1. Introduction
In the intricate tapestry of agricultural and human history, goats (
Capra hircus) have emerged as steadfast companions, which led to their domestication and modification approximately 10,500 years ago in the Fertile Crescent [
1]. Goat farming is a good option to support agricultural activities, socially and economically due to their high adaptability to diverse environments and the inclusion of technical management to achieve maximum productivity [
2]. With their primary role being meat, they are also used for milk, cheese, skins, and cashmere, providing sustenance to communities across the globe [
3].
The worldwide goat population surpassed 1 billion, with South Africa contributing around 7.8 million goats, with over 63% managed by small-scale farmers [
3,
4,
5,
6]. The South African dairy goat sector remains relatively small, with approximately 4000 goats producing roughly 1.4 million tons of milk annually [
7]. Unlike European countries where goats have long been significant milk producers, South Africa only introduced dairy goats in the 20th century from various European countries. Initially, four dairy breeds were recognized: Saanen, Toggenburg, British Alpine, and an Anglo-Nubian Swiss composite [
8,
9]. Currently, dairy production systems in South Africa include both purebred and crossbred goats derived from these initial breeds [
9].
The surge in interest in maintaining dairy goat breeds has prompted breeders to raise inquiries about the adequacy of genetic diversity within the population to sustain the expanding industry [
7]. Apart from genetic diversity, the quantity of milk, proteins, and milk fat are key economic traits, however, little is known about the genomic loci that control these important traits in goats [
10,
11]. Having genomics insights into these genetic loci will aid in the understanding of related genes and molecular mechanisms that play a role in goat milk compositions especially those that are associated with its healing properties.
Goat milk and other goat-derived products contain several bioactive compounds that might be useful in patients suffering from a variety of chronic diseases and skin conditions such as eczema and psoriasis [
12,
13]. Rich in proteins like casein, lactoferrin, and immunoglobulins, goat milk exhibits antibacterial, antioxidant, and immunomodulatory effects, enhancing skin health and offering various cosmetic benefits, etc. [
13]. While anecdotal evidence suggests its efficacy in managing conditions like eczema and psoriasis, a scientific understanding of its genomic basis and molecular mechanisms remains limited. Further research is needed to elucidate these aspects and harness goat milk's full potential for dermatological applications [
13]. This review aligns with the pressing need to unravel the genomic insights, providing a scientific foundation for the development of innovative cosmetic and medicinal applications. Understanding the medicinal properties of goat milk in treating eczema and psoriasis could pave the way for natural and sustainable skincare alternatives, addressing the challenges posed by these prevalent skin conditions.
Studying the genomics and medicinal potential of goat milk, especially concerning its role in skin conditions like eczema and psoriasis, is vital for improving milk production and developing therapies. Genome-wide SNP genotyping and milk protein sequencing are key methods for uncovering the genetic mechanisms underlying goat milk's healing properties. These approaches provide insights into SNPs, genes, and skin-protein interactions relevant to therapeutic development. The review aims to elucidate genetic factors impacting goat milk composition and its effects on the skin, emphasizing genes and molecular mechanisms crucial for skin health and their potential application in treating dermatological conditions.
2. South African Dairy Goat Breeds
Goats rank among the earliest domesticated animals, with archaeological evidence suggesting a symbiotic connection with humans primarily for milk production [
14]. However, some individuals raise them to generate income through the production of both meat and milk. The South African dairy goat population is small with 45 registered herds representing 16,561 animals, where the remainder of the goats are used in commercial milk operations and only 16 of the 45 herds (approximately 1217 goats) participate in official recording [
15]. Dairy goat breeds historically originated in developed countries, although today goats in these countries constitute only about 5% of the world’s goat population; with Europe, producing 20.7% of the world’s goat milk while maintaining only 2.5% of the total goat population [
9]. The South African dairy goat population, established in the early 1900s with imports primarily from Switzerland and the United Kingdom, has remained relatively isolated from global goat production centers due to logistical challenges, with minimal additional imports in recent decades [
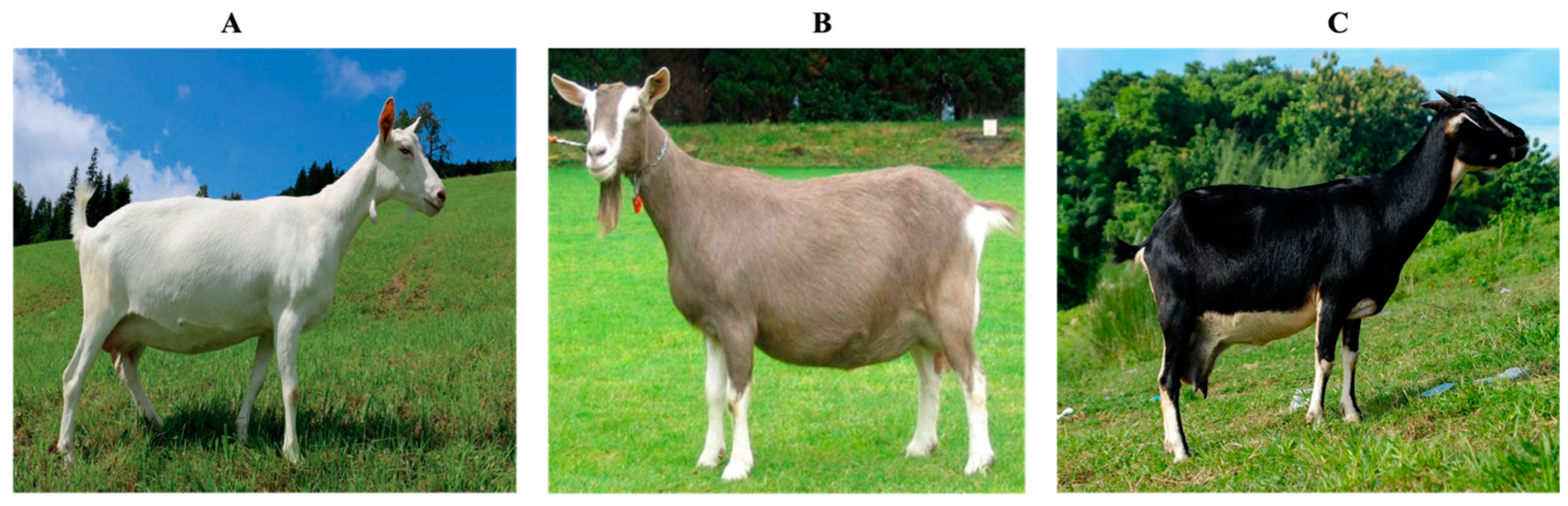
7]. Currently, the South African dairy goat industry is an important role player in the agricultural sector with animals representing the three main breeds i) Saanen ii) Toggenburg and iii) British Alpine (
Figure 1).
2.1. The Saanen
In 1898, the Saanen breed became the first milk goat breed introduced to South Africa, recognized for its exceptional milk production capabilities [
15]. The Saanen breed is characterized by its solid white coat without any markings, though the presence of black spots on the nose, eyelids, and ears is considered advantageous as it helps distinguish the animals from the undesired albino type. It is worth noting that the fair-skinned Saanen may be susceptible to the development of skin carcinomas on the udder due to intense exposure to the harsh South African sun [
16].
The current South African Saanen originated from one male and one female imported from Germany in 1923, and other importations from Switzerland, England, and Germany between 1923 and 1976. Over the years, various breeders registered these goats, however, there was no official traceability of registrations until 1947. This occurred when PR du Preez initiated the first official inquiry to register his herd. At that time, the sole breeder with registered goats possessed only two does. Du Preez acquired one of them and imported a ram from England [
16]. From 1958 to 1985, the SA Stud Book and Livestock Improvement Association (SASBLIA) registered 2388 Saanen females and 530 Saanen males, whilst 139 females and 42 males were registered from 1985 to 1991. Over the past decade, 432 Saanen females and 120 Saanen males were registered in South Africa (
http://www.milkgoats.co.za/p2/the-breed/milk-goat-breeds.html, accessed 05 February 2024)— also 15 Saanen breeders and 1 712 animals have been registered [
16].
Although participation in official animal recording is limited, the opportunity is available to record milk yield, milk composition and linear traits for selection and improvement. Heritability estimates have been reported for the SA Saanen for milk yield (0.23), butter fat yield (0.22) and protein yield (0.20). Protein and butterfat percentages had a heritability of 0.44 and 0.21, respectively [
15].
2.2. The Toggenburg
The Toggenburg dairy goat originates from the Toggenburg Valley of Switzerland in the Swiss Alps, where its breeding was strictly regulated for purity and type. It was widely exported and has a presence in many countries, including Australia, Great Britain, and the United States of America (
https://dairygoats.org.au/dairy-goat-breeds/toggenburg/, accessed 05 February 2024). In South Africa, Toggenburg goats were introduced in the early 20th century, however, for nearly two decades, they were not officially registered, leading to the disappearance of the original bloodlines. In 1951, WH Morris of Cape Town changed this by importing a Toggenburg ram and two pregnant does from Switzerland. Between 1934 and 1951, only two Toggenburg does, and 18 rams were registered directly with SA Studbook. Additionally, Toggenburg semen was later imported from New Zealand and Australia. Presently, there are 10 active Toggenburg breeders registered with the SA Milch Goat Breeders’ Society [
16].
The Toggenburg’s hair colour ranges from light fawn to dark chocolate, accentuated by accurate white or cream markings similar to those of the British Alpine (
http://www.milkgoats.co.za/p2/the-breed/milk-goat-breeds.html, accessed 05 February 2024). Its inherent robustness renders it a resilient goat capable of self-preservation. While it thrives best in moderate regions, it adapts to diverse climates, aided by its dark skin, which offers effective protection against skin cancer. Alternatively, does may display a black hue with appropriate white or cream markings. Their ears are erect and positioned forward, with the bridge of the nose either straight or dished. Toggenburgs hold the distinction of being among the earliest purebred dairy goats imported and registered in the United States (
https://adga.org/breed-standards/, accessed 05 February 2024). The Toggenburg is somewhat smaller than its Alpine counterparts, with ewes weighing a minimum of 55 kg. On average, ewes stand at about 79 cm tall at the withers, while rams typically reach 90 cm. Their teats are expected to be of moderate size, positioned squarely, and pointing slightly forward. The milk yield is around 2 kg per day, with milk containing approximately 3.2% to 3.7% fat and 2.7% protein. (
https://www.farmersweekly.co.za/farming-basics/how-to-livestock/a-sturdy-breed-some-toggenburg-basics/, accessed 16 April 2024).
2.3. The British Alpine
The British Alpine dairy goat characterized by its well-defined and neatly structured physique, originated in the early 1900s through the crossbreeding of the British Toggenburg, incorporating Nubian genetics. This resulted in a substantial, tall, and lean goat, typically black with distinctive white Swiss markings [
16]. However, its sleek black coat may lighten if there is insufficient copper in its diet, as copper is a vital mineral for all goats. British Alpines are characterized by erect ears and are available in various colors and combinations. Their hair is typically medium to short, and the bridge of the nose appears straight. Renowned for their robustness and adaptability, Alpines thrive in diverse climates, exhibiting excellent health and productivity. (
https://adga.org/breed-standards/, accessed 05 February 2024).
British Alpines are renowned for their exceptional milk production and well-sized teats, allowing easier milking. Tracey Preece achieved a significant milestone in South Africa by becoming the first successful British Alpine breeder. His goat, Darkie, born in August 1923, was officially registered in 1926, marking the breed's introduction in the country. Since then, efforts by various breeders to revive the breed have been underway, utilizing artificial insemination for greater success in adapting to the local climate [
16]. The British Alpine goat, classified as a medium to heavy milk producer, is renowned for its substantial milk yield, reaching up to 4 kg per day, characterized by superior quality. With an average butterfat content ranging between 3.5% and 3.7% and a protein content of approximately 2.74%, it stands out for its consistent performance over an extended lactation period of up to 18 months (
https://journals.co.za/doi/pdf/10.10520/ejc-ac_dm_v29_n6_a19, accessed 16 April 2024).
3. Benefits of Goat Milk
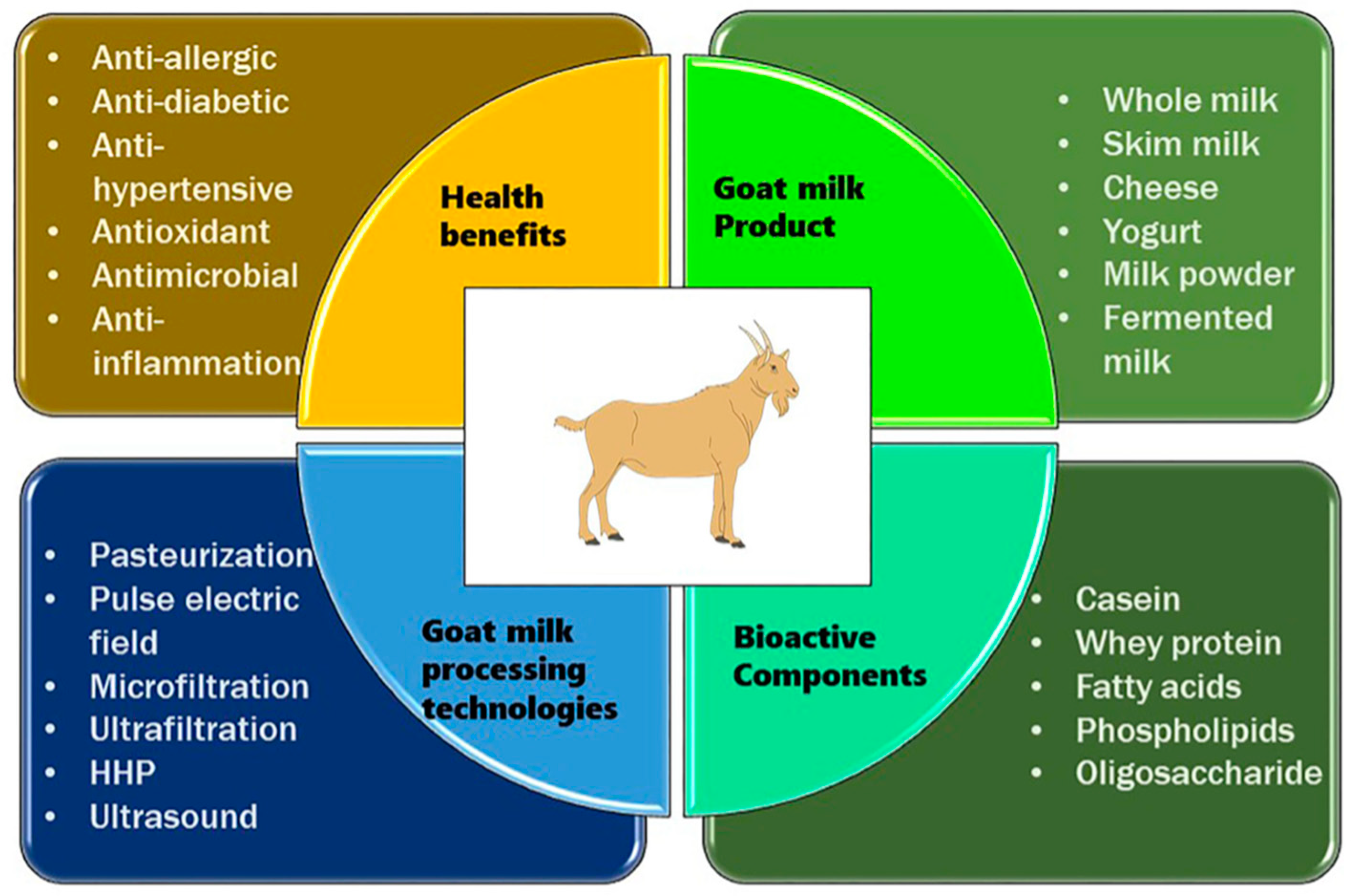
Goat milk is a notable dairy alternative and a valuable source of essential nutrients, bioactive compounds, and potential health benefits. Compared to cow's milk, goat milk exhibits unique characteristics such as smaller fat globules and different protein compositions, which contribute to its distinctive properties [
17]. Several studies have demonstrated that goat milk is rich in essential nutrients including calcium, phosphorus, potassium, vitamin A, vitamin B2 (riboflavin), and vitamin D, with potential implications for bone health, immune function, and overall well-being [
18,
19,
20]. Additionally, bioactive components present in goat milk, such as oligosaccharides and fatty acids, have been studied for their potential anti-inflammatory and immunomodulatory effects [
20]. The lower allergenicity of goat milk, attributed to reduced levels of alpha-S1 casein protein, makes it a promising alternative for individuals with cow's milk allergies [
21]. Furthermore, the application of goat milk in skincare products underscores its potential benefits for skin health, particularly in conditions such as eczema and psoriasis [
22,
23]. This highlights the scientific significance of goat milk and advances further exploration into its physiological effects and therapeutic applications. Goat milk offers several health benefits compared to cow milk, making it a valuable dietary option for many individuals.
The therapeutic and hypoallergenic properties of goat milk in human health and nutrition have long been recognized, suggesting that caprine milk may contain unique metabolically active and bioactive components specific to this species [
18,
20]. The hypoallergenic properties of goat milk, attributed to its bioactive milk components, are of paramount importance in human health and medical applications [
24]
Figure 2. shows a detailed description of the processing of goat milk, its bioactive compounds, and its health benefits to better shed light on the understanding of goat milk, its products, and the benefits thereof [
24].
With the increasing interest in goat milk for innovative cosmetic and medicinal purposes, there is a critical need to unravel medicinal properties and insights into goat milk to substantiate its healing attributes and facilitate its effective utilization in skin care and skin disease management.
Goat milk has high biological potential, and due to their natural origin and non-toxicity, they have many uses in cosmetics and dermatology. These natural products are especially rich in proteins, such as casein, β-lactoglobulin, α-lactalbumin, lactoferrin, immunoglobulins, lactoperoxidase, lysozyme, and growth factors, and possess various antibacterial, antifungal, antiviral, anticancer, antioxidant, immunomodulatory properties, etc. [
13]. Dairy derivatives find extensive application in dermatological therapies, aiding in the recovery of persistent wounds, accelerating tissue renewal, and managing conditions like acne vulgaris or plaque psoriasis. Recognized as potent components, these products contribute to enhancing skin health by diminishing acne lesions and blackheads, balancing sebum production, alleviating inflammation, and offering various benefits such as moisturization, protection, toning, smoothing, anti-irritation, whitening, soothing, and anti-ageing effects. Although milk is known to be used as a raw material in the food industry, it is also widely used in the pharmaceutical and cosmetic industries due to its considerable biological potential. It has also been the subject of detailed analyses and discussions of its components and their properties [
13].
Eczema and psoriasis represent pervasive dermatological issues, necessitating holistic and sustainable solutions. Goat milk has shown promise in promoting skin health, with anecdotal evidence suggesting its efficacy in soothing and managing these skin conditions.
4. Medicinal Properties of Goat Milk and Its Potential Use in Skin Therapies
Due to its nutritional richness and unique composition, goat milk has gained increasing scientific interest due to its potential medicinal properties (
Table 1). Emerging research suggests that goat milk may hold promise as a therapeutic agent for various skin conditions, including eczema and psoriasis. This proves a need to explore the scientific evidence supporting the use of goat milk in skin therapies, focusing on its potential benefits in managing eczema and psoriasis.
The medicinal properties of goat milk, including its anti-inflammatory, barrier-enhancing, antimicrobial, and immunomodulatory effects, hold significant therapeutic potential in the management of eczema and psoriasis. From its anti-inflammatory and antimicrobial activities to its moisturizing and antioxidant effects, goat milk offers a holistic approach to managing these chronic skin conditions. Further research is warranted to unravel the specific mechanisms of action underlying goat milk's efficacy in skin therapy and to optimize its formulation and delivery for clinical applications including the integration of goat milk genomics and delivery of goat milk-based skincare products for maximum efficacy in eczema and psoriasis management. Some genes have been associated with the medicinal properties of goat milk, and these pose a great potential for the use of goat milk in skin therapies (
Table 2).
Several candidate genes associated with the healing properties of goat milk, specifically in the context of treating skin conditions such as lactoferrin (LTF), lysozyme (LYZ), and β-casein (CSN2), are known for their antimicrobial, immunomodulatory, and anti-inflammatory effects, respectively have been identified. Understanding the genetic factors governing milk-skin interactions may provide valuable insights into the molecular pathways involved in healing skin conditions like eczema and psoriasis, ultimately contributing to the development of targeted goat milk-based skincare products for maximum efficacy in management.
5. Goat Milk Genomics and Genomic Tools
Advances in the genomics era have shown that more can be explored and achieved in terms of using genomics to reveal and aid in the understanding of genetic mechanisms in various organisms. Approximately 271 goat genes are associated with economic traits such as milk, fiber and meat production, disease resistance, reproduction, and growth in goats; where five genes were specifically associated with milk [
25,
26]. Understanding the genetic basis of milk traits is essential for dairy goat breeding programs aimed at improving milk production efficiency and dairy product quality. Due to the significant economic impact of goat dairy products, studies on genes associated with milk yield and quality have received more attention. Some of the genes influencing meat quality and yield have also been reported to affect milk yield and content traits, indirectly influencing the quality of milk and derived products [
27]. For instance, the insulin-like Growth Factor 1 gene (IGF-1), which is linked to body growth, development, and metabolism has also been reported to play a role in regulating the expression of milk protein and fat [
27,
28].
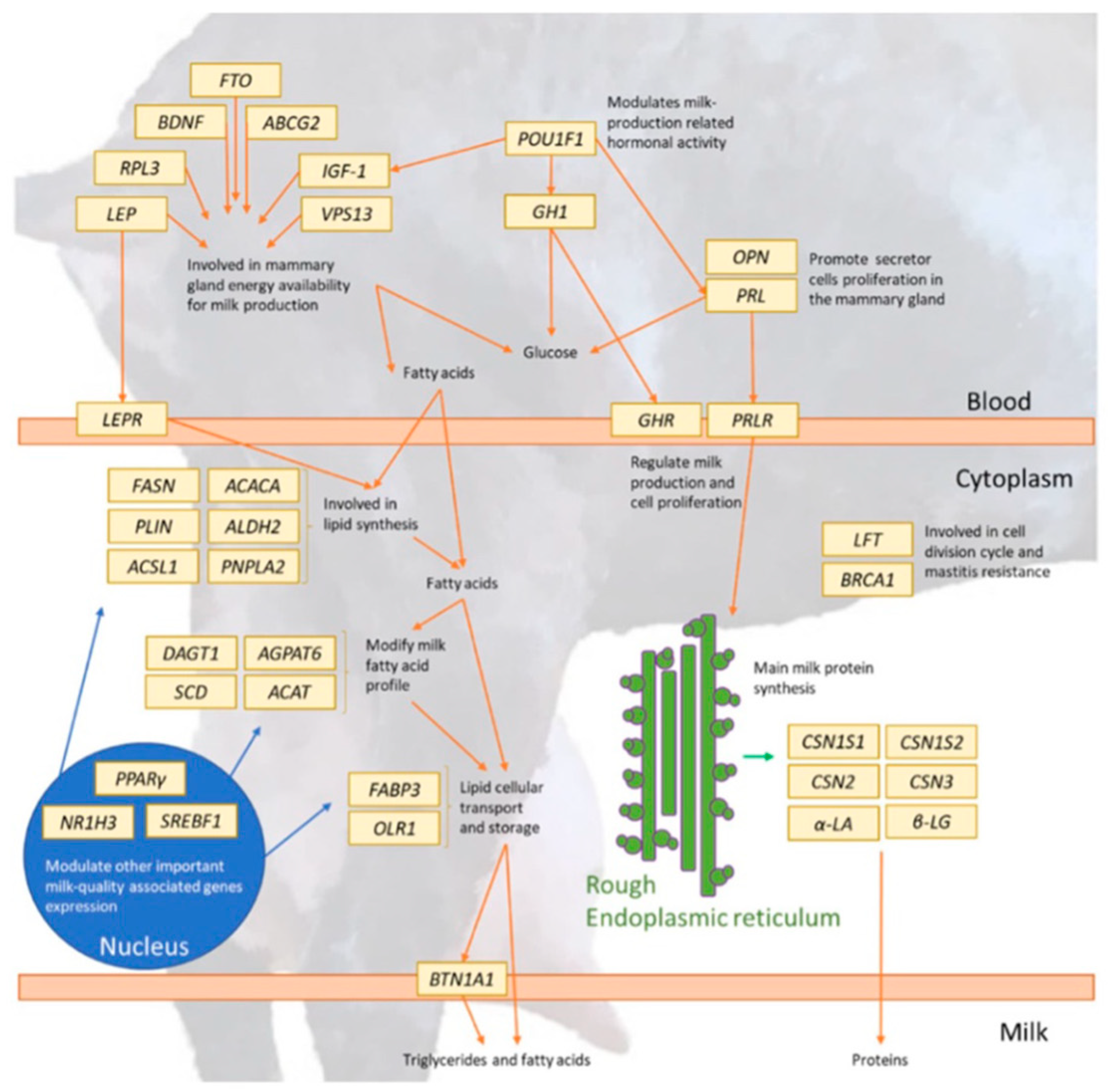
Figure 3 [
27] and
Table 3 show a representation of dairy goat candidate genes and their various roles.
Milk, proteins, and milk fat quantity are particularly important traits in dairy breed and milk production and not much is known about the area of the genome that controls these important traits in goats. A comparative analysis of signatures of selection for milk traits revealed candidate genes for milk composition and yield such as ABCG2 and NCAM2 [
10]. Some of these genes show potential for use in optimizing milk quality for production purposes. For example, the Growth Hormone 1 gene (GH1) has shown its usefulness in stimulating udder development in transgenic goats and, therefore, increasing their milk yield [
27].
Genetic studies investigating the healing properties of goat milk, particularly in the context of skin diseases, have shed light on the underlying mechanisms and identified key genes associated with these beneficial effects. While research in this area is still emerging, several studies have provided valuable insights into the genetic basis of goat milk's therapeutic properties for skin health.
Through RNA-sequencing technology [
29], Belfeki et al., (2018) explored the transcriptomic profile of goat mammary gland epithelial cells during lactation, revealing the expression of genes involved in immune modulation, wound healing, and antimicrobial defense. This study identified specific genes, such as lactoferrin (LTF) and lysozyme (LYZ), known for their antimicrobial and immunomodulatory properties, as being highly expressed in goat mammary epithelial cells, highlighting their potential role in conferring healing properties to goat milk [
29]. While specific genetic studies directly linking genes to the healing properties of goat milk for skin diseases are limited, ongoing research in this area holds promise for uncovering novel genetic pathways and biomarkers associated with goat milk's therapeutic effects. Further studies leveraging advanced genetic techniques, such as genome-wide association studies and transcriptomic analysis, may unravel the genetic mechanisms underlying goat milk's healing properties and pave the way for the development of innovative therapies for skin diseases and the next-generation genomic tools have proven to have a great potential in this regard.
The next-generation era offers a range of tools that can generate high-quality data in a short period (
www.illumina.com, accessed 05 February 2024). This data can then be used to study informative genes that can assist in bringing about useful information and hence allow for a better understanding of the dairy goats, milk genetics and more especially genes that are associated with the healing properties of goat milk. Their potential can hence be ascertained and developed for their improvement. Several tools can be used in goat genomics studies. One such tool is Single Nucleotide Polymorphisms (SNPs) which can explain most of the genetic discrepancies evident between animals [
30]. Single-nucleotide polymorphisms (SNPs) are single base-pair modifications within the genome [
31], comprising non-synonymous and synonymous [
32] as well as coding and non-coding variations. Coding SNPs are found near the coding regions of various genes such as the mitochondrial DNA [
30]. Next-generation sequencing (NGS) technologies have revolutionized the field of goat milk genomics by enabling high-throughput, cost-effective, and comprehensive analysis of the goat genome, transcriptome, and microbiome. NGS techniques, such as whole-genome sequencing (WGS), RNA sequencing (RNA-seq), and metagenomic sequencing, have been instrumental in advancing our understanding of the genetic basis of milk composition, quality, and bioactive properties in goats. Several techniques have been developed to study SNPs, which include SNP arrays, whole-genome sequencing (WGS) and targeted gene sequencing [
32], these technologies can be used to unravel the genomics potential of goat milk and its use for medicinal purposes.
5.1. The Illumina Goat SNP Chip
The progression of genetic technologies has instigated a surge of interest in genome-wide investigations [
26]. The Illumina goat SNP50K is a high-density SNP chip consisting of markers evenly spaced across the goat genome and is a useful tool for population genetics, genetic diversity, and genomic association studies [
33]. It was developed using six goat breeds and has been validated to be suitable for use in a variety of goat breeds [
33,
34,
35]. This technology has been used in various studies in South African goats. One such study was performed on Angora goats, where the suitability of the Illumina goat SNP50K Bead Chip for specific breeds was explored [
34]. Lashmar et al., (2015) [
34], also suggested that the chip could be used as a tool for genome-wide association, genetic variation, signatures of selection and genomic selection studies as well as for parentage verification. The Illumina goat SNP50K chip was used in the analysis of the population structure of South African goat populations [
36]. In this study, Mdladla et al. (2016) [
36], reported the feral Tankwa population to be a genetically distinct population with South African goat populations clustering according to their historical origins. This study further described the usefulness of the Illumina goat SNP50K in the investigation of population structure, genetic diversity, and relationships between the SA non-descript goats and the feral Tankwa populations. The Illumina goat SNP50K chip also revealed valuable information on the demographic history of South African goat populations [
35]. The Illumina goat SNP50K Bead chip was used for body morphological traits in Sudanese goats [
37].
Using this chip, a study on Canadian dairy goats revealed 189 unique and significant SNPs corresponding to 271 unique positional candidate genes within 50 kb up-and-downstream, across breeds and traits [
38]. This study further provides evidence for the economic importance of several candidate genes (e.g., CSN1S1, CSN2, CSN1S2, CSN3, DGAT1, and ZNF16) that have been associated with milk quality and production traits in the Canadian Alpine and Saanen populations [
38]. In the past, the use of very few breeds (six) to develop the goat SNP50K [
33], posed limitations in that, the breeds that were used were not of African origin and there may be some genetic variations due to ascertainment bias [
39]. While the Bead-Chip development initially focused on specific breeds, subsequent observations have revealed substantial genomic variations in excluded breeds, leading to broader applicability of the SNP50K Bead Chip across various goat breeds [
39]. The latest iteration, Goat_IGGC_65K_v2, represents an updated version containing 59,727 SNPs, enhancing genomic analysis capabilities [
39], and making it a suitable tool to use in unravelling the genomic potential of goat milk.
5.2. Next Generation Sequencing (NGS)
Next-generation sequencing (NGS) is a high-throughput technology based on massively parallel sequencing, where millions of sequences are deciphered concurrently [
40]. The technology originated in the early 2000s and it was proven a success through the complete sequencing of the human genome [
40]. NGS consists of platforms that have proven to be highly quantitative and adaptive with the promise to eliminate micro-array limitations [
41]. It can identify transcriptomes without knowledge of a particular gene, therefore, providing insights on alternative splicing and alternative variations in identified genes, and it also gives complete coverage in a relatively short period [
41,
42]. Platforms such as the Illumina HiSeq 2500 can produce about 500Gb and NovaSeq 6000 can produce about 3 TB of sequence data in one run in less than 10 days.
5.3. Whole Genome Sequencing
Whole-genome sequencing (WGS) is a method whereby the whole genetic makeup of an organism or animal is sequenced (
www.illumina.com, accessed 16 January 2024). Whole-genome sequencing in practice does, however, not necessarily cover all components of the genome (
www.acmg.net, accessed 16 January 2024). For sufficiency in data generated, a large number of sequence reads is needed (approx. 6.3 million) and multiplexing is also a challenge [
43]. These alone make sequence costs per sample very high, thereby placing limitations on sample numbers [
43].
WGS studies done on the South African feral Tankwa goat revealed variants associated with cellular pathways, defense, and immunity [
44]. SNPs that show a potential role in goat growth performance, diseases and adaptation processes have also been identified [
44]. This technology has also been used in the identification of selection of signatures in important traits such as coat colour, growth, reproduction, and high-altitude adaptation in goats [
45,
46]. The number of thoracic vertebrae is one of the important economic traits that influences carcass length and meat production in livestock and WGS revealed candidate genes that were associated with thoracic vertebrate number in sheep [
47]. Through WGS in Pakistan cattle breeds, it was revealed that most of the altered genes are significantly enriched in economically important biological processes such as heat tolerance, immune response, development, and sensory perceptions [
48]. However, as mentioned above, the main limiting factor of this technology is the high cost per sample making it too expensive, especially in the case of multiple samples.
Whole genome sequencing has been very useful in the development of goat genomic studies especially in the sequencing of goat reference genomes. The first draft genome achieved through WGS was from the female domestic goats Yunnan black goats with a genomic size of approximately 2.66 Gb [
44]. This was followed by other goat genome sequences that have been successfully used in the identification of CNVs, SNPs and other genomic variants [
44].
5.4. RNA-Sequencing
RNA-Seq is a high-throughput sequencing method for gene expression profiling widely used for mapping and quantifying transcriptomes and analyzing gene expressions in various tissues [
49]; providing more accurate levels of transcripts used to measure transcriptome composition and to discover new exons or genes [
42].
RNA-seq is one powerful approach that is widely used. Transcriptomic studies have identified genes encoding milk proteins, including caseins (e.g., CSN1S1, CSN1S2, CSN2, CSN3) and whey proteins (e.g., LALBA, LGB, LTF) [
50,
51]. These genes are highly expressed in mammary gland tissue during lactation and play crucial roles in determining milk protein composition and quality [
50]. RNA-Seq technology has also proven its usefulness in gene expression profiling of the intramuscular muscle in Nellore cattle where several genes that were associated with lipid metabolism and fatty acid composition were identified [
52]. These demonstrate the potential for RNA-Seq as a tool to be used for gene expression profiling as there are associations that can be linked to dairy goats’ traits and the identification of genes associated with the healing properties of goat milk.
While specific research on the use of genomics tools for goat milk-based therapies in eczema and psoriasis is limited, existing evidence suggests promising avenues for further investigation. By elucidating the genetic basis of goat milk's healing properties and understanding its interactions with skin biology, genomics research holds the potential to drive innovation in skincare and dermatological treatments.
6. Conclusion and Recommendations
In conclusion, this review highlights the pivotal role of genomic tools in elucidating the medicinal properties of goat milk and its potential applications in skincare and healthcare, particularly in the management of key skin diseases such as eczema and psoriasis. Through genomic analysis, researchers have identified key genes and molecular pathways underlying the bioactive constituents of goat milk, including peptides, proteins, and lipids, which contribute to its therapeutic efficacy. The integration of genomic insights with functional studies has provided valuable insights into goat milk's immunomodulatory, antimicrobial, and antioxidant activities, offering promising avenues for managing skin conditions characterized by immune dysregulation and inflammation. Furthermore, genomic approaches have enabled targeted breeding strategies to enhance the production of bioactive compounds in goat milk, facilitating the development of personalized skincare formulations tailored to individual genetic profiles. Moving forward, continued advancements in genomic technologies, combined with interdisciplinary research efforts, hold promise for further harnessing the therapeutic potential of goat milk and its derivatives in skincare and dermatological care. By leveraging genomic tools, goat milk emerges as a promising source of bioactive compounds with diverse applications in promoting skin health and wellness, ultimately contributing to the advancement of personalized medicine and the management of dermatological conditions such as eczema and psoriasis.
Author Contributions
Conceptualization, writing of the original draft, and editing K.T. Ncube; Supervision, conceptualization and editing B. Mtileni.; editing and structure of the content A.K. Nephawe, T.J. Mpofu and CM Modiba. All authors have read and agreed to the published version of the manuscript.
Funding
The project is supported and funded by the Tshwane University of Technology, Department of Animal Sciences.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
There were no new data analyzed for this review.
Acknowledgments
Tshwane University of Technology, Department of Animal Sciences.
Conflicts of Interest
The authors declare that there are no conflicts of interest.
References
- Gipson TA. Recent advances in breeding and genetics for dairy goats. Asian-Australas J Anim Sci. 2019; 32:1275–83. [CrossRef]
- Scopinich-Cisternas J, Strahsburger E. Goat type: The key factor to produce goat milk with economic profitable purpose in arid and desert zones. Idesia (Arica). 2019; 37:122–3. [CrossRef]
- Ncube KT, Dzomba EF, Hadebe K, Soma P, Frylinck L, Muchadeyi FC. Carcass Quality Profiles and Associated Genomic Regions of South African Goat Populations Investigated Using Goat SNP50K Genotypes. Animals. 2022; 12:364. [CrossRef]
- Ncube KT, Mdladla K, Dzomba EF, Muchadeyi FC. Targeted high-throughput growth hormone 1 gene sequencing reveals high within-breed genetic diversity in South African goats. Anim Genet. 2016; 47:382–5. [CrossRef]
- Dzomba EF, Mdladla K, Ncube KT, Muchadeyi FC. Genomics for the improvement of productivity and robustness of South African goat breeds. In: Sustainable Goat Production in Adverse Environments. 2018.
- Andre Mataveia G, Visser C, Sitoe A. Smallholder Goat Production in Southern Africa: A Review. In: Goat Science - Environment, Health and Economy. IntechOpen; 2023.
- Bosman L, van Marle-Köster E, Visser C. Genetic diversity of South African dairy goats for genetic management and improvement. Small Ruminant Research. 2015; 123:224–31. [CrossRef]
- Donkin E, Boyazoglu P. Diseases and mortality of goat kids in a South African milk goat herd. In: 8th International Conference on Goats. South African Journal of Animal Science; 2004. p. 258–61.
- Muller C. Genetic parameter estimation and breeding plans for the South African dairy goat herd. Thesis. University of Stellenbosch; 2005.
- Amiri Ghanatsaman Z, Ayatolahi Mehrgardi A, Asadollahpour Nanaei H, Esmailizadeh A. Comparative genomic analysis uncovers candidate genes related with milk production and adaptive traits in goat breeds. Sci Rep. 2023; 13:8722. [CrossRef]
- Martin P, Palhière I, Maroteau C, Bardou P, Canale-Tabet K, Sarry J, et al. A genome scan for milk production traits in dairy goats reveals two new mutations in Dgat1 reducing milk fat content. Sci Rep. 2017; 7:1872. [CrossRef]
- Lima MJR, Teixeira-Lemos E, Oliveira J, Teixeira-Lemos LP, Monteiro AMC, Costa JM. Nutritional and Health Profile of Goat Products: Focus on Health Benefits of Goat Milk. In: Goat Science. InTech; 2018.
- Kazimierska K, Kalinowska-Lis U. Milk Proteins—Their Biological Activities and Use in Cosmetics and Dermatology. Molecules. 2021; 26:3253. [CrossRef]
- Singh S, Kaur G, Brar RPS, Preet GS. Goat milk composition and nutritional value: A review. The Pharma Innovation Journal. 2021; 10:536–40.
- Visser C, Marle-Köster E van. The Development and Genetic Improvement of South African Goats. In: Goat Science. 2018.
- The South African Milch Goat Breeders’ Society. Preserving Genetics. 2015;34–9.
- Haenlein GFW. Goat milk in human nutrition. Small Ruminant Research. 2004; 51:155–63. [CrossRef]
- Park YW. Goat Milk – Chemistry and Nutrition. In: Handbook of Milk of Non-Bovine Mammals. Wiley; 2017. p. 42–83.
- Turkmen N. The Nutritional Value and Health Benefits of Goat Milk Components. In: Nutrients in Dairy and their Implications on Health and Disease. Elsevier; 2017. p. 441–9.
- Robin Badal, Rohit Singh, Arushi, Anjali Upadhyay. An Insight into Therapeutic and Nutritional Profile of Aja Dugdha (Goat’s Milk): A Review. International Journal of Ayurveda and Pharma Research. 2023;87–96. [CrossRef]
- Bahna SL. Cow’s milk allergy versus cow milk intolerance. Annals of Allergy, Asthma & Immunology. 2002; 89:56–60. [CrossRef]
- Kiskini A, Difilippo E. Oligosaccharides in goat milk: structure, health effects and isolation. Cell Molecular Biology. 2013; 59:25–30.
- van Leeuwen SS, te Poele EM, Chatziioannou AC, Benjamins E, Haandrikman A, Dijkhuizen L. Goat Milk Oligosaccharides: Their Diversity, Quantity, and Functional Properties in Comparison to Human Milk Oligosaccharides. J Agric Food Chem. 2020; 68:13469–85. [CrossRef]
- Nayik GA, Jagdale YD, Gaikwad SA, Devkatte AN, Dar AH, Dezmirean DS, et al. Recent Insights Into Processing Approaches and Potential Health Benefits of Goat Milk and Its Products: A Review. Front Nutr. 2021;8. [CrossRef]
- Supakorn C. The Important Candidate Genes in Goats - A Review. Journal of Science and technology. 2009. [CrossRef]
- Ncube KN. Gene Expression Profiling of South African Indigenous Goat Breeds using RNA-Seq Technologies In Search of Genes Associated With Growth and Carcass Quality Traits. University of Kwa-Zulu Natal; 2020.
- Salgado Pardo JI, Delgado Bermejo JV, González Ariza A, León Jurado JM, Marín Navas C, Iglesias Pastrana C, et al. Candidate Genes and Their Expressions Involved in the Regulation of Milk and Meat Production and Quality in Goats (Capra hircus). Animals. 2022;12:988. [CrossRef]
- Banos G, Woolliams JA, Woodward BW, Forbes AB, Coffey MP. Impact of Single Nucleotide Polymorphisms in Leptin, Leptin Receptor, Growth Hormone Receptor, and Diacylglycerol Acyltransferase (DGAT1) Gene Loci on Milk Production, Feed, and Body Energy Traits of UK Dairy Cows. J Dairy Sci. 2008; 91:3190–200. [CrossRef]
- Belfeki L, Torres D, Salemi S, Pesenti ME, Trumtel S, Boukadiri A. Transcriptomic profile of goat mammary gland epithelial cells using single-cell RNA-sequencing. Scientific Reports, 8(1), 1-12. 2018;8:1–12.
- Salem M, Vallejo RL, Leeds TD, Palti Y, Liu S, Sabbagh A, et al. RNA-seq identifies SNP markers for growth traits in rainbow trout. PLoS One. 2012;7. [CrossRef]
- Dough B. Single nucleotide polymorphisms (SNPs). Computational Molecular Biology Biochemistry. 2010. Accessed 11 Sep 2019.
- Sharma U, Banerjee P, Joshi J, Kumar R. Identification of SNPs in Goats (Capra hircus) using RNA-Seq Analysis. Int J Anim Vet Adv. 2012;4:272–83.
- Tosser-Klopp G, Bardou P, Bouchez O, Cabau C, Crooijmans R, Dong Y, et al. Design and characterization of a 52K SNP chip for goats. PLoS One. 2014. [CrossRef]
- Lashmar SF, Visser C, Van Marle-Köster E. Validation of the 50k Illumina goat SNP chip in the South African Angora goat. South African Journal of Animal Sciences. 2015;45:56–9. [CrossRef]
- Mdladla K, Dzomba EF, Muchadeyi FC. The potential of landscape genomics approach in the characterization of adaptive genetic diversity in indigenous goat genetic resources: A South African perspective. Small Ruminant Research. 2017. [CrossRef]
- Mdladla K, Dzomba EF, Huson HJ, Muchadeyi FC. Population genomic structure and linkage disequilibrium analysis of South African goat breeds using genome-wide SNP data. Anim Genet. 2016;47:471–82. [CrossRef]
- Rahmatalla SA, Arends D, Reissmann M, Wimmers K, Reyer H, Brockmann GA. Genome-wide association study of body morphological traits in Sudanese goats. Anim Genet. 2018. [CrossRef]
- Massender E, Brito LF, Maignel L, Oliveira HR, Jafarikia M, Baes CF, et al. Single-step genomic evaluation of milk production traits in Canadian Alpine and Saanen dairy goats. J Dairy Sci. 2022;105:2393–407. [CrossRef]
- Magoro AM, Mtileni B, Hadebe K, Zwane A. Assessment of Genetic Diversity and Conservation in South African Indigenous Goat Ecotypes: A Review. Animals. 2022;12:3353. [CrossRef]
- Franklin WA, Aisner DL, Post MD, Bunn PA, Garcia MV. Pathology, Biomarkers, and Molecular Diagnostics. In: Abeloff’s Clinical Oncology: Fifth Edition. 2013.
- Malatji DP. A genomic insight into the diversity and pathogenesis of village chickens in a Ascaridia galli infested environment. University of Pretoria; 2017.
- Wang GRÆX, Zhang ÆXTFÆCL. Analysis of caprine pituitary specific transcription factor-1 gene polymorphism in indigenous Chinese goats. 2009;705–9. [CrossRef]
- Boon EMJ, Faas BHW. Benefits and limitations of whole genome versus targeted approaches for noninvasive prenatal testing for fetal aneuploidies. Prenatal Diagnosis. 2013. [CrossRef]
- Mohlatlole RP, Dzomba EF, Muchadeyi FC. Addressing production challenges in goat production systems of South Africa: The genomics approach. Small Ruminant Research. 2015. [CrossRef]
- Guo J, Tao H, Li P, Li L, Zhong T, Wang L, et al. Whole-genome sequencing reveals selection signatures associated with important traits in six goat breeds. Sci Rep. 2018. [CrossRef]
- Wang X, Liu J, Zhou G, Guo J, Yan H, Niu Y, et al. Whole-genome sequencing of eight goat populations for the detection of selection signatures underlying production and adaptive traits. Sci Rep. 2016. [CrossRef]
- Li C, Li M, Li X, Ni W, Xu Y, Yao R, et al. Whole-Genome Resequencing Reveals Loci Associated With Thoracic Vertebrae Number in Sheep. Front Genet. 2019. [CrossRef]
- Iqbal N, Liu X, Yang T, Huang Z, Hanif Q, Asif M, et al. Genomic variants identified from wholegenome resequencing of indicine cattle breeds from Pakistan. PLoS One. 2019. [CrossRef]
- Cánovas, Angela., Rincon, Gonzalo., Alma Islas-Trejo, Medrano., Saumya W and JF. SNP discovery in the bovine milk transcriptome using RNA-Seq technology. Mammalian Genome. 2010;21:592–598. [CrossRef]
- Xuan R, Wang J, Zhao X, Li Q, Wang Y, Du S, et al. Transcriptome Analysis of Goat Mammary Gland Tissue Reveals the Adaptive Strategies and Molecular Mechanisms of Lactation and Involution. Int J Mol Sci. 2022;23:14424. [CrossRef]
- Guan D, Landi V, Luigi-Sierra MG, Delgado JV, Such X, Castelló A, et al. Analyzing the genomic and transcriptomic architecture of milk traits in Murciano-Granadina goats. J Anim Sci Biotechnol. 2020;11:35. [CrossRef]
- Berton MP, Fonseca LFS, Gimenez DFJ, Utembergue BL, Cesar ASM, Coutinho LL, et al. Gene expression profile of intramuscular muscle in Nellore cattle with extreme values of fatty acid. BMC Genomics. 2016;17. [CrossRef]
- Mohamad Razlan Abd Rahman, Zaiton Hassan, Mohd Sukri Hassan, Roshada Hashim, Wong Ling Shing, Sharifah Hazirah Syd Jaafar. Multi-Nutrient Milk Quality Analysis Applying Chemometrics: A Supplementation-based Approach using Dairy Goats. Journal of Advanced Research in Applied Sciences and Engineering Technology. 2022;28:123–43. [CrossRef]
- Balakrishnan G, Agrawal R. Antioxidant activity and fatty acid profile of fermented milk prepared by Pediococcus pentosaceus. J Food Sci Technol. 2014;51:4138–42. [CrossRef]
- Kober M-M, Bowe WP. The effect of probiotics on immune regulation, acne, and photoaging. Int J Womens Dermatol. 2015;1:85–9. [CrossRef]
- Triprisila L, Suharjono S, Christianto A, Fatchiyah F. The Comparing of Antimicrobial Activity of CSN1S2 Protein of Fresh Milk and Yoghurt Goat Breed Ethawah Inhibited the Pathogenic Bacteria. Materia Socio Medica. 2016;28:244. [CrossRef]
- Singh A, Duche RT, Wandhare AG, Sian JK, Singh BP, Sihag MK, et al. Milk-Derived Antimicrobial Peptides: Overview, Applications, and Future Perspectives. Probiotics Antimicrob Proteins. 2023;15:44–62. [CrossRef]
- Oftedal OT. The evolution of milk secretion and its ancient origins. Animal. 2012; 6:355–68. [CrossRef]
- Crisà A, Claps S, Moioli B, Marchitelli C. Identification of the complete coding cDNAs and expression analysis of B4GALT1, LALBA, ST3GAL5, ST6GAL1 in the colostrum and milk of the Garganica and Maltese goat breeds to reveal possible implications for oligosaccharide biosynthesis. BMC Vet Res. 2019; 15:457. [CrossRef]
- Behrouz S, Saadat S, Memarzia A, Sarir H, Folkerts G, Boskabady MH. The Antioxidant, Anti-Inflammatory and Immunomodulatory Effects of Camel Milk. Front Immunol. 2022;13. [CrossRef]
- Franzoni G, Mecocci S, De Ciucis CG, Mura L, Dell’Anno F, Zinellu S, et al. Goat milk extracellular vesicles: immuno-modulation effects on porcine monocyte-derived macrophages in vitro. Front Immunol. 2023;14. [CrossRef]
- Zervas G, Tsiplakou E. Goat Milk. In: Milk and Dairy Products in Human Nutrition. Wiley; 2013. p. 498–518.
- Kazimierska K, Kalinowska-Lis U. Milk Proteins—Their Biological Activities and Use in Cosmetics and Dermatology. Molecules. 2021;26:3253. [CrossRef]
- Wang H. A Review of the Effects of Collagen Treatment in Clinical Studies. Polymers (Basel). 2021; 13:3868. [CrossRef]
- Getaneh M, Alemayehu K. Candidate genes associated with economically important traits in dairy goats. Cogent Food Agric. 2022;8. [CrossRef]
- Nayik GA, Jagdale YD, Gaikwad SA, Devkatte AN, Dar AH, Dezmirean DS, et al. Recent Insights Into Processing Approaches and Potential Health Benefits of Goat Milk and Its Products: A Review. Front Nutr. 2021;8. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).