sinkeldam, R. W.; Tor, Y. Visibly Emissive and Responsive Extended 6-Aza-Uridines. Org. Lett. 2014, 16, 5290– 5293. [CrossRef]
- Ardhapure, A.V.; Gayakhe, V.; Bhilare, S.; Kapdi, A.R.; Bag, S.S.; Sanghvi, Y.S.; Gunturu, K.C. Extended fluorescent uridine analogues: synthesis, photophysical properties and selective interaction with BSA protein. New J. Chem. 2020, 44, 14744–14754. [Google Scholar] [CrossRef]
- Greco, N. J.; Tor, Y. Simple Fluorescent Pyrimidine Analogs Detect the Presence of DNA Abasic Sites. J. Am. Chem. Soc. 2005, 127, 10784–10785. [Google Scholar] [CrossRef]
- Bzowska A, Kulikowska E. Shugar D. Purine nucleoside phosphorylases: Properties, functions, and clinical aspects. Pharmacol. Therap. 2000, 88, 349-425. [CrossRef]
- Rodwell, V.W. Metabolism of purine and pyrimidine nucleotides. In: Harper’s Illustrated Biochemistry, 30-th edition, McGraw & Hill, 2008, pp. 347-358.
- a) Grunebaum, E.; Cohen, A.; Roifman, C.M. Recent advances in understanding and managing adenosine deaminase and purine nucleoside phosphorylase deficiencies. Curr. Opin. Allergy Clin. Immunol. 2013, 13, 630–638. DOI: 10.1097/ACI.0000000000000006; b) Fox, T.A.; Booth, C. Gene therapy for primary immunodeficiencies. British Journ. Haematol. 2021, 193, 1044-1059. DOI: 10.1111/bjh.17269.
- Ardiani, A.; Johnson, A.J.; Ruan, H.M.; Sanchez-Bonilla, M.; Serve, K.; Black, M.E. Enzymes To Die For: Exploiting Nucleotide Metabolizing Enzymes for Cancer Gene Therapy. Current Gene Therap. 2012, 12, 77–91. [Google Scholar] [CrossRef]
- Schramm, V.L. Enzymatic Transition States and Drug Design. Chem. Revs. 2018, 118, 11194–11258. [Google Scholar] [CrossRef]
- a) Lewis, D.J.; Duvic, M. Forodesine in the treatment of cutaneous T-cell lymphoma. Expert Op Investig Drugs 2017, 26, 771-775. DOI: 10.1080/13543784.2017.1324569; b) Makita S.; Maeshima A.M.; Maruyama D.; Izutsu K.; Tobinai K. "Forodesine in the treatment of relapsed/refractory peripheral T-cell lymphoma: an evidence-based review". OncoTargets & Therapy 2018, 11, 2287–2293. doi:10.2147/OTT.S140756. PMC 5916385. PMID 29719411.
- Ho, M.C.; Shi, W.; Rinaldo-Matthis, A.; Tyler, P.C.; Evans, G.B.; Clinch, K.; Almo, S.C.; Schramm, V.L. Four generations of transition-state analogues for human purine nucleoside phosphorylase. Proc. Natl. Acad. Sci. USA 2010, 107, 4805–4812. [Google Scholar] [PubMed]
- Mikhailopulo, I.A. Biotechnology of Nucleic Acid Constituents – State of the Art and Perspectives. Curr. Org. Chem. 2007, 11, 317–335. [Google Scholar] [CrossRef]
- Kamel, S.; Yehia, H.; Neubauer, P.; Wagner, A. Enzymatic Synthesis of Nucleoside Analogues by Nucleoside Phosphorylases. In: Enzymatic and Chemical Synthesis of Nucleic Acid Derivatives; Fernandez-Lucas, J., Camarasa-Rius, M.J., Eds.; Wiley-VCH, New York, 2019. Chapter 1.
- Yehia, H.; Kamel, S.; Paulick, K.; Neubauer, P.; Wagner, A. Substrate Spectra of Nucleoside Phosphorylases and their Potential in the Production of Pharmaceutically Active Compounds. Curr. Pharm. Des. 2017, 23, 6913–6935. [Google Scholar] [CrossRef]
- Westarp, S.; Kaspar, F.; Neubauer, P.; Kurreck, A. Industrial potential of the enzymatic synthesis of nucleoside analogs: existing challenges and perspectives. Current Op. Biotech. 2022, 78, 102829. [Google Scholar] [CrossRef]
- Hennen, W.J.; Wong, C.-H. A new method for the enzymic synthesis of nucleosides using purine nucleoside phosphorylase. J. Org. Chem. 1989, 4692–4695. [Google Scholar] [CrossRef]
- Krenitsky, T.A.; Koszalka, G.W.; Tuttle, J.V. Purine nucleoside synthesis: An efficient method employing nucleoside phosphorylases. Biochemistry 1981, 20, 3615–3621. [Google Scholar] [CrossRef]
- Stoeckler, J.D.; Poirot, A.F.; Smith, R.M.; Parks, R.E., Jr.; Ealick, S.E.; Takabayashi, K.; Erion, M.D. Purine-nucleoside phosphorylase. 3. Reversal of purine base specificity by site-directed mutagenesis. Biochemistry1997, 36, 11749–11756. [CrossRef]
- Kamel, S.; Thiele, I.; Neubauer, P., Wagner, A. Thermophilic nucleoside phosphorylases: Their properties, characteristics and applications. Biochim. Biophys. Acta-Proteins Proteomics 2020, 1868, 140304. [CrossRef]
- Zhou, X.R.; Mikhailopulo, I.A.; Bournazou, M.N.C.; Neubauer, P. Immobilization of thermostable nucleoside phosphorylases on MagReSyn® epoxide microspheres and their application for the synthesis of 2,6-dihalogenated purine nucleosides. J. Mol. Catal. B 2015, 115, 119-127. [CrossRef]
- a) Bzowska, A.; Kulikowska, E.; Poopeiko, N.E.; Shugar, D. Kinetics of phosphorolysis of 3-(β-D-ribofuranosyl)adenine and 3-(β-D-ribofuranosyl)hypoxanthine, non-conventional substrates of purine-nucleoside phosphorylase. Eur. J. Biochem. 1996, 239, 229–234. DOI: 10.1111/j.1432-1033.1996.0229u.x; b) Bzowska, A.; Ananiev, A.V.; Ramzaeva, N.; Alksnis, E.; Maurins, J.A.; Kulikowska, E.; Shugar, D. Purine nucleoside phosphorylase: Inhibition by purine N(7)- and N(9)-acyclonucleosides; and substrate properties of 7-β-D-ribofuranosylguanine and 7-β-D-ribofuranosylhypoxanthine. Biochem. Pharmacol. 1994, 48, 937–947.
- Fernandez-Lucas, J. Multienzymatic synthesis of nucleic acid derivatives: a general perspective. Appl Microbiol Biotechnol 2015, 99, 4615–4627. [CrossRef]
- Del Arco, J.; Acosta, J.; Fernandez-Lucas, J. New trends in the biocatalytic production of nucleosidic active pharmaceutical ingredients using 2’-deoxyribosyl transferases. Biotechnol. Advances 2021, 51, 107701. [Google Scholar] [CrossRef] [PubMed]
- Del Arco, J.; Fernandez-Lucas, J. Purine and pyrimidine phosphoribosyl transferases: a versatile tool for enzymatic synthesis of nucleoside-5’-mono phosphates, Curr. Pharm. Des. 2017, 23, 6898 – 6912. [CrossRef]
- Wierzchowski J., Stachelska-Wierzchowska, A., Wielgus-Kutrowska, B. Bzowska, A. 1,N6-ethenoadenine and other fluorescent nucleobase analogues as substrates for purine-nucleoside phosphorylases: spectroscopic and kinetic studies. Curr. Pharmaceut. Design 2017, 23, 6972-6990. [CrossRef]
- Wierzchowski, J.; Wielgus-Kutrowska, B.; Shugar, D. Fluorescence emission properties of 8- azapurines and their nucleosides, and application to the kinetics of the reverse synthetic reaction of PNP. Biochim Biophys Acta 1996, 1290, 9–17. [Google Scholar] [CrossRef]
- Albert, A. Chemistry of 8-azapurines. Adv Heterocycl Chem 1986, 39, 117–178. [Google Scholar] [CrossRef]
- Giorgi, I.; Scartoni, V. 8-Azapurine nucleus: a versatile scaffold for different targets. Mini-Reviews Medicinal Chemistry 2009, 9 1367–1378. [CrossRef]
- Wierzchowski J., Antosiewicz J.M., Shugar D. 8-Azapurines as isosteric purine fluorescent probes for nucleic acid and enzymatic research. Molecular BioSystems 2014, 10, 2756-2774. [CrossRef]
- Seela F., Jawalekar A.M., Münster, I. Replacement of Canonical DNA Nucleobases by Benzotriazole and 1,2,3-Triazolo[4,5-d]pyrimidine: Synthesis, Fluorescence, and Ambiguous Base Pairing. Helvetica Chimica Acta 2005, 88, 751-765. [CrossRef]
- Cottrell, J.W, Scott, L.G., Fedor, M.J. The pH dependence of hairpin ribozyme catalysis reflects ionization of an active site adenine, J. Biol. Chem. 2011, 286, 17658–17664. [CrossRef]
- Viladoms, J., Scott, L.G., Fedor, M.J. An active-site guanine participates in glms ribozyme catalysis in its protonated state. J. Amer. Chem. Soc. 2011, 133, 18388–18396. [CrossRef]
- a) Wierzchowski, J.; Sepiol, J.; Sulikowski, D.; Kierdaszuk, B.; Shugar, D. Fluorescence emission properties of 8-azaxanthine and its N-alkyl derivatives: Excited-state proton transfer, and potential applications in enzymology. J. Potochem. Photobiol. A-Chemistry 2006, 179, 276-282. DOI10.1016/j.jphotochem.2005.08.027; b) Wierzchowski, J.; Smyk, B. Excited-State Proton Transfer in 8-Azapurines I: A Kinetic Analysis of 8-Azaxanthine Fluorescence. Molecules 2020, 25, #2740.
- Dandanell, G.; Szczepanowski, R.H.; Kierdaszuk, B.; Shugar, D.; Bochtler, M. Escherichia coli purine nucleoside phosphorylase II, the product of the xapA gene. J. Mol. Biol. 2005, 348, 113–125. [Google Scholar] [CrossRef]
- Albertini, R.J. Albertini, R.J. HPRT mutations in humans: biomarkers for mechanistic studies. Mutat. Res. 489, 1-16. Mutation Res., 2001, 489, 1–16. [CrossRef]
- Stachelska-Wierzchowska, A; Wierzchowski, J; Wielgus-Kutrowska B.; Mikleusevic, G. Enzymatic Synthesis of Highly Fluorescent 8-Azapurine Ribosides Using a Purine Nucleoside Phosphorylase Reverse Reaction: Variable Ribosylation Sites. Molecules 2013, 18, 12587–12598. [CrossRef]
- Stachelska-Wierzchowska, A.; Wierzchowski, J.; Bzowska, A.; Wielgus-Kutrowska, B. Site-selective ribosylation of fluorescent nucleobase analogs using purine-nucleoside phosphorylase as a catalyst: effects of point mutations. Molecules 2016, 21: 44. [CrossRef]
- Wierzchowski, J.; Bzowska, A.; Stępniak, K.; Shugar, D. Interactions of Calf Spleen Purine Nucleoside Phosphorylase with 8-Azaguanine, and a Bisubstrate Analogue Inhibitor: Implications for the Reaction Mechanism. Z. Naturforsch C. 2004, 59, 713–725. [Google Scholar] [CrossRef]
- Wierzchowski, J.; Stępniak, K.; Bzowska, A.; Shugar, D. Spectroscopic and kinetic studies of interactions of calf spleen purine nucleoside phosphorylase with 8-azaguanine and its 9-(2-phosphonylmethoxyethyl) derivative. Nucleosides, Nucleotides & Nucl Acids 2005, 24, 459-464. [CrossRef]
- Stepchenko, V.A.; Seela, F.; Esipov, R.S.; Miroshnikov, A.I.; Sokolov, Y.A.; Mikhailopulo, I.A. Enzymatic Synthesis of 2’-Deoxy-β-D-ribonucleosides of 8-Azapurines and 8-Aza-7-deazapurines. Synlett 2012, 10, 1541-45. [CrossRef]
- Wierzchowski, J.; Ogiela, M.; Iwańska, B.; Shugar, D. Selective fluorescent and fluorogenic substrates for purine-nucleoside phosphorylases from various sources, and direct fluorimetric determination of enzyme levels in human and animal blood. Analytica Chimica Acta 2002, 472, 63–74. [Google Scholar] [CrossRef]
- Stachelska-Wierzchowska, A.; Wierzchowski, J. Non-typical nucleoside analogs as fluorescent and fluorogenic indicators of purine-nucleoside phosphorylase activity in biological samples. Analytica Chimica Acta 2020, 1139, 119–128. [Google Scholar] [CrossRef]
- Pyrka, M.; Maciejczyk, M. Why Purine Nucleoside Phosphorylase Ribosylates 2,6-Diamino-8-azapurine in Noncanonical Positions? A Molecular Modeling Study. J. Chem. Inf. Model. 2020, 60, 1595–1606. [Google Scholar] [CrossRef]
- a) Holy, A. Phosphonomethoxyalkyl analogs of nucleotides. Curr. Pharm. Design 2003, 9, 2567-2592; DOI: 10.2174/1381612033453668 b) De Clercq, E.; Holy, A. Acyclic nucleoside phosphonates: A key class of antiviral drugs. Nature Revs Drug Disc. 2005, 4, 928-940. https://doi.org/10.1038/nrd1877.
- Wierzchowski, J.; Kulikowska, E.; Bzowska, A.; Holy, A.; Magnowska, L.; Shugar, D. Interactions of purine nucleoside phosphorylase with antiviral acyclic nucleoside phosphonate inhibitors – kinetics and emission studies. Nucleosides & Nucleotides 1999, 18, 875-876. [CrossRef]
- Jiang, D.W.; Seela, F. Oligonucleotide Duplexes and Multistrand Assemblies with 8-Aza-2′-deoxyisoguanosine: A Fluorescent isoGd Shape Mimic Expanding the Genetic Alphabet and Forming Ionophores. J. Amer. Chem. Soc. 2010, 132, 4016–4024. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Jiang, D.W.; Schäfer, A.H.; Seela, F. 8-Aza-2′-deoxyisoguanosine Forms Fluorescent Hydrogels whereas 8-Aza-2′-deoxyguanosine Assembles into Nucleoside Nanotubes. ChemPlusChem 2017, 82, 778-784. [CrossRef]
- Emmerson, B.T. Drug therapy - The management of gout. New England J. Med. 1996, 334, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Fateev, I.V.; Kharitonova, M.I.; Antonov, K.V.; Konstantinova, I.D.; Stepanenko, V.N.; Esipov, R.S.; Seela, F.; Temburnikar, K.W.; Seley-Radtke, K.L.; Stepchenko, V.A.; et al. Recognition of Artificial Nucleobases by E. coli Purine Nucleoside Phosphorylase versus its Ser90Ala Mutant in the Synthesis of Base-Modified Nucleosides. Chem. Eur. J. 2015, 21, 13401–13419. [CrossRef]
- Seela, F.; Becher, G. Synthesis, base pairing, and fluorescence properties of oligonucleotides containing 1H-pyrazolo[3,4-d]pyrimidin-6-amine (8-aza-7-deazapurin-2-amine) as an analogue of purin-2-amine. Helvetica Chim. Acta 2000, 83, 928-942. https://doi.org/10.1002/(SICI)1522-2675(20000510)83:5<928::AID-HLCA928>3.0.CO;2-5; b) Kondhare, D.; Leonard, P.; Seela, F. Isoguanine (2-Hydroxyadenine) and 2-Aminoadenine Nucleosides with an 8-Aza-7-deazapurine Skeleton: Synthesis, Functionalization with Fluorescent and Clickable Side Chains, and Impact of 7-Substituents on Physical Properties. J.Org. Chem. 2021, 86, 14461-14475. DOI10.1021/acs.joc.1c01283.
- De Clercq, E. C-Nucleosides To Be Revisited. J. Med. Chem. 2016, 59, 2301–2311. [Google Scholar] [CrossRef]
- Wierzchowski, J.; Shugar, D. Luminescence studies on formycin, its aglycone, and their N-methyl derivatives: tautomerism, sites of protonation and phototautomerism. Photochem. Photobiol. 1982, 35, 445–458. [Google Scholar] [CrossRef]
- Karlish, S.J.D. Use of formycin nucleotides, intrinsic protein fluorescence, and... FLUORESCEIN ISOTHIOCYANATE-labeled enzymes for measurement of conformational states of NA+,K+-ATPase. Methods Enzymol. 1988, 156, 271-277. [CrossRef]
- Kierdaszuk, B.; Modrak-Wojcik, A.; Wierzchowski, J.; Shugar, D. Formycin A and its N-methyl analogues, specific inhibitors of E-coli purine nucleoside phosphorylase (PNP): induced tautomeric shifts on binding to enzyme, and enzyme -> ligand fluorescence resonance energy transfer. Biochim. Biophys. Acta - Protein Struct. Mol. Enzymol. 2000, 1476, 109-128. [CrossRef]
- Wlodarczyk, J.; Galitonov, G.S.; Kierdaszuk, B. Identification of the tautomeric form of formycin A in its complex with Escherichia coli purine nucleoside phosphorylase based on the effect of enzyme-ligand binding on fluorescence and phosphorescence. Europ. Biophys. J. Biophys. Lett. 2004, 33, 377-385. [CrossRef]
- Konstantinova, I.D.; Selezneva, O.M.; Fateev, I.V.; Balashova, T.A.; Kotovskaya, S.K.; Baskakova, Z.M.; Charushin, V.N.; Baranovsky, A.V.; Miroshnikov, A.I.; Balzarini, J.; Mikhailopulo, I.A. Chemo-Enzymatic Synthesis and Biological Evaluation of 5,6-Disubstituted Benzimidazole Ribo- and 2′-Deoxyribonucleosides. Synthesis-Stuttgart 2013, 45, 272-280. [CrossRef]
- Kharitonova, M.I.; Fateev, I.V.; Kayushin, A.L.; Konstantinova, I.D.; Kotovskaya, S.K.; Andronova, V.L.; Galegov, G.A.; Charushin, V.N.; Miroshnikov, A.I. Chemoenzymatic Synthesis and Antiherpes Activity of 5-Substituted 4,6-Difluorobenzimidazoles Ribo- and 2′-Deoxyribonucleosides. Synthesis-Stuttgart 2016, 48, 394-406. [CrossRef]
- Leonard, N.J. Etheno-substituted nucleotides and coenzymes: Fluorescence and biological activity. Crit. Revs. Biochem. 1984, 15, 125–199. [Google Scholar] [CrossRef] [PubMed]
- Leonard, N.J. Adenylates: bound and unbound. Biopolymers 1985, 24, 9–28. [Google Scholar] [CrossRef] [PubMed]
- Secrist, J. A.; Barrio, J. R.; Leonard, N. J.; Weber, G. Fluorescent modifications of adenosine containing coenzymes. Biological activities and spectroscopic properties. Biochemistry 1972, 11, 3499– 3506. [CrossRef]
- Jahnz-Wechmann, Z.; Framski, G.R.; Januszczyk, P.A.; Boryski, J. Bioactive fused heterocycles: Nucleoside analogs with an additional ring. Eur. J. Med. Chem. 2015, 97, 388–396. [Google Scholar] [CrossRef] [PubMed]
- Jahnz-Wechmann, Z.; Framski, G.R.; Januszczyk, P.A.; Boryski, J. Base-modified nucleosides: Etheno derivatives. Frontiers Chem. 2016, 4, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Stachelska-Wierzchowska, A.; Wierzchowski, J.; Bzowska, A.; Wielgus-Kutrowska, B. Tricyclic nitrogen base, 1,N6-ethenoadenine, and its ribosides, as substrates for purine-nucleoside phosphorylases: spectroscopic and kinetic studies. Nucleos. Nucleot. Nucleic Acids 2018, 37, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Kuśmierek, J.T.; Jensen, D.E.; Spengler, S.J.; Stolarski, R.; Singer, B. Synthesis and properties of N2,3-ethenoguanosine and N2,3-erthenoguanosine 5’diphosphate. J. Org. Chem. 1987, 52, 2374–2378. [Google Scholar] [CrossRef]
- Boryski, J. 1,N-2-ethenoguanosine – 3 methods of synthesis. Nucl. Nucleot. 1990, 9, 803–813. [Google Scholar] [CrossRef]
- Stachelska-Wierzchowska, A.; Wierzchowski, J.; Górka, M.; Bzowska, A.; Wielgus-Kutrowska, B. Tricyclic nucleobase analogs and their ribosides as substrates of purine-nucleoside phosphorylases. II. Guanine and isoguanine derivatives, Molecules 2019 24, 1493. [CrossRef]
- Virta, P.; Holmstrom, T.; Roslund, M.U.; Mattjus, P.; Kronberg, L.; Sjoholm, R. New nucleoside analogs from 2-amino-9-(β-d-ribofuranosyl)-purine. Org. Biomol. Chem. 2004, 2, 821–827. [Google Scholar] [CrossRef] [PubMed]
- Stachelska-Wierzchowska, A.; Wierzchowski, J.; Górka, M.; Bzowska, A.; Stolarski, R.; Wielgus-Kutrowska, B. Tricyclic nucleobase analogs and their ribosides as substrates and inhibitors of purine-nucleoside phosphorylases III. Aminopurine derivatives, Molecules 2020, 25, 681. [CrossRef]
- Von Sonntag, C.; Schuchmann, H.P. Radical-mediated damege of DNA in the presence of oxygen. Methods. Enzymol. 1990, 186, 511–520. [Google Scholar] [CrossRef]
- Horejsí, K; Pohl, R and Holy, A. Tricyclic purine analogs derived from 2-amino-6-chloropurine and 2,6-diaminopurine and their methylated quaternary salts. Coll. Czechosl. Chem. Comm. 2006, 71, 77-90. [CrossRef]
- Tsou, K.C.; Yip, K.-F., Miller, E.E., Lo, K.W. Synthesis of l,N6-etheno-2-aza-adenosine (2-aza-ε-adenosine): a new cytotoxic fluorescent nucleoside. Nucleic Acid Res. 1974, 1, 518-538. [CrossRef]
- Budow, S.; Seela, F. 2-Azapurine Nucleosides: Synthesis, Properties, and Base Pairing of Oligonucleotides. Chemistry and Biodiversity 2010, 7, 2145–2190. [Google Scholar] [CrossRef]
- Westarp, S.; Brandt, F.; Neumair, L.; Betz, C.; Dagane, A.; Kemper, S.; Jacob, C.R.; Neubauer, P.; Kurreck, A.; Kaspar, F. Nucleoside phosphorylases make N7-xanthosine. Nature Comm. 2024, 15, 3625. [Google Scholar] [CrossRef] [PubMed]
- Liu, Gaofei; Wang, Jialing; Chu, Jianlin; Jiang, Tianyue; Qin, Song; Gao, Zhen; He, Bingfang. Engineering Substrate Promiscuity of Nucleoside Phosphorylase Via an Insertions-Deletions Strategy. JACS AU, 2024, 4, 454-464. [CrossRef]
- Lovelock SL, Crawshaw R, Basler S, Levy C, Baker D, Hilvert D, Green AP: The road to fully programmable protein catalysis. Nature 2022, 606, 49–58. [CrossRef] [PubMed]
- Nyhan, W.L. Lesch-Nyhan Disease and Related Disorders of Purine Metabolism. TZU CHI Medical J. 2007, 19, 105–108. [Google Scholar] [CrossRef]
- De Bruyn, C.H.M.M. Hypoxanthine-guanine phosphoribosyl transferase deficiency. Human Genet. 1976, 31, 127–150. [Google Scholar] [CrossRef] [PubMed]
| Compound | excitation: λmax [nm] |
emission: λmax [nm] |
φ | τ [ns] |
|---|---|---|---|---|
| 2-aminopurine riboside | 305 | 380 | 0.68 | ~8 |
| 8-azaxanthosine* | 290 | 440 | >0.1 | nd |
| 8-azaguanosine (neutral) | 260 | 347 | <0.01 | |
| 8-azaguanosine (anionic) | 278 | 362 | 0.55 | 5.6 |
| 8-azaguanine-N7-β-D-riboside (anionic) | 304 | 420 | 0.03 | nd |
| 8-azaguanine-N8-β-D-riboside | ~290 | nd | >0.1 | nd |
| 8-azaadenosine | 278 | 352 | 0.068 | 0.8 |
| 8-azainosine (anionic) | 275 | 357 | 0.018 | nd |
| 2,6-diamino-8-azapurine- N9-β-D-riboside |
285 | 365 | 0.9 | ~6 |
| 2,6-diamino-8-azapurine- N7-β-D-riboside |
314 | 420 | 0.063 | 1.5;0.45 |
| 2,6-diamino-8-azapurine- N8-β-D-riboside |
313 | 430 | 0.41 | 10.5 |
| 8-aza-7-deaza-isoguanine | 288 | 370 | nd | nd |
| 8-aza-isoguanosine (anionic) | 285 | 370 | ~0.1 | nd |
| 8-aza-7-deaza-isoguanosine* | 285 | 430? | nd | nd |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





