1. Introduction
Human immunodeficiency virus type 1 (HIV-1) continues to pose significant challenges worldwide, with an estimated 39.0 million people living with the virus in 2022 [
1]. While HIV-1 treatment has been revolutionized by antiretroviral therapy, many people with HIV still present with a range of cognitive and neurologic symptoms termed HIV-associated neurocognitive disorders (HAND) [
2]. HAND symptoms are generally less severe in people using antiretroviral therapy, but treatment does not fully prevent the more subtle forms of HAND that still disrupt quality of life [
3]. Thus, HAND remains an important and unmet medical need that is increasing in prevalence as people with HIV live longer with the virus [
4,
5].
Previous work has highlighted factors that likely contribute to HAND symptoms, including but not limited to chronic low-level inflammation [
6,
7], continued expression of neurotoxic viral proteins [
8,
9], defective proviruses [
10,
11], and possibly long-term exposure to select ART regimens [
12,
13,
14]. Further, HAND symptoms can be aggravated by substance use [
15], particularly opioids [
16,
17,
18] and stimulants [
19,
20], which is more common in people with HIV [
21,
22]. This creates a complicated picture where HAND may be driven by many factors that interact in nuanced ways in the human brain. Much of our current knowledge of HAND comes from a combination of clinical studies of people with HIV and laboratory studies that use cellular and animal model systems of HIV-1 [
23]. Clinical studies provide valuable insights into the structural and functional changes in the brain associated with HIV-1 infection but lack the ability to study disease mechanisms outside clinical trials. On the contrary, cellular and animal model systems of HIV-1 allow robustly controlled studies of the underlying disease mechanisms associated with HIV-1 infection of the brain, but they do not capture the full complexity of the human brain [
24]. This disconnect could be part of the reason why we currently lack an effective adjuvant therapy to treat HAND [
25,
26].
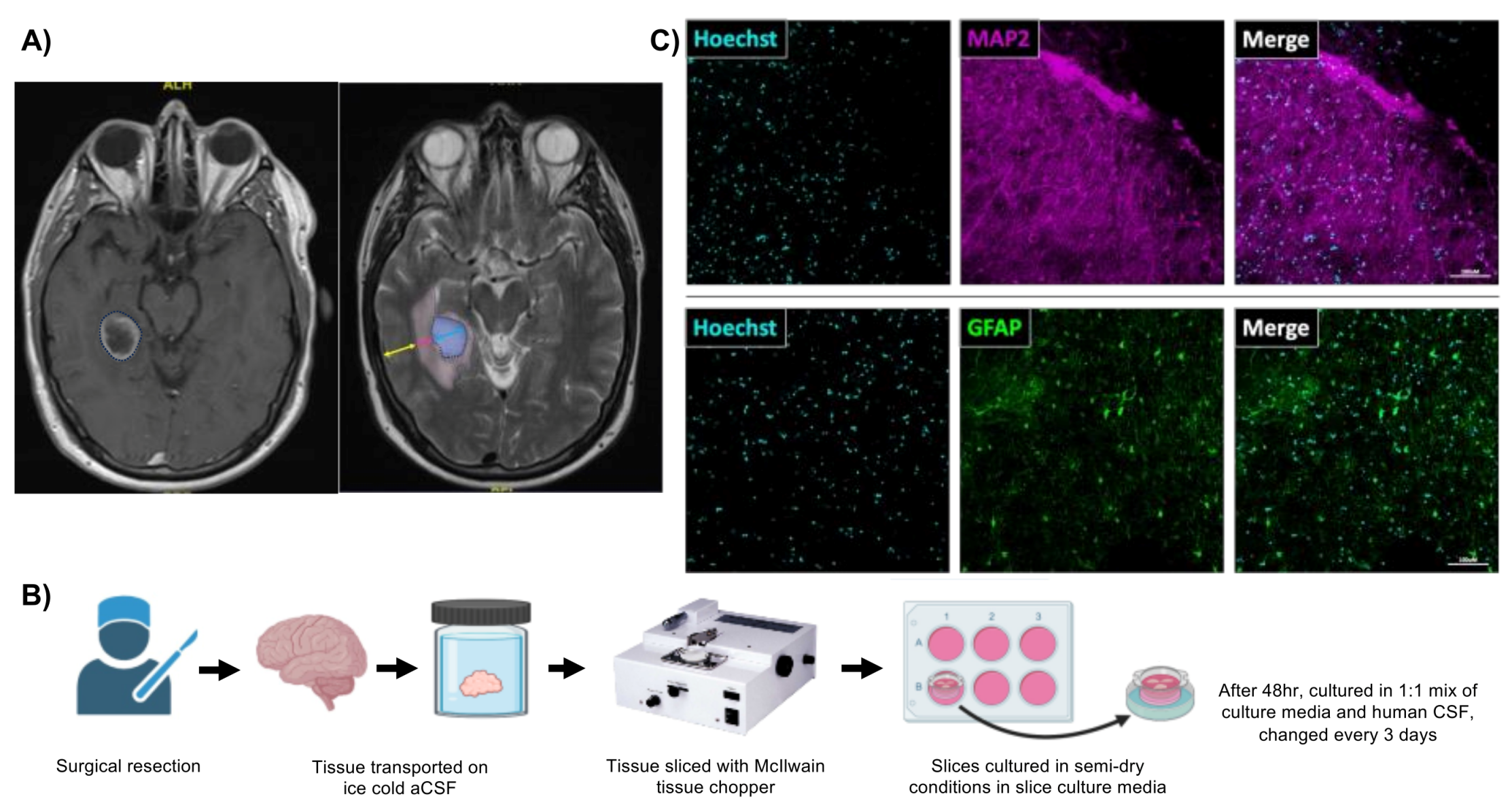
We aimed to address this disconnect by developing an adult human brain slice culture model of HIV-1 infection, which would enable controlled studies of disease mechanisms in a more physiologically relevant context. The model was developed using donated neurosurgical tissue resections and donor-matched peripheral blood from two hospitals in the Greater Philadelphia region. Importantly, our brain slice culture system only used healthy brain tissues from the supratentorial neocortical parenchyma that were removed to access deeper lesions that required surgery. The slice cultures preserved the brain’s architecture and maintained high cellular viability for several weeks, allowing for studies of HIV-1 infection over time in the same brain slice. Our model used a unique HIV-1 infection strategy where slice cultures were infected using the same tissue donor’s peripheral immune cells, thereby reflecting the trojan horse strategy [
27,
28] by which HIV-1 can enter the brain. We collected whole blood at the time of surgery and isolated T-cells and macrophages, infected these cells with a fluorescent HIV-1
in vitro, and seeded these cells directly on the donor-matched slice cultures. This strategy successfully infected cultures with HIV-1 and allowed us to measure viral replication and cellular-level infection via different techniques. Further, antiretroviral therapy strongly suppressed infection in the brain slice cultures, suggesting this system can model HIV-1 brain infection in the modern era. Although still at an early stage of development, this model offers an impactful tool to study the mechanisms of neuronal injury in HIV-1 infected brain, examine the contributions of antiretrovirals and/or HIV-1 comorbidities, monitor infection of resident cells, and test potential neuroprotective agents.
2. Materials and Methods
2.1. Patient Recruitment
All patient participation was voluntary and obtained only after extensive discussion and then signing IRB sanctioned written consent forms. Chain of Custody Tissue Tracking documentation traveled with the sample from the operating room to the laboratory. This study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the IRB committees from both participating hospitals: Crozer-Keystone Health System (IRB number #00006457) and Trinity Health Mid-Atlantic (IRB number #00012775 [active]). Patients that were positive for HIV, SARS-Cov-2, or other pathogens were not considered as potential donors in this study. All tissue samples are accompanied by de-identified demographic information and other relevant medical history.
2.2. Collection of Brain Tissue, Blood, and Cerebrospinal Fluid
Human brain tissue and blood samples were collected during normally scheduled brain surgeries at two participating hospitals. Brain tissues selected for our study consisted exclusively of supratentorial neocortical parenchyma and did not use cerebellar tissue. Patients with cortical lesions were excluded. Although surgeries were performed to remove brain tumors, our studies used brain tissues from the incision path rather than around the tumor to better reflect healthy tissues. Further, the vascularity of all tissue samples was meticulously preserved until the very moment they were removed, and resected brain tissues were submerged in ice-cold artificial cerebrospinal fluid (aCSF) within 10 seconds of resection for transport to the laboratory. Blood samples from each patient were collected into EDTA tubes (BD vacutainer 366643) and stored at 25°C until processing. Both sets of tissues were transported to Drexel University College of Medicine within 30-60 minutes from the collection time. Human cerebrospinal fluid (hCSF) used in this study was sourced from one patient (a case of benign hydrocephalus not recruited for tissue donation), who donated hCSF over two days (processed separately, ie. 24h/each). This hCSF was constantly kept on ice during collection and was then centrifuged at 4000rpm for 10 minutes at 4°C and sterile filtered using a 0.2μm pore size filter. Sterile hCSF was stored in aliquots of 15-50ml at−80°C until use in the slice culture medium.
2.3. Human Brain Slice Culture
All tools used to prepare slice cultures were sterilized using either an autoclave or UV light. All solutions/reagents were prepared before tissues arrived for processing. aCSF cutting solution (280mM sucrose, 5mM KCl, 2mM MgCl2*6H20, 1mM CaCl2, 20mM glucose, 10mM HEPES, 5mM Na-pyruvate, 3mM thiourea, 2mM Na-ascorbate) was prepared and stored in aliquots of 50mL at -20°C for no longer than 1-2 months. Human slice culture media (hSCM) (neurobasal medium, 2% B27 supplement, 1% N2 supplement, 1% L-Glutamine (x100), 0.5% glucose, 1% antibiotic/antimycotic (x100) was prepared fresh the day before surgery and then on a weekly basis.
When brain tissues arrived at the laboratory, we removed capillaries, blood clots, debris, and any visibly damaged tissue. The remaining tissue was divided into several parts (depending on tissue size) while submerged in ice-cold cutting solution. Each part was then sliced into 200μm-thick sections using a tissue chopper (McIlwain tissue chopper 800 series MCT/2), maximizing the amount of gray matter in each slice. Slices were then placed in a dish containing ice-cold aCSF, gently swirled, and carefully separated using needles. Slices with clear margins and no nicks were transferred to another dish filled with ice-cold aCSF and then rinsed in hSCM. Slices were then transferred into culture membrane inserts (0.4μM, PTFE, hydrophilic membrane inserts Millipore #PICM03050), placed in a 6-well plate, and kept at 37°C and 5% CO2. This produces a semi-dry culture that maintains slices at the air/media interface with the hSCM in the bottom of the well. Up to five slices were placed in each insert, ensuring that slices did not overlap. The slice cultures were maintained in hSCM for the first two days. After 48 hours, the hSCM was replaced with a 1:1 mixture of hSCM and hCSF. The media was changed every other day, and the slices were maintained in culture up to four weeks.
2.4. Tissue Clearing and Staining
Organotypic human brain slice cultures were cleared using a modification of the EZClear protocol [
29]. Slices were fixed in situ with 4% paraformaldehyde for 30 minutes above and below the membrane support. After fixation, slices were rinsed 3x for 1 hour in phosphate-buffered saline (PBS) with gentle shaking at 25°C. Slices were then removed from the membrane supports with a scalpel and transferred to a glass liquid scintillation vial. Tissue lipids were removed by incubation with 50% (v/v) tetrahydrofuran (THF) (with 250 ppm butylated hydroxytoluene (BHT), Millipore-Sigma, 186562) prepared in sterile Milli-Q H
2O for 16 hours. Slices were then rinsed with sterile PBS 4x for 1 hour each at 25°C to remove any trace of THF. Next, slices were moved to individual wells of a 24 well tissue culture dish for immunostaining as free-floating slices. Slices were stained with chicken anti-GFAP (Abcam Cat#ab4674, RRID:AB_304558, 1:500), or rabbit anti-MAP2 (Millipore Cat#ab5622, RRID:AB_91939, 1:500) in a PBS solution with 0.4% Triton X-100 and 5% goat serum. Slices were then stained with appropriate secondary antibodies, including goat anti-chicken Alexa Fluor 488 (Thermo Fisher Scientific Cat# A-11039, RRID:AB_2534096, 1:400) or goat anti-rabbit Alexa Fluor 546 (Thermo Fisher Scientific Cat# A-11035, RRID:AB_2534093, 1:400). Slices were incubated for up to 1 week in either secondary or primary antibodies. After antibody staining, slices were counterstained overnight with Hoechst 33342 solution (BD Pharmingen Cat#561908, 10µg/mL) and transferred to glass slides. We formed wells around each slice using a hydrophobic spacer (SecureSeal, ThermoFisher Cat# S24737) to ensure slices were not deformed during mounting. Each well received a Refractive Index matching solution (RI; EZView (80% Nycodenz (Accurate Chemical & Scientific 100334-594), 7M urea, 0.05% sodium azide prepared in 0.02M sodium phosphate buffer)) and the slices equilibrated for 24 hours prior to imaging.
2.5. Cell Culture
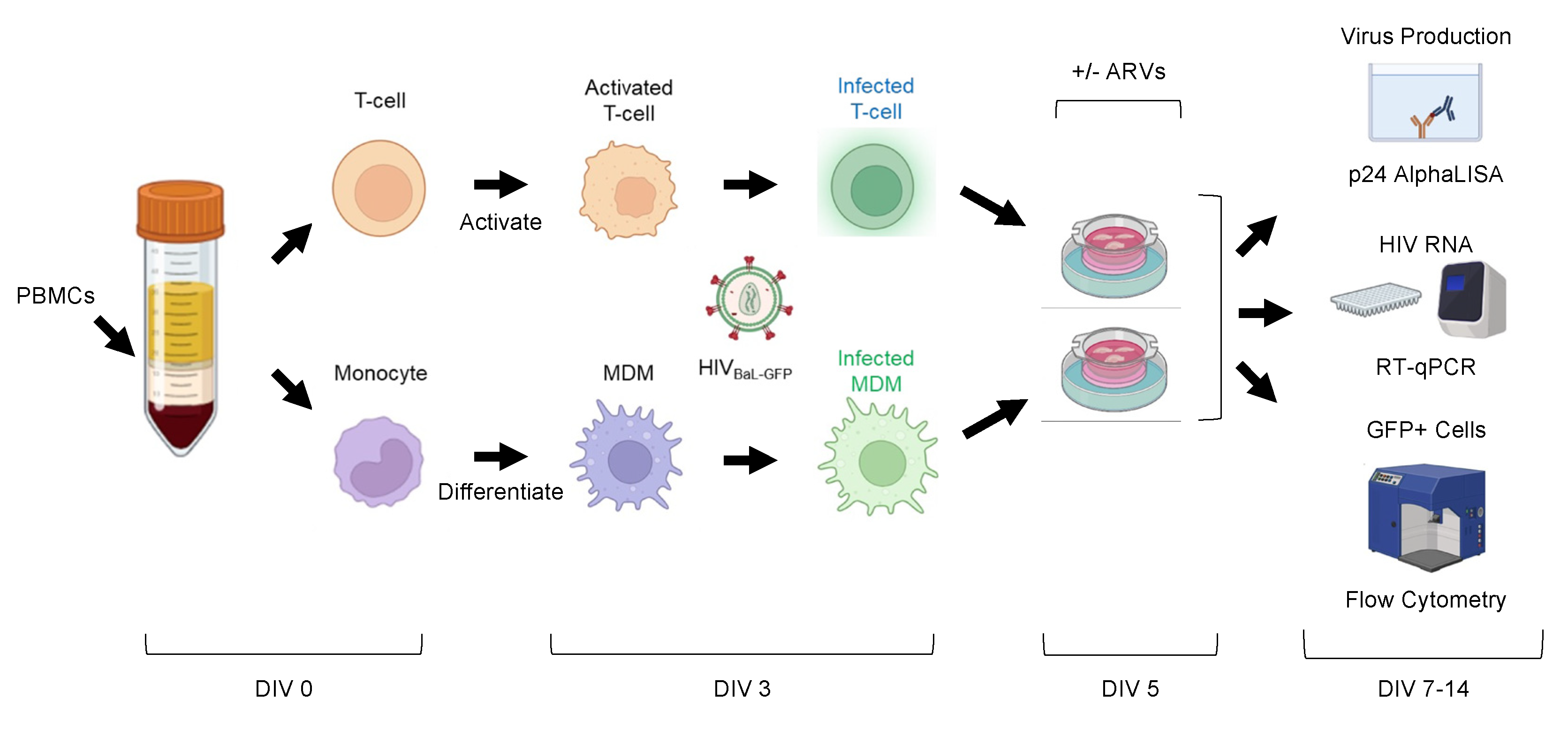
HEK293T cells (ATCC #CRL-3216) were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Corning #15-013-CV) supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (Gibco #16140071) and 1x penicillin/streptomycin/L-glutamine (PSG) (ThermoFisher Scientific #10-378-016) at 37°C and 5% CO2. Purified CD4+ T-cells were cultured in RPMI-1640 (Roswell Park Memorial Institute, Corning #15040CV) supplemented with 20% (vol/vol) fetal bovine serum, 1x PSG, and 5U/mL human rIL-2 (NIH AIDS Reagent Program), RPMI-20/IL-2. CD4+ T-cell isolates were stimulated with 5μg/mL phytohemagglutinin-P (PHA-P) (Sigma #L1668) for 48-72 hours at 37°C and 5% CO2 and subsequently cultured in RPMI-20/IL-2. Monocyte-derived macrophages (MDMs) were differentiated and cultured in a macrophage media consisting of DMEM supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum, 1x PSG, 5% (vol/vol) human AB serum (Gemini Bio-Products #100-512), 10mM HEPES (N-2-hydroxyethylpiperazine-N’-2-ethanesulphonic acid) (Corning #25-060-Cl), and 10ng/mL macrophage-colony stimulating factor (M-CSF) (Peprotech # 300-25) at 37°C and 5% CO2. MDMs were allowed to attach and differentiate by plastic adherence for 48-72 hours, then unbound cells were washed away with macrophage wash media consisting of DMEM with 10mM HEPES.
2.6. PBMC Isolation, CD4+ T-Cell Purification, and MDM Differentiation
Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood samples using Ficoll (Fisher Scientific #45-001-751) density centrifugation (1.078g/mL). Briefly, whole blood was spun at 1500rpm for 5 minutes at 25°C. Plasma was collected and stored in 1mL aliquots at -80C. Plasma volume was replaced with PBS without Ca2+/Mg2+ and diluted blood (15mL) was overlaid onto (13mL) Ficoll. Samples were spun at 300x g for 30 minutes at 25°C with no braking. PBMCs were collected from the interphase and washed with 2x volumes of PBS without Ca2+/Mg2+. PBMCs were spun at 1200rpm for 5 minutes at 4°C and counted. 1x106 PBMCs were reserved for characterization by flow cytometry. The remainder of the PBMCs were used to isolate CD4+ T-cells.
CD4+ T-cells were purified from PBMCs using MACS positive isolation beads (Miltenyi #130-045-101) according to manufacturer’s recommendations. We also collected the negatively selected flow-through (CD4-depleted PBMCs), which contained monocytes. Less than 1x106 cells were reserved from each population to determine isolate purity by flow cytometry. CD4+ T-cell isolates were cultured for 48-72 hours at 1x106 cells/well in RPMI-20/IL-2 media + PHA-P as indicated above.
CD4-depleted PBMCs were assumed to contain approximately 15% monocytes and were plated in macrophage media such that 1x106 monocytes were plated per well of a 6-well plate and allowed to attach via plastic adherence. 48-72hr post-plating, CD4+ T-cells were washed, counted, and replated in RPMI-20/IL-2 at approximately 1x106 cells/well. At the same time, unbound CD4-depleted PBMCs were removed, attached MDMs were washed with macrophage wash media, and fresh macrophage media was added. Cells were then either uninfected or infected with HIV-1 as indicated.
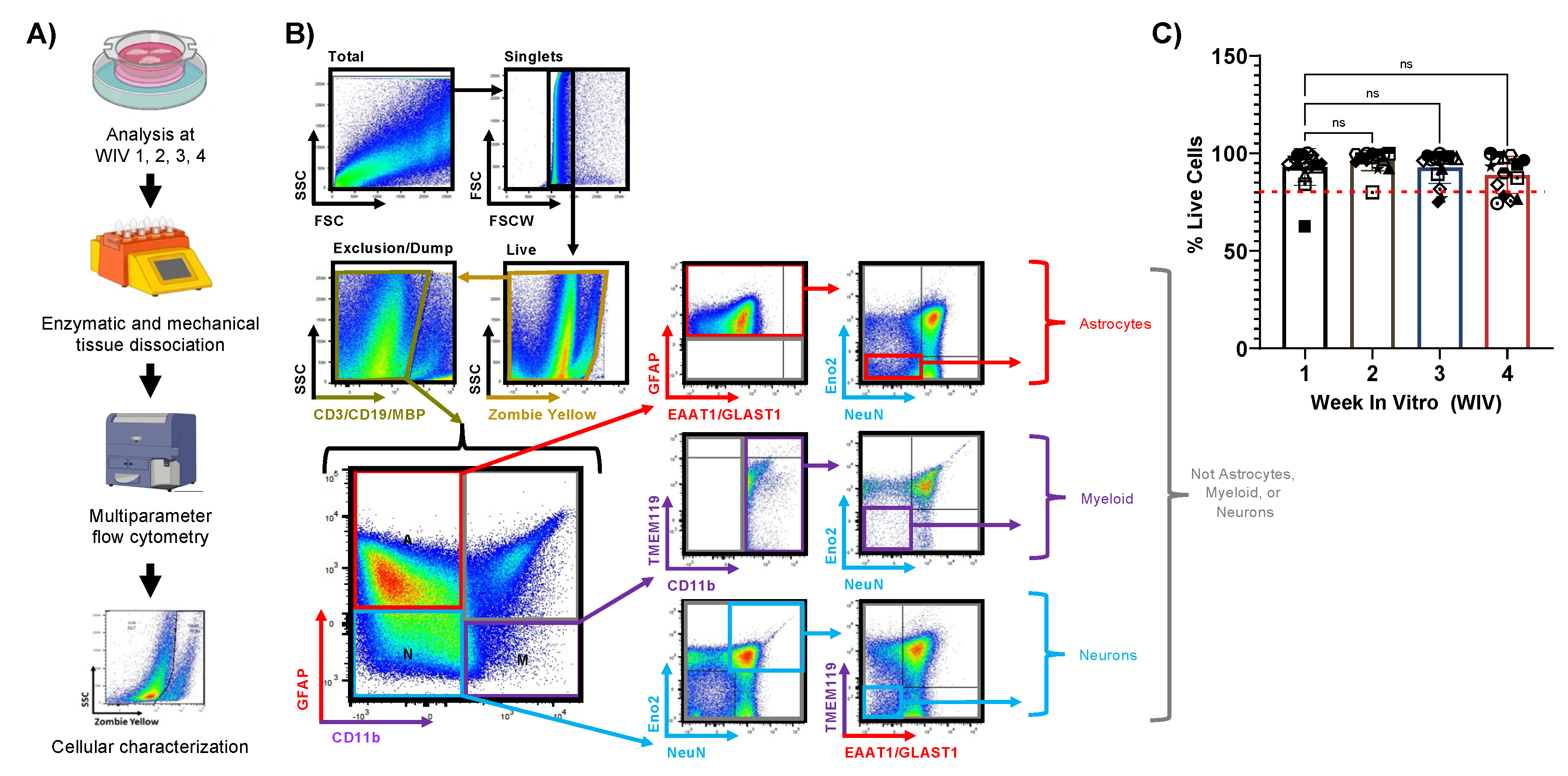
2.7. Mechanical and Enzymatic Dissociation of Tissue Slices
At the indicated time points in culture or post-infection, tissue slices were collected from the culture insert using a wet paintbrush and were processed using the Adult Brain Dissociation Kit, mouse and rat (Miltenyi #130-107-677) on the gentleMACS Octo Dissociator with Heaters (Miltenyi #130-096-427) according to manufacturer’s recommendations. Volumes were scaled down to accommodate small tissue. Though the above dissociation kit is intended for mouse/rat brain, we have used it successfully for human and mouse with no discernable differences. Briefly, tissues were incubated with two enzymes and were mechanically dissociated using the gentleMACS program “37C_ABDK_02” for 30 minutes. Cells were then washed in PBS with Ca2+/Mg2+, filtered using a 70μm filter, and pelleted. Cells were subjected to density gradient centrifugation to remove debris and myelin contamination. Lastly, residual red blood cells were lysed, cells were washed, and samples were either fixed in 1% formaldehyde or stained for flow cytometry.
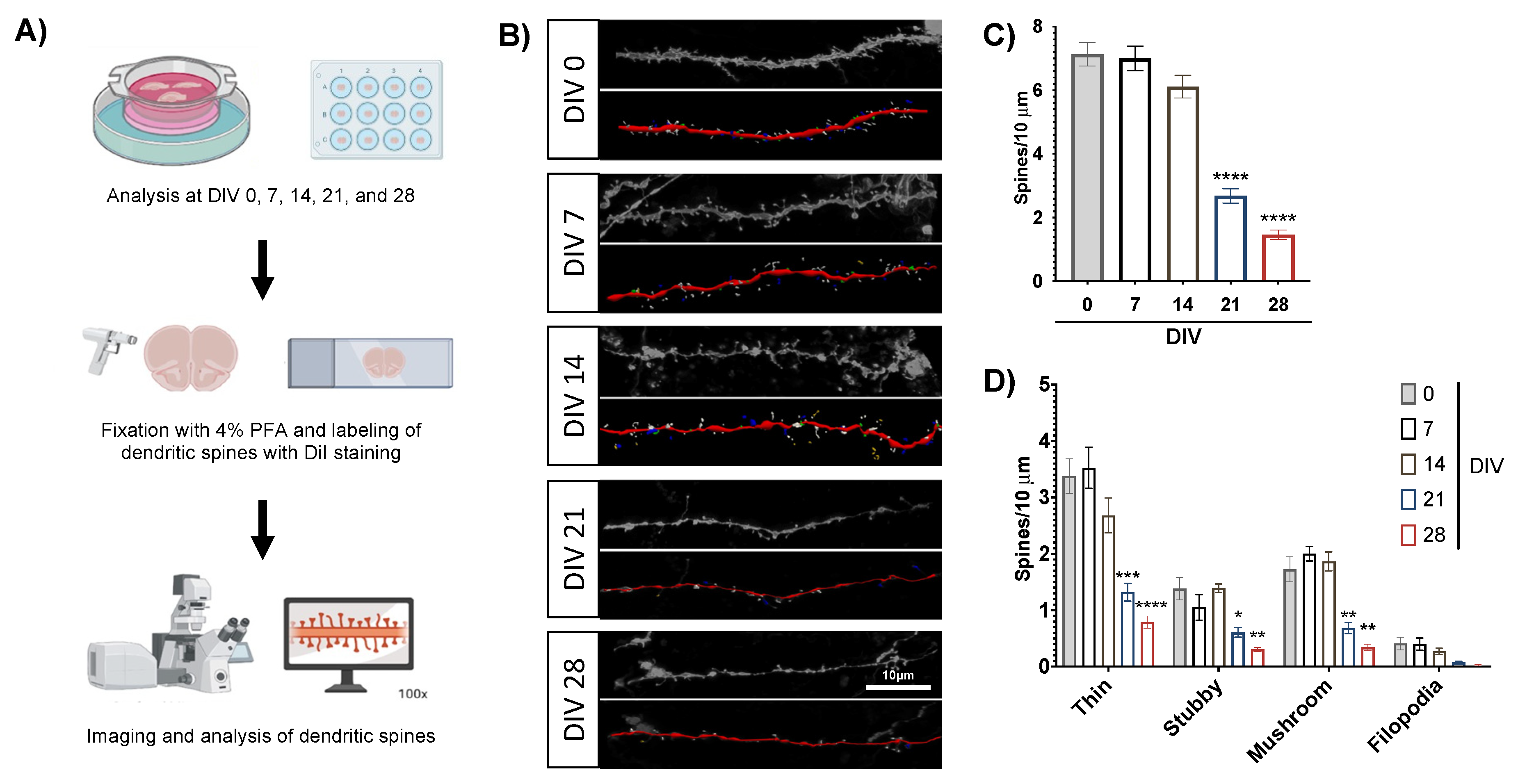
2.8. Dendritic Spine Staining and Analysis
Slice cultures used for dendritic spine analysis were sectioned at 200μm thickness and cultured for up to 4 weeks. Select cultures were fixed at days in vitro (DIV) 0, 7, 14, 21, and 28 with 4% paraformaldehyde for 10 minutes at 25°C, then rinsed 3x in PBS and stored at 4°C until beginning the staining protocol. We labeled neurons and dendritic spines using DiI stain, which was prepared as previously described [
30]. To begin, 300mg of tungsten beads (Bio-Rad 1652269) were suspended in 99.5% pure methylene chloride (Fisher Scientific D37) and sonicated in a water bath for 1 hour. Then we created the DiI solution by dissolving crystalized DiI (14.5mg, Invitrogen D282) in methylene chloride under protection from light. Tungsten beads were coated with DiI by first placing 100μl of the bead solution on a glass slide and adding 100μl of DiI solution on top with slow mixing via micropipette. The dried bead/dye mixture was scraped onto weighing paper with a razor blade, placed into a 15mL conical tube with 3mL distilled and deionized water, and sonicated in a water bath for 20 minutes. Then the mixture was drawn into Tezfel tubing coated with polyvinylpyrrolidone (Fisher Scientific BP431-100) and dried with nitrogen gas for 1 hour. Dry tubing was cut into 13mm cartridges and loaded into the Helios Gene Gun (Bio-Rad), which delivered DiI-coated beads to slices through a 3μm pore filter paper using pressurized helium gas (120 PSI). After delivery, slices were quickly washed 3x with PBS and stored overnight at 4°C to allow DiI to diffuse through the tissue. Stained slices were mounted the next day using ProLong Gold Antifade Mountant (Invitrogen P36930) and stored in the dark at 4°C until imaging.
Dendrites from human brain slices were imaged with an Olympus FLUOVIEW FV3000 confocal microscope using a 100X objective at 0.3μm per Z-step. We imaged 3-4 human brain slices at each experimental timepoint and analyzed at least 3 dendrites per slice to average as single data point. Analyzed dendrites were at least 100µm long and included both basal and apical dendrites. Dendrite micrographs were analyzed using Neurolucida 360 software, which quantified dendritic spines and classified them into predefined morphologies (thin, mushroom, stubby, filopodia) [
31].
2.9. Multielectrode Array (MEA) Electrophysiology
All solutions were oxygenated with carbogen (95% oxygen and 5% carbon dioxide) for at least 30 minutes before use with brain tissue. Immediately upon arrival to the laboratory, a small piece of brain tissue was removed for acute MEA recording. This tissue was washed in an oxygenated sucrose cutting solution composed of 150mM sucrose, 40mM NaCl, 4mM KCl, 1.25mM NaH2PO4 · H2O, 0.5mM CaCl2 · 2H2O, 7mM MgCl2*6H2O, 10mM glucose, and 26mM NaHCO3. We created a flat surface on the tissue with a razor and applied cyanoacrylate-based superglue (Gorilla Glue, Cincinnati, OH) to adhere the flat surface to a chilled vibratome stage (PELCO easiSlicer, Pelco, Redding, CA). Additional sucrose cutting solution was used to submerge the brain tissue, which was then sliced into 300μm sections that were transferred to a holding chamber with ice-cold, oxygenated artificial cerebrospinal fluid (aCSF) containing 125mM NaCl, 3.5mM KCl, 1.2mM NaH2PO4*H2O, 2.4mM CaCl2*2H2O, 1.3mM MgCl2*6H2O, 25mM glucose, and 26mM NaHCO3. The holding chamber with slices was then placed in a 35°C water bath and attached to carbogen to be continuously oxygenated.
Individual slices were transferred from the holding chamber and placed on a perforated multielectrode array (MEA) (Multi Channel Systems MCS GmbH), followed by suction of 0.1mL/minute using a peristaltic pump (Multi Channel Systems). The MEA was continuously perfused with fresh aCSF buffer using a gravity perfusion system (VC-6-pinch, Warner Instruments) at 35oC with a heated perfusion cannula (Multi Channel Systems). Recordings were made using Multi Channel Experimenter (2.0, Multi Channel Systems) at 25kHz. Treatments included a depolarizing version of the above aCSF with increased potassium chloride and no magnesium chloride (NaCl 125mM, KCl 10mM, NaH2PO4*H2O 1.2mM, CaCl2 *2H2O 2.4mM, glucose 25mM, and NaHCO3 26mM), and regular aCSF with a bolus dose of tetrodotoxin (TTX, 10nM). Each brain slice was habituated in the MEA and recorded for 30 minutes during normal aCSF perfusion (baseline), 30 minutes during perfusion with the depolarizing aCSF intended to elicit activity, and a final 30 minutes after a bolus dose of TTX at a final concentration of 10nM and washout/perfusion with regular aCSF. The last 20 minutes of each treatment period were analyzed using Multi Channel Analyzer v2.0 (Multi Channel Systems) to allow the slices 10 minutes to stabilize and respond to treatments. Spikes were defined as drops in voltage at least 5 standard deviations below the average for each electrode.
2.10. Cloning and Plasmids
pBR-NL43-IRES-eGFP-nef+ (pBR43IeG) is a proviral vector containing a HIV molecular clone that expresses HIV-1 Nef and eGFP from a single bicistronic RNA and expresses a wildtype X4-tropic NL4-3 Env (obtained from the NIH AIDS Reagent Program #11349, Accession #M19921, contributed by Jan Münch, Michael Schindler, and Frank Kirchhoff [
32,
33]). p81A-4 is a full-length, HIV-1 infectious NL4-3 based molecular clone which expresses the V1-V3 regions of BaL Env (Obtained from the NIH AIDS Reagent Program #11440, contributed by Bruse Chesebro [
34,
35,
36,
37]). We generated the R5-tropic, BaL Env expressing reporter virus BR43IeG-BaL (hereafter referred to as HIV
BaL-GFP) by cloning the
AgeI-BsaBI fragment of p81A-4 containing the BaL Env sequence into the
AgeI-BsaBI cut backbone of pBR43IeG (
Figure S1). Plasmid DNA was large-scale purified using MidiPrep Kits (Qiagen #12143) and verified by sequencing (GeneWiz).
2.11. Preparation of Virus Stocks
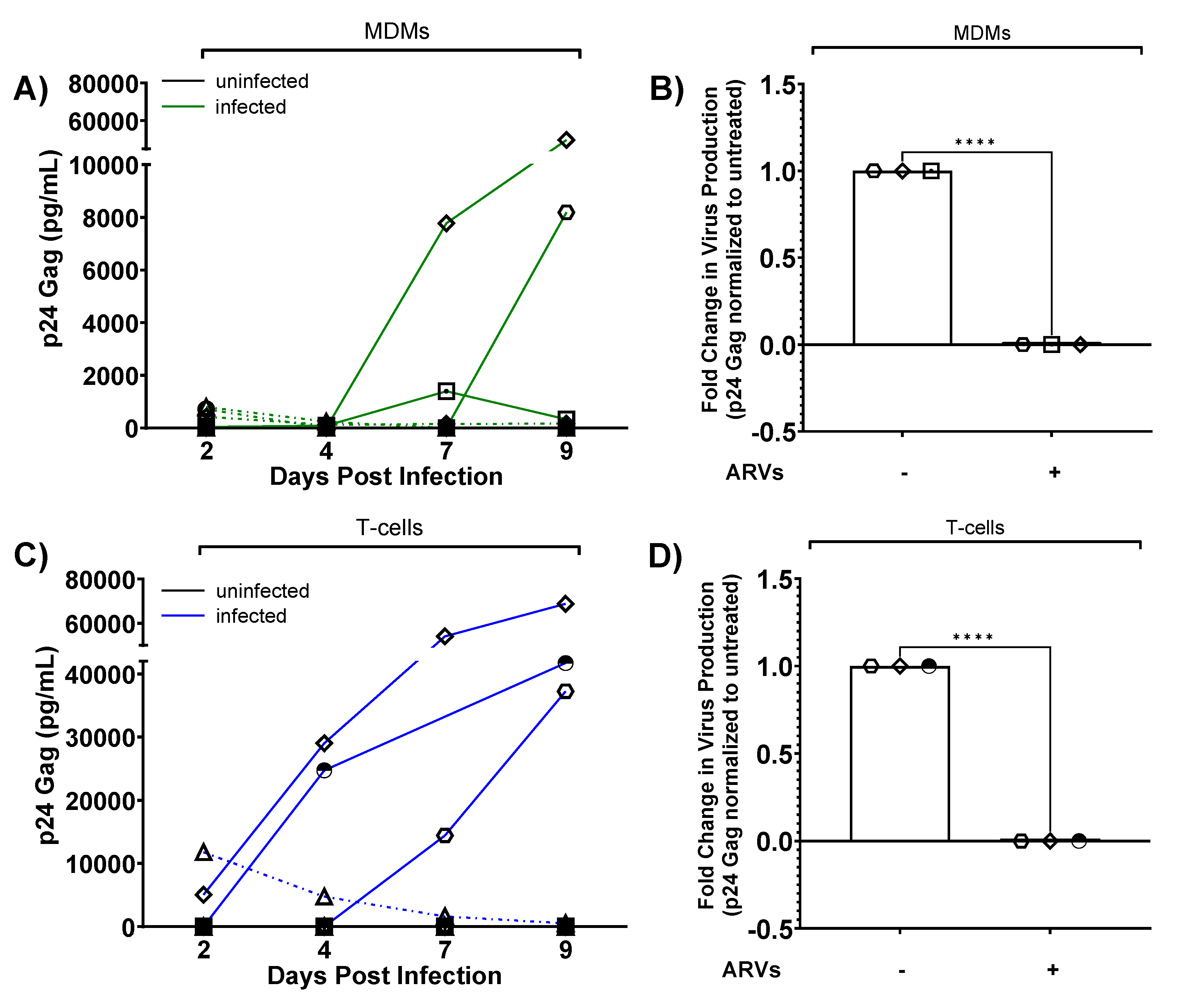
HEK293T cells were transfected with HIV-1 proviral DNA using Lipofectamine 3000 (Invitrogen #L3000001) according to manufacturer’s recommendations. Virus-containing supernatants were collected at 48 and 72 hours post-transfection, filtered through a 0.45μm filter, and concentrated 10x by volume using Retro-Concentin reagent (System Biosciences #RV100A-1) according to manufacturer’s recommendations. Concentrated stocks were aliquoted and frozen at -80°C. Virus stocks were quantified by measuring p24 Gag protein by p24 alphaLISA (Revvity #AL291C) and were titered by infecting MDMs from healthy donors at two-fold serial dilutions and determining percent GFP+ cells by flow cytometry.
2.12. Infection of Producer Cells and Inoculation of Tissue Slices
At 48-72 hours post-activation and differentiation, approximately 1x106 CD4+ T-cells or MDMs, respectively, were infected with 0.1μg or 7.2μg p24 HIVBaL-GFP virus stock, with the higher concentration being equivalent to the volume of virus that infected 10-25% of MDMs (data not shown). At 48 hours post-infection, uninfected and infected CD4+ T-cells were collected, washed, and counted, and we reserved approximately 0.1x106 cells for flow cytometry. Uninfected and infected MDMs were washed and removed from the plate by incubation with cell dissociation buffer [PBS without Ca2+/Mg2+ + 2mM EDTA (ethylenediaminetetraacetic acid, Fisher Scientific # BP2482100)] at 37°C for 30 minutes, allowing us to count and reserve approximately 0.1x106 cells for flow cytometry. T-cells and MDMs were resuspended in human slice culture media such that slices were inoculated with 10μl of producer cells each, overlaid on top of the slice. Slices were distributed in wells as follows: 4x slices per well, per condition to determine GFP+ cells by flow cytometry (1x slice was processed per timepoint); 1x slice per well to quantify p24 and extract RNA.
Where indicated, slices were cultured in the presence of antiretrovirals (ARVs) added to the culture media at the time of cell-associated virus inoculation. We used combination ARVs that comprise the first line therapy Biktarvy [
38]: 33nM bictegravir (BIC) (MedChemExpress #HY-17605), 350nM emtricitabine (FTC) (NIH AIDS Reagent Program #10071), and 5.4μM tenofovir alafenamide (TAF) (MedChemExpress #HY-15232B). These concentrations represent the 4xEC
95, which inhibit viral replication without toxicity [
39,
40]. ARVs were replenished in the slice culture media every two to three days, corresponding to collection days.
2.13. Quantification of Spreading Infection by p24 alphaLISA
p24 gag was examined from slice culture media over time using p24 AlphaLISA according to manufacturer’s recommendations. Culture media was collected and replenished every two to three days post-infection for nine days total, and frozen until the end of the experiment. If interpolated p24 values fell below the standard curve, we assigned a value of “0.3,” corresponding to the lowest concentration of the standard curve. Inoculum-matched, uninfected background signal was subtracted from infected values per timepoint, per case. If this resulted in a value less than “0,” then we assigned “0”. Where indicated, values from infected slices cultured in the presence of ARVs were normalized to untreated slices. However, if both values were “0.3,” they were removed from the analysis. Values were excluded if they did not have a matched, uninfected control or if the viability performed in parallel indicated less than 80% viable cells.
2.14. RNA Extraction, cDNA Synthesis, and qPCR from Tissue Slices
Viral RNA from tissue slices was measured at day 9 post-infection. Slices were collected using a wet paintbrush and immediately lysed in Buffer RLT (Qiagen RNeasy Kit #74106) containing 10μl/mL β-mercaptoethanol (Sigma #M3143) with vigorous vortexing. Lysates were either frozen at -80°C or processed immediately according to manufacturer’s recommendations. Purified RNA was quantified by NanoDrop, normalized, and used to generate cDNA (Invitrogen SuperScript III First-Strand Synthesis System #18080051). cDNA was diluted 1:5 and 5μl was used as template for qPCR in triplicate with SYBR Green Master Mix (ThermoFisher Scientific #4385612) and the following primers:
HIV-1 Gag (0.8μM final): 5′ GGTGCGAGAGCGTCAGTATTAAG 3′
5′ AGCTCCCTGCTTGCCCATA 3′
GAPDH (0.4μM final): 5′ GCTCACTGGCATGGCCTTCCGTGT 3′
5′ TGGAGGAGTGGGTGTCGCTGTTGA 3′
Samples were run on QuantStudio 7 real-time qPCR (Applied Biosystems). If cycle numbers returned as “Undetermined,” we assigned a value of “40,” corresponding to the highest cycle number. ΔCt values were calculated from averages of technical triplicates of Ct values (Gag-GAPDH). ΔΔCt values were calculated as 2- (ΔCt infected – ΔCt uninfected). Where indicated, ΔCt values from infected slices cultured in the presence of ARVs were normalized to untreated slices. Values were excluded if the infected values were less than the uninfected control or if the viability performed in parallel indicated less than 80% viable cells.
2.15. Flow Cytometry
All cells were stained for viability using the live/dead stain, Zombie Yellow (Biolegend # 423103), according to manufacturer’s recommendations, unless otherwise indicated (i.e., unstained). Briefly, cells were incubated with Zombie Yellow (1:1000) in PBS without Ca2+/Mg2+ for 10 min in the dark at 4°C. The stain was diluted with PBS without Ca2+/Mg2+, cells spun at 1200rpm for 5 min at 4°C and either fixed in 1% formaldehyde (Sigma 50-185-4042) or continued to be stained with extracellular and/or intracellular antibodies.
For PBMC characterization, cells were stained with antibodies against extracellular markers prepared in PBS without Ca2+/Mg2+: CD3 (AF700, BD Biosciences Cat# 557917, RRID:AB_396938), CD4 (PerCP-Cy 5.5, BD Biosciences Cat# 552838, RRID:AB_394488), CD8 (BV786, BD Biosciences Cat# 563823, RRID:AB_2687487), CD14 (BV711, BD Biosciences Cat# 563372 (also 563373), RRID:AB_2744290), CD19 (AF647, Novus Biologicals #NBP2-61908AF647), and CD56 (PE-CF594, BD Biosciences Cat# 562328, RRID:AB_11153852) for 20 minutes in the dark at 4°C. The antibodies and cells were diluted with PBS without Ca2+/Mg2+, spun at 1200rpm for 5 minutes at 4°C and fixed in 1% formaldehyde. Data are calculated as the % of Singlets and Live cells.
For isolate purity, CD4+ T-cells and CD4-depleted PBMCs were stained with Zombie Yellow and antibodies against extracellular markers: CD3 (AF700, BD #557917) and CD4 (PerCP-Cy 5.5, BD #552838) as above. Cells were fixed in 1% formaldehyde. Data were calculated as the % of Singlets, Live, CD3+, CD4+ cells.
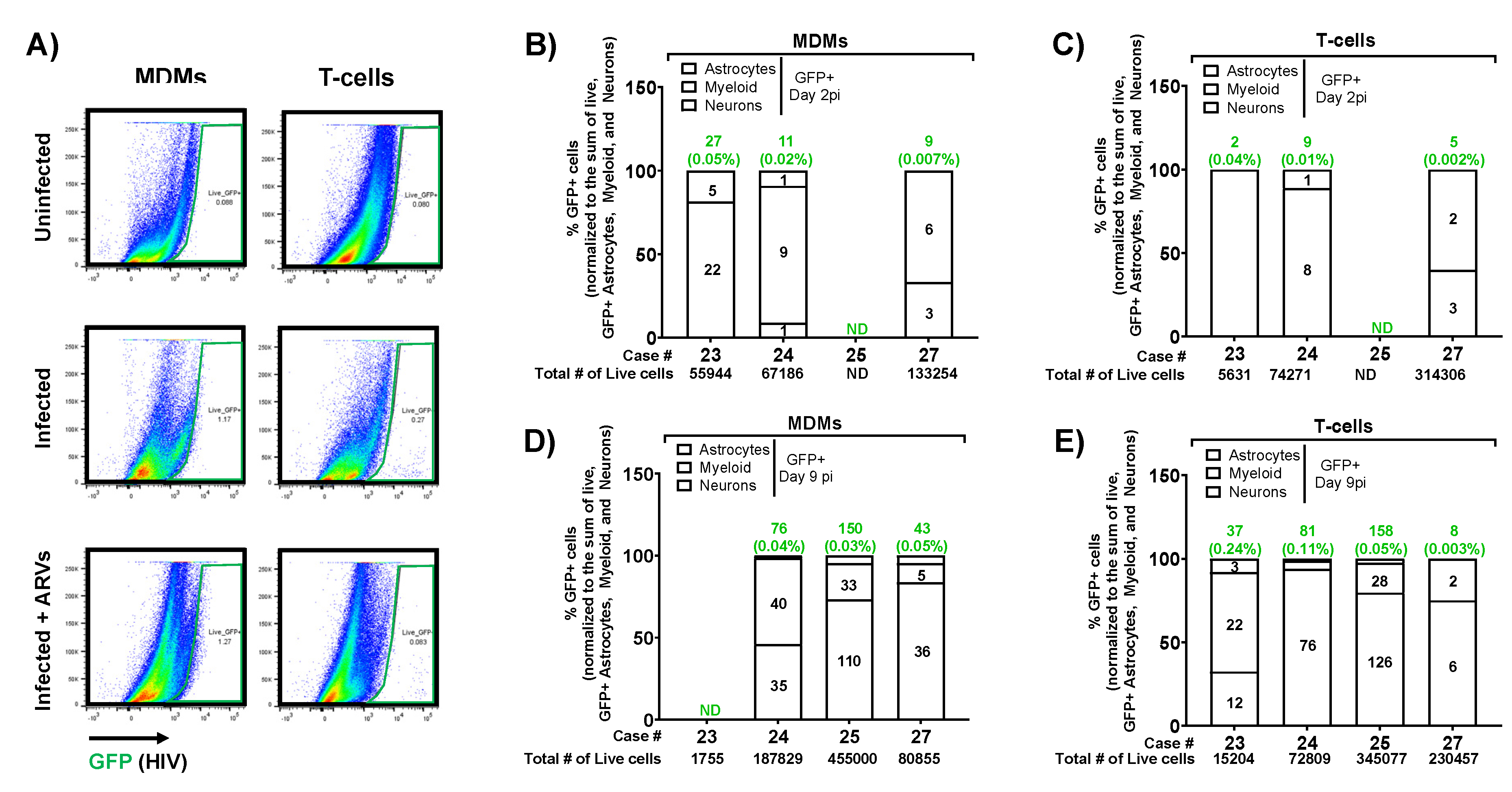
For determination of infected producer cells, CD4+ T-cells and/or MDMs were stained with Zombie Yellow as above and fixed in 1% formaldehyde. Data are calculated as the % of Singlets, Live, GFP+ cells gated based on uninfected controls. Signals from uninfected samples were subtracted from infected samples.
For viability determination of brain tissue at indicated timepoints, dissociates were stained with Zombie Yellow and antibodies against extracellular markers: CD45 (APC-H7, BD Biosciences Cat# 560274, RRID:AB_1645480), EAAT1/GLAST1 (AF405, Novus Biologicals #NB100-1869AF405), CD11b (BV711, BD Biosciences Cat# 568229, RRID:AB_2916857), TMEM119 (AF647, Novus Biologicals #FAB10313R-100ug), CD3 (AF700, BD Biosciences Cat# 557917, RRID:AB_396938), CD19 (AF700, BD Biosciences Cat# 561031, RRID:AB_10562552), and MBP (AF700, Novus Biologicals #NBP2-22121AF700) as above. Cells were then fixed and permeabilized using the Cytofix/Cytoperm kit (BD #554714) according to manufacturer’s recommendations. Briefly, cells were incubated in Fix/Perm buffer for 20 minutes in the dark at 4°C. Cells were then washed 3x with 1x Perm/Wash buffer and were stained with antibodies against intracellular antibodies prepared in 1x Perm/Wash buffer: Histone H3 (FITC, Novus Biologicals #NB500-171F) or Ki67 (FITC, BD Biosciences Cat# 556026, RRID:AB_396302), NeuN (PE, Novus Cat# NBP1-92693PE, RRID:AB_11036146), GFAP (PerCP, Novus Biologicals #NBP2-34413PCP), Eno2 (PE-ATTO594, Novus Biologicals #NBP2-54452PEATT594), and β-III-Tubulin (PE/Cy7, BioLegend Cat# 801217 (also 801218), RRID:AB_2876747) for 20 minutes in the dark at 4°C. Cells were washed twice in 1x Perm/Wash buffer and fixed in 1% formaldehyde.
To measure infection of brain tissue at indicated days post-inoculation, dissociates were stained with Zombie Yellow and antibodies against extracellular and intracellular markers as for the viability determination with the following exceptions: GFP+ cells were measured instead of Histone H3 or Ki67 and cells were stained for HIV-1 Gag (PE, HIV Reagent Program #13449) instead of NeuN.
Cells were collected and characterized by the Cytek FACSort DxP12 flow cytometer and data were analyzed using FlowJo v10.8.1 software.
2.16. Gating Strategy and Population Calculations
Populations were gated based on unstained controls collected in parallel. The total live population is defined as Zombie Yellow negative, singlets. The “dump” or “exclusion” channel eliminates CD3+ T-cells, CD19+ B-cells, and MBP+ Oligodendrocytes from our analysis. Astrocytes are defined as the population of cells that are GFAP+/CD11b-/Eno2-/NeuN- and CD3-/CD19-/MBP-. Myeloid cells are defined as the population of cells that are GFAP-/CD11b+/Eno2-/NeuN- and CD3-/CD19-/MBP-. Neurons are defined as the population of cells that are NeuN+/Eno2+/GFAP-/CD11b-/TMEM119-/EAAT1/GLAST1- and CD3-/CD19-/MBP-. The population of cells that are not astrocytes, myeloid cells or neurons were defined using Boolean “not gates” for each of the cell-type specific populations as well as all other undefined populations (GFAP+/CD11b+, GFAP+/Eno2+, GFAP+/NeuN+, GFAP+/Eno2+/NeuN+, CD11b+/Eno2+, CD11b+/NeuN+, CD11b+/Eno2+/NeuN+, GFAP-/CD11b-/Eno2-/NeuN-/TMEM119+, GFAP-/CD11b-/Eno2-/NeuN-/EAAT1/GLAST1+, and GFAP-/CD11b-/Eno2-/NeuN-/TMEM119+/GLAST1+).
The gating strategy to determine infectivity of subcellular populations from infected, dissociated brain tissue is based on
Figure 2B except for neurons being defined as the population of cells that are Eno2+/TUBB3+/GFAP-/CD11b-/TMEM119-/EAAT1/GLAST1-. Populations were initially gated based on unstained controls collected in parallel per timepoint. GFP+ cells were then determined based on gates set on the inoculum matched uninfected controls set to < 0.1% positive. Normalized GFP+ cells per case per timepoint per inoculum are calculated as the number of events within the GFP+ subpopulation divided by the sum of GFP+ astrocytes, myeloid cells, and neurons * 100.
2.17. Experimental Design and Statistical Analysis
This study used brain tissues from human donors of both sexes, which were pooled together in final analyses. Donor tissues were excluded from downstream analyses if we measured less than 80% total viable cells in flow cytometry studies. Biological replicates are values from different donors, while technical replicates are values from slice cultures made from the same donor tissue. Reported N values represent biological replicates (displayed on graphs as unique symbols) except for multi-electrode array experiments that used individual slices as N values due to the lower number of donor tissues examined. Figure legends report the exact N for each experiment and the statistical analyses used for each experiment. Data were compiled and analyzed using GraphPad PRISM 10.1.2. One case and one condition from
Figure 7A were identified as outliers by the ROUT method and excluded from the final analysis. Experiments comparing means of two independent groups were analyzed using an unpaired two-tailed t-test, while experiments comparing means of three or more groups were analyzed using one-way ANOVA with Dunnett’s post hoc. Experiments with two independent variables were analyzed using two-way ANOVA with Tukey’s post hoc. Multi-electrode array studies were analyzed by repeated measures one-way ANOVA with Tukey’s post hoc.
4. Discussion
This study is the first to report a tractable model of HIV-1 brain infection based entirely on healthy, intact adult human brain tissues in organotypic slice culture. This new model is a significant advance in the field, as it has the potential to overcome long-standing issues with animal models that lack the structure and complexity of the human brain and to better identify therapeutic approaches for HAND in the preclinical stage. The model is made possible by tissue donors undergoing brain surgeries, which provide us radiographically normal, healthy tissues that are distant from the targeted surgical lesion. Importantly, resected brain tissues are immediately preserved in cold aCSF and transferred to the laboratory, thus maximally preserving tissue structure and health. The brain slice cultures are viable for several weeks, they contain all the major CNS cell types and preserve their biological complexity, and they demonstrate both structural and functional aspects of healthy neuronal networks. We also provide one of the first reports of a highly stringent human CNS-specific flow cytometry panel to characterize cells from human brain tissue. Most importantly, we can infect brain slice cultures with a GFP-expressing HIV-1 using same tissue donor’s peripheral immune cells as trojan horses, thereby reflecting how HIV-1 likely infects the brain in vivo. This approach produces a spreading infection that can be measured by multiple techniques (including within individual cells), and the infection is fully suppressed by current first-line antiretroviral therapy. Overall, this adult, human brain-based model of neuroHIV is an important step forward that will facilitate future studies on mechanistic drivers of HAND/neuroHIV, drug target discovery, and preclinical treatment efficacy in human tissues.
The brain slice culture protocol we employed was informed by work from another group that optimized and characterized human brain slice cultures using similar surgical resections from epilepsy patients. This group substantially improved culture viability by adding hCSF to the culture media [
48] and robustly characterized neuronal health and activity via electrophysiology and other approaches [
41], lending confidence that optimized human brain slice cultures indeed preserve the complexity and function of human cortical tissues. We also included hCSF in our culture media and achieved high cellular viability and neuronal dendritic spine density over several weeks, further validating the human brain slice culture system. Notably, our study only used hCSF from a single donor, which eliminated this aspect as a source of variability. Our system could also detect local field potentials from these cultures using multi-electrode arrays, in line with another recent methods paper [
42]. Together, these data suggest that human brain resections from the incision path of a surgery produce healthy, functional, long-term slice cultures regardless of the underlying patient pathology (epilepsy or a deep brain tumor).
Since our tissue donors present with pathologies that require different surgical paths and must meet selection criteria, one limitation of our system is that we receive heterogeneous cortical areas to use for slice cultures. However, as studies proceed and our sample size increases, these heterogeneous tissues may turn into an advantage as the outcomes from various brain regions could reveal important region-specific mechanistic differences that may also be relevant to HAND or other CNS pathologies. Future neuroHIV studies in this model should include cultures with detectable supernatant HIV p24 and tissue associated viral RNA, as this strongly verifies the infection. In our study, three of the four slice cultures that produced detectable supernatant p24 were from the frontal cortex, suggesting a possible brain region susceptibility to infection ex vivo. However, cultures treated with antiretrovirals had suppressed viral RNA regardless of brain region or supernatant p24 levels, suggesting we achieved at least a low-level infection in all cases.
Additionally, pooled data from our slice culture system is well equipped to detect common physiological and disease mechanisms that are preserved throughout the brain, which could translate to more broadly relevant CNS drug targets. Importantly, our system only used healthy cortical tissue that was distant from deeper pathologies and radiographically normal, which limits confounds that may be present in pathology-adjacent tissues. Retrospective analyses of frozen tissue from these specimens showed negligible to no infiltration of inflammatory cells (data not shown), further suggesting our cultures lack this major confounding variable.
Our work to validate and adapt adult human brain slice cultures to model HIV-1 infection is a critical contribution to preclinical HIV research. Though valuable, most preclinical studies of HIV-1 brain infection currently rely on either small animal models that only express a subset of viral proteins, humanized mice that don’t fully reconstitute human CNS cells and architecture, or SIV-infected macaques, which don’t fully reflect HIV-associated neurocognitive pathologies [
49,
50,
51,
52]. Some HIV-1 studies take advantage of brain organoids made of human CNS cells [
53,
54,
55], but these systems fail to recapitulate the structure of the human brain and lack its cellular diversity [
56]. Further, they require addition of microglia from an external source or derive microglia from induced pluripotent stem cells [
57], which is an important caveat since microglia remain as HIV reservoirs in people taking antiretroviral therapies [
58]. Other groups have studied postmortem brain tissues from people with HIV, and although these retrospective studies provided important insights, they have so far mostly examined tissues from people with AIDS or HIV encephalitis that are less relevant to the modern era [
59]. Modern neuroimaging approaches overcome some of these limitations [
60], but they are not yet sensitive enough to measure subtle structural changes in neurons that likely contribute to impairment, for example dendritic spine density [
23]. To date, the field has lacked a physiological model of HIV-1 infection in live human CNS tissue, with the exception of one study that created a co-culture system using an HIV-infected cell line and human fetal brain slice cultures [
61]. However, our system is much more relevant to the modern pathology, as we use adult human brain tissues, we infect the tissues with the same donor’s primary immune cells instead of a cell line, we detect HIV infection in the brain tissue itself, and we can suppress the infection using antiretrovirals. No other experimental systems in the field provide the same level of experimental opportunities while directly modeling the presumed route of brain infection and the overall complex structure of the human brain. Therefore, our slice culture model of neuroHIV is a very promising approach with strong potential to move the field forward.
A few other groups have published adult human brain slice culture models of viral infections, though they each have important caveats. For example, one group published a free-floating slice culture model to study Oropouche virus infection with human brain surgical resections [
62], but their approach only maintained culture viability for a few days, therefore limiting the model’s utility. Another group recently published an approach to culture postmortem adult human cortical tissue slices as a model to study Tahyna virus infection [
63]. Though this system allows investigators to choose the cortical region they wish to culture and study, the downside is the postmortem interval before collecting tissue ranged between 2-12 hours. This long interval of little to no oxygenation likely changes the physiological nature of the brain [
64], which may confound the results and reduce the benefits of culturing slices from similar cortical areas. Our slice culture system overcomes these downsides, as we immediately preserve live surgical resections in cold aCSF, we slice and culture all tissues within 2-3 hours of resection, and the cultures remain highly viable for weeks using an optimized protocol. Therefore, our system is more likely to model the normal state of the brain and will allow longer-term investigations of viral infection and progression in healthy tissue, which will likely be important to accurately model disease mechanisms and discover relevant drug targets. In the future, we expect adult human brain slice cultures to be further adapted and developed to study a variety of pathologies and brain infections [
65].
To conclude, our human brain slice model of HIV-1 infection is a significant step towards improving the experimental models available for preclinical neuroHIV studies as well as our understanding of the molecular and cellular mechanisms that drive disease in human tissues. The model also provides an unmatched approach for interventional studies in human tissues, which would typically only occur during clinical trials. Future work using this system is poised to overcome the long-standing limitations of animal models used in neuroHIV research and possibly discover a much-needed adjuvant strategy to treat modern forms of HAND that present despite viral suppression by antiretroviral therapies.
Author Contributions
Conceptualization, O.M., Z.A.K., A.S., J.G.J.; Methodology, E.I., R.V.D., J.G.J. J.A.J., A.M.G., B.N., A.L., E.V.O., L.M. A.S.; Software / Validation: N/A; Formal Analysis, E.I., R.V.D., J.A.J., B.N.; Investigation, E.I., R.V.D., J.A.J.; Resources, O.M., Z.A.K, A.S., J.G.J.; Data Curation, E.I., R.V.D., J.A.J., A.M.G., A.L., E.V.O., L.M., B.N.; Writing – Original Draft Preparation, R.V.D., E.I., Z.A.K., O.M., B.N.; Writing – Review & Editing, O.M, B.N., Z.A.K.; Visualization: all authors; Supervision, O.M., Z.A.K., J.G.J.; Project Administration, O.M. Z.A.K, A.S.; Funding Acquisition, O.M., Z.A.K., J.G.J.