Submitted:
23 April 2024
Posted:
23 April 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Genetic Identification of Bacterial Strains by Phylogenetic Analysis of 16S rRNA Encoding Gene Sequence
2.1.1. PCR Amplification of the 16S rRNA Gene
2.1.2. Nucleotide Sequence Analysis
2.1.3. Data Availability and Nucleotide Sequences Accession Numbers
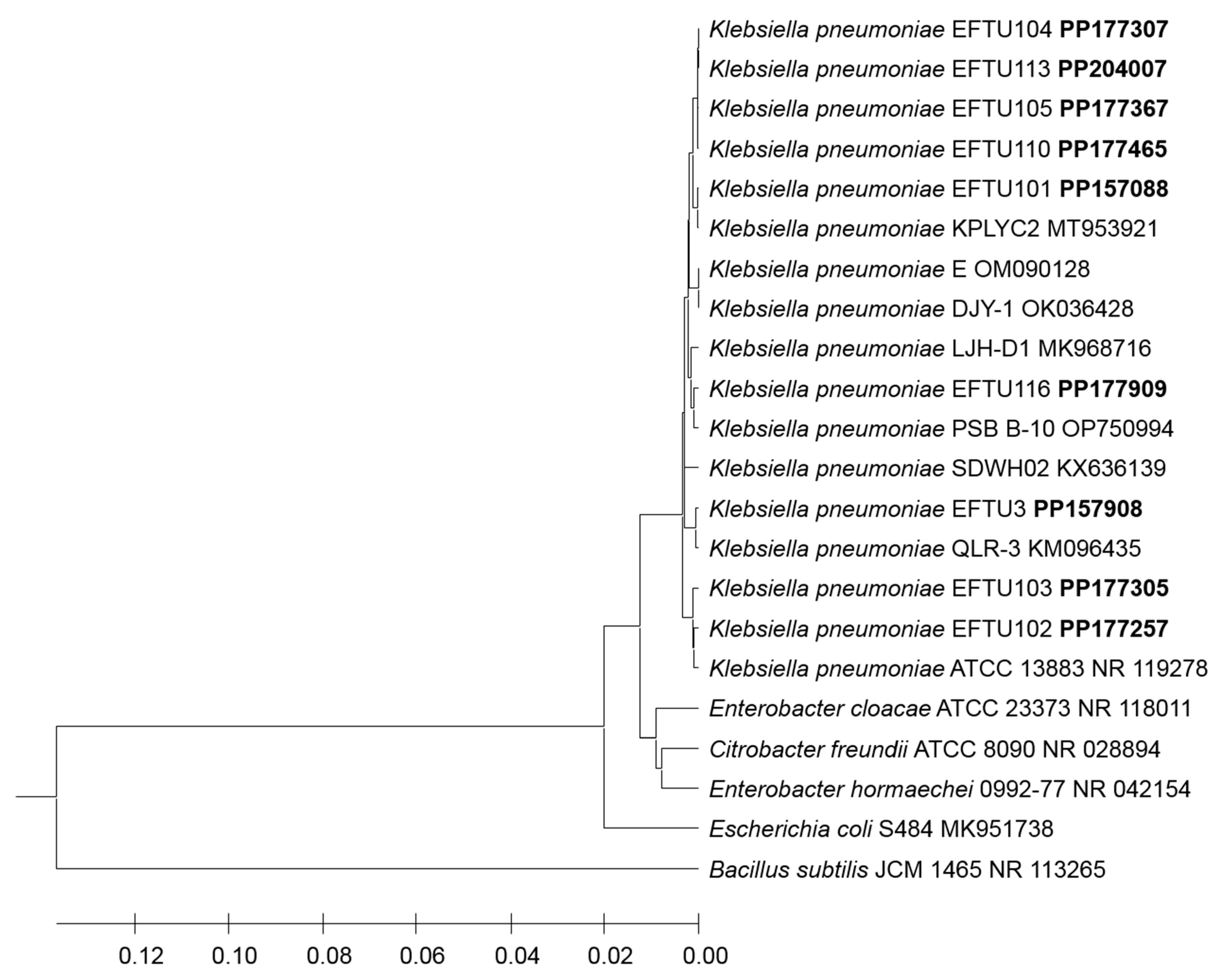
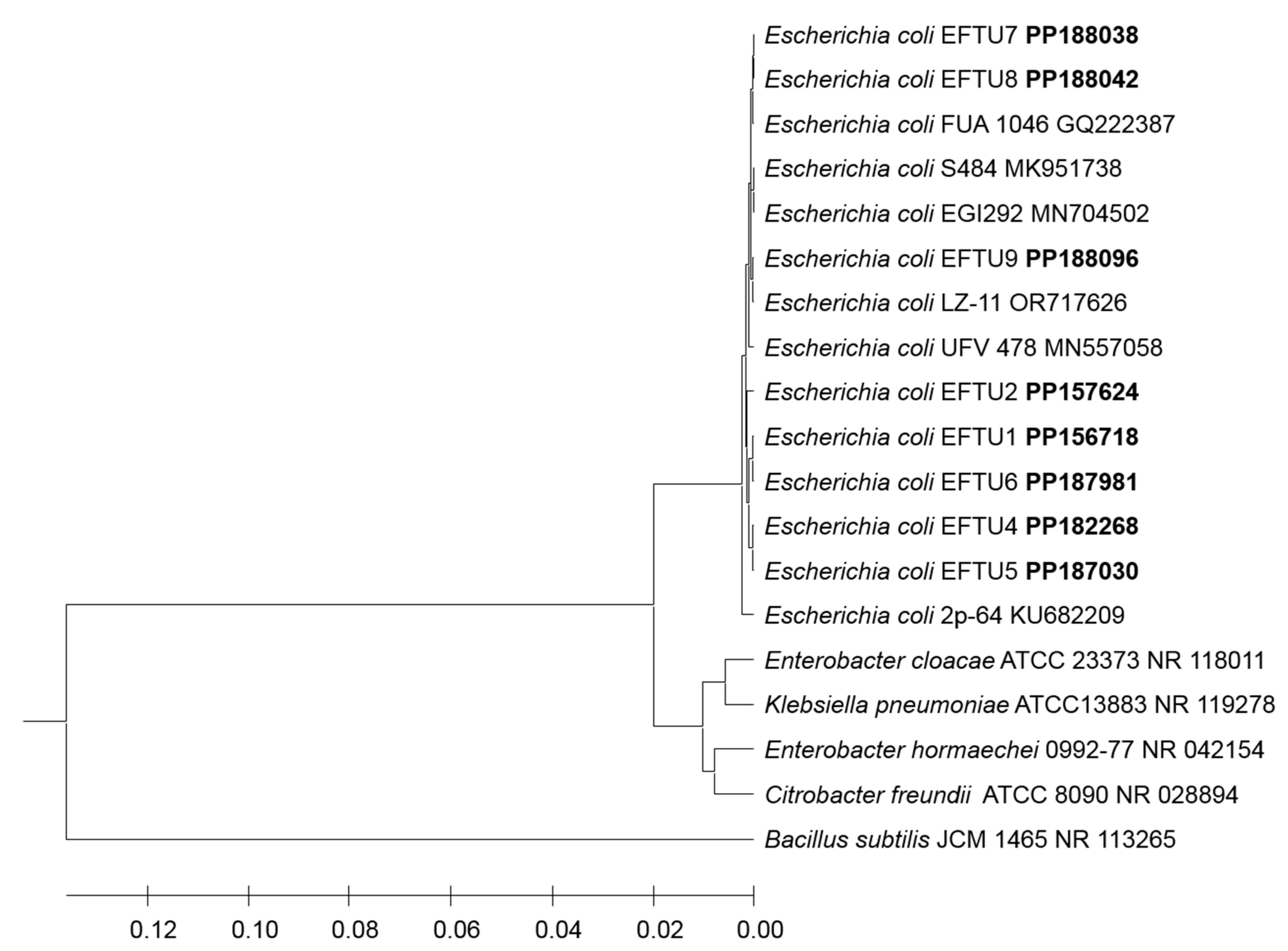
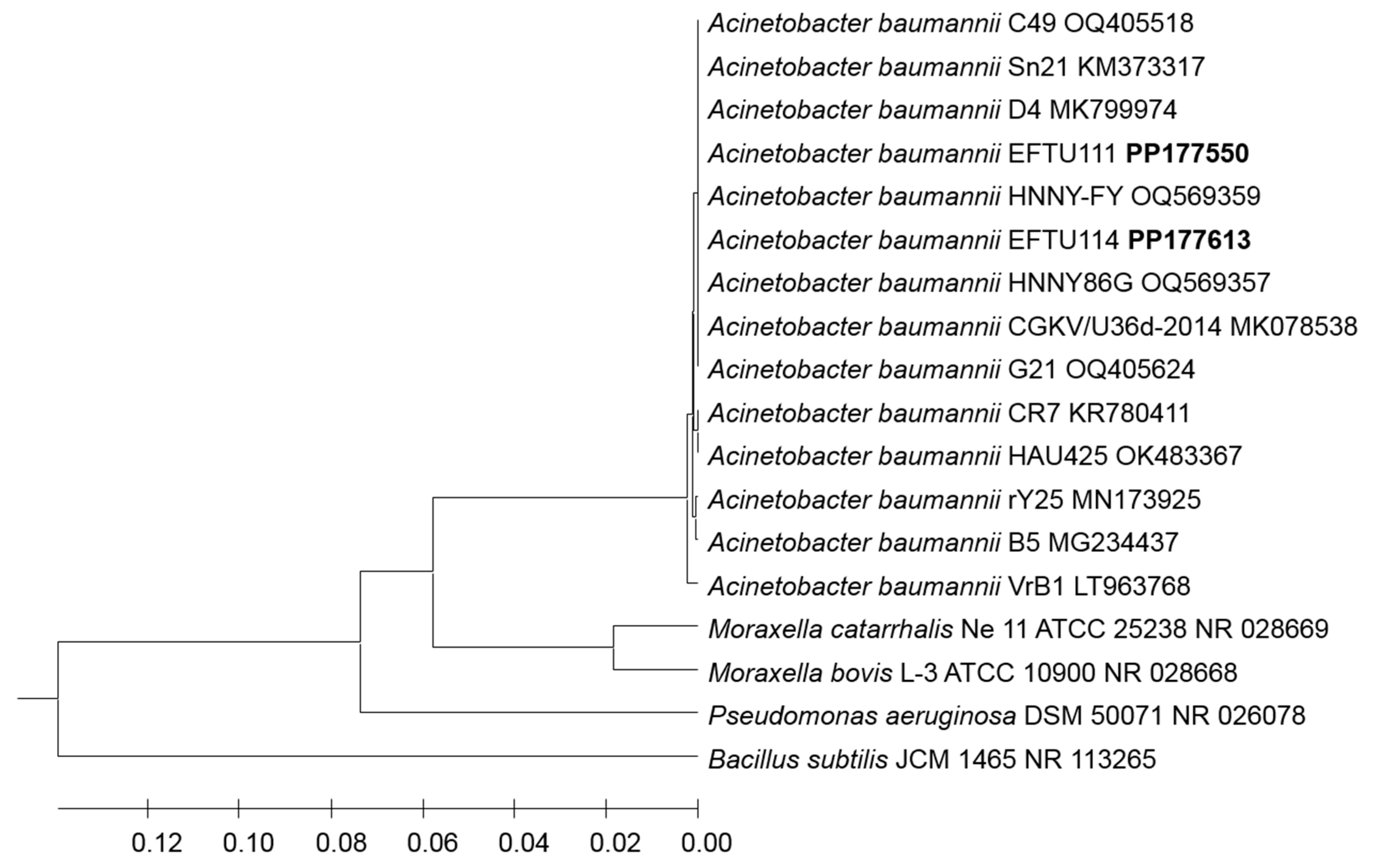
3. Results
4. Discussion
Conflicts of Interest Statement
Acknowledgments
References
- Nordmann, P.; Dortet, L.; Poirel, L. Carbapenem resistance in Enterobacteriaceae: Here is the storm! Trends Mol. Med. 2012, 18, 263–272. [Google Scholar] [CrossRef]
- Ellappan, K.; Belgode Narasimha, H.; Kumar, S. Coexistence of multidrug resistance mechanisms and virulence genes in carbapenem-resistant Pseudomonas aeruginosa strains from a tertiary care hospital in South India. J. Glob. Antimicrob. Resist. 2018, 12, 37–43. [Google Scholar] [CrossRef]
- Lee, Y.; Bradley, N. Overview and insights into Carbapenem allergy. Pharmacy 2019, 7, 110–116. [Google Scholar] [CrossRef]
- Elshamy, A.A.; Aboshanab, K.M. A review on bacterial resistance to carbapenems: epidemiology, detection and treatment options. Future Sci. OA 2020, 6, FSO438. [Google Scholar] [CrossRef]
- Mohamed, A.; Daef, E.; Nafie, A.; Shaban, L.; Ibrahim, M. Characteristics of Carbapenem-resistant Gram-negative Bacilli in patients with ventilator-associated pneumonia. Antibiotics 2021, 10, 1325. [Google Scholar] [CrossRef]
- Pitout, J.D.D.; Nordmann, P.; Poirel, L. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob. Agents Chemother. 2015, 59, 5873–5884. [Google Scholar] [CrossRef]
- Grönlund, M.M.; Arvilommi, H.; Kero, P.; Lehtonen, O.P.; Isolauri, E. Importance of intestinal colonisation in the maturation of humoral immunity in early infancy: a prospective follow up study of healthy infants aged 0–6 months. Arch. Dis. Child. Fetal. Neonatal. Ed. 2000, 83, F186–F192. [Google Scholar] [CrossRef]
- MacPherson, A.J.; Harris, N.L. Interactions between commensal intestinal bacteria and the immune system. Nat. Rev. Immunol. 2004, 4, 478–485. [Google Scholar] [CrossRef]
- Stavropoulou, E.; Bezirtzoglou, E. Human microbiota in aging and infection: a review. Crit Rev Food Sci Nutr 2019, 59, 537–545. [Google Scholar] [CrossRef]
- Fanaro, S.; Chierici, R.; Guerrini, P.; Vigi, V. Intestinal microflora in early infancy: composition and development. Acta Paediatr. Suppl. 2003, 91, 48–55. [Google Scholar] [CrossRef]
- Faith, J.J.; Guruge, J.L.; Charbonneau, M.; Subramanian, S.; Seedorf, H.; Goodman, A.L.; Clemente, J.C.; Knight, R.; Heath, A.C.; Leibel, R.L.; et al. The long term stability of the human gut microbiota. Science 2013, 341, 1237439. [Google Scholar] [CrossRef]
- Sommer, F.; Anderson, J.M.; Bharti, R.; Raes, J.; Rosenstiel, P. The resilience of the intestinal microbiota influences health and disease. Nat. Rev. Microbiol. 2017, 15, 630–638. [Google Scholar] [CrossRef]
- Al-Balawi, M.; Morsy, F.M. Enterococcus faecalis is a better competitor than other lactic acid bacteria in the initial colonization of colon of healthy newborn babies at first week of their life. Front. Microbiol. 2020, 11, 2017. [Google Scholar] [CrossRef]
- Al-Balawi, M.; Morsy, F.M. Prenatal versus postnatal initial colonization of healthy neonates’ colon ecosystem by the enterobacterium Escherichia coli. Microbiol. Spectrum 2021, 9, e00379-21. [Google Scholar] [CrossRef]
- Bocci, V. The neglected organ: bacterial flora has a crucial immunostimulatory role. Perspect. Biol. Med. 1992, 35, 251–260. [Google Scholar] [CrossRef]
- O’Hara, A.; Shanahan, F. The gut flora as a forgotten organ. EMBO Rep. 2006, 7, 688–693. [Google Scholar] [CrossRef]
- Guinane, C.M.; Cotter, P.D. Role of the gut microbiota in health and chronic gastrointestinal disease: understanding a hidden metabolic organ. Therap. Adv. Gastroenterol. 2013, 6, 295–308. [Google Scholar] [CrossRef]
- Van de Vliet, M.; Joossens, M. The resemblance between bacterial gut colonization in pigs and humans. Microorganisms 2022, 10, 1831. [Google Scholar] [CrossRef]
- Flint, H.J.; Scott, K.P.; Louis, P.; Duncan, S.H. The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 577–589. [Google Scholar] [CrossRef]
- Tamburini, S.; Shen, N.; Wu, H.C.; Clemente, J.C. The microbiome in early life: Implications for health outcomes. Nat. Med. 2016, 22, 713–722. [Google Scholar] [CrossRef]
- Gentile, C.L.; Weir, T.L. The gut microbiota at the intersection of diet and human health. Science 2018, 362, 776–780. [Google Scholar] [CrossRef]
- Shanahan, F. The host–microbe interface within the gut. Best Pract. Res. Clin. Gastroenterol. 2002, 16, 915–931. [Google Scholar] [CrossRef]
- Belkaid, Y.; Hand, T.W. Role of the microbiota in immunity and inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef]
- Ley, R.E.; Peterson, D.A.; Gordon, J.I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 2006, 124, 837–848. [Google Scholar] [CrossRef]
- Sousa, S.A.; Feliciano, J.R.; Pita, T.; Soeiro, C.F.; Mendes, B.L.; Alves, L.G.; Leitão, J.H. Bacterial nosocomial infections: Multidrug resistance as a trigger for the development of novel antimicrobials. Antibiotics 2021, 10, 942. [Google Scholar] [CrossRef]
- Coque, T.M.; Cantón, R.; Pérez-Cobas, A.E.; Fernández-de-Bobadilla, M.D.; Baquero, F. Antimicrobial Resistance in the Global Health Network: Known Unknowns and Challenges for Efficient Responses in the 21st Century. Microorganisms 2023, 11, 1050. [Google Scholar] [CrossRef]
- Freitas, A.R.; Werner, G. Nosocomial Pathogens and Antimicrobial Resistance: Modern Challenges and Future Opportunities. Microorganisms 2023, 11, 1685. [Google Scholar] [CrossRef]
- Duan, Z.; Li, X.; Li, S.; Zhou, H.; Hu, L.; Xia, H.; Xie, L.; Xie, F. Nosocomial surveillance of multidrug-resistant Acinetobacter baumannii: a genomic epidemiological study. Microbiol Spectr. 2024, 12, e0220723. [Google Scholar] [CrossRef]
- Valzano, F.; Coda, A.R.D.; Liso, A.; Arena, F. Multidrug-resistant bacteria contaminating plumbing components and sanitary installations of hospital restrooms. Microorganisms 2024, 12, 136. [Google Scholar] [CrossRef]
- Hocquet, D.; Muller, A.; Bertrand, X. What Happens in Hospitals Does Not Stay in Hospitals: Antibiotic-Resistant Bacteria in Hospital Wastewater Systems. J. Hosp. Infect. 2016, 93, 395–402. [Google Scholar] [CrossRef]
- Kizny Gordon, A.E.; Mathers, A.J.; Cheong, E.Y.L.; Gottlieb, T.; Kotay, S.; Walker, A.S.; Peto, T.E.A.; Crook, D.W.; Stoesser, N. The Hospital Water Environment as a Reservoir for Carbapenem-Resistant Organisms Causing Hospital-Acquired Infections-A Systematic Review of the Literature. Clin. Infect. Dis. 2017, 64, 1435–1444. [Google Scholar] [CrossRef]
- Park, S.C.; Parikh, H.; Vegesana, K.; Stoesser, N.; Barry, K.E.; Kotay, S.M.; Dudley, S.; Peto, T.E.A.; Crook, D.W.; Walker, A.S.; et al. Risk Factors Associated with Carbapenemase-Producing Enterobacterales (CPE) Positivity in the Hospital Wastewater Environment. Appl. Environ. Microbiol. 2020, 86, e01715–e01720. [Google Scholar] [CrossRef]
- MacFaddin, J. Media for isolation-cultivation-identification-maintenance of medical bacteria, Baltimore, Maryland: Williams and Wilkins; 1985, vol. 1.
- Brenner, D.J.; Krieg, N.R.; Staley, J.T.; Garrity, G.M. Bergey’s manual of systematic bacteriology. 2005, 2nd edition, Vol. 2 (The Proteobacteria), part C (The Alpha-, Beta-, Delta-, and Epsilonproteobacteria), Springer, New York, NY.
- Lane, D.J. 16S/23S rRNA sequencing, 1991, p 115–175. In Stackebrandt E, Goodfellow M (ed), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, NewYork, NY.
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-termination inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef]
- Thompson, D.; Gibson, J.; Plewinak, F.; Jeanmougin, F.; Higgins, G. The ClastalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nuc. Acids Res. 1997, 25, 4867–4887. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 38,3022–3027. [Google Scholar] [CrossRef]
- Bhattacharyya, D.K.; Kwon, O.; Meganathan, R. Vitamin K2 (menaquinone) biosynthesis in Escherichia coli: evidence for the presence of an essential histidine residue in o-succinylbenzoyl coenzyme A synthetase. J. Bacteriol. 1997, 179, 6061–6065. [Google Scholar] [CrossRef]
- Gao, Y.D.; Zhao, Y.; Huang, J. 2014. Metabolic modeling of common Escherichia coli strains in human gut microbiome. Biomed Res Int 2014, 694967. [Google Scholar] [CrossRef]
- Delmas, J.; Dalmasso, G.; Bonnet, R. Escherichia coli: the good, the bad and the ugly. Clin. Microbiol.2015, 4:2.
- Cools, P. The role of Escherichia coli in reproductive health: state of the art. Res. Microbiol. 2017, 168, 892–901. [Google Scholar] [CrossRef]
- Christofi, T.; Panayidou, S.; Dieronitou, I.; Michael, C.; Apidianakis, Y. Metabolic output defines Escherichia coli as a health-promoting microbe against intestinal Pseudomonas aeruginosa. Sci Rep 2019, 9, 14463. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Cao, Y.; Guo, Z.F.; Chen, M.; Chen, X.; Guo, Z. Menaquinone biosynthesis in Escherichia coli: identification of 2-succinyl-5-enolpyruvyl-6- hydroxy-3-cyclohexene-1-carboxylate as a novel intermediate and re-evaluation of MenD activity. Biochemistry 2007, 46, 10979–10989. [Google Scholar] [CrossRef] [PubMed]
- Leonardi, R.; Fairhurst, S.A.; Kriek, M.; Lowe, D.J.; Roach, P.L. Thiamine biosynthesis in Escherichia coli: isolation and initial characterization of the ThiGH complex. FEBS Lett. 2003, 539, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.; Wang, H.; Xie, J. Thiamin (vitamin B1) biosynthesis and regulation: a rich source of antimicrobial drug targets? Int. J. Biol. Sci. 2011, 7, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Jacobi, C.A.; Malfertheiner, P. Escherichia coli Nissle 1917 (Mutaflor): new insights into an old probiotic bacterium. Dig Dis. 2011, 29, 600–607. [Google Scholar] [CrossRef]
- Sonnenborn, U. Escherichia coli strain Nissle 1917-from bench to bedside and back: history of a special Escherichia coli strain with probiotic properties. FEMS Microbiol. Lett. 2016, 363, fnw212. [Google Scholar] [CrossRef] [PubMed]
- Beimfohr, C. A review of research conducted with probiotic E. coli marketed as Symbioflor. Int. J. Bacteriol. 2016, 2016, 3535621. [Google Scholar] [CrossRef]
- Escribano-Vazquez, U.; Beimfohr, C.; Bellet, D.; Thomas, M.; Zimmermann, K.; Langella, P.; Cherbuy, C. Symbioflor2® Escherichia coli genotypes enhance ileal and colonic gene expression associated with mucosal defense in gnotobiotic mice. Microorganisms 2020, 8, 512. [Google Scholar] [CrossRef] [PubMed]
- Altenhoefer, A.; Oswald, S.; Sonnenborn, U.; Enders, C.; Schulze, J.; Hacker, J.; Oelschlaeger, T.A. The probiotic Escherichia coli strain Nissle 1917 interferes with invasion of human intestinal epithelial cells by different enteroinvasive bacterial pathogens. FEMS Immunol. Med. Microbiol. 2004, 40, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Westendorf, A.M.; Gunzer, F.; Deppenmeier, S.; Tapadar, D.; Hunger, J.K.; Schmidt, M.A.; Buer, J.; Bruder, D. Intestinal immunity of Escherichia coli NISSLE 1917: a safe carrier for therapeutic molecules. FEMS Immunol. Med. Microbiol. 2005, 43, 373–384. [Google Scholar] [CrossRef]
- Ukena, S.N.; Singh, A.; Dringenberg, U.; Engelhardt, R.; Seidler, U.; Hansen, W.; Bleich, A.; Bruder, D.; Franzke, A.; Rogler, G.; et al. Probiotic Escherichia coli Nissle 1917 inhibits leaky gut by enhancing mucosal integrity. PLoS ONE 2007, 2, e1308. [Google Scholar] [CrossRef]
- Mohsin, M.; Guenther, S.; Schierack, P.; Tedin, K.; Wieler, L.H. Probiotic Escherichia coli Nissle 1917 reduces growth, Shiga toxin expression, release and thus cytotoxicity of enterohemorrhagic Escherichia coli. Int. J. Med. Microbiol. 2015, 305, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Wassenaar, T.M. Insights from 100 years of research with probiotic. Eur. J. Microbiol. Immunol. 2016, 2016. 6, 147–161. [Google Scholar] [CrossRef]
- Zimmer, C.; Dorea, C. Enumeration of Escherichia coli in probiotic products. Microorganisms 2019, 7, 437. [Google Scholar] [CrossRef] [PubMed]
- Rebai, Y.; Wagner, L.; Gnaien, M.; Hammer, M.L.; Kapitan, M.; Niemiec, M.J.; Mami, W.; Mosbah, A.; Messadi, E.; Mardassi, H.; et al. Escherichia coli Nissle 1917 Antagonizes Candida albicans Growth and Protects Intestinal Cells from C. albicans-Mediated Damage. Microorganisms 2023, 11, 1929. [Google Scholar] [CrossRef]
- Zschüttig, A.; Auerbach, C.; Meltke, S.; Eichhorn, C.; Brandt, M.; Blom, J.; Goesmann, A.; Jarek, M.; Scharfe, M.; Zimmermann, K.; et al. Complete sequence of probiotic symbioflor 2 Escherichia coli strain G3/10 and draft sequences of symbioflor 2 E. coli strains G1/2, G4/9, G5, G6/7, and G8. Genome Announc. 2015, 3, e01330-14. [Google Scholar] [CrossRef]
- Miller, T.L.; Wolin, M.J. Fermentation by the human large intestine microbial community in an in vitro semi-continuous culture system. Appl. Environ. Microbiol. 1981, 42, 400–407. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, G.T.; Macfarlane, S. Fermentation in the human large intestine: its physiologic consequences and the potential contribution of prebiotics. J. Clin. Gastroenterol. 2011, 45, S120–S127. [Google Scholar] [CrossRef] [PubMed]
- Abat, C.; Fournier, P.E.; Jimeno, M.T.; Rolain, J.M.; Raoult, D. Extremely and pandrug-resistant bacteria extra-deaths: myth or reality? Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 1687–1697. [Google Scholar] [CrossRef]
- Gajdács, M.; Ábrók, M.; Lázár, A.; Jánvári, L.; Tóth, Á.; Terhes, G.; Burián, K. Detection of VIM, NDM and OXA-48 producing carbapenem resistant Enterobacterales among clinical isolates in Southern Hungary. Acta Microbiol. Immunol. Hung. 2020, 67, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Zarrilli, R.; Bagattini, M.; Esposito, E.P.; Triassi, M. Acinetobacter Infections in Neonates. Curr. Infect. Dis. Rep. 2018, 10, 48. [Google Scholar] [CrossRef]
- Wisplinghoff, H.; Bischoff, T.; Tallent, S.M.; Seifert, H.; Wenzel, R.P.; Edmond, M.B. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 2004, 39, 309–317. [Google Scholar] [CrossRef]
- Montefour, K.; Frieden, J.; Hurst, S.; Helmich, C.; Headley, D.; Martin, M.; Boyle, D.A. Acinetobacter baumannii: an emerging multidrug-resistant pathogen in critical care. Crit. Care Nurse 2008, 28, 15–26. [Google Scholar] [CrossRef]
- Howard, A.; O'Donoghue, M.; Feeney, A.; Sleator, R.D. Acinetobacter baumannii: an emerging opportunistic pathogen. Virulence 2012, 3, 243–250. [Google Scholar] [CrossRef]
- Meatherall, B.L.; Gregson, D.; Ross, T.; Pitout, J.D.; Laupland, K.B. Incidence, risk factors, and outcomes of Klebsiella pneumoniae bacteremia. Am. J. Med. 2009, 122, 866–873. [Google Scholar] [CrossRef]
- Girometti, N.; Lewis, R.E.; Giannella, M.; Ambretti, S.; Bartoletti, M.; Tedeschi, S.; Tumietto, F.; Cristini, F.; Trapani, F.; Gaibani, P.; et al. Klebsiella pneumoniae bloodstream infection: epidemiology and impact of inappropriate empirical therapy. Medicine 2014, 93, 298–309. [Google Scholar] [CrossRef]
- Durdu, B.; Hakyemez, I.N.; Bolukcu, S.; Okay, G.; Gultepe, B.; Aslan, T. Mortality markers in nosocomial Klebsiella pneumoniae bloodstream infection. SpringerPlus 2016, 5, 1892. [Google Scholar] [CrossRef]
- Li, L.; Huang, H. Risk factors of mortality in bloodstream infections caused by Klebsiella pneumonia: A single-center retrospective study in China. Medicine 2017, 96, e7924. [Google Scholar] [CrossRef]
- Tang, L.M.; Chen, S.T. Klebsiella pneumoniae meningitis: prognostic factors. Scand. J. Infect. Dis. 1994, 26, 95–102. [Google Scholar] [CrossRef]
- Ku, Y.H.; Chuang, Y.C.; Chen, C.C.; Lee, M.F.; Yang, Y.C.; Tang, H.J.; Yu, W.L. Klebsiella pneumoniae isolates from meningitis: Epidemiology, virulence and antibiotic resistance. Sci. Rep. 2017, 7, 6634. [Google Scholar] [CrossRef]
- Abbas, R.; Chakkour, M.; Zein El Dine, H.; Obaseki, E.F.; Obeid, S.T.; Jezzini, A.; Ghssein, G.; Ezzeddine, Z. General Overview of Klebsiella pneumonia: Epidemiology and the Role of Siderophores in Its Pathogenicity. Biology 2024, 13, 78. [Google Scholar] [CrossRef]
- Mukherjee, S.; Mitra, S.; Dutta, S.; Basu, S. Neonatal Sepsis: The Impact of Carbapenem-Resistant and Hypervirulent Klebsiella pneumoniae. Front. Med. 2021, 8, 634349. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Bhadury, P.; Mitra, S.; Naha, S.; Saha, B.; Dutta, S.; Basu, S. Hypervirulent Klebsiella pneumoniae Causing Neonatal Bloodstream Infections: Emergence of NDM-1-Producing Hypervirulent ST11-K2 and ST15-K54 Strains Possessing pLVPK-Associated Markers. Microbiol. Spectr. 2023, 11, e0412122. [Google Scholar] [CrossRef]

| Preterm Neonates subject # | Age days |
Gestational age (Weeks) | Types of antibiotics Treated with |
Bacterial strains Colonizing Preterm Neonates Colon | Accession number | |
|---|---|---|---|---|---|---|
| 1 st week | Case 3 | 2 | 31 | Ampicillin 50mg/ivq12h Cefotaxim50mg ivq12/h |
ND | - |
| Case 4 1st time | 5 | 32 | Gentamicin Ampicillin50mg/iv/q12h |
ND | - | |
| Case 5 1st time | 6 | 32 | Gentamicin Ampicillin50mg/iv/q12h |
ND | - | |
| Case 9 | 2 | 30 | Merobenem80mg q12h/iv Cefotaxime 80mg ivq12h |
ND | - | |
| Case 18 | 3 | 33 | Ampicillin 90Mg iv q 12h Gentamicin 8Mg iv q 36h |
ND | - | |
| Case 19 |
5 | 26 | Ampicillin 12Mg iv q 12h CEFOTAXIME --Mg iv q 48h |
ND | - | |
| > 1 week | Case 6 1st time | 8 | 28 | Ampicillin50mg/iv/q12h Cefotaxim50mg/iv/q12h |
ND | - |
| Case 1 | 22 | 27 | Vancomycin10mg/ivq36h | Escherichia coli EFTU1 | PP156718 | |
| Case 2 | 18 | 25 | Ampicillin 50mg/ivq12h Cefotaxim50mg ivq12/h Fucanazol13mg/ivq12 |
Klebsiella pneumoniae EFTU101 |
PP157088 | |
| Case 4 2nd time |
15 | 32 | Stop AB before 5 days |
E. coli EFTU2 |
PP157624 | |
| Case 5 2nd time | 16 | 33 | Gentamicin Ampicillin50mg/iv/q12h |
Klebsiella pneumoniae EFTU102 | PP177257 | |
| Case 6 2nd time | 18 | 28 | Ampicillin50mg/iv/q12h Cefotaxim50mg/iv/q12h |
Klebsiella pneumoniae EFTU103 |
PP177305 | |
| Case 7 | 44 | 26 | Vancomycin 10mg q12h/iv Merobenem20mg q12h/iv |
Klebsiella pneumoniae EFTU104 | PP177307 | |
| Case 8 | 44 | 26 | Vancomycin 10mg q12h/iv Merobenem20mg q12h/iv |
Klebsiella pneumoniae EFTU105 | PP177367 | |
| Case 10 | 44 | 29 | He was taking vancomycin 15mg iv q 12 h and stopping before 18day | Klebsiella pneumoniae EFTU3 | PP157908 | |
| Case 11 | 24 | 31 | No antibiotic | Escherichia coli EFTU4 | PP182268 | |
| Case 13 | 21 | 25 | Merobenem27mg q12h Colitin45mg q12h/iv |
Escherichia coli EFTU5 | PP187030 | |
| Case 14 | 16 | 33 | Gentamicin8mgivq36h Ampicillin76mg/iv/q12h |
Klebsiella pneumoniae EFTU110 | PP177465 | |
| Case 15 |
46 | 27 | Vancomycin 10mg q8h/iv Merobenem20mg q8h/iv |
Escherichia coli EFTU6 | PP187981 | |
| Klebsiella pneumoniae EFTU113 | PP204007 | |||||
| Acinetobacter baumannii EFTU111 | PP177550 |
|||||
| Case 16 | 29 | 29 | Ampicillin 65Mg iv q 12h Gentamicin 65Mg iv q 48h |
Acinetobacter baumannii EFTU114 | PP177613 |
|
| Case 17 | 32 | 29 | Ampicillin 60Mg iv q 12h CEFOTAXIME 60Mg iv q 48h |
Escherichia coli EFTU7 | PP188038 | |
| Klebsiella quasipneumoniae EFTU115 | PP177908 |
|||||
| Case 20 |
64 | 31 | meropenem 50Mg iv q 8h Colistin 50Mg iv q 8h |
Escherichia coli EFTU8 | PP188042 | |
| Klebsiella pneumoniae EFTU116 | PP177909 | |||||
| Case 21 | 14 | 31 | AMPCILLIN 67MGiv q 12h Gentamicin 6,7Mg iv q 36h stopping before six day |
Escherichia coli EFTU9 |
PP188096 |
| Bacterial strains | ESBL | Antibiotic | |||||||||||||||
| AMP | Amox | Tzp | CF | FOX | CAZ | CRO | FEP | ||||||||||
| MIC | Intpr | MIC | Intpr | MIC | Intpr | MIC | Intpr | MIC | Intpr | MIC | Intpr | MIC | Intpr | MIC | Intpr | ||
| Klebsiella pneumoniae EFTU101 | P0S | >=32 | R | 4 | S | <=4 | S | >=64 | R | <=4 | S | 8 | R | >=64 | R | >=64 | R |
| Klebsiella pneumoniae EFTU102 | NEG | >=32 | R | <=2 | S | <=4 | S | <=2 | S | <=4 | S | <=1 | S | <=1 | S | <=1 | S |
| Klebsiella pneumoniae EFTU103 | NEG | >=32 | R | <=2 | S | <=4 | S | <=2 | S | <=4 | S | <=1 | S | <=1 | S | <=1 | S |
| Klebsiella pneumoniae EFTU104 | P0S | >=32 | R | 16 | i | 32 | i | >=64 | R | 16 | R | >=64 | R | >=64 | R | >=64 | R |
| Klebsiella pneumoniae EFTU105 | P0S | >=32 | R | 8 | S | <=4 | S | >=64 | R | <=4 | S | 8 | R | >=64 | R | 2 | R |
| Klebsiella pneumoniae EFTU3 | NEG | >=32 | R | 4 | S | <=4 | S | 16 | I | <=4 | S | <=1 | S | <=1 | S | <=1 | S |
| Klebsiella pneumoniae EFTU110 | NEG | >=32 | R | >=32 | R | ≥128 | R | >=64 | R | >=64 | R | 16 | R | >=64 | R | >=64 | R |
| Acinetobacter baumannii EFTU111 | NM | NM | - | NM | - | <=4 | S | NM | - | NM | - | 4 | S | 16 | R | 2 | S |
| Klebsiella pneumoniae EFTU113 | P0S | >=32 | R | >=32 | R | 3 | R | >=64 | R | <=4 | R | >=64 | R | >=64 | R | >=64 | R |
| Acinetobacter baumannii EFTU114 | NM | NM | - | NM | - | <=4 | S | NM | - | NM | - | 4 | S | 8 | R | 2 | S |
| Klebsiella quasipneumoniae EFTU115 | NM | NM | - | NM | - | <=4 | S | NM | - | NM | - | 4 | S | 8 | R | 2 | S |
| Klebsiella pneumoniae EFTU116 | NEG | >=32 | R | >=32 | R | 8 | S | >=64 | R | >=64 | R | 4 | S | <=1 | S | <=1 | S |
| Bacterial strains | Antibiotic | ||||||||||||||||
| IMI | MERO | AK | GM | CIP | TGC | FT | SXT | ||||||||||
| MIC | Intpr | MIC | Intpr | MIC | Intpr | MIC | Intpr | MIC | Intpr | MIC | Intpr | MIC | Intpr | MIC | Intpr | ||
| Klebsiella pneumoniae EFTU101 | <=0,25 | S | <=0,25 | S | <=2 | S | <=1 | S | 0, 5 | S | 1 | S | 64 | I | 320 | R | |
| Klebsiella pneumoniae EFTU102 | <=0,25 | S | <=0,25 | S | <=2 | S | <=1 | S | <=0, 25 | S | =<0,5 | S | 32 | S | 20 | S | |
| Klebsiella pneumoniae EFTU103 | <=0,25 | S | <=0,25 | S | <=2 | S | <=1 | S | <=0, 25 | S | =<0,5 | S | 32 | S | 20 | S | |
| Klebsiella pneumoniae EFTU104 | <=0,25 | S | <=0,25 | S | <=2 | S | <=1 | S | 1 | S | 1 | S | 128 | R | >=320 | R | |
| Klebsiella pneumoniae EFTU105 | <=0,25 | S | <=0,25 | S | <=2 | S | <=1 | S | 1 | S | 2 | S | 64 | I | >=320 | R | |
| Klebsiella pneumoniae EFTU3 | <=0,25 | S | <=0,25 | S | <=2 | S | <=1 | S | <=0,25 | S | <=0,5 | S | <=16 | S | <=20 | S | |
| Klebsiella pneumoniae EFTU110 | >=16 | R | >=16 | R | ≥64 | R | <=1 | S | >=4 | R | 2 | S | 256 | R | 40 | S | |
| Acinetobacter baumannii EFTU111 | <=0,25 | S | <=0,25 | S | NM | - | <=1 | S | <=0,25 | S | <=0,5 | S | NM | - | <=20 | S | |
| Klebsiella pneumoniae EFTU113 | 2 | I | 2 | I | 4 | S | 2 | S | 2 | R | 2 | S | 128 | R | <=320 | R | |
| Acinetobacter baumannii EFTU114 | <=0,25 | S | <=0,25 | S | NM | - | <=1 | S | <=0,25 | S | <=0,5 | S | NM | - | <=20 | S | |
| Klebsiella quasipneumoniae EFTU115 | <=0,25 | S | <=0,25 | S | NM | - | <=1 | S | <=0,25 | S | <=0,5 | S | NM | - | <=20 | S | |
| Klebsiella pneumoniae EFTU116 | <=0,25 | S | <=0,25 | S | <=2 | S | <=1 | S | 1 | S | 2 | S | 128 | R | 320 | R | |
| Strains | ESBL |
Antibiotics | ||||||||||||||||||||||||||||
| AMP | Amox | Tzp | CF | FOX | CAZ | CRO | FEP | |||||||||||||||||||||||
| MIC | Intpr | MIC | Intpr | MIC | Intpr | MIC | Intpr | MIC | Intpr | MIC | Intpr | MIC | Intpr | MIC | Intpr | |||||||||||||||
| E. coli EFTU 1 | NEG | >=32 | R | 4 | S | <=4 | S | 16 | I | <=4 | S | <=1 | S | <=1 | S | <=1 | S | |||||||||||||
| E. coli EFTU 2 | POS | >=32 | R | 4 | S | <=4 | S | >=64 | R | <=4 | S | 4 | R | <=64 | R | 2 | R | |||||||||||||
| E. coli EFTU 4 | NEG | >=32 | R | 4 | S | <=4 | S | 16 | I | <=4 | S | <=1 | S | <=1 | S | <=1 | S | |||||||||||||
| E. coli ETTU5 | NEG | <=2 | S | <=2 | S | <=4 | S | 4 | S | <=4 | S | <=1 | S | <=1 | S | <=1 | S | |||||||||||||
| E. coli EFTU6 | NEG | >=32 | R | 16 | I | <=4 | S | 16 | I | <=4 | S | <=1 | S | <=1 | S | <=1 | S | |||||||||||||
| E. coli EFTU7 | P0S | >=32 | R | 4 | S | <=4 | S | >=64 | R | <=4 | S | <=1 | R | >=64 | R | <=1 | R | |||||||||||||
| E. coli EFTU8 | P0S | >=32 | R | 4 | S | <=4 | S | >=64 | R | <=4 | S | 16 | R | >=64 | R | 8 | R | |||||||||||||
| E. coli EFTU9 | P0S | >=32 | R | 4 | S | <=4 | S | >=64 | R | <=4 | S | 16 | R | >=64 | R | >=64 | R | |||||||||||||
| Strains | Antibiotics | |||||||||||||||||||||||||||||
| IMI | MERO | AK | GM | CIP | TGC | FT | SXT | |||||||||||||||||||||||
| MIC | Intpr | MIC | Intpr | MIC | Intpr | MIC | Intpr | MIC | Intpr | MIC | Intpr | MIC | Intpr | MIC | Intpr | |||||||||||||||
| E. coli EFTU 1 | <=0,25 | S | <=0,25 | S | <=2 | S | <=1 | S | <=0,25 | S | <=0,5 | S | <=16 | S | <=20 | S | ||||||||||||||
| E. coli EFTU 2 | <=0,25 | S | <=0,25 | S | <=2 | S | <=1 | S | <=0,25 | S | <=0,5 | S | <=16 | S | <=20 | S | ||||||||||||||
| E. coli EFTU 4 | <=0,25 | S | <=0,25 | S | <=2 | S | <=1 | S | <=0,25 | S | <=0,5 | S | <=16 | S | <=20 | S | ||||||||||||||
| E. coli ETTU5 | <=0,25 | S | <=0,25 | S | <=2 | S | <=1 | S | <=0,25 | S | <=0, 5 | S | <=16 | S | <=20 | S | ||||||||||||||
| E. coli EFTU6 | <=0,25 | S | <=0,25 | S | <=2 | S | <=1 | S | <=0,25 | S | <=0,5 | S | <=16 | S | >=320 | R | ||||||||||||||
| E. coli EFTU7 | <=0,25 | S | <=0,25 | S | <=2 | S | <=1 | S | <=0,25 | S | <=0, 5 | S | <=16 | S | <=20 | S | ||||||||||||||
| E. coli EFTU8 | <=0,25 | S | <=0,25 | S | <=2 | S | <=1 | S | 0, 5 | S | <=0, 5 | S | <=16 | S | <=20 | S | ||||||||||||||
| E. coli EFTU9 | <=0,25 | S | <=0,25 | S | <=2 | S | <=1 | S | 0, 5 | S | <=0, 5 | S | <=16 | S | <=20 | S | ||||||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




