Submitted:
18 January 2024
Posted:
19 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Culture Conditions of P. anserina:
2.2. Metabolic Profiling of P. anserina:
2.3. Global Proteome Profile of P. anserina:
2.4. Mouse Cerebellar Metabolome:
2.5. Mouse Metabolic Validation Study:
2.6. Statistical Analyses
3. Results
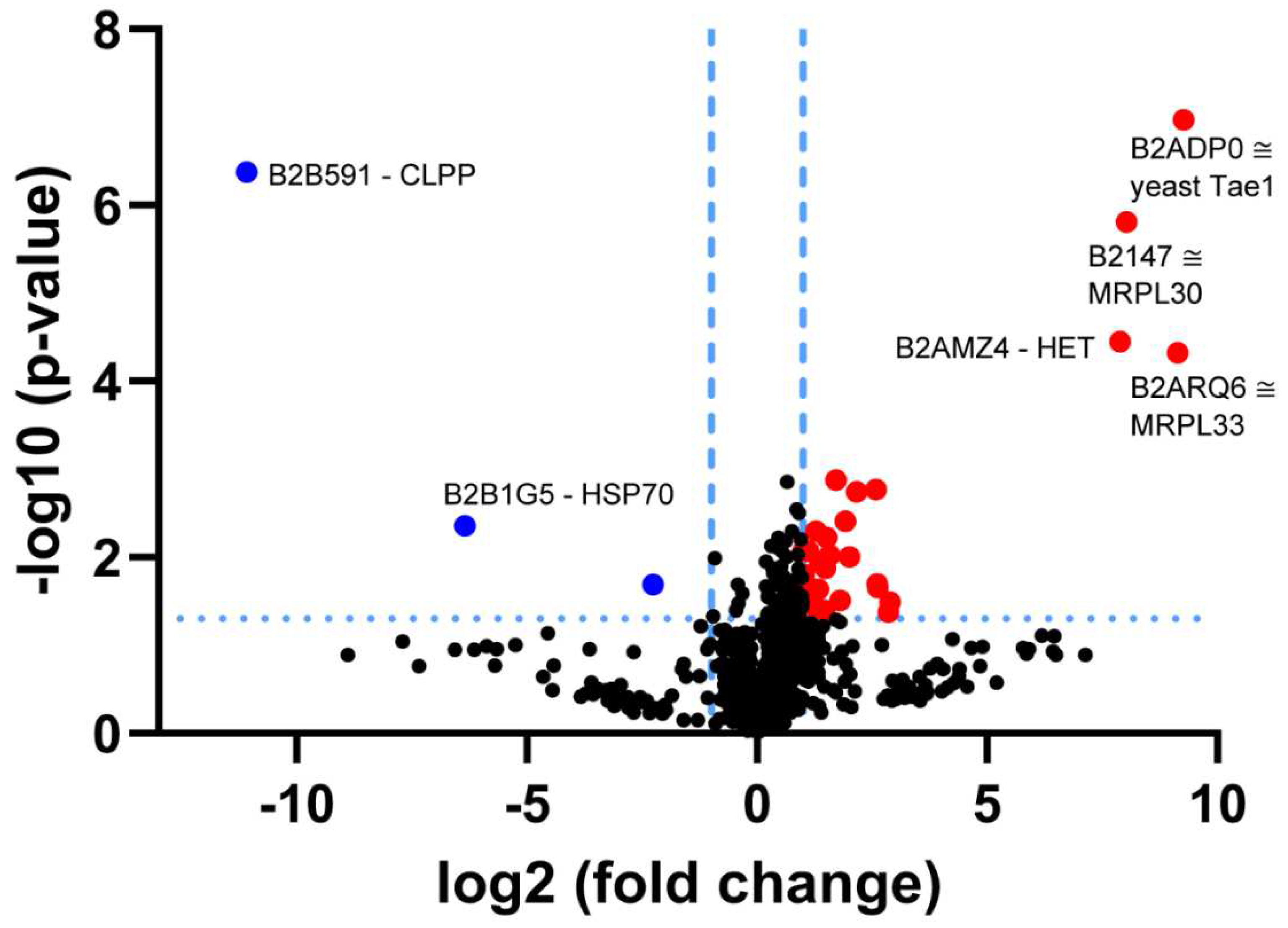
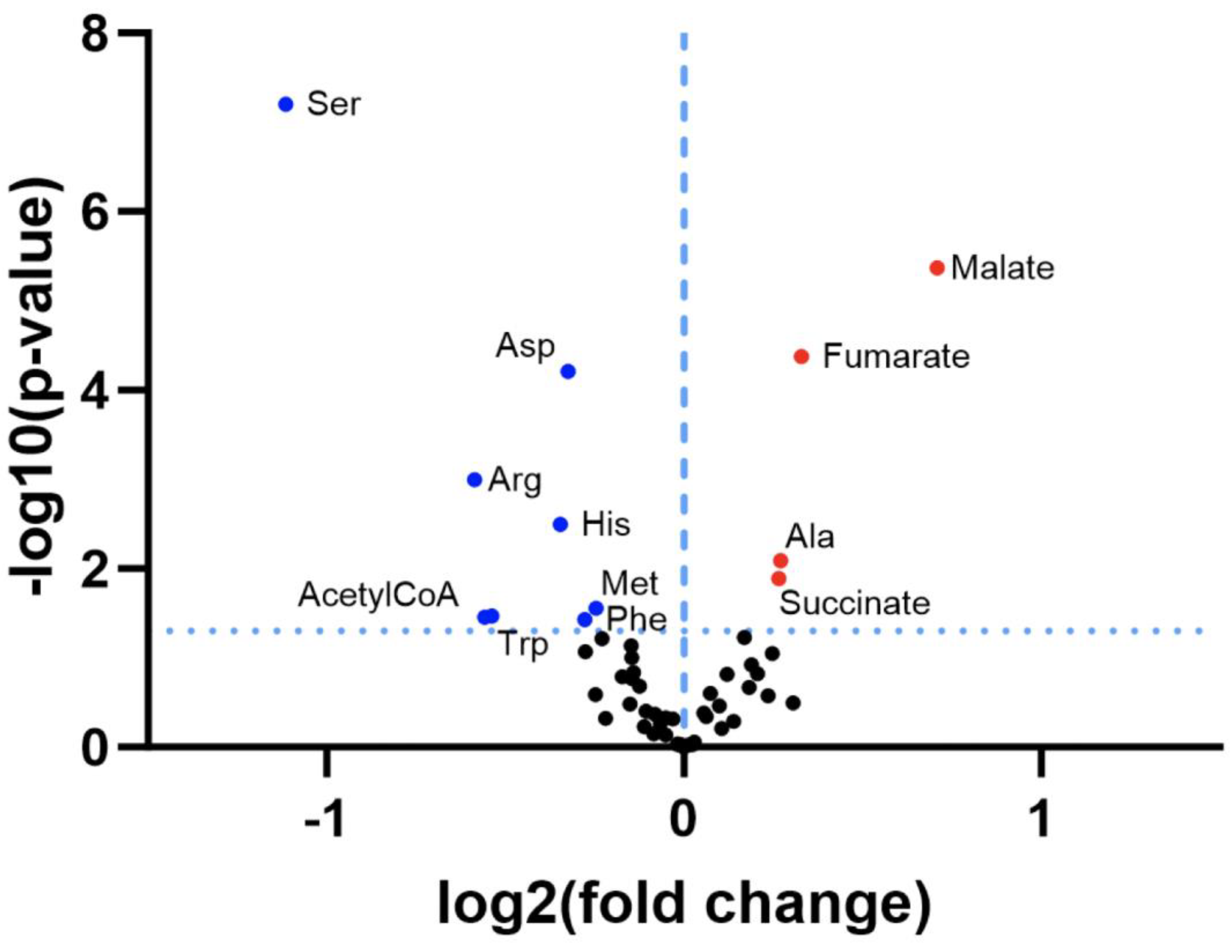
3.1. Re-Analysis of CLPP-Mutation Effects in P. anserina:
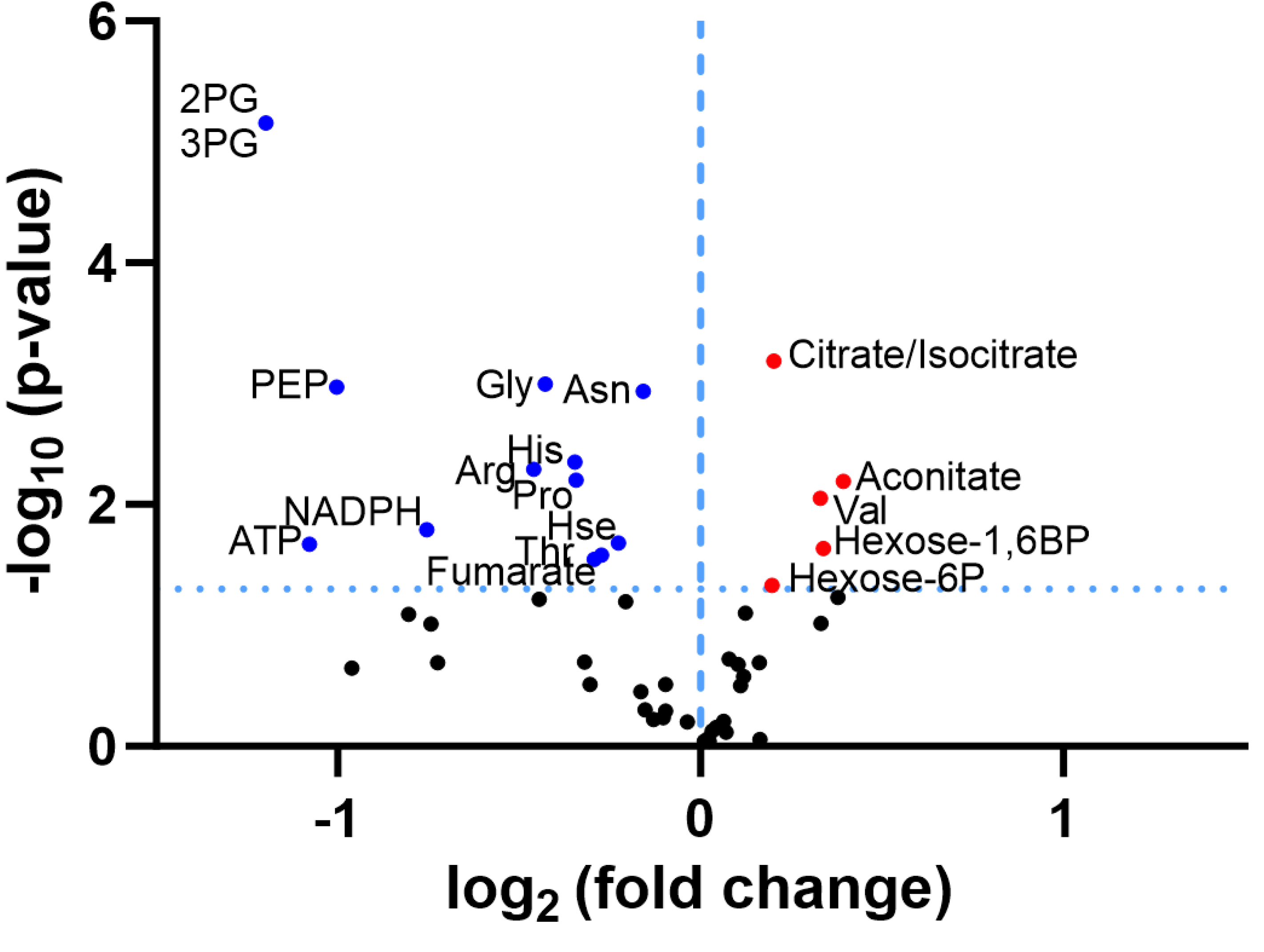
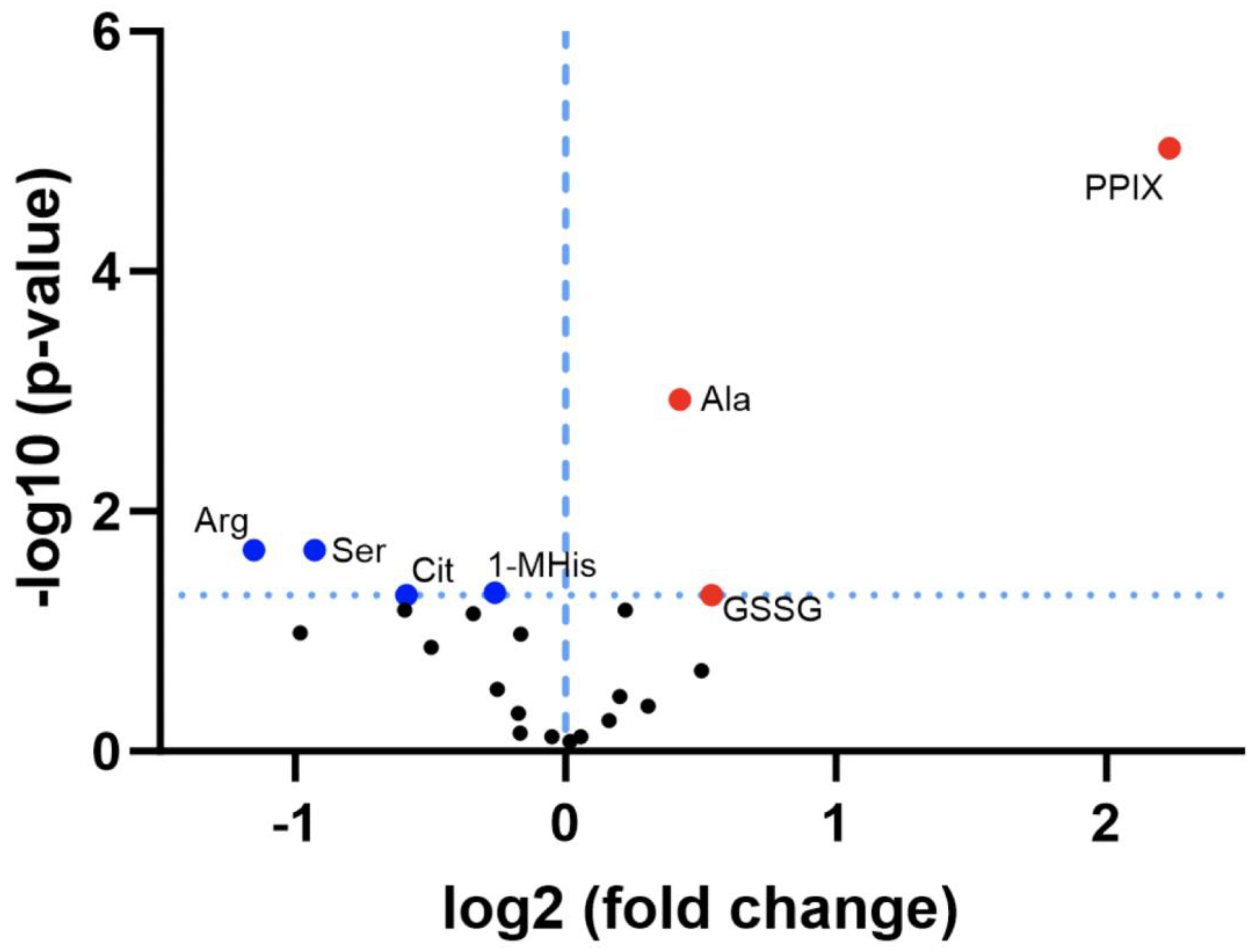
3.2. Discovery Survey of Cognate Amino Acid Levels in CLPP-Null Mouse Cerebellum:
3.3. Confirmatory Survey with Selected Non-Cognate Amino Acids in CLPP-Null Mouse Cerebellum:
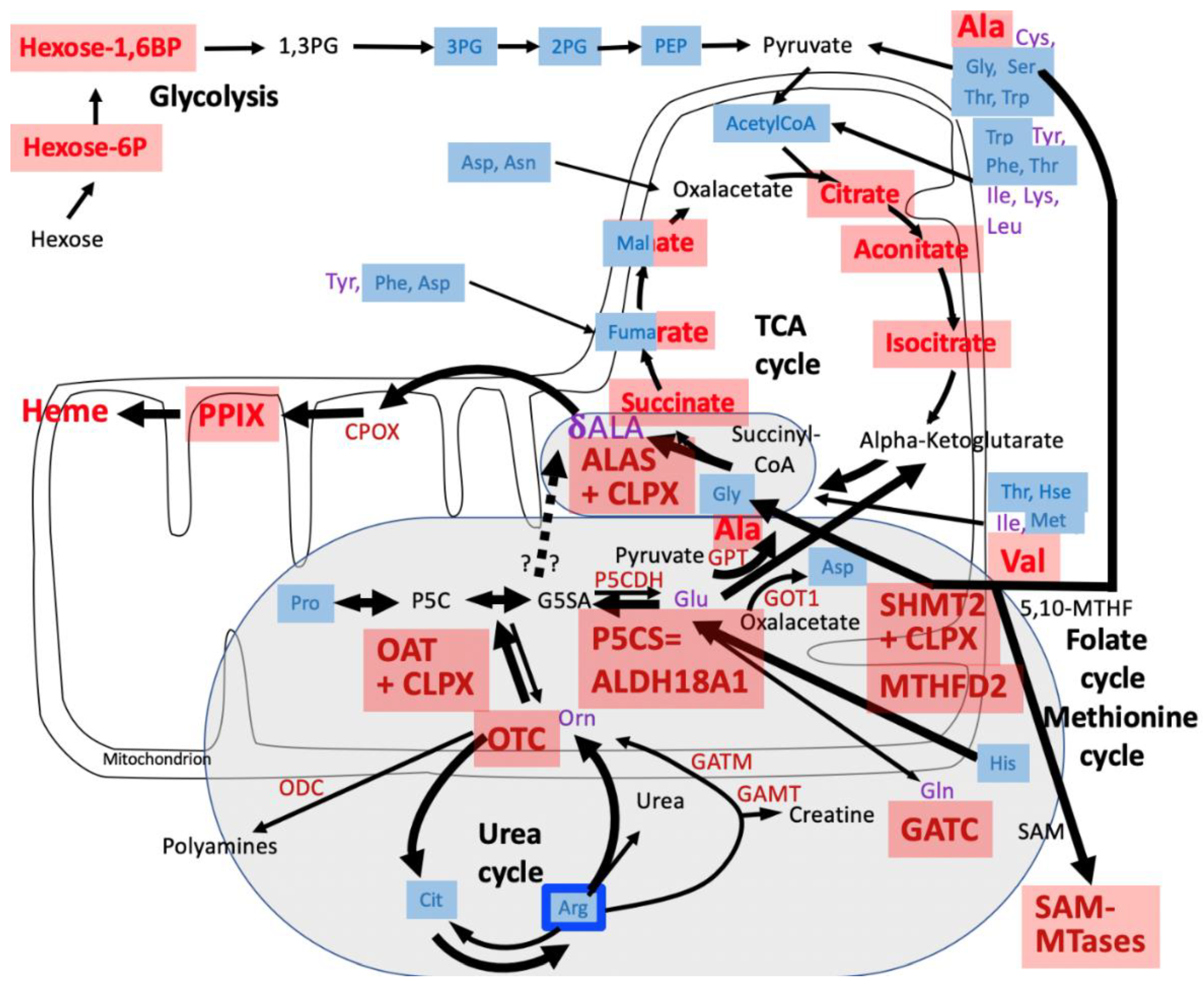
4. Discussion
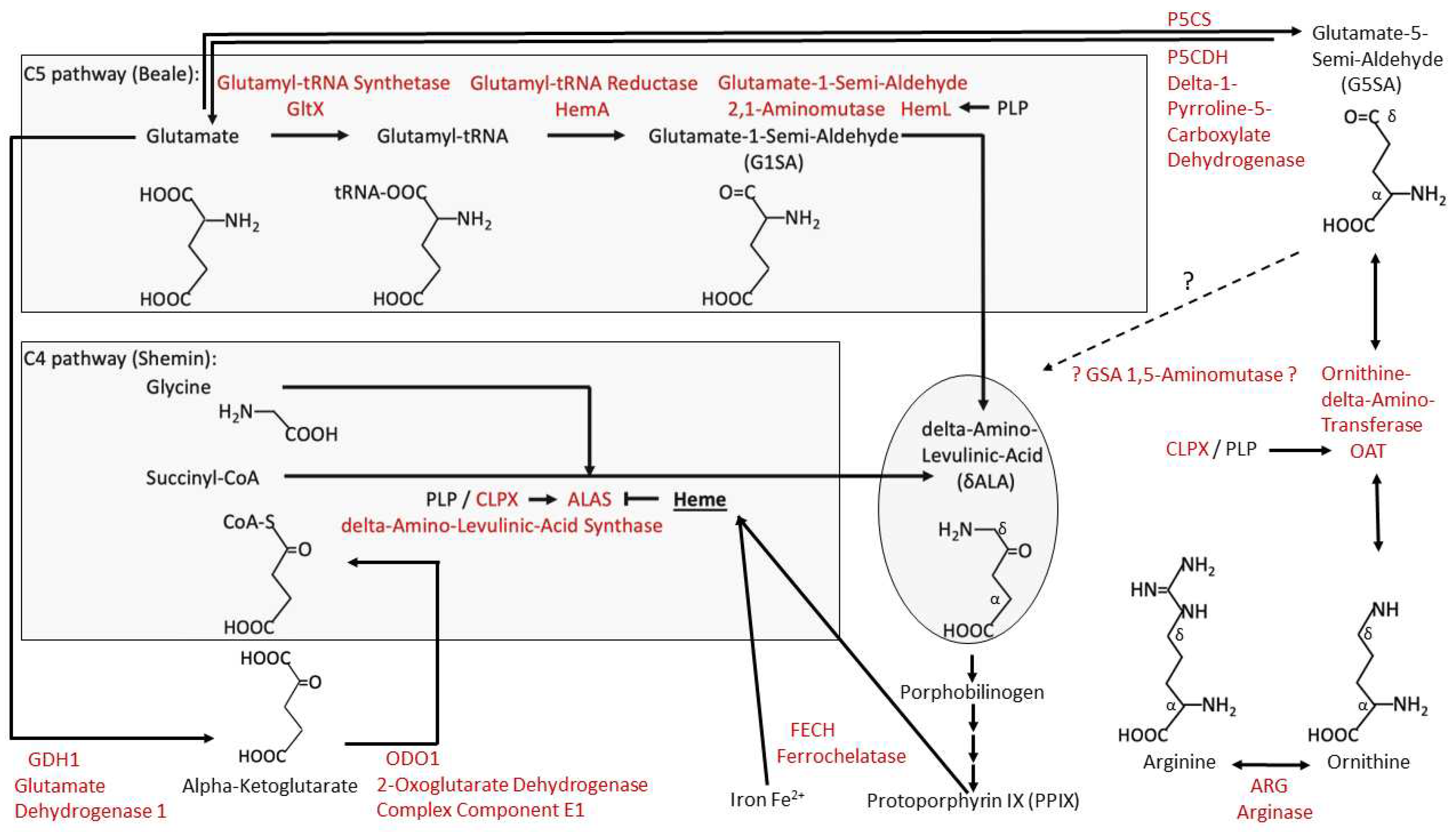
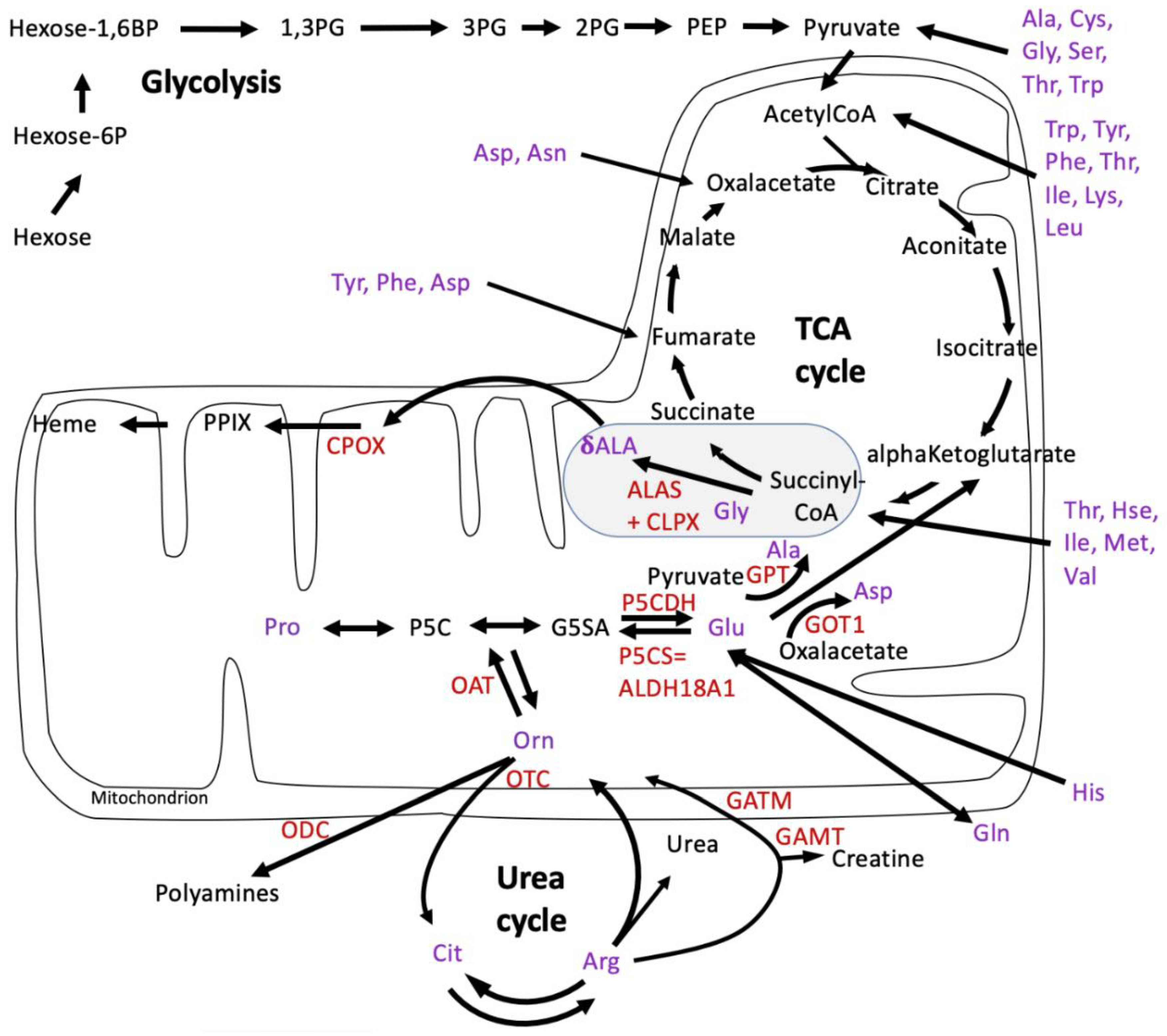
4.1. CLPP-Absence Affects Amino-Acid Metabolism
4.2. CLPP-Absence Affects Mitoribosomal LSU and Its rRNA/tRNAVal/Phe
4.3. CLPP-Absence Affects SAM-Dependent Methyltransferases
4.4. CLPP-Absence Affects Molecular Chaperones and the UPRmt
4.5. CLPP-Absence Affects the Respiratory Chain
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 2PG | 2-phosphoglycerate |
| 5S rRNA | the smallest rRNA within the eukaryotic ribosomal LSU |
| 60S subunit | eukaryotic ribosomal LSU with sedimentation at 60 Svedberg units |
| AAA+ | ATPases associated with various cellular activities |
| ABAT | 4-aminobutyrate aminotransferase |
| ABC | ammonium bicarbonate |
| AGAT | = GATM, L-arginine:glycine amidinotransferase |
| AGC | automatic gain control |
| AGXT2 | alanine-glyoxylate aminotransferase 2 |
| ALA | = deltaALA , delta-amino-levulinic-acid |
| Ala | alanine |
| ALAS | delta-amino-levulinic-acid synthase, generic |
| ALAS1 | delta-amino-levulinic-acid synthase 1, non-specific |
| ALAS2 | delta-amino-levulinic-acid synthase 1, erythroid-specific |
| ALDH18A1 | aldehyde dehydrogenase 18 family member A1 |
| Arg | arginine |
| ASMT | acetylserotonin O-methyltransferase |
| Asn | asparagine |
| Asp | aspartate |
| ATF4 | activating transcription factor 4 |
| ATP | adenosine trisphosphate |
| BAG-domain | Bcl-2-associated athanogene-domain |
| BCAT2 | branched chain amino acid transaminase 2 |
| BCL2 | BCL2 apoptosis regulator |
| C1/4/5/6 | chain composed of 1/4/5/6 carbons |
| C57BL/6 | inbred substrain 6 generated by C.C. Little, from Abbie Lathrop’s mouse 57, with nearly black coat |
| CI | respiratory chain complex I |
| CIV | respiratory chain complex IV |
| Cit | citrulline |
| CLPA-E | caseinolytic mitochondrial matrix peptidase chaperone subunit A-E |
| CLPP | caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| CLPX | caseinolytic mitochondrial matrix peptidase chaperone subunit X |
| CM liquid medium | complete medium containing glucose monohydrate |
| Co2+ | elemental cobalt as divalent cation |
| CoA | coenzyme A |
| COX2 | mitochondrially encoded cytochrome C oxidase II |
| COX7A | cytochrome C oxidase subunit 7A1 |
| COXFA4 | = NDUFA4; cytochrome C oxidase subunit FA4 |
| CPOX | coproporphyrinogen oxidase |
| CPT | carnitine palmitoyltransferase 2 |
| CTP | cytidine trisphosphate |
| Cys | cysteine |
| DNA | desoxyribonucleic acid |
| DnaK | E. coli chaperone protein |
| DTT | dithiothreitol |
| EARS2 | glutamyl-tRNA synthetase 2, mitochondrial |
| eIF6 | eukaryotic translation initiation factor 6 |
| ETNPPL | ethanolamine-phosphate phospho-lyase |
| ESI | electrospray ionization |
| ESSS | = NDUFB11, NADH-ubiquinone oxidoreductase subunit B11 |
| FAD | flavin adenine dinucleotide |
| FC | fold-change |
| FDR | false discovery rate |
| Fe2+ | ferrous iron = iron(II), elemental iron as divalent cation |
| FECH | ferrochelatase |
| FeS clusters | iron-sulfur clusters |
| G1SA | =GSA, glutamate-1-semialdehyde |
| G5SA | glutamate-5’-semialdehyde |
| GABA | gamma-amino-butyric-acid |
| GAMT | guanidinoacetate N-methyltransferase |
| GAR1 | Gar1 Ribonucleoprotein Homolog |
| GATC | glutaminyl-tRNA-synthase subunit C, mitochondrial |
| GATM | glycine amidinotransferase |
| GDH1 | glutamate decarboxylase 1 |
| GFM1/2 | translation elongation factor G, mitochondrial, variant 1/2 |
| GH | growth hormone |
| GLDC | glycine decarboxylase |
| Gln | glutamine |
| GltX | glutamate-tRNA ligase |
| Glu | glutamate |
| Gly | glycine |
| GO-term | gene ontology term |
| GOT1/2 | glutamic-oxaloacetic transaminase 1/2 |
| GPT | glutamic-pyruvic transaminase |
| GSSG | glutathione disulfide |
| GTP | guanosine triphosphate |
| H/ACA | sequence motifs H box (consensus ANANNA) and ACA box (ACA) |
| HARS2 | histidine-tRNA ligase, mitochondrial |
| HemA | glutamyl-tRNA reductase |
| HemL | glutamate-1-semialdehyde 2,1-aminomutase |
| Hexose-1,6BP | hexose-1,6-bisphosphate |
| Hexose-6P | hexose-6-phosphate |
| His | histidine |
| Hsc70 | heat shock cognate 71 kDa protein |
| Hse | homoserine |
| HSP70 | heat shock protein family A (Hsp70) member 4 |
| i-AAA | mitochondrial intermembrane space AAA+ protease |
| Ile | isoleucine |
| kDa | kiloDalton (molecular weight unit) |
| L7/L12 stalk | stalk structure in the mitoribosomal LSU, composed of proteins 7 / 12 |
| LARS2 | leucyl-tRNA synthetase 2, mitochondrial |
| LC/MS | liquid chromatography / mass spectrometry |
| Leu | leucine |
| LMBRD1 | lysosomal cobalamin transport escort protein LMBR1 |
| LonP | Lon peptidase 1 homolog, mitochondrial |
| Lsm7 | like-SM domain containing protein 7, cytosolic |
| LSU | mitoribosomal large subunit |
| LYRM6 | protein 6 with conserved tripeptide (LYR) motif |
| m3G | 2,2,7-trimethyl guanosine |
| MCX1 | yeast CLPX homolog |
| Met | methionine |
| Mg2+ | elemental magnesium as divalent cation |
| 1-MHis | 1-methylhistamine |
| MRM | multiple reaction mode |
| MRPP1 | = TRMT10C; mitochondrial ribonuclease P protein 1 |
| MRPP3 | = PRORP; mitochondrial ribonuclease P protein 3 |
| MS | mass spectrometry |
| MRPL30 | large ribosomal subunit protein uL30m |
| MRPL33 | large ribosomal subunit protein bL33m |
| MTBE | methyl-tert-butyl ester |
| mt-CO3 | mitochondrially encoded cytochrome C oxidase III |
| mtDNA | mitochondrial DNA, nucleoid |
| 5,10-MTHF | 5,10-methylenetetrahydrofolate |
| MTHFD2 | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase. |
| NADPH | nicotinamide adenine dinucleotide phosphate |
| NDUFA1 | NADH:ubiquinone oxidoreductase subunit A1 |
| NDUFA2 | NADH:ubiquinone oxidoreductase subunit A2 |
| NDUFA2 | NADH:ubiquinone oxidoreductase subunit A3 |
| NDUFA4 | NADH:ubiquinone oxidoreductase subunit A4 |
| NDUFA6 | NADH:ubiquinone oxidoreductase subunit A6 |
| NDUFB2 | NADH:ubiquinone oxidoreductase subunit B2 |
| NDUFB11 | NADH:ubiquinone oxidoreductase subunit B11 |
| NDUFS6 | NADH:ubiquinone oxidoreductase subunit S6 |
| NFS1 | nitrogen fixing bacteria S-like protein 1, cysteine desulfurase |
| NOP10 | homolog of yeast Nop10p |
| NTMT1/2 | N-terminal Xaa-Pro-Lys N-methyltransferase 1/2 |
| OAT | ornithine aminotransferase |
| ODC | = ODC1, ornithine decarboxylase 1 |
| ODO1 | 2-oxoglutarate dehydrogenase complex component E1 |
| ONC201 | = dordaviprone: 11-benzyl-7-[(2-methylphenyl)methyl]-2,5,7,11-tetrazatricyclo[7.4.0.02,6]trideca-1(9),5-dien-8-one |
| Orn | ornithine |
| OTC | ornithine transcarbamylase |
| P5C | delta-1-pyrroline-5-carboxylate |
| P5CDH | pyrroline-5-carboxylate dehydrogenase |
| P5CS | pyrroline-5-carboxylate synthetase = ALDH18A1 |
| P5P | pyridoxal-5’-phosphate |
| Pa | Podospora anserina fungus |
| PADI | peptidylarginine-deiminases |
| PAGE | polyacrylamide gel electrophoresis |
| PaIap | = B2B020 in UniProt; mitoch. intermembrane space AAA+ protease |
| PaSnf1 | = B2B4C1 in UniProt; sucrose non-fermenting complex, catalytic 1 |
| PEP | phosphoenolpyruvate |
| 1,3PG | 1,3-bisphospho-glycerate |
| 2PG | 2-phosphoglycerate |
| 3PG | 3-phosphoglycerate |
| PHD | prolyl-3-hydroxlase domain |
| Phe | phenylalanine |
| PHYKPL | 5-phosphohydroxy-L-lysine phospho-lyase |
| PLP | pyridoxal-5’-phosphate |
| PodAns | Podospora anserina fungus |
| PP/PE | polypropylene/polyehtylene |
| PPIX | protoporphyrinogen IX |
| PRLTS3 | Perrault syndrome type 3 |
| Pro | proline |
| PRORP | protein only RNase P catalytic subunit |
| RMND1 | required for meiotic nuclear division 1 homolog |
| RNA | ribonucleic acid |
| rRNA | ribosomal RNA |
| SAM | S-adenosyl-methionine |
| SAM-MTases | S-adenosyl-methionine-dependent methyltransferases |
| SAP domain | DNA-binding 35-residue motif, named after SAF-A/B, Acinus and PIAS, three proteins known to contain it |
| SDS | sodium dodecyl sulfate |
| Ser | serine |
| SHMT2 | serine hydroxymethyltransferase 2, mitochondrial |
| Sm domain | occur in Sm proteins, named in honor of patient Stephanie Smith |
| SmG-like | domain spliceosomal core protein SmG, binding to AU-dinucleotide |
| snoRNA | small nucleolar RNA |
| snRNA | small nuclear RNA |
| snRNP | small nuclear ribonucleoprotein |
| SSU | ribosomal small subunit |
| STRING | search tool for the retrieval of interacting genes/proteins |
| Tae1 | alpha N-terminal protein methyltransferase 1 |
| TCA cycle | tricarboxylic acid cycle |
| Thr | threonine |
| TRMT10C | tRNA methyltransferase 10C, mitochondrial RNase P mubunit |
| tRNA | transfer RNA |
| tRNAVal/Phe | transfer RNA for valine or phenylalanine |
| Trp | tryptophan |
| TWNK | twinkle |
| Tyr | tyrosine |
| UniProt | public database about proteins, unifying nomenclature |
| UniProt-ID | UniProt protein identifier number |
| UPRmt | mitochondrial unfolded protein response |
| UTP | uridine triphosphate |
| v/v | volume per volume |
| WT | wildtype |
| ZFE | Zentrale orschungs-Einrichtung |
References
- Olivares, A.O., T.A. Baker, and R.T. Sauer, Mechanistic insights into bacterial AAA+ proteases and protein-remodelling machines. Nat Rev Microbiol, 2016. 14(1): p. 33-44. [CrossRef]
- Huang, S., J. Petereit, and A.H. Millar, Loss of conserved mitochondrial CLPP and its functions lead to different phenotypes in plants and other organisms. Plant Signal Behav, 2020. 15(12): p. 1831789. [CrossRef]
- Auburger, G., J. Key, and S. Gispert, The Bacterial ClpXP-ClpB Family Is Enriched with RNA-Binding Protein Complexes. Cells, 2022. 11(15). [CrossRef]
- Mabanglo, M.F., V. Bhandari, and W.A. Houry, Substrates and interactors of the ClpP protease in the mitochondria. Curr Opin Chem Biol, 2022. 66: p. 102078. [CrossRef]
- Key, J., et al., Global Proteome of LonP1(+/-) Mouse Embryonal Fibroblasts Reveals Impact on Respiratory Chain, but No Interdependence between Eral1 and Mitoribosomes. Int J Mol Sci, 2019. 20(18). [CrossRef]
- Chandu, D. and D. Nandi, Comparative genomics and functional roles of the ATP-dependent proteases Lon and Clp during cytosolic protein degradation. Res Microbiol, 2004. 155(9): p. 710-9. [CrossRef]
- Maurizi, M.R., et al., Sequence and structure of Clp P, the proteolytic component of the ATP-dependent Clp protease of Escherichia coli. J Biol Chem, 1990. 265(21): p. 12536-45. [CrossRef]
- Gispert, S., et al., Loss of mitochondrial peptidase Clpp leads to infertility, hearing loss plus growth retardation via accumulation of CLPX, mtDNA and inflammatory factors. Hum Mol Genet, 2013. 22(24): p. 4871-87. [CrossRef]
- Froese, D.S., B. Fowler, and M.R. Baumgartner, Vitamin B(12) , folate, and the methionine remethylation cycle-biochemistry, pathways, and regulation. J Inherit Metab Dis, 2019. 42(4): p. 673-685. [CrossRef]
- Kardon, J.R., et al., Mitochondrial ClpX Activates a Key Enzyme for Heme Biosynthesis and Erythropoiesis. Cell, 2015. 161(4): p. 858-67. [CrossRef]
- van der Vorm, L.N. and B.H. Paw, Studying disorders of vertebrate iron and heme metabolism using zebrafish. Methods Cell Biol, 2017. 138: p. 193-220. [CrossRef]
- Yien, Y.Y., et al., Mutation in human CLPX elevates levels of delta-aminolevulinate synthase and protoporphyrin IX to promote erythropoietic protoporphyria. Proc Natl Acad Sci U S A, 2017. 114(38): p. E8045-E8052. [CrossRef]
- Ducamp, S., et al., A mutation in the iron-responsive element of ALAS2 is a modifier of disease severity in a patient suffering from CLPX associated erythropoietic protoporphyria. Haematologica, 2021. 106(7): p. 2030-2033. [CrossRef]
- Shemin, D., An illustration of the use of isotopes: the biosynthesis of porphyrins. Bioessays, 1989. 10(1): p. 30-5. [CrossRef]
- Beale, S.I. and P.A. Castelfranco, The Biosynthesis of delta-Aminolevulinic Acid in Higher Plants: II. Formation of C-delta-Aminolevulinic Acid from Labeled Precursors in Greening Plant Tissues. Plant Physiol, 1974. 53(2): p. 297-303. [CrossRef]
- Iida, K., I. Mimura, and M. Kajiwara, Evaluation of two biosynthetic pathways to delta-aminolevulinic acid in Euglena gracilis. Eur J Biochem, 2002. 269(1): p. 291-7. [CrossRef]
- Petricek, M., et al., Occurrence of two 5-aminolevulinate biosynthetic pathways in Streptomyces nodosus subsp. asukaensis is linked with the production of asukamycin. J Bacteriol, 2006. 188(14): p. 5113-23. [CrossRef]
- Key, J., et al., Translation fidelity and respiration deficits in CLPP-deficient tissues: Mechanistic insights from mitochondrial complexome. Int J Mol Sci, 2023. 24(24): p. 17503. [CrossRef]
- Koper, K., et al., Evolutionary origin and functional diversification of aminotransferases. J Biol Chem, 2022. 298(8): p. 102122. [CrossRef]
- Cellini, B., et al., The chaperone role of the pyridoxal 5'-phosphate and its implications for rare diseases involving B6-dependent enzymes. Clin Biochem, 2014. 47(3): p. 158-65. [CrossRef]
- Liang, J., et al., Current Advances on Structure-Function Relationships of Pyridoxal 5'-Phosphate-Dependent Enzymes. Front Mol Biosci, 2019. 6: p. 4. [CrossRef]
- Shin, B.S., et al., Amino acid substrates impose polyamine, eIF5A, or hypusine requirement for peptide synthesis. Nucleic Acids Res, 2017. 45(14): p. 8392-8402. [CrossRef]
- Pegg, A.E., Toxicity of polyamines and their metabolic products. Chem Res Toxicol, 2013. 26(12): p. 1782-800. [CrossRef]
- di Salvo, M.L., N. Budisa, and R. Contestabile, PLP-dependent Enzymes: a Powerful Tool for Metabolic Synthesis of Non-canonical Amino Acids. https://www.beilstein-institut.de/download/65/plp-dependent_enzymes_a_powerful_tool_for_metabolic_synthesis_of_non-canonical_amino_acids_.pdf, 2012.
- Obermaier, S. and M. Muller, Ibotenic Acid Biosynthesis in the Fly Agaric Is Initiated by Glutamate Hydroxylation. Angew Chem Int Ed Engl, 2020. 59(30): p. 12432-12435. [CrossRef]
- Chen, M., C.T. Liu, and Y. Tang, Discovery and Biocatalytic Application of a PLP-Dependent Amino Acid gamma-Substitution Enzyme That Catalyzes C-C Bond Formation. J Am Chem Soc, 2020. 142(23): p. 10506-10515. [CrossRef]
- Haynes, C.M., et al., ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev Cell, 2007. 13(4): p. 467-80. [CrossRef]
- Zhou, Z., et al., The mitochondrial unfolded protein response: A multitasking giant in the fight against human diseases. Ageing Res Rev, 2022. 81: p. 101702. [CrossRef]
- Al-Furoukh, N., et al., ClpX stimulates the mitochondrial unfolded protein response (UPRmt) in mammalian cells. Biochim Biophys Acta, 2015. 1853(10 Pt A): p. 2580-91. [CrossRef]
- Levchenko, I., et al., A specificity-enhancing factor for the ClpXP degradation machine. Science, 2000. 289(5488): p. 2354-6. [CrossRef]
- Lytvynenko, I., et al., Alanine Tails Signal Proteolysis in Bacterial Ribosome-Associated Quality Control. Cell, 2019. 178(1): p. 76-90 e22. [CrossRef]
- Wu, Z., et al., MISTERMINATE Mechanistically Links Mitochondrial Dysfunction with Proteostasis Failure. Mol Cell, 2019. 75(4): p. 835-848 e8. [CrossRef]
- Huter, P., et al., Structural Basis for Polyproline-Mediated Ribosome Stalling and Rescue by the Translation Elongation Factor EF-P. Mol Cell, 2017. 68(3): p. 515-527 e6. [CrossRef]
- Konovalova, S., et al., Exposure to arginine analog canavanine induces aberrant mitochondrial translation products, mitoribosome stalling, and instability of the mitochondrial proteome. Int J Biochem Cell Biol, 2015. 65: p. 268-74. [CrossRef]
- Ou, X., et al., Errors in translational decoding: tRNA wobbling or misincorporation? PLoS Genet, 2019. 15(3): p. e1008017. [CrossRef]
- Jenkinson, E.M., et al., Perrault syndrome is caused by recessive mutations in CLPP, encoding a mitochondrial ATP-dependent chambered protease. Am J Hum Genet, 2013. 92(4): p. 605-13. [CrossRef]
- Newman, W.G., et al., Perrault Syndrome, in GeneReviews((R)), M.P. Adam, et al., Editors. 1993: Seattle (WA).
- Theunissen, T.E., et al., Specific MRI Abnormalities Reveal Severe Perrault Syndrome due to CLPP Defects. Front Neurol, 2016. 7: p. 203. [CrossRef]
- Key, J., et al., Inactivity of Peptidase ClpP Causes Primary Accumulation of Mitochondrial Disaggregase ClpX with Its Interacting Nucleoid Proteins, and of mtDNA. Cells, 2021. 10(12). [CrossRef]
- Faridi, R., et al., New insights into Perrault syndrome, a clinically and genetically heterogeneous disorder. Hum Genet, 2022. 141(3-4): p. 805-819. [CrossRef]
- Hochberg, I., et al., Bi-allelic variants in the mitochondrial RNase P subunit PRORP cause mitochondrial tRNA processing defects and pleiotropic multisystem presentations. Am J Hum Genet, 2021. 108(11): p. 2195-2204. [CrossRef]
- Bhandari, V., et al., The Role of ClpP Protease in Bacterial Pathogenesis and Human Diseases. ACS Chem Biol, 2018. 13(6): p. 1413-1425. [CrossRef]
- Prabhu, V.V., et al., ONC201 and imipridones: Anti-cancer compounds with clinical efficacy. Neoplasia, 2020. 22(12): p. 725-744. [CrossRef]
- Zhang, R., et al., Assessment of the structure-activity relationship and antileukemic activity of diacylpyramide compounds as human ClpP agonists. Eur J Med Chem, 2023. 258: p. 115577. [CrossRef]
- Key, J., et al., CLPP Depletion Causes Diplotene Arrest; Underlying Testis Mitochondrial Dysfunction Occurs with Accumulation of Perrault Proteins ERAL1, PEO1, and HARS2. Cells, 2022. 12(1). [CrossRef]
- Bhaskaran, S., et al., Loss of mitochondrial protease ClpP protects mice from diet-induced obesity and insulin resistance. EMBO Rep, 2018. 19(3). [CrossRef]
- Becker, C., et al., CLPP deficiency protects against metabolic syndrome but hinders adaptive thermogenesis. EMBO Rep, 2018. 19(5). [CrossRef]
- Szczepanowska, K., et al., CLPP coordinates mitoribosomal assembly through the regulation of ERAL1 levels. EMBO J, 2016. 35(23): p. 2566-2583. [CrossRef]
- Key, J., et al., Loss of mitochondrial ClpP, Lonp1, and Tfam triggers transcriptional induction of Rnf213, a susceptibility factor for moyamoya disease. Neurogenetics, 2020. 21(3): p. 187-203. [CrossRef]
- Torres-Odio, S., et al., Loss of Mitochondrial Protease CLPP Activates Type I IFN Responses through the Mitochondrial DNA-cGAS-STING Signaling Axis. J Immunol, 2021. 206(8): p. 1890-1900. [CrossRef]
- Maletzko, A., et al., Increased presence of nuclear DNAJA3 and upregulation of cytosolic STAT1 and of nucleic acid sensors trigger innate immunity in the ClpP-null mouse. Neurogenetics, 2021. 22(4): p. 297-312. [CrossRef]
- Osiewacz, H.D., Mitochondrial quality control in aging and lifespan control of the fungal aging model Podospora anserina. Biochem Soc Trans, 2011. 39(5): p. 1488-92. [CrossRef]
- Osiewacz, H.D., A. Hamann, and S. Zintel, Assessing Organismal Aging in the Filamentous Fungus Podospora anserina, in Cell Senescence: Methods and Protocols, Methods in Molecular Biology, L. Galluzzi, et al., Editors. 2013, Humana: Totowa, NJ. [CrossRef]
- Fischer, F., et al., Human CLPP reverts the longevity phenotype of a fungal ClpP deletion strain. Nat Commun, 2013. 4: p. 1397. [CrossRef]
- Heinz, D., et al., Simultaneous Ablation of the Catalytic AMPK alpha-Subunit SNF1 and Mitochondrial Matrix Protease CLPP Results in Pronounced Lifespan Extension. Front Cell Dev Biol, 2021. 9: p. 616520. [CrossRef]
- Fischer, F., J.D. Langer, and H.D. Osiewacz, Identification of potential mitochondrial CLPXP protease interactors and substrates suggests its central role in energy metabolism. Sci Rep, 2015. 5: p. 18375. [CrossRef]
- Szczepanowska, K., et al., A salvage pathway maintains highly functional respiratory complex I. Nat Commun, 2020. 11(1): p. 1643. [CrossRef]
- Petereit, J., et al., Mitochondrial CLPP2 Assists Coordination and Homeostasis of Respiratory Complexes. Plant Physiol, 2020. 184(1): p. 148-164. [CrossRef]
- Meierhofer, D., et al., Ataxin-2 (Atxn2)-Knock-Out Mice Show Branched Chain Amino Acids and Fatty Acids Pathway Alterations. Mol Cell Proteomics, 2016. 15(5): p. 1728-39. [CrossRef]
- Gielisch, I. and D. Meierhofer, Metabolome and proteome profiling of complex I deficiency induced by rotenone. J Proteome Res, 2015. 14(1): p. 224-35. [CrossRef]
- Funck, D., B. Stadelhofer, and W. Koch, Ornithine-delta-aminotransferase is essential for arginine catabolism but not for proline biosynthesis. BMC Plant Biol, 2008. 8: p. 40. [CrossRef]
- Du, J., et al., Proline metabolism and transport in retinal health and disease. Amino Acids, 2021. 53(12): p. 1789-1806. [CrossRef]
- Monne, M., et al., Mitochondrial transport and metabolism of the major methyl donor and versatile cofactor S-adenosylmethionine, and related diseases: A review(dagger). IUBMB Life, 2022. 74(7): p. 573-591. [CrossRef]
- Rebelo-Guiomar, P., et al., A late-stage assembly checkpoint of the human mitochondrial ribosome large subunit. Nat Commun, 2022. 13(1): p. 929. [CrossRef]
- Koditz, J., et al., Oxygen-dependent ATF-4 stability is mediated by the PHD3 oxygen sensor. Blood, 2007. 110(10): p. 3610-7. [CrossRef]
- Stranska, J., et al., Ornithine delta-aminotransferase: An enzyme implicated in salt tolerance in higher plants. Plant Signal Behav, 2008. 3(11): p. 929-35. [CrossRef]
- Llacer, J.L., I. Fita, and V. Rubio, Arginine and nitrogen storage. Curr Opin Struct Biol, 2008. 18(6): p. 673-81. [CrossRef]
- Pencharz, P.B., Assessment of protein nutritional status in children. Pediatr Blood Cancer, 2008. 50(2 Suppl): p. 445-6; discussion 451. [CrossRef]
- Sodero, G., et al., Growth hormone responses during arginine and clonidine stimulation test: Correlations with patients' auxological and metabolic parameters in a single centre study. Growth Horm IGF Res, 2023. 68: p. 101522. [CrossRef]
- Stefani, G.P., et al., Resistance training and L-arginine supplementation are determinant in genomic stability, cardiac contractility and muscle mass development in rats. PLoS One, 2018. 13(9): p. e0204858. [CrossRef]
- Vanderniet, J.A., et al., Barth syndrome with severe dilated cardiomyopathy and growth hormone resistance: a case report. J Pediatr Endocrinol Metab, 2021. 34(7): p. 951-955. [CrossRef]
- Brown, A., et al., Structure of the large ribosomal subunit from human mitochondria. Science, 2014. 346(6210): p. 718-722. [CrossRef]
- Miluzio, A., et al., Eukaryotic initiation factor 6 mediates a continuum between 60S ribosome biogenesis and translation. EMBO Rep, 2009. 10(5): p. 459-65. [CrossRef]
- Gartmann, M., et al., Mechanism of eIF6-mediated inhibition of ribosomal subunit joining. J Biol Chem, 2010. 285(20): p. 14848-14851. [CrossRef]
- Weis, F., et al., Mechanism of eIF6 release from the nascent 60S ribosomal subunit. Nat Struct Mol Biol, 2015. 22(11): p. 914-9. [CrossRef]
- Jaako, P., et al., eIF6 rebinding dynamically couples ribosome maturation and translation. Nat Commun, 2022. 13(1): p. 1562. [CrossRef]
- Pesce, E., et al., Discovery and Preliminary Characterization of Translational Modulators that Impair the Binding of eIF6 to 60S Ribosomal Subunits. Cells, 2020. 9(1). [CrossRef]
- Balogh, E., et al., Pseudouridylation defect due to DKC1 and NOP10 mutations causes nephrotic syndrome with cataracts, hearing impairment, and enterocolitis. Proc Natl Acad Sci U S A, 2020. 117(26): p. 15137-15147. [CrossRef]
- De Zoysa, M.D. and Y.T. Yu, Posttranscriptional RNA Pseudouridylation. Enzymes, 2017. 41: p. 151-167. [CrossRef]
- Jakubowski, H., Misacylation of tRNALys with noncognate amino acids by lysyl-tRNA synthetase. Biochemistry, 1999. 38(25): p. 8088-93. [CrossRef]
- Bauerle, M.R., E.L. Schwalm, and S.J. Booker, Mechanistic diversity of radical S-adenosylmethionine (SAM)-dependent methylation. J Biol Chem, 2015. 290(7): p. 3995-4002. [CrossRef]
- Webb, K.J., et al., Identification of protein N-terminal methyltransferases in yeast and humans. Biochemistry, 2010. 49(25): p. 5225-35. [CrossRef]
- Bhatta, A., et al., Structural basis of RNA processing by human mitochondrial RNase P. Nat Struct Mol Biol, 2021. 28(9): p. 713-723. [CrossRef]
- Metodiev, M.D., et al., Recessive Mutations in TRMT10C Cause Defects in Mitochondrial RNA Processing and Multiple Respiratory Chain Deficiencies. Am J Hum Genet, 2016. 98(5): p. 993-1000. [CrossRef]
- Averbeck, N.B., et al., Identification and characterization of PaMTH1, a putative O-methyltransferase accumulating during senescence of Podospora anserina cultures. Curr Genet, 2000. 37(3): p. 200-8. [CrossRef]
- Kunstmann, B. and H.D. Osiewacz, Over-expression of an S-adenosylmethionine-dependent methyltransferase leads to an extended lifespan of Podospora anserina without impairments in vital functions. Aging Cell, 2008. 7(5): p. 651-62. [CrossRef]
- Kunstmann, B. and H.D. Osiewacz, The S-adenosylmethionine dependent O-methyltransferase PaMTH1: a longevity assurance factor protecting Podospora anserina against oxidative stress. Aging (Albany NY), 2009. 1(3): p. 328-34. [CrossRef]
- Knab, B. and H.D. Osiewacz, Methylation of polyphenols with vicinal hydroxyl groups: A protection pathway increasing organismal lifespan. Cell Cycle, 2010. 9(17): p. 3387-8. [CrossRef]
- Chatterjee, D., et al., Structure and Biophysical Characterization of the S-Adenosylmethionine-dependent O-Methyltransferase PaMTH1, a Putative Enzyme Accumulating during Senescence of Podospora anserina. J Biol Chem, 2015. 290(26): p. 16415-30. [CrossRef]
- Madeo, F., et al., Spermidine in health and disease. Science, 2018. 359(6374). [CrossRef]
- Gailus, S., et al., Insights into lysosomal cobalamin trafficking: lessons learned from cblF disease. J Mol Med (Berl), 2010. 88(5): p. 459-66. [CrossRef]
- Balsa, E., et al., NDUFA4 is a subunit of complex IV of the mammalian electron transport chain. Cell Metab, 2012. 16(3): p. 378-86. [CrossRef]
- Kadenbach, B., Regulation of Mammalian 13-Subunit Cytochrome c Oxidase and Binding of other Proteins: Role of NDUFA4. Trends Endocrinol Metab, 2017. 28(11): p. 761-770. [CrossRef]
- Yagil, C., R. Varadi-Levi, and Y. Yagil, A novel mutation in the NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 4 (Ndufa4) gene links mitochondrial dysfunction to the development of diabetes in a rodent model. Dis Model Mech, 2018. 11(11). [CrossRef]
- Kadenbach, B., Complex IV - The regulatory center of mitochondrial oxidative phosphorylation. Mitochondrion, 2021. 58: p. 296-302. [CrossRef]
- Sorouri, M., et al., Signatures of host-pathogen evolutionary conflict reveal MISTR-A conserved MItochondrial STress Response network. PLoS Biol, 2020. 18(12): p. e3001045. [CrossRef]
- Hock, D.H., D.R.L. Robinson, and D.A. Stroud, Blackout in the powerhouse: clinical phenotypes associated with defects in the assembly of OXPHOS complexes and the mitoribosome. Biochem J, 2020. 477(21): p. 4085-4132. [CrossRef]
- Letts, J.A., K. Fiedorczuk, and L.A. Sazanov, The architecture of respiratory supercomplexes. Nature, 2016. 537(7622): p. 644-648. [CrossRef]
- Guerrero-Castillo, S., et al., The Assembly Pathway of Mitochondrial Respiratory Chain Complex I. Cell Metab, 2017. 25(1): p. 128-139. [CrossRef]
- Signes, A. and E. Fernandez-Vizarra, Assembly of mammalian oxidative phosphorylation complexes I-V and supercomplexes. Essays Biochem, 2018. 62(3): p. 255-270. [CrossRef]
- Formosa, L.E., et al., Building a complex complex: Assembly of mitochondrial respiratory chain complex I. Semin Cell Dev Biol, 2018. 76: p. 154-162. [CrossRef]
- Giachin, G., et al., Dynamics of Human Mitochondrial Complex I Assembly: Implications for Neurodegenerative Diseases. Front Mol Biosci, 2016. 3: p. 43. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




