Submitted:
29 April 2024
Posted:
01 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1 Population Analyzed in the Study
2.2. DNA Extraction and Exome Analysis
2.3. Gene Selection
2.4. Statistical and Bioinformatic Analysis
2.5. Inclusion Criteria
3. Results
4. Discussion
Supplementary Materials
Conflicts of Interest
References
- Shiota M, Narita S, Akamatsu S, et al. Association of Missense Polymorphism in HSD3B1 With Outcomes Among Men With Prostate Cancer Treated With Androgen-Deprivation Therapy or Abiraterone. JAMA Netw Open. 2019;2(2):e190115. doi:10.1001/jamanetworkopen.2019.0115. [CrossRef]
- Alex AB, Pal SK, Agarwal N. CYP17 inhibitors in prostate cancer: latest evidence and clinical potential. Ther Adv Med Oncol. 2016 Jul;8(4):267-75. doi: 10.1177/1758834016642370. Epub 2016 Apr 19. PMID: 27482286; PMCID: PMC4952018. [CrossRef]
- Hearn JWD, Xie W, Nakabayashi M, et al. Association of HSD3B1 Genotype With Response to Androgen-Deprivation Therapy for Biochemical Recurrence After Radiotherapy for Localized Prostate Cancer. JAMA Oncol. 2018;4(4):558–562. doi:10.1001/jamaoncol.2017.3164. [CrossRef]
- Almassi N, Reichard C, Li J, et al. HSD3B1 and Response to a Nonsteroidal CYP17A1 Inhibitor in Castration-Resistant Prostate Cancer. JAMA Oncol. 2018;4(4):554–557. doi:10.1001/jamaoncol.2017.3159. [CrossRef]
- Sai Harisha Rajanala, Anna Plym, Jane B Vaselkiv, Ericka M Ebot, Konstantina Matsoukas, Zhike Lin, Goutam Chakraborty, Sarah C Markt, Kathryn L Penney, Gwo-Shu M Lee, Lorelei A Mucci, Philip W Kantoff, Konrad H Stopsack, SLCO1B3 and SLCO2B1 genotypes, androgen deprivation therapy, and prostate cancer outcomes: a prospective cohort study and meta-analysis, Carcinogenesis, Volume 45, Issue 1-2, January-February 2024, Pages 35–44, https://doi.org/10.1093/carcin/bgad075. [CrossRef]
- Andrew W Hahn et al., Germline variant alleles in rs12422149 of SLCO2B1 and response to abiraterone acetate (AA) in men with metastatic castration-resistant prostate cancer (mCRPC).. JCO 36, 5076-5076(2018). DOI:10.1200/JCO.2018.36.15_suppl.5076. [CrossRef]
- Fujimoto N, Kubo T, Inatomi H, Bui HT, Shiota M, Sho T, Matsumoto T. Polymorphisms of the androgen transporting gene SLCO2B1 may influence the castration resistance of prostate cancer and the racial differences in response to androgen deprivation. Prostate Cancer Prostatic Dis. 2013 Dec;16(4):336-40. doi: 10.1038/pcan.2013.23. Epub 2013 Jul 30. PMID: 23896625. [CrossRef]
- Fujimoto N, Kubo T, Inatomi H, Bui HT, Shiota M, Sho T, Matsumoto T. Polymorphisms of the androgen transporting gene SLCO2B1 may influence the castration resistance of prostate cancer and the racial differences in response to androgen deprivation. Prostate Cancer Prostatic Dis. 2013 Dec;16(4):336-40. doi: 10.1038/pcan.2013.23. Epub 2013 Jul 30. PMID: 23896625. [CrossRef]
- Wilborn, Teresa & Lang, Nicholas & Johnson, Michelle & Meleth, Sreelatha & Falany, Charles. (2006). Association of SULT2A1 allelic variants with plasma adrenal androgens and prostate cancer in African American men. The Journal of steroid biochemistry and molecular biology. 99. 209-14. 10.1016/j.jsbmb.2006.01.006. [CrossRef]
- Kurogi K, Rasool MI, Alherz FA, El Daibani AA, Bairam AF, Abunnaja MS, Yasuda S, Wilson LJ, Hui Y, Liu MC. SULT genetic polymorphisms: physiological, pharmacological and clinical implications. Expert Opin Drug Metab Toxicol. 2021 Jul;17(7):767-784. doi: 10.1080/17425255.2021.1940952. Epub 2021 Jun 30. PMID: 34107842; PMCID: PMC8369464. [CrossRef]
- Teresa W. Wilborn, Nicholas P. Lang, Michelle Smith, Sreelatha Meleth, Charles N. Falany, Association of SULT2A1 allelic variants with plasma adrenal androgens and prostate cancer in African American men, The Journal of Steroid Biochemistry and Molecular Biology, Volume 99, Issues 4–5, 2006, Pages 209-214, ISSN 0960-0760, https://doi.org/10.1016/j.jsbmb.2006.01.006. [CrossRef]
- Laverdière I, Flageole C, Audet-Walsh É, Caron P, Fradet Y, Lacombe L, Lévesque É, Guillemette C. The UGT1 locus is a determinant of prostate cancer recurrence after prostatectomy. Endocr Relat Cancer. 2015 Feb;22(1):77-85. doi: 10.1530/ERC-14-0423. Epub 2014 Dec 1. PMID: 25452636. [CrossRef]
- Amrousy YM, Haffez H, Abdou DM, Atya HB. Role of single nucleotide polymorphisms of the HSD3B1 gene (rs6203 and rs33937873) in the prediction of prostate cancer risk. Mol Med Rep. 2022 Aug;26(2):271. doi: 10.3892/mmr.2022.12787. Epub 2022 Jul 7. PMID: 35795973; PMCID: PMC9309536. [CrossRef]
- National Library of Medicine, National Center for Biotechnology information, acessed 16 March 2024 <https://www.ncbi.nlm.nih.gov/gene/1576>.
- World Health Organization, Health Topics, acessed 12 March 2024 https://www.who.int/news-room/fact-sheets/detail/cancer.
- Juliana Carla Gomes Rodrigues et al., Identification of pharmacogenomic variants associated with oncology treatments in Brazilian Amazonian Amerindians. JCO 40, e15082-e15082(2022). DOI:10.1200/JCO.2022.40.16_suppl.e15082. [CrossRef]
- National Comprehensive Cancer Network, NCCN Guidelines, Prostate Cancer, acessed 18 March 2024 https://www.nccn.org/guidelines/guidelines-detail?category=1&id=1459.
- Rodrigues, J.C.G.; Fernandes, M.R.; Ribeiro-dos-Santos, A.M.; de Araújo, G.S.; de Souza, S.J.; Guerreiro, J.F.; Ribeiro-dos-Santos, Â.; de Assumpção, P.P.; Santos, N.P.C.d.; Santos, S. Pharmacogenomic Profile of Amazonian Amerindians. J. Pers. Med. 2022, 12, 952. https://doi.org/10.3390/jpm12060952. [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring: Harbor, NY, USA, 1989.
- M. Whirl-Carrillo1, R. Huddart1, L. Gong, K. Sangkuhl, C.F. Thorn, R. Whaley and T.E. Klein. "An evidence-based framework for evaluating pharmacogenomics knowledge for personalized medicine" Clinical Pharmacology & Therapeutics (2021) online ahead of print.In Progress: Abiraterone Pathway, Pharmacokinetics. acessed 20 March 2024 https://www.pharmgkb.org/pathway/PA166310681.
- Jason W D Hearn*, Ghada AbuAli*, Chad A Reichard, Chandana A Reddy, Cristina Magi-Galluzzi, Kai-Hsiung Chang, Rachel Carlson,Laureano Rangel, Kevin Reagan, Brian J Davis, R Jeffrey Karnes, Manish Kohli, Donald Tindall, Eric A Klein, Nima Sharif. HSD3B1 and resistance to androgen-deprivation therapy in prostate cancer: a retrospective, multicohort study. www.thelancet.com/oncology Published online August 26, 2016 http://dx.doi.org/10.1016/S1470-2045(16)30227-3. [CrossRef]
- Cohen-Paes, A.d.N.; de Carvalho, D.C.; Pastana, L.F.; Dobbin, E.A.F.; Moreira, F.C.; de Souza, T.P.; Fernandes, M.R.; Leal, D.F.d.V.B.; de Sá, R.B.A.; de Alcântara, A.L.; et al. Characterization of PCLO gene in Amazonian Native American populations.Genes 2022, 13, 499.Author 1, A.; Author 2, B. Title of the chapter. In Book Title, 2nd ed.; Editor 1, A., Editor 2, B., Eds.; Publisher: Publisher Location, Country, 2007; Volume 3, pp. 154–196.
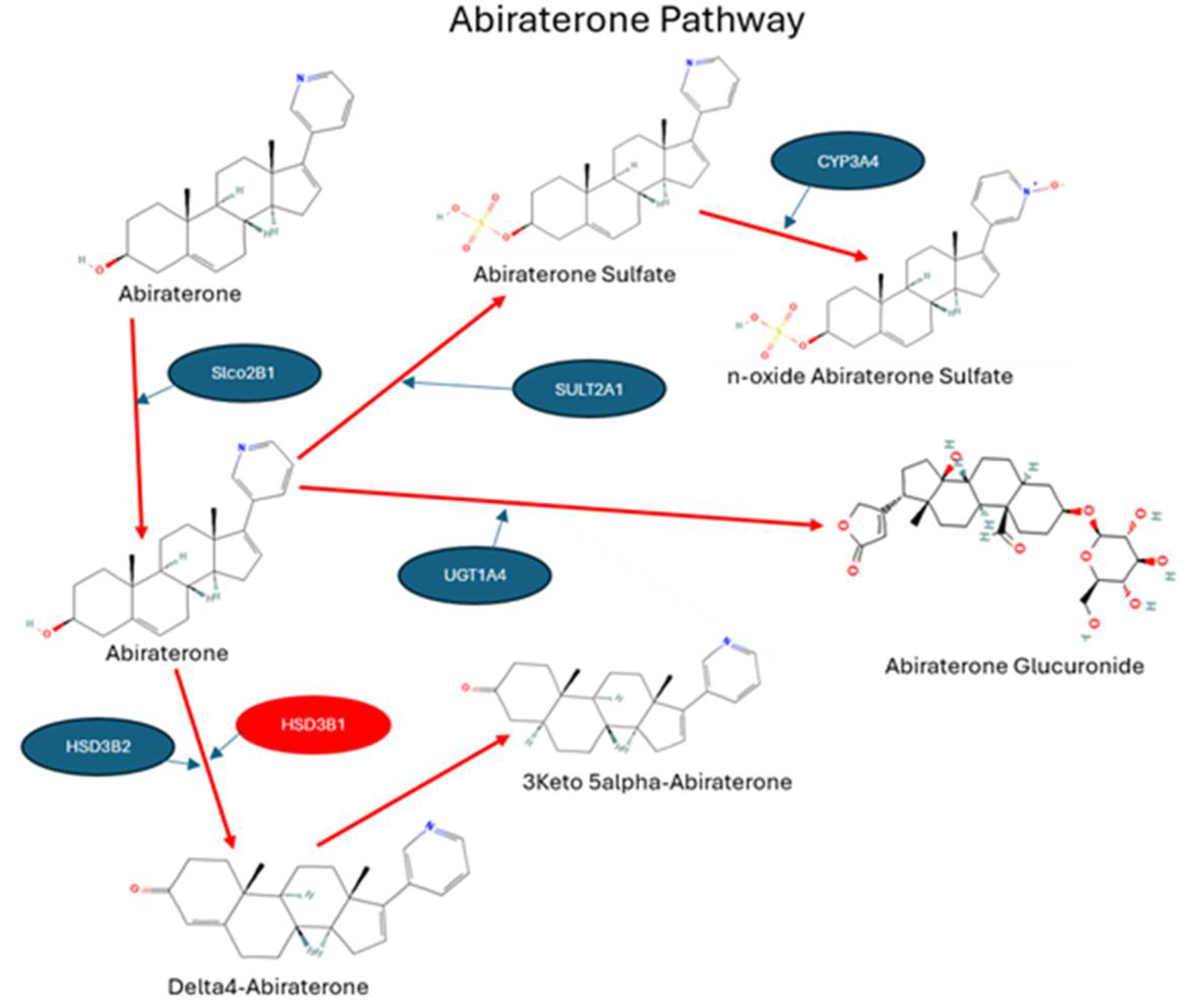
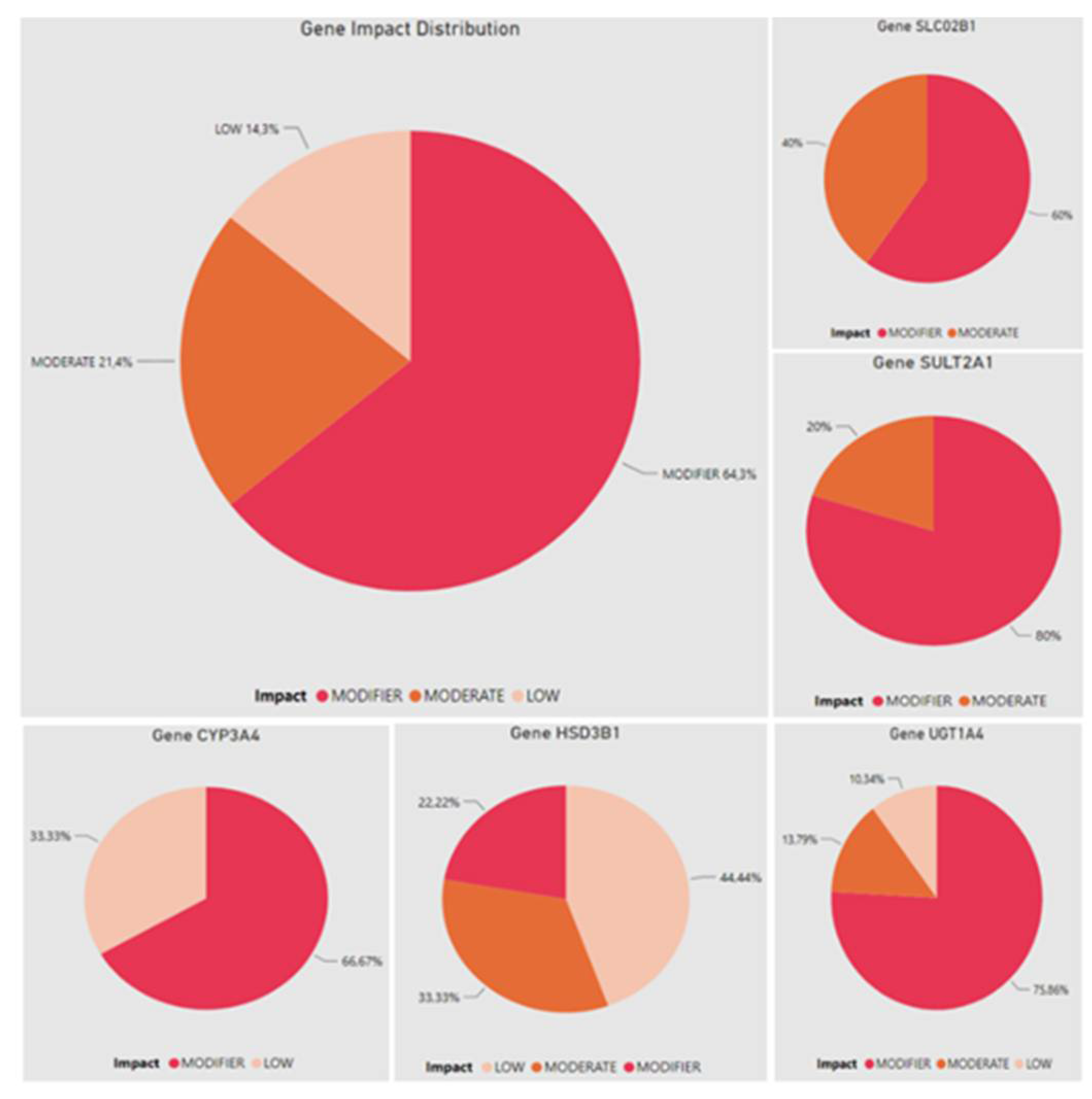
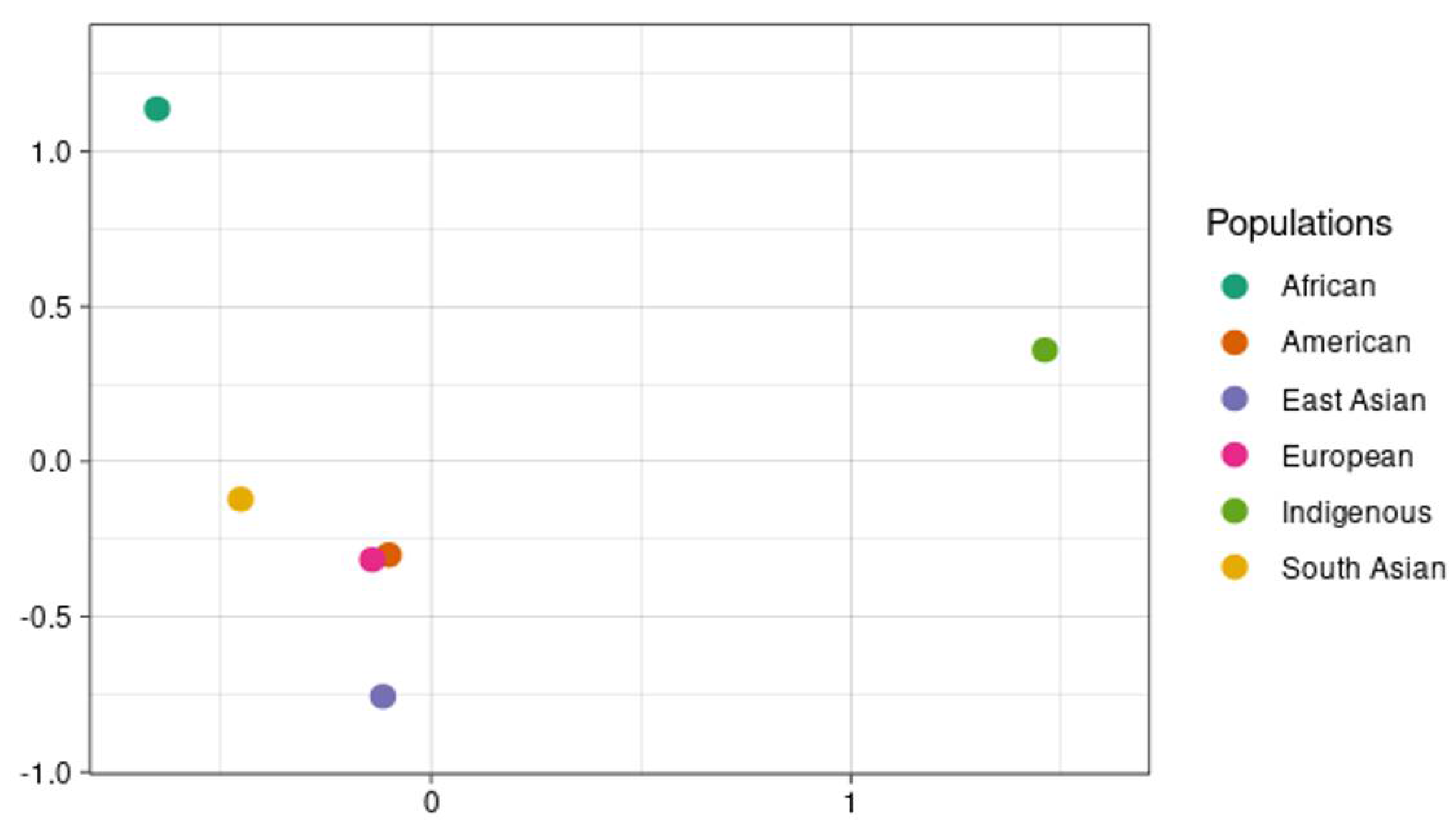
| Gene | SNP ID | Region | Impact | IND | AFR | AMR | EAS | EUR | SAS |
|---|---|---|---|---|---|---|---|---|---|
| CYP3A4 | . | cds | low | 0,0179 | 0 | 0 | 0 | 0 | 0 |
| CYP3A4 | rs12721620 | Intronic | Modifier | 0,0161 | 0,2671 | 0,0118 | 0 | 0,0006 | 0,008 |
| CYP3A4 | rs2687116 | Intronic | Modifier | 0 | 0,3782 | 0,9165 | 0,9982 | 0,9624 | 0,9629 |
| HSD3B1 | rs1047303 | Cds | Moderate | 0 | 0,8897 | 0,8599 | 0,9376 | 0,6797 | 0,8118 |
| HSD3B1 | rs6201 | Cds | Moderate | 0 | 0,33 | 0,0794 | 0,0412 | 0,003 | 0,0557 |
| HSD3B1 | rs6205 | Cds | Moderate | 0,0391 | 0,39 | 0,0844 | 0,0428 | 0,0034 | 0,0561 |
| HSD3B1 | rs6671149 | Intronic | Modifier | 0 | 0,151 | 0,046 | 0,055 | 0,003 | 0,049 |
| HSD3B1 | rs6673653 | Intronic | Modifier | 0 | 0,1 | 0,04 | 0,054 | 0,002 | 0,05 |
| SLCO2B1 | rs12422149 | Cds | Moderate | 0,6667 | 0,0953 | 0,4573 | 0,3336 | 0,1094 | 0,2231 |
| SLCO2B1 | rs149765874 | Cds | Moderate | 0 | 0 | 0,0001 | 0 | 0,0002 | 0,001 |
| SLCO2B1 | rs2306168 | Cds | Moderate | 0,1349 | 0,3431 | 0,0972 | 0,2308 | 0,0266 | 0,0619 |
| SLCO2B1 | rs60113013 | Cds | Moderate | 0 | 0,0038 | 0,0618 | 0,1107 | 0,0211 | 0,0379 |
| SLCO2B1 | rs12287059 | Intronic | Modifier | 0,0135 | 0,1543 | 0,0074 | 0 | 0,001 | 0,0002 |
| SLCO2B1 | rs1944612 | 5utr | Modifier | 1 | 0,9999 | 0,9994 | 0,8861 | 0,9992 | 0,9961 |
| SLCO2B1 | rs2851069 | 5utr | Modifier | 0,0676 | 0,144 | 0,372 | 0,268 | 0,622 | 0,555 |
| SLCO2B1 | rs7125268 | Intronic | Modifier | 0,1429 | 0,4186 | 0,5222 | 0,3276 | 0,2235 | 0,3283 |
| SLCO2B1 | rs74885054 | Intronic | Modifier | 0,1087 | 0,089 | 0,386 | 0,311 | 0,116 | 0,302 |
| SLCO2B1 | rs995893327 | Intronic | Modifier | 0 | 0 | 0,0002 | 0 | 0 | 0 |
| SULT2A1 | rs11569679 | Cds | Moderate | 0 | 0,1248 | 0,0048 | 0 | 0,0003 | 0,0007 |
| SULT2A1 | rs11569678 | 3utr | Modifier | 0,0161 | 0,049 | 0,049 | 0 | 0 | 0 |
| SULT2A1 | rs2547238 | Intronic | Modifier | 0,0833 | 0,026 | 0,367 | 0,506 | 0,284 | 0,355 |
| SULT2A1 | rs62531056 | Intronic | Modifier | 0,0405 | 0,0007 | 0,0627 | 0,0003 | 0 | 0,0007 |
| SULT2A1 | rs767511533 | Intronic | Modifier | 0 | 0 | 0,0001 | 0 | 0 | 0 |
| UGT1A4 | rs2011425 | cds | Moderate | 0 | 0,0986 | 0,1275 | 0,2065 | 0,0907 | 0,1996 |
| UGT1A4 | rs3892221 | cds | Moderate | 0 | 0,0429 | 0,0011 | 0,005 | 0,0011 | 0,0006 |
| UGT1A4 | rs45540231 | cds | Moderate | 0 | 0,0641 | 0,0032 | 0,0001 | 0,0002 | |
| UGT1A4 | rs6755571 | cds | Moderate | 0 | 0,0162 | 0,0215 | 0,0001 | 0,0553 | 0,0102 |
| UGT1A4 | rs10929301 | Intronic | Modifier | 0 | 0,7048 | 0,4579 | 0,3323 | 0,4511 | 0,6108 |
| UGT1A4 | rs12466997 | Intronic | Modifier | 0,0811 | 0,288 | 0,107 | 0,225 | 0,069 | 0,201 |
| UGT1A4 | rs12471326 | Intronic | Modifier | 0,1111 | 0,0489 | 0,1074 | 0,0038 | 0,0296 | 0,0174 |
| UGT1A4 | rs199892897 | Intronic | Modifier | 0 | 0,086 | 0,004 | 0 | 0 | 0 |
| UGT1A4 | rs2011219 | Intronic | Modifier | 0 | 0,0519 | 0,1106 | 0,1979 | 0,0598 | 0,1759 |
| UGT1A4 | rs2302538 | Intronic | Modifier | 0 | 0,425 | 0,097 | 0,052 | 0,128 | 0,157 |
| UGT1A4 | rs2361501 | Intronic | Modifier | 0 | 0,8134 | 0,4585 | 0,2834 | 0,4469 | 0,6085 |
| UGT1A4 | rs28898618 | Intronic | Modifier | 0,0208 | 0,0172 | 0,0005 | 0 | 0 | 0,0001 |
| UGT1A4 | rs28900402 | Intronic | Modifier | 0 | 0,104 | 0,006 | 0 | 0 | 0,066 |
| UGT1A4 | rs34547608 | Intronic | Modifier | 0 | 0,099 | 0,007 | 0,001 | 0 | 0 |
| UGT1A4 | rs34622615 | Intronic | Modifier | 0 | 0,0519 | 0,0165 | 0,0017 | 0,0307 | 0,0194 |
| UGT1A4 | rs34650714 | Intronic | Modifier | 0 | 0,1218 | 0,0064 | 0,0012 | 0,0008 | 0,0004 |
| UGT1A4 | rs377453564 | Intronic | Modifier | 0 | 0,0001 | 0,0001 | 0,0001 | 0,0002 | 0 |
| UGT1A4 | rs3821242 | Intronic | Modifier | 0,3684 | 0,6401 | 0,4537 | 0,3342 | 0,4453 | 0,61 |
| UGT1A4 | rs4148323 | Intronic | Modifier | 0,0172 | 0,0009 | 0,026 | 0,1524 | 0,0036 | 0,019 |
| UGT1A4 | rs45449995 | Intronic | Modifier | 0,0159 | 0,0485 | 0,0165 | 0,0018 | 0,0306 | 0,0193 |
| UGT1A4 | rs570829500 | Intronic | Modifier | 0 | 0,0003 | 0,0002 | 0,0001 | 0 | 0 |
| UGT1A4 | rs62191918 | Intronic | Modifier | 0,1016 | 0,0946 | 0,1192 | 0,2103 | 0,0615 | 0,1818 |
| UGT1A4 | rs6431625 | Intronic | Modifier | 0,4531 | 0,6698 | 0,3398 | 0,1206 | 0,3867 | 0,4283 |
| UGT1A4 | rs6706232 | Intronic | Modifier | 0,582 | 0,7644 | 0,4583 | 0,3345 | 0,4473 | 0,61 |
| UGT1A4 | rs7574296 | Intronic | Modifier | 0,5192 | 0,7646 | 0,4587 | 0,3322 | 0,4478 | 0,6098 |
| UGT1A4 | rs77053267 | Intronic | Modifier | 0 | 0,0149 | 0,0004 | 0,0001 | 0,0001 |
| Gene | Refseq | dbSNP | Var Type | Impact | Region Detailed |
|---|---|---|---|---|---|
| CYP3A4 | NM_017460.5 | . | SNV | LOW | SYNON_COD |
| SULT2A1 | NM_003167.3 | rs11569678 | SNV | MODIFIER | UTR_3_PRIME |
| SLCO2B1 | NM_007256.4 | rs12287059 | SNV | MODIFIER | INTRON |
| SLCO2B1 | NM_007256.4 | rs12422149 | SNV | MODERATE | N_SYNON_COD |
| UGT1A4 | NM_007120.2 | rs12466997 | SNV | MODIFIER | INTRON |
| UGT1A4 | NM_007120.2 | rs12471326 | SNV | MODIFIER | INTRON |
| CYP3A4 | NM_017460.5 | rs12721620 | SNV | MODIFIER | INTRON |
| SLCO2B1 | NM_007256.4 | rs1944612 | SNV | MODIFIER | UTR_5_PRIME |
| UGT1A4 | NM_007120.2 | rs2011404 | SNV | LOW | SYNON_COD |
| SLCO2B1 | NM_007256.4 | rs2306168 | SNV | MODERATE | N_SYNON_COD |
| SULT2A1 | NM_003167.3 | rs2547238 | SNV | MODIFIER | INTRON |
| SLCO2B1 | NM_007256.4 | rs2851069 | SNV | MODIFIER | UTR_5_PRIME |
| UGT1A4 | NM_007120.2 | rs28898618 | SNV | MODIFIER | INTRON |
| HSD3B1 | NM_000862.2 | rs33937873 | SNV | LOW | SYNON_COD |
| UGT1A4 | NM_007120.2 | rs3732217 | SNV | LOW | SYNON_COD |
| UGT1A4 | NM_007120.2 | rs3821242 | SNV | MODIFIER | INTRON |
| UGT1A4 | NM_007120.2 | rs4148323 | SNV | MODIFIER | INTRON |
| UGT1A4 | NM_007120.2 | rs45449995 | SNV | MODIFIER | INTRON |
| HSD3B1 | NM_000862.2 | rs6203 | SNV | LOW | SYNON_COD |
| HSD3B1 | NM_000862.2 | rs6205 | SNV | MODERATE | N_SYNON_COD |
| UGT1A4 | NM_007120.2 | rs62191918 | SNV | MODIFIER | INTRON |
| SULT2A1 | NM_003167.3 | rs62531056 | SNV | MODIFIER | INTRON |
| UGT1A4 | NM_007120.2 | rs6431625 | SNV | MODIFIER | INTRON |
| UGT1A4 | NM_007120.2 | rs6706232 | SNV | MODIFIER | INTRON |
| SLCO2B1 | NM_007256.4 | rs7125268 | SNV | MODIFIER | INTRON |
| SLCO2B1 | NM_007256.4 | rs74885054 | INDEL | MODIFIER | INTRON |
| UGT1A4 | NM_007120.2 | rs7574296 | SNV | MODIFIER | INTRON |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





