Submitted:
11 May 2024
Posted:
13 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Selection of Kombucha Beverages
2.2. Viable Culture Analysis
2.3. Analytical Techniques
2.4. Molecular Identification of Select Kombucha Colony Isolates
2.5. Amplicon Profiling of Kombucha Beverages
3. Results
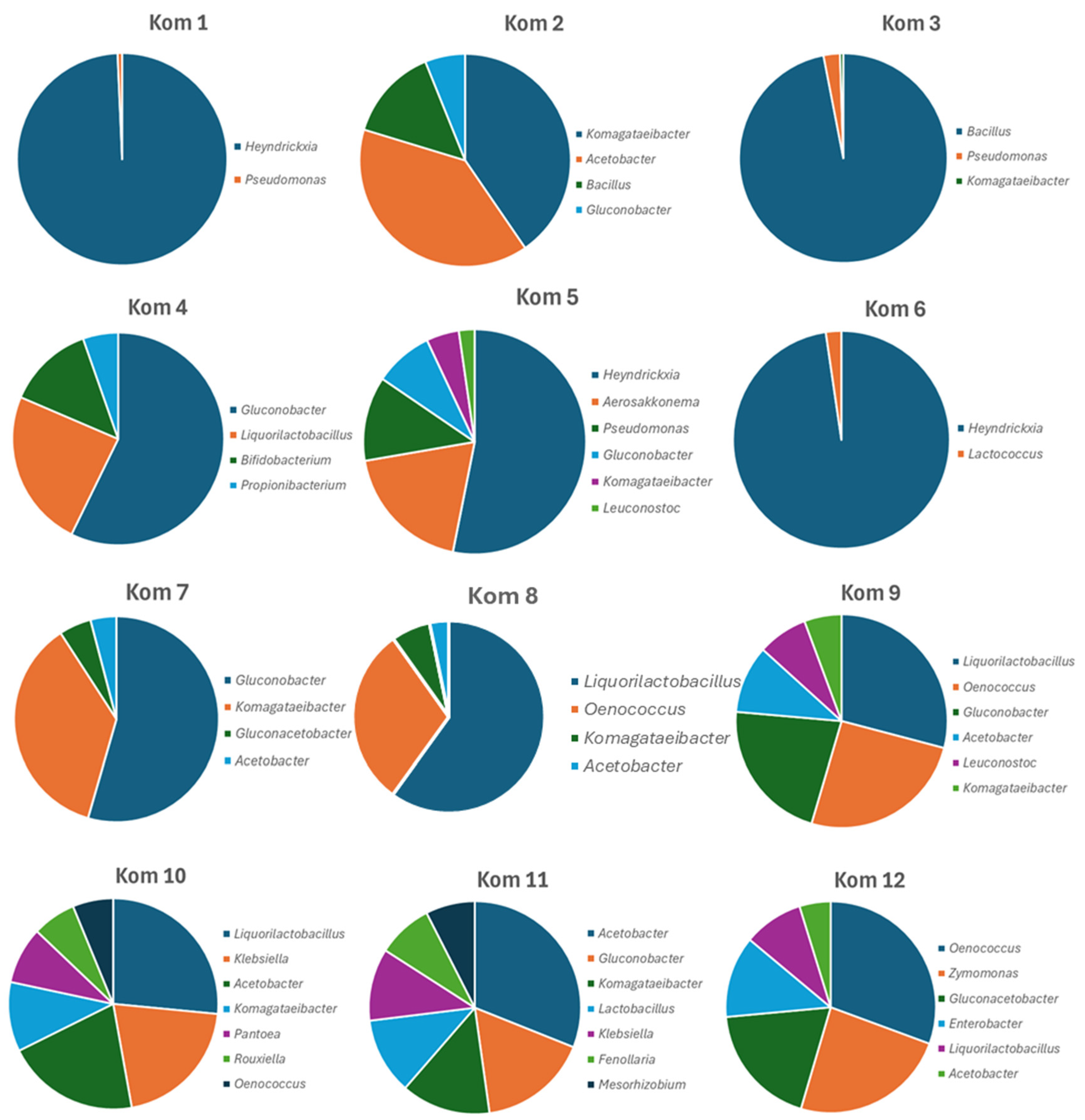
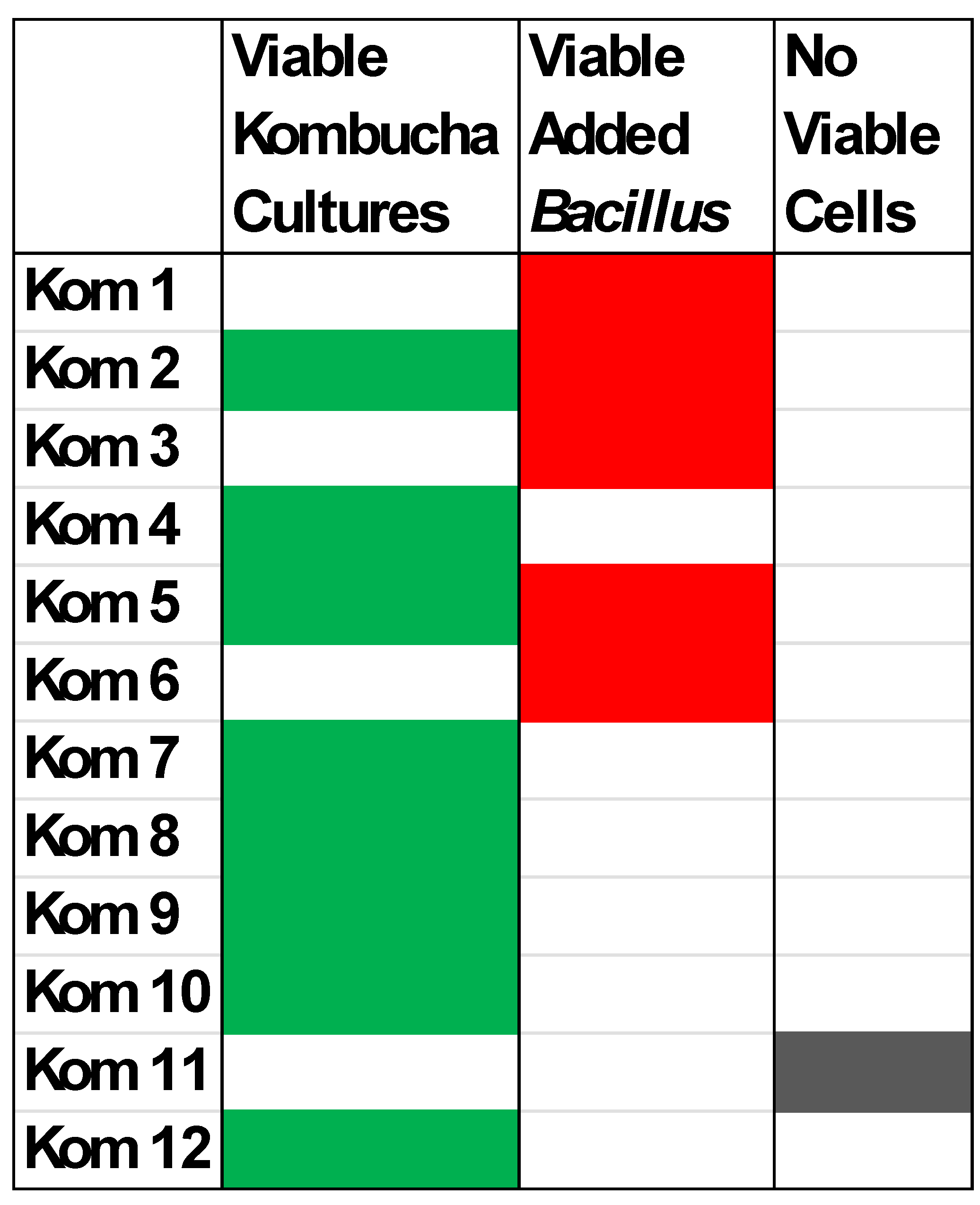
3.1. Viable Culture Analysis of Kom 1 – Kom 12
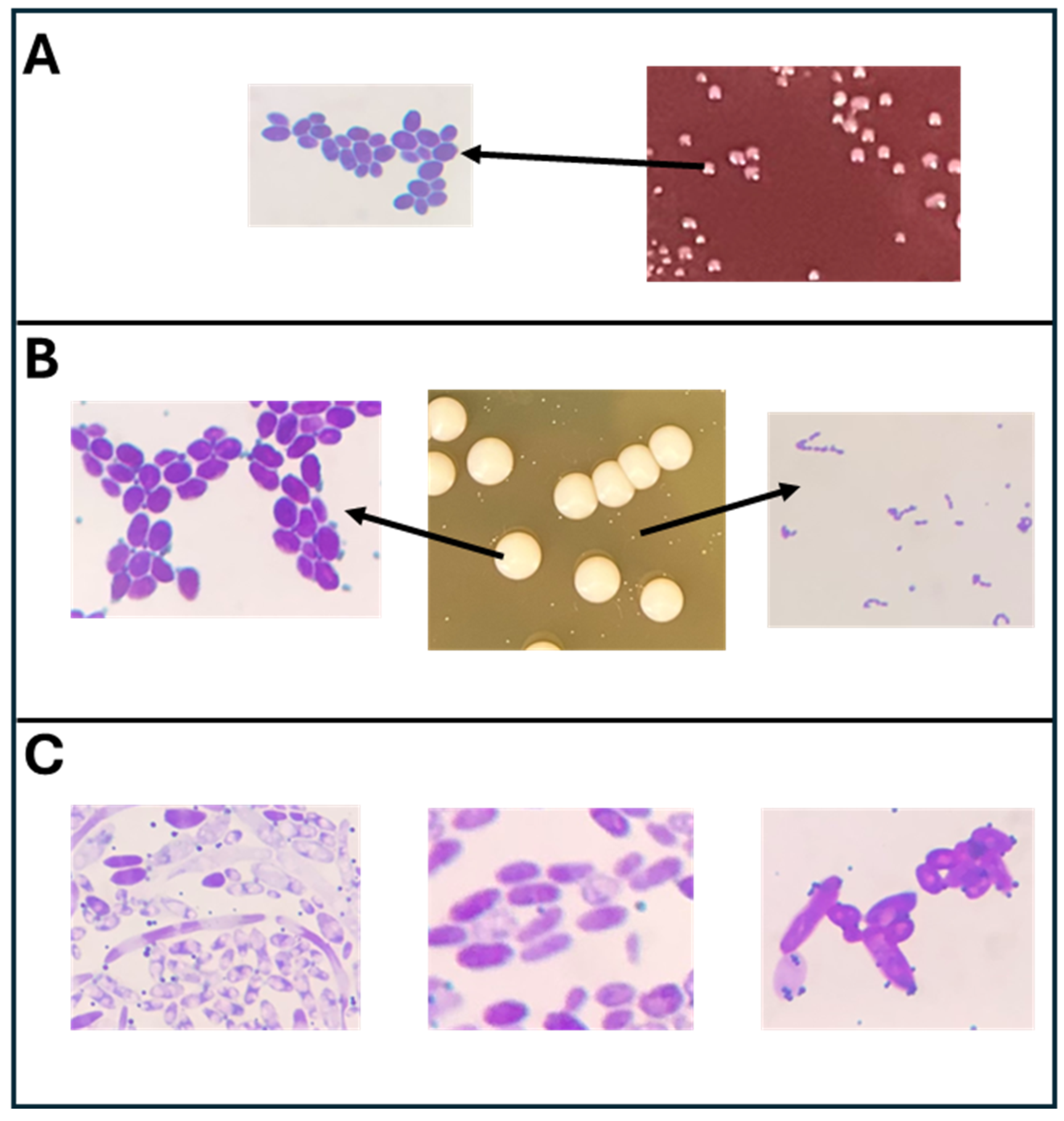
3.2. Microscopic Analysis of Different Cultures Isolated from the Selective Media
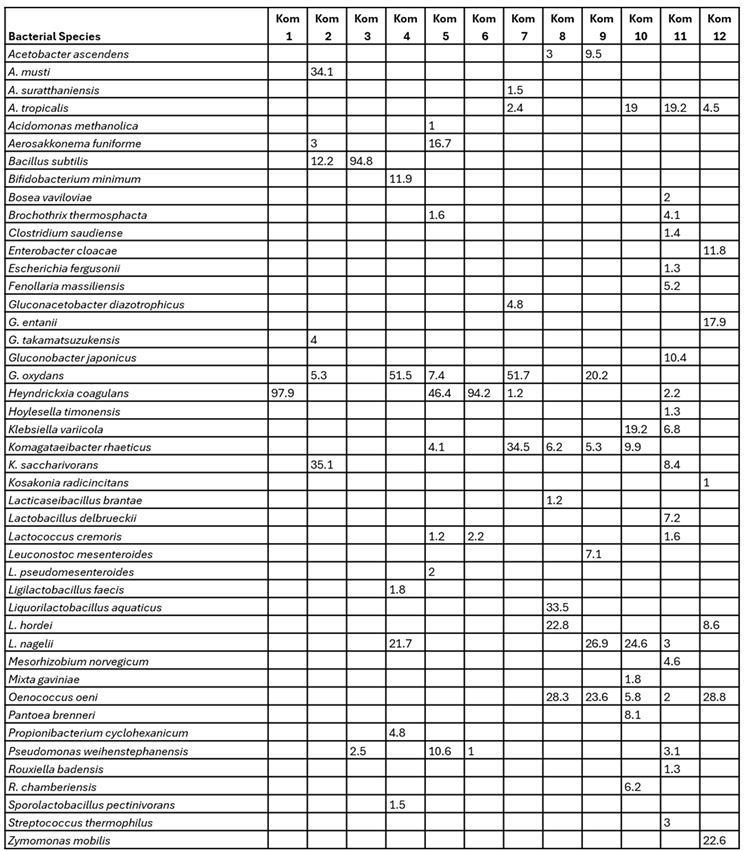
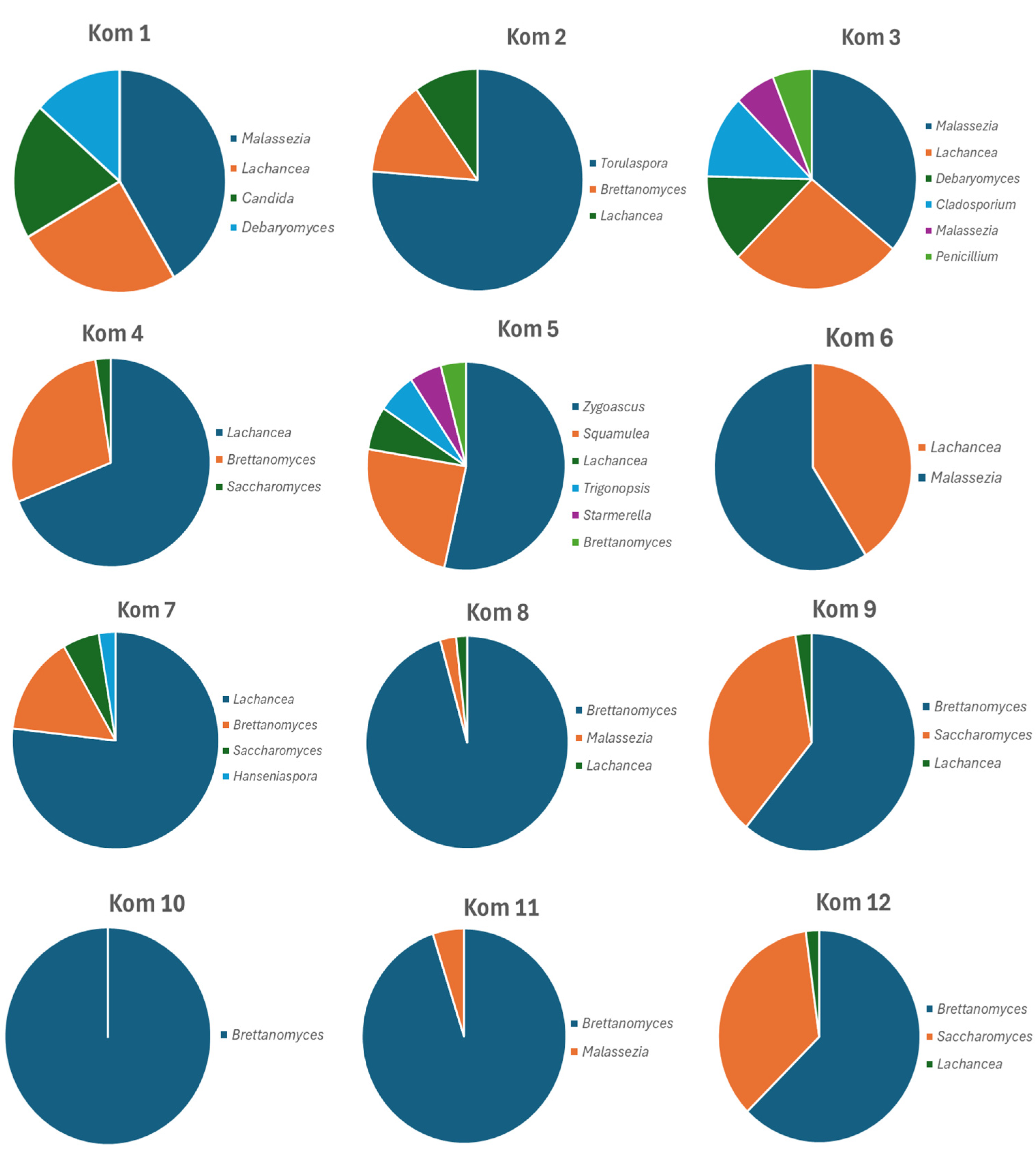
3.3. Bacterial and Fungal Diversity Obtained from Direct Amplicon Profiling of Each Kombucha
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- de Oliveira:, P.V.; da Silva Júnior, A.H.; de Oliveira, C.R.S.; Assumpção, C.F.; Ogeda, C.H. Kombucha Benefits, Risks and Regulatory Frameworks: A Review. Food Chemistry Advances 2023, 2, 100288. [Google Scholar] [CrossRef]
- Mayser, P.; Fromme, S.; Leitzmann, G.; Gründer, K. The Yeast Spectrum of the ‘Tea Fungus Kombucha’: Das Hefespektrum Des ‘Teepilzes Kombucha. ’ Mycoses 1995, 38, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Blanc, P.J. Characterization of the Tea Fungus Metabolites. Biotechnol Lett 1996, 18, 139–142. [Google Scholar] [CrossRef]
- Cambell-Platt, G. G. Campbell-Platt: Fermented Foods of the World. A Dictionary and Guide. 291 Seiten. Butterworth, London, Boston, Durban u. a. 1987. Preis: 35,— £ (Hardcover). Food/Nahrung 1989, 33, 304–304. [Google Scholar] [CrossRef]
- Jayabalan, R.; Malbaša, R.V.; Lončar, E.S.; Vitas, J.S.; Sathishkumar, M. A Review on Kombucha Tea—Microbiology, Composition, Fermentation, Beneficial Effects, Toxicity, and Tea Fungus. Comprehensive Reviews in Food Science and Food Safety 2014, 13, 538–550. [Google Scholar] [CrossRef]
- Kombucha Market Size, Share & Trends Report, 2022-2030 Available online: https://www.grandviewresearch.com/industry-analysis/kombucha-market (accessed on 18 April 2024).
- Chong, A.Q.; Lau, S.W.; Chin, N.L.; Talib, R.A.; Basha, R.K. Fermented Beverage Benefits: A Comprehensive Review and Comparison of Kombucha and Kefir Microbiome. Microorganisms 2023, 11, 1344. [Google Scholar] [CrossRef]
- Fabricio, M.F.; Vargas, B.K.; Tischer, B.; Wagner, R.; Ribeiro, S.R.; Cordeiro, N.; Flôres, S.H.; Záchia Ayub, M.A. Revamping Kombucha Production: Achieving Consistency and Probiotic Potential through a Tailor-Made Microbial Consortium. International Journal of Gastronomy and Food Science 2023, 34, 100844. [Google Scholar] [CrossRef]
- Coton, M.; Pawtowski, A.; Taminiau, B.; Burgaud, G.; Deniel, F.; Coulloumme-Labarthe, L.; Fall, A.; Daube, G.; Coton, E. Unraveling Microbial Ecology of Industrial-Scale Kombucha Fermentations by Metabarcoding and Culture-Based Methods. FEMS Microbiology Ecology 2017, 93, fix048. [Google Scholar] [CrossRef] [PubMed]
- Marsh, A.J.; O’Sullivan, O.; Hill, C.; Ross, R.P.; Cotter, P.D. Sequence-Based Analysis of the Bacterial and Fungal Compositions of Multiple Kombucha (Tea Fungus) Samples. Food Microbiology 2014, 38, 171–178. [Google Scholar] [CrossRef]
- Wang, B.; Rutherfurd-Markwick, K.; Zhang, X.-X.; Mutukumira, A.N. Isolation and Characterisation of Dominant Acetic Acid Bacteria and Yeast Isolated from Kombucha Samples at Point of Sale in New Zealand. Current Research in Food Science 2022, 5, 835–844. [Google Scholar] [CrossRef]
- Harrison, K.; Navarro, R.; Jensen, K.; Cayler, W.; Nielsen, T.; Curtin, C. Live, Probiotic, or Neither? Microbial Composition of Retail-Available Kombucha and “Hard” Kombucha in the Pacific Northwest of the United States. Beverages 2023, 9, 59. [Google Scholar] [CrossRef]
- Hesseltine, C.W. A Millennium of Fungi, Food, and Fermentation. Mycologia 1965, 57, 149–197. [Google Scholar] [CrossRef]
- De Roos, J.; De Vuyst, L. Acetic Acid Bacteria in Fermented Foods and Beverages. Curr Opin Biotechnol 2018, 49, 115–119. [Google Scholar] [CrossRef]
- Kapp, J.M.; Sumner, W. Kombucha: A Systematic Review of the Empirical Evidence of Human Health Benefit. Annals of Epidemiology 2019, 30, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Morales, D. Biological Activities of Kombucha Beverages: The Need of Clinical Evidence. Trends in Food Science & Technology 2020, 105, 323–333. [Google Scholar] [CrossRef]
- Rasouli, L.; Aryaeian, N.; Gorjian, M.; Nourbakhsh, M.; Amiri, F. Evaluation of Cytotoxicity and Anticancer Activity of Kombucha and Doxorubicin Combination Therapy on Colorectal Cancer Cell Line HCT-116. J Educ Health Promot 2021, 10, 376. [Google Scholar] [CrossRef]
- Xu, S.; Wang, Y.; Wang, J.; Geng, W. Kombucha Reduces Hyperglycemia in Type 2 Diabetes of Mice by Regulating Gut Microbiota and Its Metabolites. Foods 2022, 11, 754. [Google Scholar] [CrossRef]
- Wang, P.; Feng, Z.; Sang, X.; Chen, W.; Zhang, X.; Xiao, J.; Chen, Y.; Chen, Q.; Yang, M.; Su, J. Kombucha Ameliorates LPS-Induced Sepsis in a Mouse Model. Food Funct. 2021, 12, 10263–10280. [Google Scholar] [CrossRef]
- Costa, M.A. de C.; Vilela, D.L. de S.; Fraiz, G.M.; Lopes, I.L.; Coelho, A.I.M.; Castro, L.C.V.; Martin, J.G.P. Effect of Kombucha Intake on the Gut Microbiota and Obesity-Related Comorbidities: A Systematic Review. Crit Rev Food Sci Nutr 2023, 63, 3851–3866. [Google Scholar] [CrossRef]
- Arıkan, M.; Mitchell, A.L.; Finn, R.D.; Gürel, F. Microbial Composition of Kombucha Determined Using Amplicon Sequencing and Shotgun Metagenomics. J Food Sci 2020, 85, 455–464. [Google Scholar] [CrossRef]
- Yamada, Y.; Yukphan, P.; Vu, H.T.L.; Muramatsu, Y.; Ochaikul, D.; Nakagawa, Y. Subdivision of the Genus Gluconacetobacter Yamada, Hoshino and Ishikawa 1998: The Proposal of Komagatabacter Gen. Nov., for Strains Accommodated to the Gluconacetobacter Xylinus Group in the α-Proteobacteria. Ann Microbiol 2012, 62, 849–859. [Google Scholar] [CrossRef]
- Yang, J.; Lagishetty, V.; Kurnia, P.; Henning, S.M.; Ahdoot, A.I.; Jacobs, J.P. Microbial and Chemical Profiles of Commercial Kombucha Products. Nutrients 2022, 14, 670. [Google Scholar] [CrossRef] [PubMed]
- Gohl, D.M.; Vangay, P.; Garbe, J.; MacLean, A.; Hauge, A.; Becker, A.; Gould, T.J.; Clayton, J.B.; Johnson, T.J.; Hunter, R.; et al. Systematic Improvement of Amplicon Marker Gene Methods for Increased Accuracy in Microbiome Studies. Nat Biotechnol 2016, 34, 942–949. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- de Boer, E. Update on Media for Isolation of Enterobacteriaceae from Foods. Int J Food Microbiol 1998, 45, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-S.; Kim, Y.-J.; Chon, J.-W.; Kim, D.-H.; Kim, K.-Y.; Seo, K.-H. Citrobacter Braakii: A Major Cause of False-Positive Results on MacConkey and Levine’s Eosin Methylene Blue Selective Agars Used for the Isolation of Escherichia Coli from Fresh Vegetable Samples. Journal of Food Safety 2016, 36, 33–37. [Google Scholar] [CrossRef]
- Hong, S.-M.; Kwon, H.-J.; Park, S.-J.; Seong, W.-J.; Kim, I.; Kim, J.-H. Genomic and Probiotic Characterization of SJP-SNU Strain of Pichia Kudriavzevii. AMB Express 2018, 8, 80. [Google Scholar] [CrossRef] [PubMed]
- Trček, J.; Mira, N.P.; Jarboe, L.R. Adaptation and Tolerance of Bacteria against Acetic Acid. Appl Microbiol Biotechnol 2015, 99, 6215–6229. [Google Scholar] [CrossRef] [PubMed]
- Diguță, C.F.; Nițoi, G.D.; Matei, F.; Luță, G.; Cornea, C.P. The Biotechnological Potential of Pediococcus Spp. Isolated from Kombucha Microbial Consortium. Foods 2020, 9, 1780. [Google Scholar] [CrossRef]
- Guzzo, J.; Coucheney, F.; Pierre, F.; Fortier, L.-C.; Delams, F.; Divies, C.; Tourdot-Maréchal, R. Acidophilic Behaviour of the Malolactic Bacterium Oenococcus Oeni. Sciences Des Aliments - SCI ALIMENT 2002, 22, 107–111. [Google Scholar] [CrossRef]
- Balmaseda, A.; Lorentzen, M.; Dutilh, L.; Bauduin, R.; Guichard, H.; Ollivier, S.; Miot-Sertier, C.; Lucas, P.M. Alcoholic Fermentation Drives the Selection of Oenococcus Oeni Strains in Wine but Not in Cider. International Journal of Food Microbiology 2023, 400, 110276. [Google Scholar] [CrossRef] [PubMed]
- Lorentzen, M.P.; Campbell-Sills, H.; Jorgensen, T.S.; Nielsen, T.K.; Coton, M.; Coton, E.; Hansen, L.; Lucas, P.M. Expanding the Biodiversity of Oenococcus Oeni through Comparative Genomics of Apple Cider and Kombucha Strains. BMC Genomics 2019, 20, 330. [Google Scholar] [CrossRef] [PubMed]
- Villarreal, P.; O’Donnell, S.; Agier, N.; Muñoz-Guzman, F.; Benavides-Parra, J.; Urbina, K.; Peña, T.A.; Solomon, M.; Nespolo, R.F.; Fischer, G.; et al. Domestication Signatures in the Non-Conventional Yeast Lachancea Cidri. mSystems 9, e01058-23. [CrossRef]
- Bellut, K.; Krogerus, K.; Arendt, E.K. Lachancea Fermentati Strains Isolated From Kombucha: Fundamental Insights, and Practical Application in Low Alcohol Beer Brewing. Front Microbiol 2020, 11, 764. [Google Scholar] [CrossRef]
- Chakravorty, S.; Bhattacharya, S.; Chatzinotas, A.; Chakraborty, W.; Bhattacharya, D.; Gachhui, R. Kombucha Tea Fermentation: Microbial and Biochemical Dynamics. International Journal of Food Microbiology 2016, 220, 63–72. [Google Scholar] [CrossRef]
- Harrison, K.; Curtin, C. Microbial Composition of SCOBY Starter Cultures Used by Commercial Kombucha Brewers in North America. Microorganisms 2021, 9, 1060. [Google Scholar] [CrossRef]
- Barbosa, C.D.; Trovatti Uetanabaro, A.P.; Rodrigues Santos, W.C.; Caetano, R.G.; Albano, H.; Kato, R.; Cosenza, G.P.; Azeredo, A.; Góes-Neto, A.; Rosa, C.A.; et al. Microbial–Physicochemical Integrated Analysis of Kombucha Fermentation. LWT 2021, 148, 111788. [Google Scholar] [CrossRef]
- Kaashyap, M.; Cohen, M.; Mantri, N. Microbial Diversity and Characteristics of Kombucha as Revealed by Metagenomic and Physicochemical Analysis. Nutrients 2021, 13, 4446. [Google Scholar] [CrossRef]
- Prohic, A.; Jovovic Sadikovic, T.; Krupalija-Fazlic, M.; Kuskunovic-Vlahovljak, S. Malassezia Species in Healthy Skin and in Dermatological Conditions. Int J Dermatol 2016, 55, 494–504. [Google Scholar] [CrossRef]
- Panesar, P.S.; Marwaha, S.S.; Kennedy, J.F. Zymomonas Mobilis: An Alternative Ethanol Producer. Journal of Chemical Technology & Biotechnology 2006, 81, 623–635. [Google Scholar] [CrossRef]
- Escalante-Minakata, P.; Blaschek, H.P.; Barba de la Rosa, A.P.; Santos, L.; De León-Rodríguez, A. Identification of Yeast and Bacteria Involved in the Mezcal Fermentation of Agave Salmiana. Lett Appl Microbiol 2008, 46, 626–630. [Google Scholar] [CrossRef]
- Mas, P.; Tran, T.; Verdier, F.; Martin, A.; Alexandre, H.; Grandvalet, C.; Tourdot-Maréchal, R. Evolution in Composition of Kombucha Consortia over Three Consecutive Years in Production Context. Foods 2022, 11, 614. [Google Scholar] [CrossRef] [PubMed]
- Ruíz-Ramírez, Y.; Guadarrama-Mendoza, P.C.; Escalante, A.; Giles-Gómez, M.; Valadez-Blanco, R. Probiotic Activity Traits in Vitro and Production of Antimicrobial Peptides by Lactobacillaceae Isolates from Pulque Using Lactobacillus Acidophilus NCFM as Control. Braz J Microbiol 2022, 53, 921–933. [Google Scholar] [CrossRef] [PubMed]
- Fuller, R. Probiotics in Man and Animals. J Appl Bacteriol 1989, 66, 365–378. [Google Scholar]
- O’Sullivan, D.J. Genomics Can Advance the Potential for Probiotic Cultures to Improve Liver and Overall Health. Curr Pharm Des 2008, 14, 1376–1381. [Google Scholar] [CrossRef] [PubMed]
| Kombucha beverage | State | Relevant ingredients listed regarding microbial fermentation | Relevant label claims |
|---|---|---|---|
| Kom 1b | CA | Kombucha cultures, Bacillus coagulans MTCC5856, cane sugar, black tea, green tea, lime juice and grape juice, | “Bubbly probiotic tea” |
| Kom 2b | WA | Kombucha culture, Bacillus subtilis, cane sugar, tea blend, ginger juice, whole lemon puree | “2 billion CFU Probiotic Cultures” |
| Kom 3b | OR | Kombucha cultures, Bacillus subtilis, green tea, black tea, cane sugar, apple, white grape and passion fruit juice, mango puree | “Probiotic Kombucha” “2 billion Probiotic Cultures” |
| Kom 4b | OR | Kombucha culture, currant, Oolong tea, Elderberry, Hibiscus, Blueberry, Goji Berry, Strawberry, Raspberry, Cane Sugar | “Fizzy Probiotic Tea” |
| Kom 5b | CA | Kombucha culture, black tea, green tea, kiwi and ginger juice, Bacillus coagulans GBI-306086 | “9 billion Living Probiotics” |
| Kom 6b | CA | Kombucha culture, Bacillus coagulans MTCC 5856, black and green tea, cane sugar, stevia leaf extract, green coffee bean extract | “Live Probiotics” |
| Kom 7b | IL | Black tea, cane sugar, cherry and lemon juice concentrate | “Live Probiotics & Enzymes” “Heirloom cultures” |
| Kom 8c | MN | Black and green tea, cane sugar, apple juice | “Live Probiotics” “Active Kombucha Culture” |
| Kom 9c | WI | Kombucha culture, green tea, hibiscus flowers, cane sugar | “Forage Kombucha is Alive” “Kombucha Culture” |
| Kom 10b | MN | Peach oolong tea, mango turmeric tea, cane sugar | “Non-pasteurized” “Fresh Brewed” |
| Kom 11b | TX | Kombucha culture, sugar, stevia extract | “Live Sparkling Probiotic Kombucha" |
| Kom 12b | WI | Kombucha culture, black tea, safflower, cane sugar | “Contains millions of living probiotic cultures” “Live Kombucha Culture” |
| Kombucha Sample | pH Reading | Total CFU BHI (cfu/ml) |
Total CFU MRS (cfu/ml) | Total CFU ABS (cfu/ml) |
Total CFU PDA-Y (cfu/ml) |
Total CFU MAC (cfu/ml) |
|---|---|---|---|---|---|---|
| Kom 1 | 3.2 | 3.2 × 105 | 3.5 × 105 | nc1 | nc | nc |
| Kom 2 | 3.5 | 3.4 × 106 | 3.1 × 106 | 1.7 × 105 | 1.7 × 105 | 1.0 × 104 |
| Kom 3 | 3.7 | 5.0 × 105 | 2.5 × 106 | nc | nc | nc |
| Kom 4 | 3.3 | 3.0 × 103 | 5.5 × 103 | 1.2 × 104 | 1.2 × 104 | nc |
| Kom 5 | 3.1 | 1.1 × 104 | 3.3 × 103 | 7.0 × 103 | 1.1 × 104 | nc |
| Kom 6 | 3.2 | 1.0 × 103 | 1.0 × 103 | nc | nc | nc |
| Kom 7 | 3.3 | 5.5 × 105 | 9.7 × 105 | 8.5 × 105 | 4.7 × 105 | 3.0 × 103 |
| Kom 8 | 3.6 | 2.0 × 105 | 6.3 × 105 | 5.2 × 106 | 1.5 × 106 | nc |
| Kom 9 | 3.0 | 2.4 × 106 | 1.5 × 106 | 1.6 × 106 | 2.0 × 106 | nc |
| Kom 10 | 3.4 | 1.1 × 106 | 2.2 × 106 | 3.8 × 106 | 6.0 × 106 | nc |
| Kom 11 | 3.5 | nc | nc | nc | nc | nc |
| Kom 12 | 3.8 | 2.3 × 106 | 1.9 × 106 | 3.1 × 106 | 2.1 × 106 | nc |
| Fungal Species | Kom 1 | Kom 2 | Kom 3 | Kom 4 | Kom 5 | Kom 6 | Kom 7 | Kom 8 | Kom 9 | Kom 10 | Kom 11 | Kom 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allodekkera sacchari | 7.1 | |||||||||||
| Brettanomyces anomalus | 14.7 | 4.1 | 11.8 | 3.5 | 35.2 | |||||||
| B. bruxellensis | 12.2 | 26.5 | 3.8 | 88.7 | 48.9 | 95.1 | 94.5 | 26.6 | ||||
| Candida mesenterica | 5 | 1.6 | ||||||||||
| C. parapsilosis | 14 | |||||||||||
| Cladosporium angustiherbarum | 3 | 11.5 | ||||||||||
| Cyberlindnera jadinii | 3.5 | |||||||||||
| Debaryomyces psychrosporus | 13 | 12.2 | ||||||||||
| Gibellulopsis serrae | 1.2 | |||||||||||
| Hanseniaspora valbyensis | 2.6 | |||||||||||
| Lachancea cidri | 24.1 | 8.5 | 25 | 67.8 | 6.2 | 38.5 | 75.5 | 1.7 | 2.5 | 2.1 | ||
| Malassezia arunalokei | 14.5 | 1.6 | 38.5 | |||||||||
| M. globosa | 5.9 | 14.4 | 2.5 | |||||||||
| M. restricta | 25.2 | 33.8 | 2.4 | 2.4 | 2 | |||||||
| M. slooffiae | 3 | |||||||||||
| Penicillium hordei | 5.7 | |||||||||||
| Pichia cecembensis | 1.8 | |||||||||||
| Saccharomyces bayanus | 2.5 | |||||||||||
| S. cerevisiae | 2.1 | 5.6 | 36.5 | 34.8 | ||||||||
| Squamulea flakusii | 22.2 | |||||||||||
| Starmerella davenportii | 4.8 | |||||||||||
| Torulaspora microellipsoides | 66.5 | 2 | ||||||||||
| Trigonopsis variabilis | 5.8 | |||||||||||
| Zygoascus hellenicus | 49.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




