Submitted:
25 April 2024
Posted:
26 April 2024
You are already at the latest version
Abstract
Keywords:
1. CLPP is a Key Modifier of Growth and Lifespan, But Its Substrates Remain Unclear
2. Novel Evidence on CLPP and CLPX Functions from Clpp-KO Mice and PRLTS3 Patients
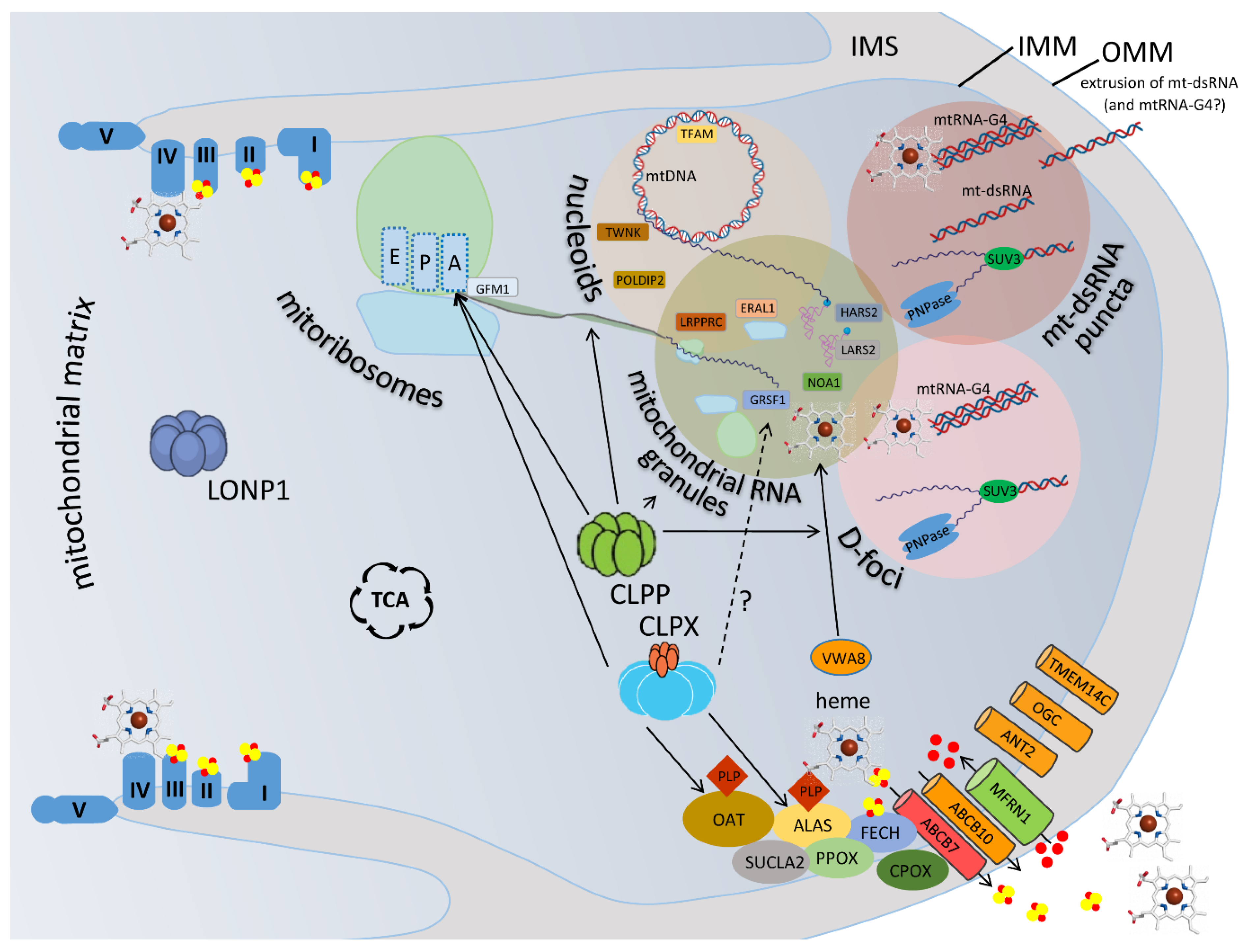
2.1. Prominent Impact of CLPP Absence and CLPX Excess on Mitochondrial Nucleoids
2.2. Prominent Impact of CLPP Absence and CLPX Excess on Mitoribosomes, Mostly on the tRNA-/mRNA-Associated and rRNA-Containing mtSSU
2.3. Prominent Impact of CLPP Absence and CLPX Excess on Mitochondrial RNA Processing Granules
2.4. Prominent Impact of CLPP Absence and CLPX Excess on Mitochondrial D-foci where RNA Degradation, Extrusion and Innate Immunity Activation are Decided
2.5. Prominent Impact of CLPP Absence via CLPX Accumulation on Heme Biosynthesis and Incorporation into the Complex-IV of the Respiratory Chain
2.6. Prominent Impact of CLPP Absence on the Fe-S Cluster Containing Peripheral Arm of Respiratory Complex-I
3. Phase-Separated Condensates in Mitochondria and the Cytosol
4. Proposal
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 4Fe–4S | iron sulfur clusters consisting of two interleaved 4Fe- and 4S-tetrahedra |
| AAA+ | ATPases Associated with diverse cellular Activities, and other ring-shaped P-loop NTPases |
| ABCB7 | ATP Binding Cassette Subfamily B Member 7 |
| ABCB10 | ATP Binding Cassette Subfamily B Member 10 |
| ALAS | delta-Amino-Levulinic Acid Synthase |
| ALAS2 | delta-Amino-Levulinic Acid Synthase 2, erythroid-specific |
| ALDH18A1 | Aldehyde Dehydrogenase 18 family member A1, = P5CS, delta-1-Pyrroline-5-Carboxylate Synthase |
| ANT2 | Adenine Nucleotide Translocator 2, = SLC25A25 |
| ATP | Adenosine Tri-Phosphate |
| ATPase | Adenosine Tri-Phosphatase |
| CAT-tail | C-terminal alanine and threonine tail |
| cGAS | cyclic GMP-AMP Synthase |
| CLPP | Caseinolytic Mitochondrial Matrix Peptidase Proteolytic Subunit |
| CLPX | Caseinolytic Mitochondrial Matrix Peptidase Chaperone Subunit X |
| CODAS | multiple anomalies syndrome with Cerebral, Ocular, Dental, Auricular and Skeletal anomalies |
| Cox1 | mitochondrially encoded Cytochrome C Oxidase I, mRNA |
| COX15 | Cytochrome C Oxidase assembly homolog COX15 |
| CPOX | Copro-Porphyrinogen OXidase |
| D-foci | degradosome granules in mitochondrial matrix |
| D-loop | displacement loop within the mtDNA |
| DARS2 | Aspartyl-tRNA Synthetase 2, mitochondrial |
| deltaALA | delta-aminolevulinic acid |
| DNA | Deoxyribo-Nucleic Acid |
| DNAzyme | catalytically active DNA sequences |
| dsRNA | double-stranded RNA |
| EF-G | Elongation Factor G |
| EFG1 | G Elongation Factor, mitochondrial 1 |
| ERAL1 | Era (E. coli) -Like 12S mitochondrial rRNA chaperone 1 |
| Fe2+ | divalent cation of iron, ferrous iron |
| FECH | Ferrochelatase |
| Fe-S | iron-sulfur |
| G4 | Guanine-quadruplex where RNA or DNA acquires four-stranded conformation |
| GFM1 | G elongation Factor Mitochondrial 1 |
| GRSF1 | G-rich RNA Sequence binding Factor 1 |
| GSA | Glutamate-5-Semi-Aldehyde |
| GTP | Guanosine-5'-triphosphate |
| HARS2 | Histidyl-tRNA Synthetase 2, mitochondrial |
| hsp-6 | heat shock protein family B (small) member 6 |
| hsp-60 | heat shock protein family D (hsp60) member 1, = human HSPD1 |
| HSPD1 | Heat Shock Protein family D (hsp60) member 1, = human HSP60, chaperonin |
| HTRA2 | High Temperature Requirement A serine peptidase 2 |
| i-AAA | ATP-dependent zinc metalloprotease YME1 (S. cerevisiae) -Like 1 |
| IDR | Intrinsically Disordered Region |
| IMM | Inner Membrane of Mitochondria |
| ISC | Iron-Sulfur-Cluster |
| KO | Knock-Out |
| LARS2 | Leucyl-tRNA Synthetase 2, mitochondrial |
| lncRNA | long non-coding RNA |
| LLPS | liquid-liquid phase separation |
| LONP1 | Lon Peptidase 1, Mitochondrial |
| LRPPRC | Leucine Rich Pentatrico-Peptide Repeat Containing |
| m-AAA | AFG3-like matrix AAA peptidase subunit 2 and SPG7 matrix AAA peptidase subunit paraplegin |
| MEFs | mouse embryonic fibroblasts |
| MFRN1 | mitoferrin 1, = SLC25A37, Solute Carrier Family 25 Member 37 |
| Mg2+ | divalent cation of magnesium |
| MPP | mitochondrial processing peptidase |
| MRPL12 | Mitochondrial Ribosomal Protein L12 |
| MRPL18 | Mitochondrial Ribosomal Protein L18 |
| MRPL37 | Mitochondrial Ribosomal Protein L37 |
| MRPL38 | Mitochondrial Ribosomal Protein L38 |
| MRPS15 | Mitochondrial Ribosomal Protein S15 |
| MRPS35 | Mitochondrial Ribosomal Protein S35 |
| mtDNA | mitochondrial DNA |
| mtHSP75 | mitochondrial heat shock protein 75, = human TRAP1 |
| mtLSU | mitoribosomal 39S large subunit |
| mtRNA | mitochondrial RNA |
| mtSSB | mitochondrial single-stranded DNA binding protein |
| mtSSU | mitoribosomal 28S small subunit |
| mt-tRNA | mitochondrial transfer RNA |
| NADH | Nicotinamide Adenine Dinucleotide, reduced form |
| NDUFS2 | NADH:Ubiquinone Oxidoreductase Core Subunit S2 |
| NDUFV1 | NADH:Ubiquinone Oxidoreductase Core Subunit V1 |
| NDUFV2 | NADH:Ubiquinone Oxidoreductase Core Subunit V2 |
| NOA1 | Nitric Oxide Associated 1 |
| NTPase | Nucleoside-Tri-Phosphatase |
| OAT | Ornithine delta-Amino-Transferase |
| OGC | 2-Oxoglutarate/Malate Carrier protein, mitochondrial |
| OMA1 | Overlapping with the M-AAA protease 1 homolog, zinc metallopeptidase |
| OMM | Outer Membrane of Mitochondria |
| PARL | Presenilin Associated Rhomboid Like |
| PcG bodies | polycomb bodies |
| PEO1 | Progressive External Ophthalmoplegia 1 protein = TWNK |
| Phe | phenylalanine |
| PLP | Pyridoxal-5’-Phosphate |
| PLPBP | PLP-binding protein |
| PNPase | Polyribo-Nucleotide Phosphorylase / Nucleotidyl-Transferase 1 = PNPT1 in human |
| PNPT1 | Polyribo-Nucleotide Phosphorylase / Nucleotidyl-Transferase 1 = PNPase |
| POLDIP2 | DNA Polymerase Delta Interacting Protein 2 |
| poly(A) tail | poly(adenine) tail of messenger RNAs |
| poly(I:C) | poly(inosinic : cytidylic) acid |
| PPOX | Proto-Porphyrinogen OXidase |
| PRLTS3 | Perrault Syndrome type 3 |
| PRORP | Protein Only RNase P catalytic subunit |
| qPCR | quantitative Polymerase Chain Reaction |
| RMND1 | Required for Meiotic Nuclear Division 1 homolog |
| RNA | Ribo-Nucleic Acid |
| RNA-G4 | RNA, guanine-rich, in quadruplex conformation |
| RNase | ribonuclease |
| RNAzyme | catalytically active RNA sequences |
| RNF213 | Ring Finger protein 213 |
| rRNA | ribosomal RNA |
| SFXN4 | Sideroflexin 4 |
| SLC25A37 | Solute Carrier family 25 member 37, Mitoferrin 1 |
| SLC25A38 | Solute Carrier Family 25 Member 38, mitochondrial glycine transporter |
| STING | STimulator of INterferon response cGAMP interactor 1 |
| SUCLA2 | Succinate-CoA Ligase ADP-forming subunit beta |
| SUPV3L1 | = SUV3, Suv3-like RNA helicase |
| TCA | Tri-Carboxylic Acid cycle, = Krebs cycle |
| TFAM | Transcription Factor A, Mitochondrial |
| TIGM | Texas Institute of Genomic Medicine |
| TMEM14C | Transmembrane Protein 14C |
| tRNA | transfer RNA |
| TWNK | Twinkle mtDNA helicase, = PEO1 |
| UPR | unfolded protein response |
| UPRmt | unfolded protein response in mitochondria |
| Val | valine |
| VWA8 | von Willebrand Factor A domain containing 8 |
References
- Key, J.; Kohli, A.; Barcena, C.; Lopez-Otin, C.; Heidler, J.; Wittig, I.; Auburger, G. Global Proteome of LonP1(+/-) Mouse Embryonal Fibroblasts Reveals Impact on Respiratory Chain, but No Interdependence between Eral1 and Mitoribosomes. Int J Mol Sci 2019, 20. [CrossRef]
- Bezawork-Geleta, A.; Brodie, E.J.; Dougan, D.A.; Truscott, K.N. LON is the master protease that protects against protein aggregation in human mitochondria through direct degradation of misfolded proteins. Sci Rep 2015, 5, 17397. [CrossRef]
- Stahlberg, H.; Kutejova, E.; Suda, K.; Wolpensinger, B.; Lustig, A.; Schatz, G.; Engel, A.; Suzuki, C.K. Mitochondrial Lon of Saccharomyces cerevisiae is a ring-shaped protease with seven flexible subunits. Proc Natl Acad Sci U S A 1999, 96, 6787-6790. [CrossRef]
- Baker, T.A.; Sauer, R.T. ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim Biophys Acta 2012, 1823, 15-28. [CrossRef]
- Kardon, J.R.; Yien, Y.Y.; Huston, N.C.; Branco, D.S.; Hildick-Smith, G.J.; Rhee, K.Y.; Paw, B.H.; Baker, T.A. Mitochondrial ClpX Activates a Key Enzyme for Heme Biosynthesis and Erythropoiesis. Cell 2015, 161, 858-867. [CrossRef]
- Fischer, F.; Weil, A.; Hamann, A.; Osiewacz, H.D. Human CLPP reverts the longevity phenotype of a fungal ClpP deletion strain. Nat Commun 2013, 4, 1397. [CrossRef]
- Dikoglu, E.; Alfaiz, A.; Gorna, M.; Bertola, D.; Chae, J.H.; Cho, T.J.; Derbent, M.; Alanay, Y.; Guran, T.; Kim, O.H.; et al. Mutations in LONP1, a mitochondrial matrix protease, cause CODAS syndrome. Am J Med Genet A 2015, 167, 1501-1509. [CrossRef]
- Strauss, K.A.; Jinks, R.N.; Puffenberger, E.G.; Venkatesh, S.; Singh, K.; Cheng, I.; Mikita, N.; Thilagavathi, J.; Lee, J.; Sarafianos, S.; et al. CODAS syndrome is associated with mutations of LONP1, encoding mitochondrial AAA+ Lon protease. Am J Hum Genet 2015, 96, 121-135. [CrossRef]
- Faridi, R.; Stratton, P.; Salmeri, N.; Morell, R.J.; Khan, A.A.; Usmani, M.A.; Newman, W.G.; Riazuddin, S.; Friedman, T.B. Homozygous novel truncating variant of CLPP associated with severe Perrault syndrome. Clin Genet 2024, 105, 584-586. [CrossRef]
- Brodie, E.J.; Zhan, H.; Saiyed, T.; Truscott, K.N.; Dougan, D.A. Perrault syndrome type 3 caused by diverse molecular defects in CLPP. Sci Rep 2018, 8, 12862. [CrossRef]
- Theunissen, T.E.; Szklarczyk, R.; Gerards, M.; Hellebrekers, D.M.; Mulder-Den Hartog, E.N.; Vanoevelen, J.; Kamps, R.; de Koning, B.; Rutledge, S.L.; Schmitt-Mechelke, T.; et al. Specific MRI Abnormalities Reveal Severe Perrault Syndrome due to CLPP Defects. Front Neurol 2016, 7, 203. [CrossRef]
- Dursun, F.; Mohamoud, H.S.; Karim, N.; Naeem, M.; Jelani, M.; Kirmizibekmez, H. A Novel Missense Mutation in the CLPP Gene Causing Perrault Syndrome Type 3 in a Turkish Family. J Clin Res Pediatr Endocrinol 2016, 8, 472-477. [CrossRef]
- Demain, L.A.; Urquhart, J.E.; O'Sullivan, J.; Williams, S.G.; Bhaskar, S.S.; Jenkinson, E.M.; Lourenco, C.M.; Heiberg, A.; Pearce, S.H.; Shalev, S.A.; et al. Expanding the genotypic spectrum of Perrault syndrome. Clin Genet 2017, 91, 302-312. [CrossRef]
- Ahmed, S.; Jelani, M.; Alrayes, N.; Mohamoud, H.S.; Almramhi, M.M.; Anshasi, W.; Ahmed, N.A.; Wang, J.; Nasir, J.; Al-Aama, J.Y. Exome analysis identified a novel missense mutation in the CLPP gene in a consanguineous Saudi family expanding the clinical spectrum of Perrault Syndrome type-3. J Neurol Sci 2015, 353, 149-154. [CrossRef]
- Jenkinson, E.M.; Rehman, A.U.; Walsh, T.; Clayton-Smith, J.; Lee, K.; Morell, R.J.; Drummond, M.C.; Khan, S.N.; Naeem, M.A.; Rauf, B.; et al. Perrault syndrome is caused by recessive mutations in CLPP, encoding a mitochondrial ATP-dependent chambered protease. Am J Hum Genet 2013, 92, 605-613. [CrossRef]
- Hochberg, I.; Demain, L.A.M.; Richer, J.; Thompson, K.; Urquhart, J.E.; Rea, A.; Pagarkar, W.; Rodriguez-Palmero, A.; Schluter, A.; Verdura, E.; et al. Bi-allelic variants in the mitochondrial RNase P subunit PRORP cause mitochondrial tRNA processing defects and pleiotropic multisystem presentations. Am J Hum Genet 2021, 108, 2195-2204. [CrossRef]
- Newman, W.G.; Friedman, T.B.; Conway, G.S.; Demain, L.A.M. Perrault Syndrome. In GeneReviews((R)), Adam, M.P., Feldman, J., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Amemiya, A., Eds.; Seattle (WA), 1993.
- Kobe, C.; Kracht, L.W.; Timmermann, L.; Bachmann, J.; Schmidt, M.C. Perrault Syndrome with progressive nervous system involvement. Clin Nucl Med 2008, 33, 922-924. [CrossRef]
- Gottschalk, M.E.; Coker, S.B.; Fox, L.A. Neurologic anomalies of Perrault syndrome. Am J Med Genet 1996, 65, 274-276. [CrossRef]
- Linssen, W.H.; Van den Bent, M.J.; Brunner, H.G.; Poels, P.J. Deafness, sensory neuropathy, and ovarian dysgenesis: a new syndrome or a broader spectrum of Perrault syndrome? Am J Med Genet 1994, 51, 81-82. [CrossRef]
- Faridi, R.; Rea, A.; Fenollar-Ferrer, C.; O'Keefe, R.T.; Gu, S.; Munir, Z.; Khan, A.A.; Riazuddin, S.; Hoa, M.; Naz, S.; et al. New insights into Perrault syndrome, a clinically and genetically heterogeneous disorder. Hum Genet 2022, 141, 805-819. [CrossRef]
- Tucker, E.J.; Rius, R.; Jaillard, S.; Bell, K.; Lamont, P.J.; Travessa, A.; Dupont, J.; Sampaio, L.; Dulon, J.; Vuillaumier-Barrot, S.; et al. Genomic sequencing highlights the diverse molecular causes of Perrault syndrome: a peroxisomal disorder (PEX6), metabolic disorders (CLPP, GGPS1), and mtDNA maintenance/translation disorders (LARS2, TFAM). Hum Genet 2020, 139, 1325-1343. [CrossRef]
- Gotta, F.; Lamp, M.; Geroldi, A.; Trevisan, L.; Origone, P.; Fugazza, G.; Fabbri, S.; Nesti, C.; Rubegni, A.; Morani, F.; et al. A novel mutation of Twinkle in Perrault syndrome: A not rare diagnosis? Ann Hum Genet 2020, 84, 417-422. [CrossRef]
- Smith, T.B.; Rea, A.; Thomas, H.B.; Thompson, K.; Olahova, M.; Maroofian, R.; Zamani, M.; He, L.; Sadeghian, S.; Galehdari, H.; et al. Novel homozygous variants in PRORP expand the genotypic spectrum of combined oxidative phosphorylation deficiency 54. Eur J Hum Genet 2023, 31, 1190-1194. [CrossRef]
- Chatzispyrou, I.A.; Alders, M.; Guerrero-Castillo, S.; Zapata Perez, R.; Haagmans, M.A.; Mouchiroud, L.; Koster, J.; Ofman, R.; Baas, F.; Waterham, H.R.; et al. A homozygous missense mutation in ERAL1, encoding a mitochondrial rRNA chaperone, causes Perrault syndrome. Hum Mol Genet 2017, 26, 2541-2550. [CrossRef]
- Rioux, A.V.; Bergeron, N.A.; Riopel, J.; Marcoux, N.; Theriault, C.; Gould, P.V.; Garneau, A.P.; Isenring, P. The ever wider clinical spectrum of RMND1-related disorders and limitedness of phenotype-based classifications. J Mol Med (Berl) 2023, 101, 1229-1236. [CrossRef]
- Neyroud, A.S.; Rudinger-Thirion, J.; Frugier, M.; Riley, L.G.; Bidet, M.; Akloul, L.; Simpson, A.; Gilot, D.; Christodoulou, J.; Ravel, C.; et al. LARS2 variants can present as premature ovarian insufficiency in the absence of overt hearing loss. Eur J Hum Genet 2023, 31, 453-460. [CrossRef]
- Ozieblo, D.; Pazik, J.; Stepniak, I.; Skarzynski, H.; Oldak, M. Two Novel Pathogenic Variants Confirm RMND1 Causative Role in Perrault Syndrome with Renal Involvement. Genes (Basel) 2020, 11. [CrossRef]
- Riley, L.G.; Rudinger-Thirion, J.; Frugier, M.; Wilson, M.; Luig, M.; Alahakoon, T.I.; Nixon, C.Y.; Kirk, E.P.; Roscioli, T.; Lunke, S.; et al. The expanding LARS2 phenotypic spectrum: HLASA, Perrault syndrome with leukodystrophy, and mitochondrial myopathy. Hum Mutat 2020, 41, 1425-1434. [CrossRef]
- Demain, L.A.M.; Gerkes, E.H.; Smith, R.J.H.; Molina-Ramirez, L.P.; O'Keefe, R.T.; Newman, W.G. A recurrent missense variant in HARS2 results in variable sensorineural hearing loss in three unrelated families. J Hum Genet 2020, 65, 305-311. [CrossRef]
- Karstensen, H.G.; Rendtorff, N.D.; Hindbaek, L.S.; Colombo, R.; Stein, A.; Birkebaek, N.H.; Hartmann-Petersen, R.; Lindorff-Larsen, K.; Hojland, A.T.; Petersen, M.B.; et al. Novel HARS2 missense variants identified in individuals with sensorineural hearing impairment and Perrault syndrome. Eur J Med Genet 2020, 63, 103733. [CrossRef]
- Demain, L.A.M.; Antunes, D.; O'Sullivan, J.; Bhaskhar, S.S.; O'Keefe, R.T.; Newman, W.G. A known pathogenic variant in the essential mitochondrial translation gene RMND1 causes a Perrault-like syndrome with renal defects. Clin Genet 2018, 94, 276-277. [CrossRef]
- Kosaki, R.; Horikawa, R.; Fujii, E.; Kosaki, K. Biallelic mutations in LARS2 can cause Perrault syndrome type 2 with neurologic symptoms. Am J Med Genet A 2018, 176, 404-408. [CrossRef]
- Pierce, S.B.; Gersak, K.; Michaelson-Cohen, R.; Walsh, T.; Lee, M.K.; Malach, D.; Klevit, R.E.; King, M.C.; Levy-Lahad, E. Mutations in LARS2, encoding mitochondrial leucyl-tRNA synthetase, lead to premature ovarian failure and hearing loss in Perrault syndrome. Am J Hum Genet 2013, 92, 614-620. [CrossRef]
- Pierce, S.B.; Chisholm, K.M.; Lynch, E.D.; Lee, M.K.; Walsh, T.; Opitz, J.M.; Li, W.; Klevit, R.E.; King, M.C. Mutations in mitochondrial histidyl tRNA synthetase HARS2 cause ovarian dysgenesis and sensorineural hearing loss of Perrault syndrome. Proc Natl Acad Sci U S A 2011, 108, 6543-6548. [CrossRef]
- Auburger, G.; Key, J.; Gispert, S. The Bacterial ClpXP-ClpB Family Is Enriched with RNA-Binding Protein Complexes. Cells 2022, 11. [CrossRef]
- Ducamp, S.; Luscieti, S.; Ferrer-Cortes, X.; Nicolas, G.; Manceau, H.; Peoc'h, K.; Yien, Y.Y.; Kannengiesser, C.; Gouya, L.; Puy, H.; et al. A mutation in the iron-responsive element of ALAS2 is a modifier of disease severity in a patient suffering from CLPX associated erythropoietic protoporphyria. Haematologica 2021, 106, 2030-2033. [CrossRef]
- Yien, Y.Y.; Ducamp, S.; van der Vorm, L.N.; Kardon, J.R.; Manceau, H.; Kannengiesser, C.; Bergonia, H.A.; Kafina, M.D.; Karim, Z.; Gouya, L.; et al. Mutation in human CLPX elevates levels of delta-aminolevulinate synthase and protoporphyrin IX to promote erythropoietic protoporphyria. Proc Natl Acad Sci U S A 2017, 114, E8045-E8052. [CrossRef]
- van der Vorm, L.N.; Paw, B.H. Studying disorders of vertebrate iron and heme metabolism using zebrafish. Methods Cell Biol 2017, 138, 193-220. [CrossRef]
- Gispert, S.; Parganlija, D.; Klinkenberg, M.; Drose, S.; Wittig, I.; Mittelbronn, M.; Grzmil, P.; Koob, S.; Hamann, A.; Walter, M.; et al. Loss of mitochondrial peptidase Clpp leads to infertility, hearing loss plus growth retardation via accumulation of CLPX, mtDNA and inflammatory factors. Hum Mol Genet 2013, 22, 4871-4887. [CrossRef]
- Seiferling, D.; Szczepanowska, K.; Becker, C.; Senft, K.; Hermans, S.; Maiti, P.; Konig, T.; Kukat, A.; Trifunovic, A. Loss of CLPP alleviates mitochondrial cardiomyopathy without affecting the mammalian UPRmt. EMBO Rep 2016, 17, 953-964. [CrossRef]
- Key, J.; Gispert, S.; Koepf, G.; Steinhoff-Wagner, J.; Reichlmeir, M.; Auburger, G. Translation Fidelity and Respiration Deficits in CLPP-Deficient Tissues: Mechanistic Insights from Mitochondrial Complexome Profiling. Int J Mol Sci 2023, 24. [CrossRef]
- Szczepanowska, K.; Maiti, P.; Kukat, A.; Hofsetz, E.; Nolte, H.; Senft, K.; Becker, C.; Ruzzenente, B.; Hornig-Do, H.T.; Wibom, R.; et al. CLPP coordinates mitoribosomal assembly through the regulation of ERAL1 levels. EMBO J 2016, 35, 2566-2583. [CrossRef]
- Arribas, J.; Castano, J.G. A comparative study of the chymotrypsin-like activity of the rat liver multicatalytic proteinase and the ClpP from Escherichia coli. J Biol Chem 1993, 268, 21165-21171.
- Mabanglo, M.F.; Wong, K.S.; Barghash, M.M.; Leung, E.; Chuang, S.H.W.; Ardalan, A.; Majaesic, E.M.; Wong, C.J.; Zhang, S.; Lang, H.; et al. Potent ClpP agonists with anticancer properties bind with improved structural complementarity and alter the mitochondrial N-terminome. Structure 2023, 31, 185-200 e110. [CrossRef]
- Prabhu, V.V.; Morrow, S.; Rahman Kawakibi, A.; Zhou, L.; Ralff, M.; Ray, J.; Jhaveri, A.; Ferrarini, I.; Lee, Y.; Parker, C.; et al. ONC201 and imipridones: Anti-cancer compounds with clinical efficacy. Neoplasia 2020, 22, 725-744. [CrossRef]
- Wong, K.S.; Houry, W.A. Chemical Modulation of Human Mitochondrial ClpP: Potential Application in Cancer Therapeutics. ACS Chem Biol 2019, 14, 2349-2360. [CrossRef]
- Key, J.; Gispert, S.; Kandi, A.R.; Heinz, D.; Hamann, A.; Osiewacz, H.D.; Meierhofer, D.; Auburger, G. CLPP-Null Eukaryotes with Excess Heme Biosynthesis Show Reduced L-arginine Levels, Probably via CLPX-Mediated OAT Activation. Biomolecules 2024, 14. [CrossRef]
- Key, J.; Gispert, S.; Koornneef, L.; Sleddens-Linkels, E.; Kohli, A.; Torres-Odio, S.; Koepf, G.; Amr, S.; Reichlmeir, M.; Harter, P.N.; et al. CLPP Depletion Causes Diplotene Arrest; Underlying Testis Mitochondrial Dysfunction Occurs with Accumulation of Perrault Proteins ERAL1, PEO1, and HARS2. Cells 2022, 12. [CrossRef]
- Becker, C.; Kukat, A.; Szczepanowska, K.; Hermans, S.; Senft, K.; Brandscheid, C.P.; Maiti, P.; Trifunovic, A. CLPP deficiency protects against metabolic syndrome but hinders adaptive thermogenesis. EMBO Rep 2018, 19. [CrossRef]
- Bhaskaran, S.; Pharaoh, G.; Ranjit, R.; Murphy, A.; Matsuzaki, S.; Nair, B.C.; Forbes, B.; Gispert, S.; Auburger, G.; Humphries, K.M.; et al. Loss of mitochondrial protease ClpP protects mice from diet-induced obesity and insulin resistance. EMBO Rep 2018, 19. [CrossRef]
- Hofsetz, E.; Demir, F.; Szczepanowska, K.; Kukat, A.; Kizhakkedathu, J.N.; Trifunovic, A.; Huesgen, P.F. The Mouse Heart Mitochondria N Terminome Provides Insights into ClpXP-Mediated Proteolysis. Mol Cell Proteomics 2020, 19, 1330-1345. [CrossRef]
- Benedetti, C.; Haynes, C.M.; Yang, Y.; Harding, H.P.; Ron, D. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics 2006, 174, 229-239. [CrossRef]
- Key, J.; Torres-Odio, S.; Bach, N.C.; Gispert, S.; Koepf, G.; Reichlmeir, M.; West, A.P.; Prokisch, H.; Freisinger, P.; Newman, W.G.; et al. Inactivity of Peptidase ClpP Causes Primary Accumulation of Mitochondrial Disaggregase ClpX with Its Interacting Nucleoid Proteins, and of mtDNA. Cells 2021, 10. [CrossRef]
- Cheng, X.; Kanki, T.; Fukuoh, A.; Ohgaki, K.; Takeya, R.; Aoki, Y.; Hamasaki, N.; Kang, D. PDIP38 associates with proteins constituting the mitochondrial DNA nucleoid. J Biochem 2005, 138, 673-678. [CrossRef]
- Strack, P.R.; Brodie, E.J.; Zhan, H.; Schuenemann, V.J.; Valente, L.J.; Saiyed, T.; Lowth, B.R.; Angley, L.M.; Perugini, M.A.; Zeth, K.; et al. Polymerase delta-interacting protein 38 (PDIP38) modulates the stability and activity of the mitochondrial AAA+ protease CLPXP. Commun Biol 2020, 3, 646. [CrossRef]
- Zhang, R.; Wang, P.; Wei, B.; Chen, L.; Song, X.; Pan, Y.; Li, J.; Gan, J.; Zhang, T.; Yang, C.G. Assessment of the structure-activity relationship and antileukemic activity of diacylpyramide compounds as human ClpP agonists. Eur J Med Chem 2023, 258, 115577. [CrossRef]
- Nikali, K.; Suomalainen, A.; Saharinen, J.; Kuokkanen, M.; Spelbrink, J.N.; Lonnqvist, T.; Peltonen, L. Infantile onset spinocerebellar ataxia is caused by recessive mutations in mitochondrial proteins Twinkle and Twinky. Hum Mol Genet 2005, 14, 2981-2990. [CrossRef]
- Munson, H.E.; De Simone, L.; Schwaede, A.; Bhatia, A.; Mithal, D.S.; Young, N.; Kuntz, N.; Rao, V.K. Axonal polyneuropathy and ataxia in children: consider Perrault Syndrome, a case report. BMC Med Genomics 2023, 16, 278. [CrossRef]
- Wei, L.; Hou, L.; Ying, Y.Q.; Luo, X.P. A Novel Missense Mutation in TWNK Gene Causing Perrault Syndrome Type 5 in a Chinese Family and Review of the Literature. Pharmgenomics Pers Med 2022, 15, 1-8. [CrossRef]
- Chen, Z.; Tang, S.; Li, H.; Xu, X.; Lyu, J. [Analysis of TWNK variant in a family affected with Perrault syndrome]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2020, 37, 739-742. [CrossRef]
- Kume, K.; Morino, H.; Miyamoto, R.; Matsuda, Y.; Ohsawa, R.; Kanaya, Y.; Tada, Y.; Kurashige, T.; Kawakami, H. Middle-age-onset cerebellar ataxia caused by a homozygous TWNK variant: a case report. BMC Med Genet 2020, 21, 68. [CrossRef]
- Fekete, B.; Pentelenyi, K.; Rudas, G.; Gal, A.; Grosz, Z.; Illes, A.; Idris, J.; Csukly, G.; Domonkos, A.; Molnar, M.J. Broadening the phenotype of the TWNK gene associated Perrault syndrome. BMC Med Genet 2019, 20, 198. [CrossRef]
- Dominguez-Ruiz, M.; Garcia-Martinez, A.; Corral-Juan, M.; Perez-Alvarez, A.I.; Plasencia, A.M.; Villamar, M.; Moreno-Pelayo, M.A.; Matilla-Duenas, A.; Menendez-Gonzalez, M.; Del Castillo, I. Perrault syndrome with neurological features in a compound heterozygote for two TWNK mutations: overlap of TWNK-related recessive disorders. J Transl Med 2019, 17, 290. [CrossRef]
- Oldak, M.; Ozieblo, D.; Pollak, A.; Stepniak, I.; Lazniewski, M.; Lechowicz, U.; Kochanek, K.; Furmanek, M.; Tacikowska, G.; Plewczynski, D.; et al. Novel neuro-audiological findings and further evidence for TWNK involvement in Perrault syndrome. J Transl Med 2017, 15, 25. [CrossRef]
- Morino, H.; Pierce, S.B.; Matsuda, Y.; Walsh, T.; Ohsawa, R.; Newby, M.; Hiraki-Kamon, K.; Kuramochi, M.; Lee, M.K.; Klevit, R.E.; et al. Mutations in Twinkle primase-helicase cause Perrault syndrome with neurologic features. Neurology 2014, 83, 2054-2061. [CrossRef]
- Yamamoto, H.; Qin, Y.; Achenbach, J.; Li, C.; Kijek, J.; Spahn, C.M.; Nierhaus, K.H. EF-G and EF4: translocation and back-translocation on the bacterial ribosome. Nat Rev Microbiol 2014, 12, 89-100. [CrossRef]
- Carbone, C.E.; Loveland, A.B.; Gamper, H.B., Jr.; Hou, Y.M.; Demo, G.; Korostelev, A.A. Time-resolved cryo-EM visualizes ribosomal translocation with EF-G and GTP. Nat Commun 2021, 12, 7236. [CrossRef]
- Koripella, R.K.; Sharma, M.R.; Bhargava, K.; Datta, P.P.; Kaushal, P.S.; Keshavan, P.; Spremulli, L.L.; Banavali, N.K.; Agrawal, R.K. Structures of the human mitochondrial ribosome bound to EF-G1 reveal distinct features of mitochondrial translation elongation. Nat Commun 2020, 11, 3830. [CrossRef]
- Kummer, E.; Ban, N. Structural insights into mammalian mitochondrial translation elongation catalyzed by mtEFG1. EMBO J 2020, 39, e104820. [CrossRef]
- Chen, Y.; Feng, S.; Kumar, V.; Ero, R.; Gao, Y.G. Structure of EF-G-ribosome complex in a pretranslocation state. Nat Struct Mol Biol 2013, 20, 1077-1084. [CrossRef]
- Moraleda, J.M.; Gonzalez, R.; Alegre, A.; Anta, J.P.; San Miguel, J.F. Bone marrow necrosis and treatment with interferon. J Clin Pathol 1986, 39, 1045. [CrossRef]
- Sitron, C.S.; Park, J.H.; Giafaglione, J.M.; Brandman, O. Aggregation of CAT tails blocks their degradation and causes proteotoxicity in S. cerevisiae. PLoS One 2020, 15, e0227841. [CrossRef]
- Wu, Z.; Tantray, I.; Lim, J.; Chen, S.; Li, Y.; Davis, Z.; Sitron, C.; Dong, J.; Gispert, S.; Auburger, G.; et al. MISTERMINATE Mechanistically Links Mitochondrial Dysfunction with Proteostasis Failure. Mol Cell 2019, 75, 835-848 e838. [CrossRef]
- Lytvynenko, I.; Paternoga, H.; Thrun, A.; Balke, A.; Muller, T.A.; Chiang, C.H.; Nagler, K.; Tsaprailis, G.; Anders, S.; Bischofs, I.; et al. Alanine Tails Signal Proteolysis in Bacterial Ribosome-Associated Quality Control. Cell 2019, 178, 76-90 e22. [CrossRef]
- Gottesman, S.; Roche, E.; Zhou, Y.; Sauer, R.T. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev 1998, 12, 1338-1347. [CrossRef]
- Gruffaz, C.; Smirnov, A. GTPase Era at the heart of ribosome assembly. Front Mol Biosci 2023, 10, 1263433. [CrossRef]
- Dennerlein, S.; Rozanska, A.; Wydro, M.; Chrzanowska-Lightowlers, Z.M.; Lightowlers, R.N. Human ERAL1 is a mitochondrial RNA chaperone involved in the assembly of the 28S small mitochondrial ribosomal subunit. Biochem J 2010, 430, 551-558. [CrossRef]
- Uchiumi, T.; Ohgaki, K.; Yagi, M.; Aoki, Y.; Sakai, A.; Matsumoto, S.; Kang, D. ERAL1 is associated with mitochondrial ribosome and elimination of ERAL1 leads to mitochondrial dysfunction and growth retardation. Nucleic Acids Res 2010, 38, 5554-5568. [CrossRef]
- Accardi, R.; Oxelmark, E.; Jauniaux, N.; de Pinto, V.; Marchini, A.; Tommasino, M. High levels of the mitochondrial large ribosomal subunit protein 40 prevent loss of mitochondrial DNA in null mmf1 Saccharomyces cerevisiae cells. Yeast 2004, 21, 539-548. [CrossRef]
- Zhang, X.; Gao, X.; Coots, R.A.; Conn, C.S.; Liu, B.; Qian, S.B. Translational control of the cytosolic stress response by mitochondrial ribosomal protein L18. Nat Struct Mol Biol 2015, 22, 404-410. [CrossRef]
- Xu, P.; Wang, L.; Peng, H.; Liu, H.; Liu, H.; Yuan, Q.; Lin, Y.; Xu, J.; Pang, X.; Wu, H.; et al. Disruption of Hars2 in Cochlear Hair Cells Causes Progressive Mitochondrial Dysfunction and Hearing Loss in Mice. Front Cell Neurosci 2021, 15, 804345. [CrossRef]
- Gong, S.; Wang, X.; Meng, F.; Cui, L.; Yi, Q.; Zhao, Q.; Cang, X.; Cai, Z.; Mo, J.Q.; Liang, Y.; et al. Overexpression of mitochondrial histidyl-tRNA synthetase restores mitochondrial dysfunction caused by a deafness-associated tRNA(His) mutation. J Biol Chem 2020, 295, 940-954. [CrossRef]
- van der Knaap, M.S.; Bugiani, M.; Mendes, M.I.; Riley, L.G.; Smith, D.E.C.; Rudinger-Thirion, J.; Frugier, M.; Breur, M.; Crawford, J.; van Gaalen, J.; et al. Biallelic variants in LARS2 and KARS cause deafness and (ovario)leukodystrophy. Neurology 2019, 92, e1225-e1237. [CrossRef]
- Perli, E.; Fiorillo, A.; Giordano, C.; Pisano, A.; Montanari, A.; Grazioli, P.; Campese, A.F.; Di Micco, P.; Tuppen, H.A.; Genovese, I.; et al. Short peptides from leucyl-tRNA synthetase rescue disease-causing mitochondrial tRNA point mutations. Hum Mol Genet 2016, 25, 903-915. [CrossRef]
- Hornig-Do, H.T.; Montanari, A.; Rozanska, A.; Tuppen, H.A.; Almalki, A.A.; Abg-Kamaludin, D.P.; Frontali, L.; Francisci, S.; Lightowlers, R.N.; Chrzanowska-Lightowlers, Z.M. Human mitochondrial leucyl tRNA synthetase can suppress non cognate pathogenic mt-tRNA mutations. EMBO Mol Med 2014, 6, 183-193. [CrossRef]
- Levinger, L.; Morl, M.; Florentz, C. Mitochondrial tRNA 3' end metabolism and human disease. Nucleic Acids Res 2004, 32, 5430-5441. [CrossRef]
- Rumyantseva, A.; Popovic, M.; Trifunovic, A. CLPP deficiency ameliorates neurodegeneration caused by impaired mitochondrial protein synthesis. Brain 2022, 145, 92-104. [CrossRef]
- Lyu, B.; Song, Q. The intricate relationship of G-Quadruplexes and bacterial pathogenicity islands. Elife 2024, 12. [CrossRef]
- Mestre-Fos, S.; Ito, C.; Moore, C.M.; Reddi, A.R.; Williams, L.D. Human ribosomal G-quadruplexes regulate heme bioavailability. J Biol Chem 2020, 295, 14855-14865. [CrossRef]
- Chang, T.; Liu, X.; Cheng, X.; Qi, C.; Mei, H.; Shangguan, D. Selective isolation of G-quadruplexes by affinity chromatography. J Chromatogr A 2012, 1246, 62-68. [CrossRef]
- Li, W.; Li, Y.; Liu, Z.; Lin, B.; Yi, H.; Xu, F.; Nie, Z.; Yao, S. Insight into G-quadruplex-hemin DNAzyme/RNAzyme: adjacent adenine as the intramolecular species for remarkable enhancement of enzymatic activity. Nucleic Acids Res 2016, 44, 7373-7384. [CrossRef]
- Grigg, J.C.; Shumayrikh, N.; Sen, D. G-quadruplex structures formed by expanded hexanucleotide repeat RNA and DNA from the neurodegenerative disease-linked C9orf72 gene efficiently sequester and activate heme. PLoS One 2014, 9, e106449. [CrossRef]
- Li, C.; Yin, Z.; Xiao, R.; Huang, B.; Cui, Y.; Wang, H.; Xiang, Y.; Wang, L.; Lei, L.; Ye, J.; et al. G-quadruplexes sense natural porphyrin metabolites for regulation of gene transcription and chromatin landscapes. Genome Biol 2022, 23, 259. [CrossRef]
- Varshney, D.; Cuesta, S.M.; Herdy, B.; Abdullah, U.B.; Tannahill, D.; Balasubramanian, S. RNA G-quadruplex structures control ribosomal protein production. Sci Rep 2021, 11, 22735. [CrossRef]
- Pietras, Z.; Wojcik, M.A.; Borowski, L.S.; Szewczyk, M.; Kulinski, T.M.; Cysewski, D.; Stepien, P.P.; Dziembowski, A.; Szczesny, R.J. Dedicated surveillance mechanism controls G-quadruplex forming non-coding RNAs in human mitochondria. Nat Commun 2018, 9, 2558. [CrossRef]
- Noh, J.H.; Kim, K.M.; Abdelmohsen, K.; Yoon, J.H.; Panda, A.C.; Munk, R.; Kim, J.; Curtis, J.; Moad, C.A.; Wohler, C.M.; et al. HuR and GRSF1 modulate the nuclear export and mitochondrial localization of the lncRNA RMRP. Genes Dev 2016, 30, 1224-1239. [CrossRef]
- Jourdain, A.A.; Koppen, M.; Wydro, M.; Rodley, C.D.; Lightowlers, R.N.; Chrzanowska-Lightowlers, Z.M.; Martinou, J.C. GRSF1 regulates RNA processing in mitochondrial RNA granules. Cell Metab 2013, 17, 399-410. [CrossRef]
- Hensen, F.; Potter, A.; van Esveld, S.L.; Tarres-Sole, A.; Chakraborty, A.; Sola, M.; Spelbrink, J.N. Mitochondrial RNA granules are critically dependent on mtDNA replication factors Twinkle and mtSSB. Nucleic Acids Res 2019, 47, 3680-3698. [CrossRef]
- Xavier, V.J.; Martinou, J.C. RNA Granules in the Mitochondria and Their Organization under Mitochondrial Stresses. Int J Mol Sci 2021, 22. [CrossRef]
- Antonicka, H.; Sasarman, F.; Nishimura, T.; Paupe, V.; Shoubridge, E.A. The mitochondrial RNA-binding protein GRSF1 localizes to RNA granules and is required for posttranscriptional mitochondrial gene expression. Cell Metab 2013, 17, 386-398. [CrossRef]
- Pietras, Z.; Wojcik, M.A.; Borowski, L.S.; Szewczyk, M.; Kulinski, T.M.; Cysewski, D.; Stepien, P.P.; Dziembowski, A.; Szczesny, R.J. Controlling the mitochondrial antisense - role of the SUV3-PNPase complex and its co-factor GRSF1 in mitochondrial RNA surveillance. Mol Cell Oncol 2018, 5, e1516452. [CrossRef]
- Wang, J.; Lu, J.; Zhu, Y.; Huang, Q.; Gu, Q.; Tian, S.; Ge, J.; Lin, X.; Sha, W. Guanine-rich RNA sequence binding factor 1 regulates neuronal ferroptosis after spinal cord injury in rats via the GPX4 signaling pathway. Brain Res 2023, 1818, 148497. [CrossRef]
- Dumoulin, B.; Heydeck, D.; Jahn, D.; Lasse, M.; Sofi, S.; Ufer, C.; Kuhn, H. Male guanine-rich RNA sequence binding factor 1 knockout mice (Grsf1(-/-)) gain less body weight during adolescence and adulthood. Cell Biosci 2022, 12, 199. [CrossRef]
- Noh, J.H.; Kim, K.M.; Pandey, P.R.; Noren Hooten, N.; Munk, R.; Kundu, G.; De, S.; Martindale, J.L.; Yang, X.; Evans, M.K.; et al. Loss of RNA-binding protein GRSF1 activates mTOR to elicit a proinflammatory transcriptional program. Nucleic Acids Res 2019, 47, 2472-2486. [CrossRef]
- Al-Furoukh, N.; Goffart, S.; Szibor, M.; Wanrooij, S.; Braun, T. Binding to G-quadruplex RNA activates the mitochondrial GTPase NOA1. Biochim Biophys Acta 2013, 1833, 2933-2942. [CrossRef]
- He, J.; Cooper, H.M.; Reyes, A.; Di Re, M.; Kazak, L.; Wood, S.R.; Mao, C.C.; Fearnley, I.M.; Walker, J.E.; Holt, I.J. Human C4orf14 interacts with the mitochondrial nucleoid and is involved in the biogenesis of the small mitochondrial ribosomal subunit. Nucleic Acids Res 2012, 40, 6097-6108. [CrossRef]
- Kolanczyk, M.; Pech, M.; Zemojtel, T.; Yamamoto, H.; Mikula, I.; Calvaruso, M.A.; van den Brand, M.; Richter, R.; Fischer, B.; Ritz, A.; et al. NOA1 is an essential GTPase required for mitochondrial protein synthesis. Mol Biol Cell 2011, 22, 1-11. [CrossRef]
- Al-Furoukh, N.; Kardon, J.R.; Kruger, M.; Szibor, M.; Baker, T.A.; Braun, T. NOA1, a novel ClpXP substrate, takes an unexpected nuclear detour prior to mitochondrial import. PLoS One 2014, 9, e103141. [CrossRef]
- Luo, M.; Ma, W.; Sand, Z.; Finlayson, J.; Wang, T.; Brinton, R.D.; Willis, W.T.; Mandarino, L.J. Von Willebrand factor A domain-containing protein 8 (VWA8) localizes to the matrix side of the inner mitochondrial membrane. Biochem Biophys Res Commun 2020, 521, 158-163. [CrossRef]
- Chen, Z.; Suzuki, H.; Kobayashi, Y.; Wang, A.C.; DiMaio, F.; Kawashima, S.A.; Walz, T.; Kapoor, T.M. Structural Insights into Mdn1, an Essential AAA Protein Required for Ribosome Biogenesis. Cell 2018, 175, 822-834 e818. [CrossRef]
- Kawashima, S.A.; Chen, Z.; Aoi, Y.; Patgiri, A.; Kobayashi, Y.; Nurse, P.; Kapoor, T.M. Potent, Reversible, and Specific Chemical Inhibitors of Eukaryotic Ribosome Biogenesis. Cell 2016, 167, 512-524 e514. [CrossRef]
- Prattes, M.; Lo, Y.H.; Bergler, H.; Stanley, R.E. Shaping the Nascent Ribosome: AAA-ATPases in Eukaryotic Ribosome Biogenesis. Biomolecules 2019, 9. [CrossRef]
- Linke, R.; Limmer, M.; Juranek, S.A.; Heine, A.; Paeschke, K. The Relevance of G-Quadruplexes for DNA Repair. Int J Mol Sci 2021, 22. [CrossRef]
- McShane, E.; Couvillion, M.; Ietswaart, R.; Prakash, G.; Smalec, B.M.; Soto, I.; Baxter-Koenigs, A.R.; Choquet, K.; Churchman, L.S. A kinetic dichotomy between mitochondrial and nuclear gene expression processes. Mol Cell 2024. [CrossRef]
- Honarmand, S.; Shoubridge, E.A. Poly (A) tail length of human mitochondrial mRNAs is tissue-specific and a mutation in LRPPRC results in transcript-specific patterns of deadenylation. Mol Genet Metab Rep 2020, 25, 100687. [CrossRef]
- Wilson, W.C.; Hornig-Do, H.T.; Bruni, F.; Chang, J.H.; Jourdain, A.A.; Martinou, J.C.; Falkenberg, M.; Spahr, H.; Larsson, N.G.; Lewis, R.J.; et al. A human mitochondrial poly(A) polymerase mutation reveals the complexities of post-transcriptional mitochondrial gene expression. Hum Mol Genet 2014, 23, 6345-6355. [CrossRef]
- Chujo, T.; Ohira, T.; Sakaguchi, Y.; Goshima, N.; Nomura, N.; Nagao, A.; Suzuki, T. LRPPRC/SLIRP suppresses PNPase-mediated mRNA decay and promotes polyadenylation in human mitochondria. Nucleic Acids Res 2012, 40, 8033-8047. [CrossRef]
- Ruzzenente, B.; Metodiev, M.D.; Wredenberg, A.; Bratic, A.; Park, C.B.; Camara, Y.; Milenkovic, D.; Zickermann, V.; Wibom, R.; Hultenby, K.; et al. LRPPRC is necessary for polyadenylation and coordination of translation of mitochondrial mRNAs. EMBO J 2012, 31, 443-456. [CrossRef]
- Pajak, A.; Laine, I.; Clemente, P.; El-Fissi, N.; Schober, F.A.; Maffezzini, C.; Calvo-Garrido, J.; Wibom, R.; Filograna, R.; Dhir, A.; et al. Defects of mitochondrial RNA turnover lead to the accumulation of double-stranded RNA in vivo. PLoS Genet 2019, 15, e1008240. [CrossRef]
- Szczesny, R.J.; Wojcik, M.A.; Borowski, L.S.; Szewczyk, M.J.; Skrok, M.M.; Golik, P.; Stepien, P.P. Yeast and human mitochondrial helicases. Biochim Biophys Acta 2013, 1829, 842-853. [CrossRef]
- Borowski, L.S.; Dziembowski, A.; Hejnowicz, M.S.; Stepien, P.P.; Szczesny, R.J. Human mitochondrial RNA decay mediated by PNPase-hSuv3 complex takes place in distinct foci. Nucleic Acids Res 2013, 41, 1223-1240. [CrossRef]
- Szczesny, R.J.; Borowski, L.S.; Malecki, M.; Wojcik, M.A.; Stepien, P.P.; Golik, P. RNA degradation in yeast and human mitochondria. Biochim Biophys Acta 2012, 1819, 1027-1034. [CrossRef]
- Clemente, P.; Pajak, A.; Laine, I.; Wibom, R.; Wedell, A.; Freyer, C.; Wredenberg, A. SUV3 helicase is required for correct processing of mitochondrial transcripts. Nucleic Acids Res 2015, 43, 7398-7413. [CrossRef]
- Jain, M.; Golzarroshan, B.; Lin, C.L.; Agrawal, S.; Tang, W.H.; Wu, C.J.; Yuan, H.S. Dimeric assembly of human Suv3 helicase promotes its RNA unwinding function in mitochondrial RNA degradosome for RNA decay. Protein Sci 2022, 31, e4312. [CrossRef]
- Toompuu, M.; Tuomela, T.; Laine, P.; Paulin, L.; Dufour, E.; Jacobs, H.T. Polyadenylation and degradation of structurally abnormal mitochondrial tRNAs in human cells. Nucleic Acids Res 2018, 46, 5209-5226. [CrossRef]
- Szczesny, R.J.; Borowski, L.S.; Brzezniak, L.K.; Dmochowska, A.; Gewartowski, K.; Bartnik, E.; Stepien, P.P. Human mitochondrial RNA turnover caught in flagranti: involvement of hSuv3p helicase in RNA surveillance. Nucleic Acids Res 2010, 38, 279-298. [CrossRef]
- Wang, D.D.; Shu, Z.; Lieser, S.A.; Chen, P.L.; Lee, W.H. Human mitochondrial SUV3 and polynucleotide phosphorylase form a 330-kDa heteropentamer to cooperatively degrade double-stranded RNA with a 3'-to-5' directionality. J Biol Chem 2009, 284, 20812-20821. [CrossRef]
- Sarkar, D.; Fisher, P.B. Human polynucleotide phosphorylase (hPNPase old-35): an RNA degradation enzyme with pleiotrophic biological effects. Cell Cycle 2006, 5, 1080-1084. [CrossRef]
- Piazza, F.; Zappone, M.; Sana, M.; Briani, F.; Deho, G. Polynucleotide phosphorylase of Escherichia coli is required for the establishment of bacteriophage P4 immunity. J Bacteriol 1996, 178, 5513-5521. [CrossRef]
- Chen, P.L. SUV3 Helicase and Mitochondrial Homeostasis. Int J Mol Sci 2023, 24. [CrossRef]
- Silva, S.; Camino, L.P.; Aguilera, A. Human mitochondrial degradosome prevents harmful mitochondrial R loops and mitochondrial genome instability. Proc Natl Acad Sci U S A 2018, 115, 11024-11029. [CrossRef]
- Carzaniga, T.; Sbarufatti, G.; Briani, F.; Deho, G. Polynucleotide phosphorylase is implicated in homologous recombination and DNA repair in Escherichia coli. BMC Microbiol 2017, 17, 81. [CrossRef]
- Tuteja, N.; Tarique, M.; Tuteja, R. Rice SUV3 is a bidirectional helicase that binds both DNA and RNA. BMC Plant Biol 2014, 14, 283. [CrossRef]
- Minczuk, M.; Piwowarski, J.; Papworth, M.A.; Awiszus, K.; Schalinski, S.; Dziembowski, A.; Dmochowska, A.; Bartnik, E.; Tokatlidis, K.; Stepien, P.P.; et al. Localisation of the human hSuv3p helicase in the mitochondrial matrix and its preferential unwinding of dsDNA. Nucleic Acids Res 2002, 30, 5074-5086. [CrossRef]
- Barbier, M.; Bahlo, M.; Pennisi, A.; Jacoupy, M.; Tankard, R.M.; Ewenczyk, C.; Davies, K.C.; Lino-Coulon, P.; Colace, C.; Rafehi, H.; et al. Heterozygous PNPT1 Variants Cause Spinocerebellar Ataxia Type 25. Ann Neurol 2022, 92, 122-137. [CrossRef]
- Eaton, A.; Bernier, F.P.; Goedhart, C.; Caluseriu, O.; Lamont, R.E.; Boycott, K.M.; Parboosingh, J.S.; Innes, A.M.; Care4Rare Canada, C. Is PNPT1-related hearing loss ever non-syndromic? Whole exome sequencing of adult siblings expands the natural history of PNPT1-related disorders. Am J Med Genet A 2018, 176, 2487-2493. [CrossRef]
- Matilainen, S.; Carroll, C.J.; Richter, U.; Euro, L.; Pohjanpelto, M.; Paetau, A.; Isohanni, P.; Suomalainen, A. Defective mitochondrial RNA processing due to PNPT1 variants causes Leigh syndrome. Hum Mol Genet 2017, 26, 3352-3361. [CrossRef]
- Sato, R.; Arai-Ichinoi, N.; Kikuchi, A.; Matsuhashi, T.; Numata-Uematsu, Y.; Uematsu, M.; Fujii, Y.; Murayama, K.; Ohtake, A.; Abe, T.; et al. Novel biallelic mutations in the PNPT1 gene encoding a mitochondrial-RNA-import protein PNPase cause delayed myelination. Clin Genet 2018, 93, 242-247. [CrossRef]
- Slavotinek, A.M.; Garcia, S.T.; Chandratillake, G.; Bardakjian, T.; Ullah, E.; Wu, D.; Umeda, K.; Lao, R.; Tang, P.L.; Wan, E.; et al. Exome sequencing in 32 patients with anophthalmia/microphthalmia and developmental eye defects. Clin Genet 2015, 88, 468-473. [CrossRef]
- von Ameln, S.; Wang, G.; Boulouiz, R.; Rutherford, M.A.; Smith, G.M.; Li, Y.; Pogoda, H.M.; Nurnberg, G.; Stiller, B.; Volk, A.E.; et al. A mutation in PNPT1, encoding mitochondrial-RNA-import protein PNPase, causes hereditary hearing loss. Am J Hum Genet 2012, 91, 919-927. [CrossRef]
- Dhir, A.; Dhir, S.; Borowski, L.S.; Jimenez, L.; Teitell, M.; Rotig, A.; Crow, Y.J.; Rice, G.I.; Duffy, D.; Tamby, C.; et al. Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature 2018, 560, 238-242. [CrossRef]
- Key, J.; Maletzko, A.; Kohli, A.; Gispert, S.; Torres-Odio, S.; Wittig, I.; Heidler, J.; Barcena, C.; Lopez-Otin, C.; Lei, Y.; et al. Loss of mitochondrial ClpP, Lonp1, and Tfam triggers transcriptional induction of Rnf213, a susceptibility factor for moyamoya disease. Neurogenetics 2020, 21, 187-203. [CrossRef]
- Maletzko, A.; Key, J.; Wittig, I.; Gispert, S.; Koepf, G.; Canet-Pons, J.; Torres-Odio, S.; West, A.P.; Auburger, G. Increased presence of nuclear DNAJA3 and upregulation of cytosolic STAT1 and of nucleic acid sensors trigger innate immunity in the ClpP-null mouse. Neurogenetics 2021, 22, 297-312. [CrossRef]
- Torres-Odio, S.; Lei, Y.; Gispert, S.; Maletzko, A.; Key, J.; Menissy, S.S.; Wittig, I.; Auburger, G.; West, A.P. Loss of Mitochondrial Protease CLPP Activates Type I IFN Responses through the Mitochondrial DNA-cGAS-STING Signaling Axis. J Immunol 2021, 206, 1890-1900. [CrossRef]
- McKay, R.; Druyan, R.; Getz, G.S.; Rabinowitz, M. Intramitochondrial localization of delta-aminolaevulate synthetase and ferrochelatase in rat liver. Biochem J 1969, 114, 455-461. [CrossRef]
- Medlock, A.E.; Shiferaw, M.T.; Marcero, J.R.; Vashisht, A.A.; Wohlschlegel, J.A.; Phillips, J.D.; Dailey, H.A. Identification of the Mitochondrial Heme Metabolism Complex. PLoS One 2015, 10, e0135896. [CrossRef]
- Ferreira, G.C.; Andrew, T.L.; Karr, S.W.; Dailey, H.A. Organization of the terminal two enzymes of the heme biosynthetic pathway. Orientation of protoporphyrinogen oxidase and evidence for a membrane complex. J Biol Chem 1988, 263, 3835-3839.
- Lim, S.; Clark, D.S. Phase-separated biomolecular condensates for biocatalysis. Trends Biotechnol 2024, 42, 496-509. [CrossRef]
- Dahmani, I.; Qin, K.; Zhang, Y.; Fernie, A.R. The formation and function of plant metabolons. Plant J 2023, 114, 1080-1092. [CrossRef]
- Kastritis, P.L.; Gavin, A.C. Enzymatic complexes across scales. Essays Biochem 2018, 62, 501-514. [CrossRef]
- Kim, H.J.; Khalimonchuk, O.; Smith, P.M.; Winge, D.R. Structure, function, and assembly of heme centers in mitochondrial respiratory complexes. Biochim Biophys Acta 2012, 1823, 1604-1616. [CrossRef]
- Maio, N.; Kim, K.S.; Holmes-Hampton, G.; Singh, A.; Rouault, T.A. Dimeric ferrochelatase bridges ABCB7 and ABCB10 homodimers in an architecturally defined molecular complex required for heme biosynthesis. Haematologica 2019, 104, 1756-1767. [CrossRef]
- Chen, W.; Dailey, H.A.; Paw, B.H. Ferrochelatase forms an oligomeric complex with mitoferrin-1 and Abcb10 for erythroid heme biosynthesis. Blood 2010, 116, 628-630. [CrossRef]
- Richardson, D.R.; Lane, D.J.; Becker, E.M.; Huang, M.L.; Whitnall, M.; Suryo Rahmanto, Y.; Sheftel, A.D.; Ponka, P. Mitochondrial iron trafficking and the integration of iron metabolism between the mitochondrion and cytosol. Proc Natl Acad Sci U S A 2010, 107, 10775-10782. [CrossRef]
- Shum, M.; Shintre, C.A.; Althoff, T.; Gutierrez, V.; Segawa, M.; Saxberg, A.D.; Martinez, M.; Adamson, R.; Young, M.R.; Faust, B.; et al. ABCB10 exports mitochondrial biliverdin, driving metabolic maladaptation in obesity. Sci Transl Med 2021, 13. [CrossRef]
- Pearson, S.A.; Cowan, J.A. Evolution of the human mitochondrial ABCB7 [2Fe-2S](GS)(4) cluster exporter and the molecular mechanism of an E433K disease-causing mutation. Arch Biochem Biophys 2021, 697, 108661. [CrossRef]
- Martinez, M.; Fendley, G.A.; Saxberg, A.D.; Zoghbi, M.E. Stimulation of the human mitochondrial transporter ABCB10 by zinc-mesoporphrin. PLoS One 2020, 15, e0238754. [CrossRef]
- Yamamoto, M.; Arimura, H.; Fukushige, T.; Minami, K.; Nishizawa, Y.; Tanimoto, A.; Kanekura, T.; Nakagawa, M.; Akiyama, S.; Furukawa, T. Abcb10 role in heme biosynthesis in vivo: Abcb10 knockout in mice causes anemia with protoporphyrin IX and iron accumulation. Mol Cell Biol 2014, 34, 1077-1084. [CrossRef]
- Chen, W.; Paradkar, P.N.; Li, L.; Pierce, E.L.; Langer, N.B.; Takahashi-Makise, N.; Hyde, B.B.; Shirihai, O.S.; Ward, D.M.; Kaplan, J.; et al. Abcb10 physically interacts with mitoferrin-1 (Slc25a37) to enhance its stability and function in the erythroid mitochondria. Proc Natl Acad Sci U S A 2009, 106, 16263-16268. [CrossRef]
- Clough, C.A.; Pangallo, J.; Sarchi, M.; Ilagan, J.O.; North, K.; Bergantinos, R.; Stolla, M.C.; Naru, J.; Nugent, P.; Kim, E.; et al. Coordinated missplicing of TMEM14C and ABCB7 causes ring sideroblast formation in SF3B1-mutant myelodysplastic syndrome. Blood 2022, 139, 2038-2049. [CrossRef]
- Harding, C.R.; Sidik, S.M.; Petrova, B.; Gnadig, N.F.; Okombo, J.; Herneisen, A.L.; Ward, K.E.; Markus, B.M.; Boydston, E.A.; Fidock, D.A.; et al. Genetic screens reveal a central role for heme metabolism in artemisinin susceptibility. Nat Commun 2020, 11, 4813. [CrossRef]
- Yien, Y.Y.; Robledo, R.F.; Schultz, I.J.; Takahashi-Makise, N.; Gwynn, B.; Bauer, D.E.; Dass, A.; Yi, G.; Li, L.; Hildick-Smith, G.J.; et al. TMEM14C is required for erythroid mitochondrial heme metabolism. J Clin Invest 2014, 124, 4294-4304. [CrossRef]
- Nilsson, R.; Schultz, I.J.; Pierce, E.L.; Soltis, K.A.; Naranuntarat, A.; Ward, D.M.; Baughman, J.M.; Paradkar, P.N.; Kingsley, P.D.; Culotta, V.C.; et al. Discovery of genes essential for heme biosynthesis through large-scale gene expression analysis. Cell Metab 2009, 10, 119-130. [CrossRef]
- Azuma, M.; Kabe, Y.; Kuramori, C.; Kondo, M.; Yamaguchi, Y.; Handa, H. Adenine nucleotide translocator transports haem precursors into mitochondria. PLoS One 2008, 3, e3070. [CrossRef]
- Kabe, Y.; Ohmori, M.; Shinouchi, K.; Tsuboi, Y.; Hirao, S.; Azuma, M.; Watanabe, H.; Okura, I.; Handa, H. Porphyrin accumulation in mitochondria is mediated by 2-oxoglutarate carrier. J Biol Chem 2006, 281, 31729-31735. [CrossRef]
- Lash, L.H. Mitochondrial glutathione transport: physiological, pathological and toxicological implications. Chem Biol Interact 2006, 163, 54-67. [CrossRef]
- Deery, E.; Schroeder, S.; Lawrence, A.D.; Taylor, S.L.; Seyedarabi, A.; Waterman, J.; Wilson, K.S.; Brown, D.; Geeves, M.A.; Howard, M.J.; et al. An enzyme-trap approach allows isolation of intermediates in cobalamin biosynthesis. Nat Chem Biol 2012, 8, 933-940. [CrossRef]
- Cheung, C.W.; Cohen, N.S.; Raijman, L. Channeling of urea cycle intermediates in situ in permeabilized hepatocytes. J Biol Chem 1989, 264, 4038-4044.
- Chen, C.; Hamza, I. Notes from the Underground: Heme Homeostasis in C. elegans. Biomolecules 2023, 13. [CrossRef]
- Chambers, I.G.; Willoughby, M.M.; Hamza, I.; Reddi, A.R. One ring to bring them all and in the darkness bind them: The trafficking of heme without deliverers. Biochim Biophys Acta Mol Cell Res 2021, 1868, 118881. [CrossRef]
- Sachar, M.; Anderson, K.E.; Ma, X. Protoporphyrin IX: the Good, the Bad, and the Ugly. J Pharmacol Exp Ther 2016, 356, 267-275. [CrossRef]
- Liou, Y.F.; Charoenkwan, P.; Srinivasulu, Y.; Vasylenko, T.; Lai, S.C.; Lee, H.C.; Chen, Y.H.; Huang, H.L.; Ho, S.Y. SCMHBP: prediction and analysis of heme binding proteins using propensity scores of dipeptides. BMC Bioinformatics 2014, 15 Suppl 16, S4. [CrossRef]
- Whitman, J.C.; Paw, B.H.; Chung, J. The role of ClpX in erythropoietic protoporphyria. Hematol Transfus Cell Ther 2018, 40, 182-188. [CrossRef]
- Fouquet, C.; Le Rouzic, M.A.; Leblanc, T.; Fouyssac, F.; Leverger, G.; Hessissen, L.; Marlin, S.; Bourrat, E.; Fahd, M.; Raffoux, E.; et al. Genotype/phenotype correlations of childhood-onset congenital sideroblastic anaemia in a European cohort. Br J Haematol 2019, 187, 530-542. [CrossRef]
- Kardon, J.R.; Moroco, J.A.; Engen, J.R.; Baker, T.A. Mitochondrial ClpX activates an essential biosynthetic enzyme through partial unfolding. Elife 2020, 9. [CrossRef]
- Brown, B.L.; Kardon, J.R.; Sauer, R.T.; Baker, T.A. Structure of the Mitochondrial Aminolevulinic Acid Synthase, a Key Heme Biosynthetic Enzyme. Structure 2018, 26, 580-589 e584. [CrossRef]
- Huang, L. An experimental study of the principle of electronic root canal measurement. J Endod 1987, 13, 60-64. [CrossRef]
- Kubota, Y.; Nomura, K.; Katoh, Y.; Yamashita, R.; Kaneko, K.; Furuyama, K. Novel Mechanisms for Heme-dependent Degradation of ALAS1 Protein as a Component of Negative Feedback Regulation of Heme Biosynthesis. J Biol Chem 2016, 291, 20516-20529. [CrossRef]
- Schiroli, D.; Peracchi, A. A subfamily of PLP-dependent enzymes specialized in handling terminal amines. Biochim Biophys Acta 2015, 1854, 1200-1211. [CrossRef]
- Tarnacka, B.; Jopowicz, A.; Maslinska, M. Copper, Iron, and Manganese Toxicity in Neuropsychiatric Conditions. Int J Mol Sci 2021, 22. [CrossRef]
- Jiang, X.; Stockwell, B.R.; Conrad, M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol 2021, 22, 266-282. [CrossRef]
- Laut, C.L.; Leasure, C.S.; Pi, H.; Carlin, S.M.; Chu, M.L.; Hillebrand, G.H.; Lin, H.K.; Yi, X.I.; Stauff, D.L.; Skaar, E.P. DnaJ and ClpX Are Required for HitRS and HssRS Two-Component System Signaling in Bacillus anthracis. Infect Immun 2022, 90, e0056021. [CrossRef]
- Farrand, A.J.; Friedman, D.B.; Reniere, M.L.; Ingmer, H.; Frees, D.; Skaar, E.P. Proteomic analyses of iron-responsive, Clp-dependent changes in Staphylococcus aureus. Pathog Dis 2015, 73. [CrossRef]
- Farrand, A.J.; Reniere, M.L.; Ingmer, H.; Frees, D.; Skaar, E.P. Regulation of host hemoglobin binding by the Staphylococcus aureus Clp proteolytic system. J Bacteriol 2013, 195, 5041-5050. [CrossRef]
- Stanne, T.M.; Sjogren, L.L.; Koussevitzky, S.; Clarke, A.K. Identification of new protein substrates for the chloroplast ATP-dependent Clp protease supports its constitutive role in Arabidopsis. Biochem J 2009, 417, 257-268. [CrossRef]
- Wang, L.; Elliott, M.; Elliott, T. Conditional stability of the HemA protein (glutamyl-tRNA reductase) regulates heme biosynthesis in Salmonella typhimurium. J Bacteriol 1999, 181, 1211-1219. [CrossRef]
- Szczepanowska, K.; Trifunovic, A. Tune instead of destroy: How proteolysis keeps OXPHOS in shape. Biochim Biophys Acta Bioenerg 2021, 1862, 148365. [CrossRef]
- Vercellino, I.; Sazanov, L.A. The assembly, regulation and function of the mitochondrial respiratory chain. Nat Rev Mol Cell Biol 2022, 23, 141-161. [CrossRef]
- Giachin, G.; Bouverot, R.; Acajjaoui, S.; Pantalone, S.; Soler-Lopez, M. Dynamics of Human Mitochondrial Complex I Assembly: Implications for Neurodegenerative Diseases. Front Mol Biosci 2016, 3, 43. [CrossRef]
- Berrisford, J.M.; Baradaran, R.; Sazanov, L.A. Structure of bacterial respiratory complex I. Biochim Biophys Acta 2016, 1857, 892-901. [CrossRef]
- Friedrich, T. On the mechanism of respiratory complex I. J Bioenerg Biomembr 2014, 46, 255-268. [CrossRef]
- Clason, T.; Ruiz, T.; Schagger, H.; Peng, G.; Zickermann, V.; Brandt, U.; Michel, H.; Radermacher, M. The structure of eukaryotic and prokaryotic complex I. J Struct Biol 2010, 169, 81-88. [CrossRef]
- Szczepanowska, K.; Senft, K.; Heidler, J.; Herholz, M.; Kukat, A.; Hohne, M.N.; Hofsetz, E.; Becker, C.; Kaspar, S.; Giese, H.; et al. A salvage pathway maintains highly functional respiratory complex I. Nat Commun 2020, 11, 1643. [CrossRef]
- Feric, M.; Demarest, T.G.; Tian, J.; Croteau, D.L.; Bohr, V.A.; Misteli, T. Self-assembly of multi-component mitochondrial nucleoids via phase separation. EMBO J 2021, 40, e107165. [CrossRef]
- Rey, T.; Zaganelli, S.; Cuillery, E.; Vartholomaiou, E.; Croisier, M.; Martinou, J.C.; Manley, S. Mitochondrial RNA granules are fluid condensates positioned by membrane dynamics. Nat Cell Biol 2020, 22, 1180-1186. [CrossRef]
- Guilhas, B.; Walter, J.C.; Rech, J.; David, G.; Walliser, N.O.; Palmeri, J.; Mathieu-Demaziere, C.; Parmeggiani, A.; Bouet, J.Y.; Le Gall, A.; et al. ATP-Driven Separation of Liquid Phase Condensates in Bacteria. Mol Cell 2020, 79, 293-303 e294. [CrossRef]
- Ladouceur, A.M.; Parmar, B.S.; Biedzinski, S.; Wall, J.; Tope, S.G.; Cohn, D.; Kim, A.; Soubry, N.; Reyes-Lamothe, R.; Weber, S.C. Clusters of bacterial RNA polymerase are biomolecular condensates that assemble through liquid-liquid phase separation. Proc Natl Acad Sci U S A 2020, 117, 18540-18549. [CrossRef]
- Muthunayake, N.S.; Tomares, D.T.; Childers, W.S.; Schrader, J.M. Phase-separated bacterial ribonucleoprotein bodies organize mRNA decay. Wiley Interdiscip Rev RNA 2020, 11, e1599. [CrossRef]
- Kato, M.; Han, T.W.; Xie, S.; Shi, K.; Du, X.; Wu, L.C.; Mirzaei, H.; Goldsmith, E.J.; Longgood, J.; Pei, J.; et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 2012, 149, 753-767. [CrossRef]
- Annunziata, O.; Asherie, N.; Lomakin, A.; Pande, J.; Ogun, O.; Benedek, G.B. Effect of polyethylene glycol on the liquid-liquid phase transition in aqueous protein solutions. Proc Natl Acad Sci U S A 2002, 99, 14165-14170. [CrossRef]
- Zhang, L.; Wang, S.; Wang, W.; Shi, J.; Stovall, D.B.; Li, D.; Sui, G. Phase-Separated Subcellular Compartmentation and Related Human Diseases. Int J Mol Sci 2022, 23. [CrossRef]
- Peran, I.; Mittag, T. Molecular structure in biomolecular condensates. Curr Opin Struct Biol 2020, 60, 17-26. [CrossRef]
- Alberti, S.; Dormann, D. Liquid-Liquid Phase Separation in Disease. Annu Rev Genet 2019, 53, 171-194. [CrossRef]
- Dutta, A.; Sepehri, A.; Lazaridis, T. Putative Pore Structures of Amyloid beta 25-35 in Lipid Bilayers. Biochemistry 2023, 62, 2549-2558. [CrossRef]
- Vendruscolo, M.; Fuxreiter, M. Sequence Determinants of the Aggregation of Proteins Within Condensates Generated by Liquid-liquid Phase Separation. J Mol Biol 2022, 434, 167201. [CrossRef]
- Stockl, M.T.; Zijlstra, N.; Subramaniam, V. alpha-Synuclein oligomers: an amyloid pore? Insights into mechanisms of alpha-synuclein oligomer-lipid interactions. Mol Neurobiol 2013, 47, 613-621. [CrossRef]
- Fuller, G.G.; Kim, J.K. Compartmentalization and metabolic regulation of glycolysis. J Cell Sci 2021, 134. [CrossRef]
- Nesterov, S.V.; Ilyinsky, N.S.; Plokhikh, K.S.; Manuylov, V.D.; Chesnokov, Y.M.; Vasilov, R.G.; Kuznetsova, I.M.; Turoverov, K.K.; Gordeliy, V.I.; Fonin, A.V.; et al. Order wrapped in chaos: On the roles of intrinsically disordered proteins and RNAs in the arrangement of the mitochondrial enzymatic machines. Int J Biol Macromol 2024, 267, 131455. [CrossRef]
- Zurita Rendon, O.; Shoubridge, E.A. LONP1 Is Required for Maturation of a Subset of Mitochondrial Proteins, and Its Loss Elicits an Integrated Stress Response. Mol Cell Biol 2018, 38. [CrossRef]
- Szczepanowska, K.; Trifunovic, A. Mitochondrial matrix proteases: quality control and beyond. FEBS J 2022, 289, 7128-7146. [CrossRef]
- Feng, Y.; Nouri, K.; Schimmer, A.D. Mitochondrial ATP-Dependent Proteases-Biological Function and Potential Anti-Cancer Targets. Cancers (Basel) 2021, 13. [CrossRef]
- Quiros, P.M.; Langer, T.; Lopez-Otin, C. New roles for mitochondrial proteases in health, ageing and disease. Nat Rev Mol Cell Biol 2015, 16, 345-359. [CrossRef]
- Bohovych, I.; Chan, S.S.; Khalimonchuk, O. Mitochondrial protein quality control: the mechanisms guarding mitochondrial health. Antioxid Redox Signal 2015, 22, 977-994. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



