Submitted:
12 April 2024
Posted:
15 April 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Material and Methods
2.1. Fusarium Strains
2.2. Plant Materials and Growth
2.3. Plant Inoculation
2.4. Assessment of Symptoms
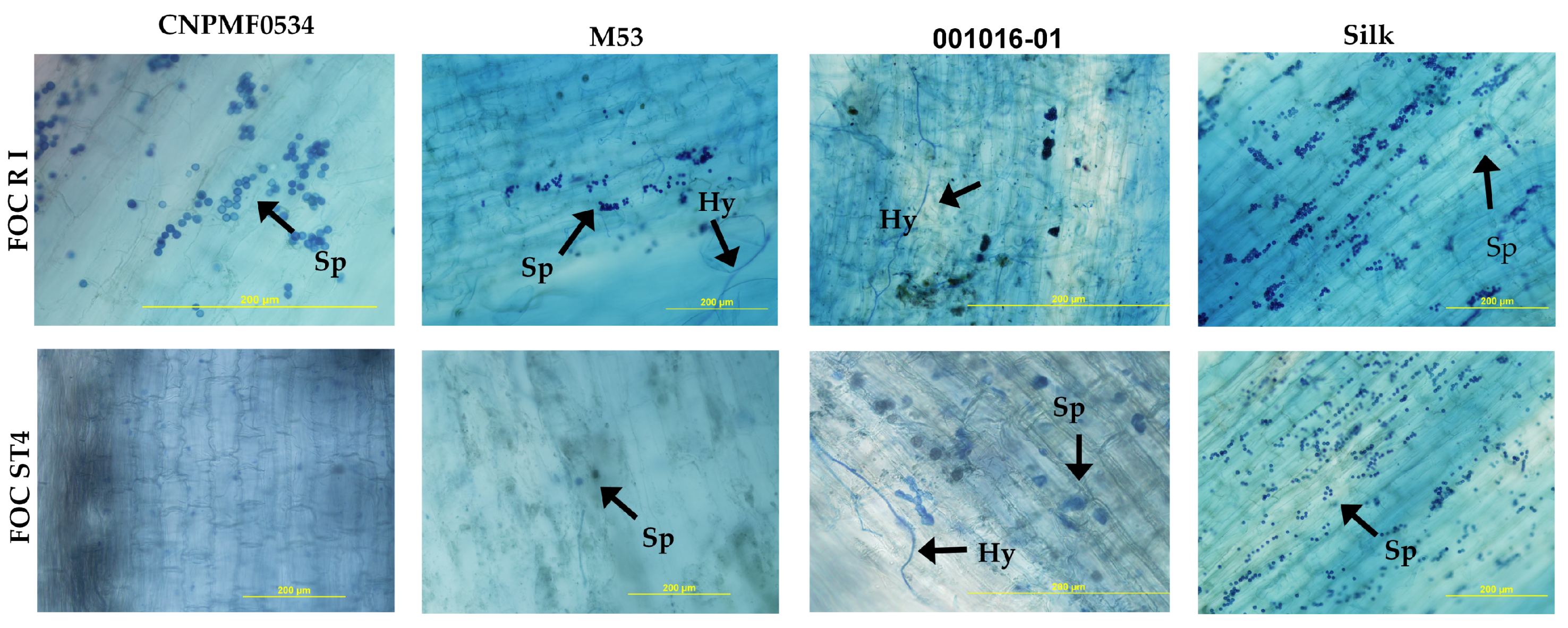
2.5. Histological and Histochemical Analysis
2.5.1. Whitening and Staining of Fungal Structures in Roots
2.5.2. Histochemical Analysis
2.5.3. Scanning Electron Microscopy
3. Results
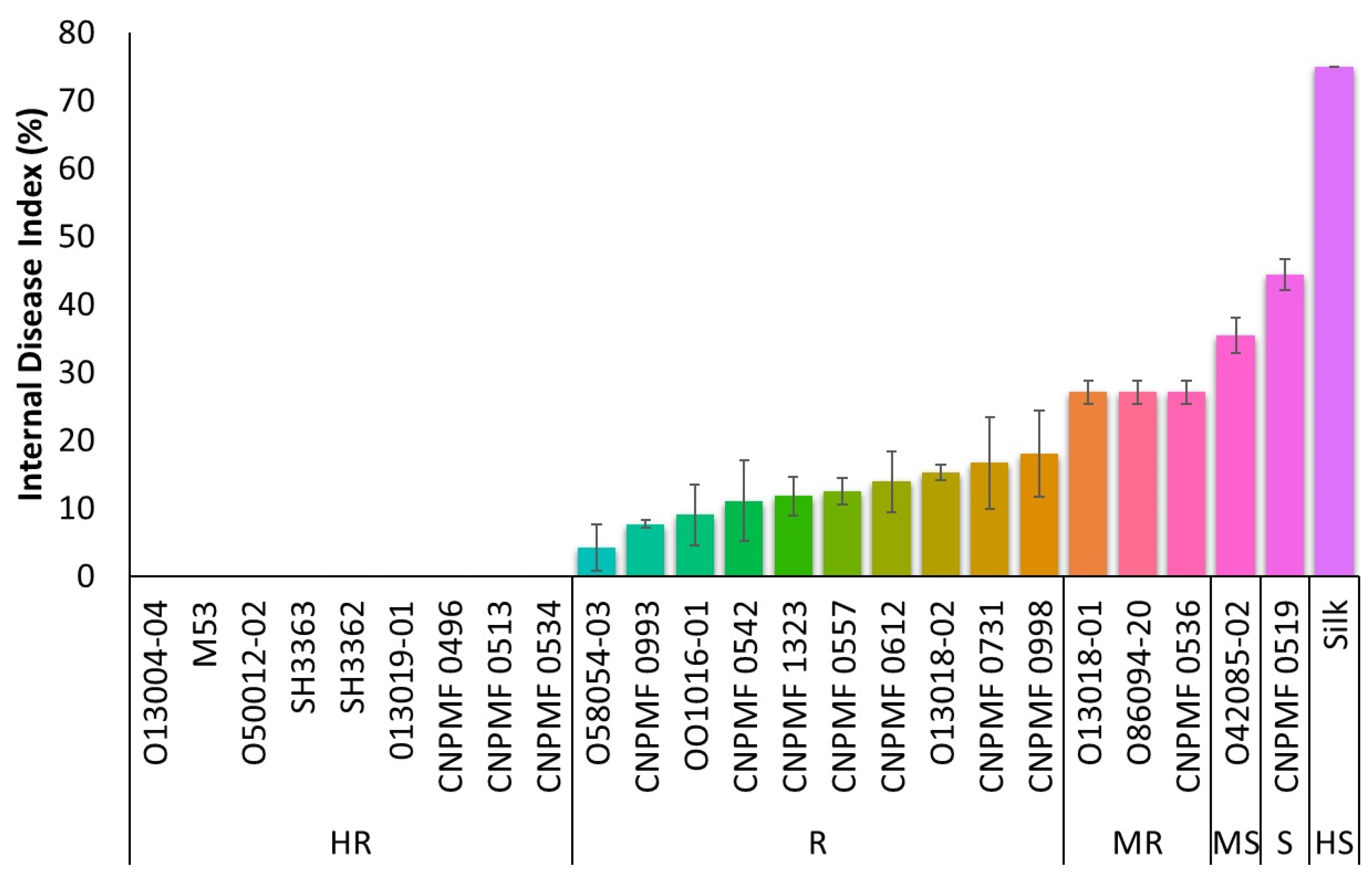
3.1. Screening for Resistance to Foc R1
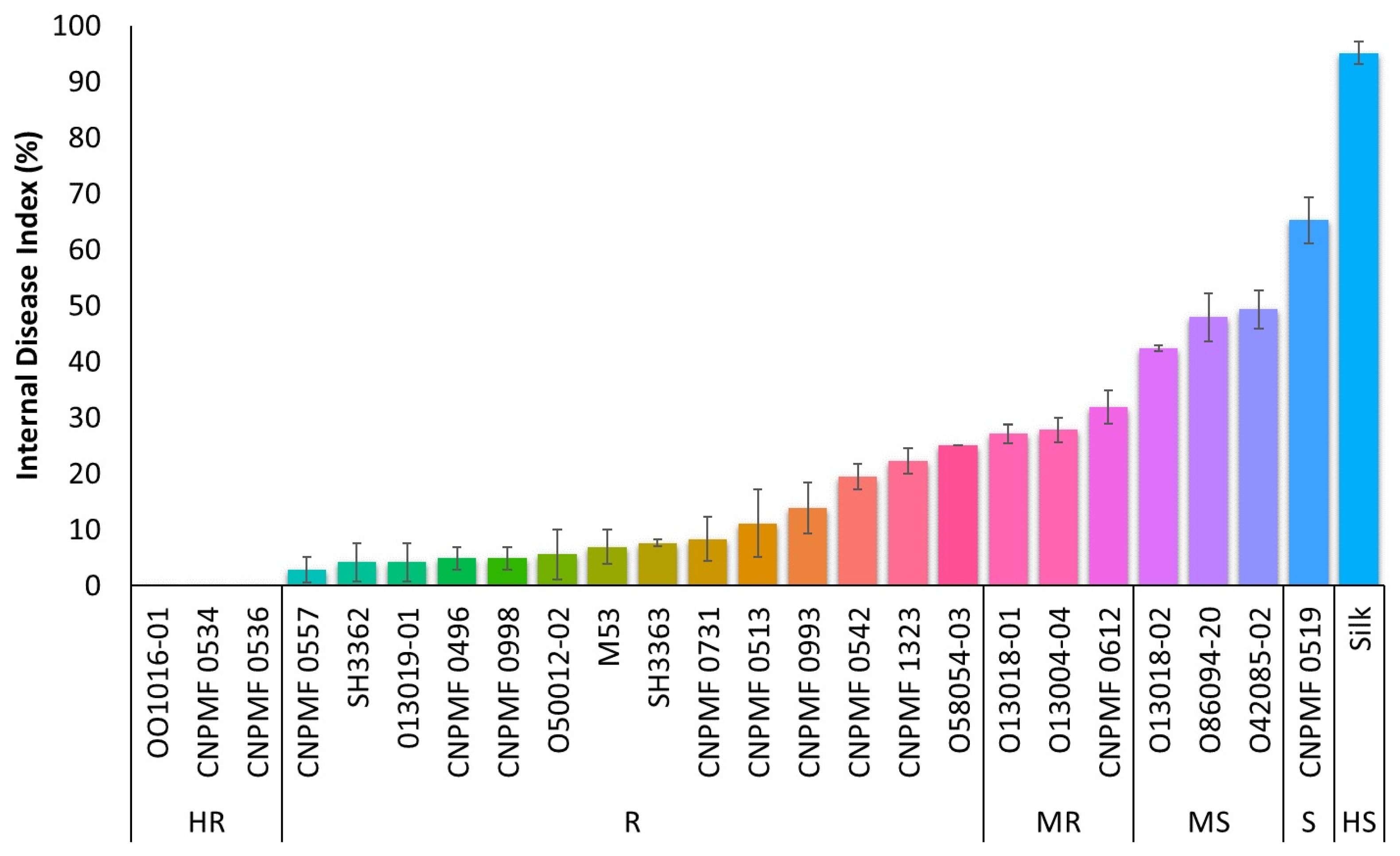
3.2. Screening for Resistance to Foc ST4
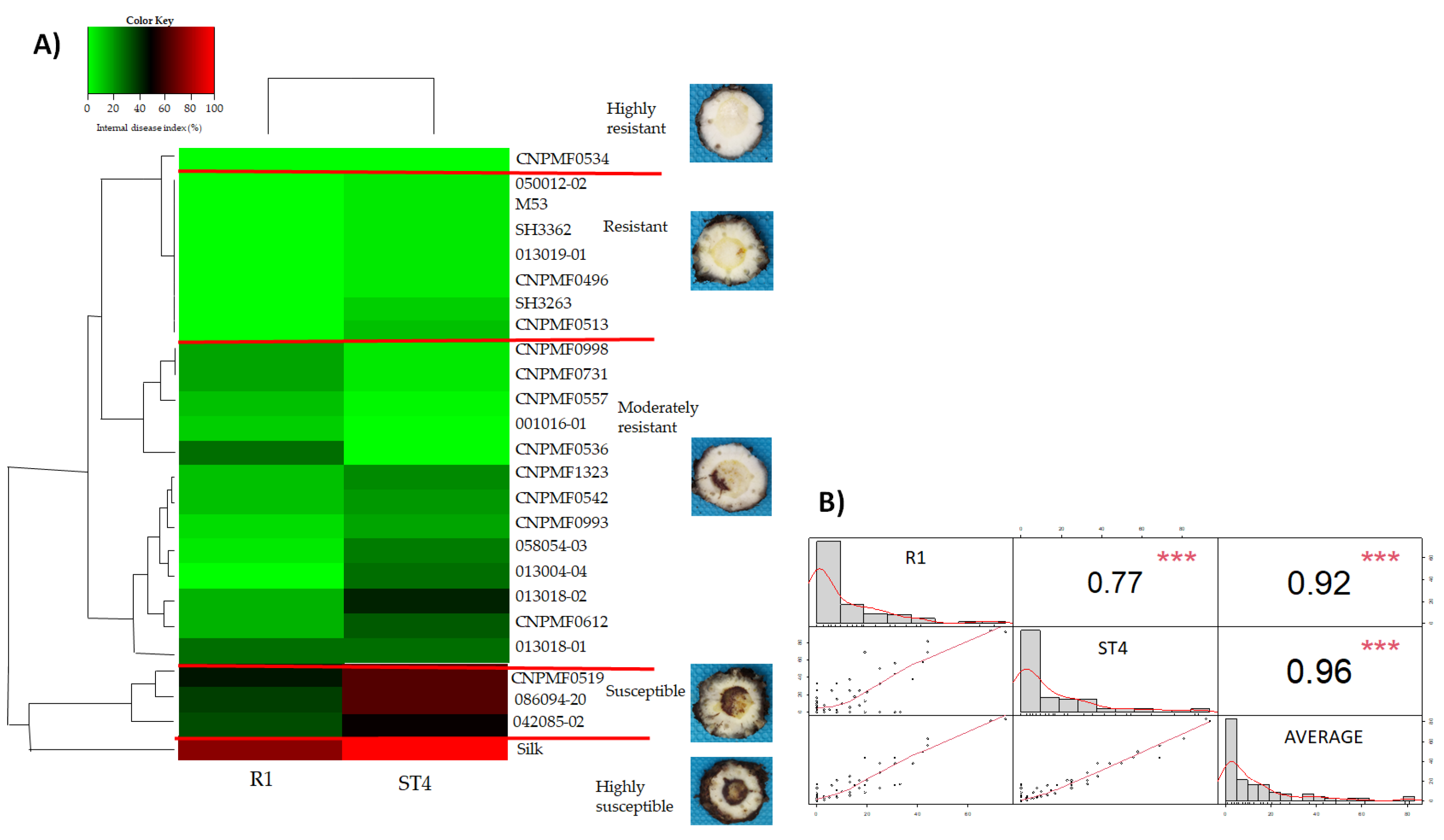
3.3. Cluster Analysis between Diploids for Responses to Foc R1 and Foc ST4
4. Discussion
5. Conclusion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAOSTAT (2024) Estatística da produção e de comércio. Disponivel em: https://www.fao.org/faostat/en/#data (Acesso em 16 de Marco de 2024).
- Mintoff, T.V.; Nguyen, C.K.; Samantha, C.M.H.; Robert, W.; Jeffrey, W. D e Lucy, T.T Tran-Nguyen. Field screening of banana cultivars for resistance to Fusarium oxysporum f.sp. Cuban tropical breed 4 in the North Shar.JL Territory. J. Fungi, 2021, 7, 627. [Google Scholar]
- Ploetz, R.C. (2015). Management of Fusarium wilt of banana: A review with special reference to tropical race 4. Crop Protection. 2015, 73, 7–15. [Google Scholar]
- Ploetz, R.C. Fusarium wilt of banana is caused by several pathogens referred to as fusarium oxyporum f. sp. Cubense. Phytopathology, Saint Paul. 2006, 96, 653–656. [Google Scholar]
- Chen, A.; Sun, J.; Matthews, A.; Armas-Egas, L.; Chen, N.; Hamill, S.; Mintoff, S.; Tran-nguyen, L.T.T.; Batley, J.; Aitken, E.A.B. Assessing variations in host resistance to Fusarium oxysporum f sp. cubense race 4 in Musa species, with a focus on the subtropical race 4. Front. Microbiol. 2019, 10, 1062. [Google Scholar]
- Dita, M.; Barquero, M.; Heck, D.; Mizubute, E.S.G.; Staver, CP. Fusarium wilt of banana: current knowledge on epidemiology and research needs toward sustainable disease management. Frontiers of Plant Science. 2018, 9, p–1468. [Google Scholar]
- Rocha, A.J.; Ferreira, M.S.; Rocha, L.S.; Oliveira, S.A.; Amorim, E.P.; Mizubuti, E.S.; Haddad, F. Interaction between Fusarium oxysporum f. sp. cubense and Radopholus similis may lead to changes in the resistance of banana cultivars to Fusarium wilt. European Journal of Plant Pathology, Dordrecht. 2020, 4, 1–15. [Google Scholar]
- Batista, I.C.A.; Heck, D.W.; Santos, A.; Alves, G.; Ferro, C.G.; Dita, M.; Haddad, F.; Michereff, S.J.; Correia, K.C.; Silva, C.F.B.; Mizubuti, E.S.G. A População de Fusarium oxysporum f. sp. cubense no Brasil Não é Estruturada por Grupo de Compatibilidade Vegetativa ou por Origem Geográfica. Fitopatologia 2022. [Google Scholar]
- Buddenhagen, I. Understanding the diversity of strains in Fusarium oxysporum f. sp. Cuban and history of introduction of the "Tropical Breed 4" to better manage banana production. Acta Hortic. 2009, 828, 193–204. [Google Scholar]
- Garcia, B.F.A.; Quintero-Vargas, J.C.; Ayala-Vasquez, M.; Schermer, T.; Seidl, M.F.; Santos-Paiva, M.; Noguera, A.M.; Aguilera-Galvez, C.; Wittenberg, A.; Hofstede, R. First report of wilt of Fusarium Tropical Breed 4 in Cavendish bananas caused by Fusarium odoratissimum in Colombia. Planta Dis. 2020. [Google Scholar]
- Martíne, G.; Olivares, B.O.; Rey, J.C.; Rojas, J.; Cardens., J.; Muentes, C.; Dawson, C. The Advance of Fusarium Wilt Tropical Race 4 in Musaceae of Latin America and the Caribbean: Current Situation. Pathogens 2023, 12, 277. [Google Scholar] [CrossRef]
- Zheng, S.J.; García-Bastidas, F.A.; Li, X.D.; Zeng, L.; Bai, T.T.; Xu, S.T.; Yin, K.S.; Li, H.X.X.; Fu, G.; Yu, Y.C.; Yang, L.; Nguyen, H.C.; Douangboupha, B.; Khaing, A.A.; Drenth, A.; Seidl, M.F.; Meijer, H.G.J.; Kema, G.H.J. New incursions of Fusarium oxysporum f. sp. cubense tropical Race 4 across the Greater Me kong Subregion. Frontier in Plant Science. 2018, 4, 208–218. [Google Scholar]
- Rocha, A.D.J.; Soares, J.M.D.S.; Nascimento, F.D.S.; Santos, A.S.; Amorim, V.B.D.O.; Ferreira, C.F.; Haddad, F.; Santos-Serejo, J.A.D.; Amorim, E.P. Improvements in the Resistance of the Banana Species to Fusarium Wilt: A Systematic Review of Methods and Perspectives. J. Fungi. 2022, 7, 249. [Google Scholar]
- Li, W.; Ge, X.; Wu, W.; Wang, W.; Hu, Y.; Mo, Y.; Xie, J. Identification of defense-related genes in banana roots infected by Fusarium oxysporum f. sp. cubense tropical race 4. Euphytica, Dordrecht. 2015, 205, n–3. [Google Scholar]
- Ferreira, M.D.S.; Moura, E.R.D.; Lino, L.S.M.; Amorim, E.P.; Santos-Serejo, J.A.D.; Haddad, F. Selection of somaclonal variants of the cultivar ‘Prata-Anã’ for resistance to Fusarium oxysporum f. sp. cubense race 1. Rev. Bras. Frutic. 2020, 42. [Google Scholar]
- Zuo, C.; Deng, G.; Li, B.; Huo, H.; Li, C.; Hu, C.; Kuang, R.; Yang, Q.; Dong, T.; Sheng, O.; et al. Germplasm screening of Musa spp. for resistance to Fusarium oxysporum f. sp. cubense tropical race 4 (Foc TR4). Eur. J. Plant. Pathol. 2018, 151, 723–734. [Google Scholar]
- Amorim, E.P. Melhoramento genético In: FERREIRA, C.F. et al (org) O agronegócio da banana 1. ed. Brasília-DF, Embrapa/ Cruz das Almas. 2016, p.173- 200.
- Gonçalves, Z.S.; Rocha, A.D.J.; Haddad, F.; Amorim, V.B.O.; Ferreira, C.F.; Amorim, E.P. Selection of Diploid and Tetraploid Banana Hybrids Resistant to Pseudocercospora fijiensis. Agronomy 2021, 11, 2483. [Google Scholar] [CrossRef]
- Rocha, A.D.J.; Soares, J.M.D.S.; Nascimento, F.D.S.; Santos, A.S.; Amorim, V.B.D.O.; Ferreira, C.F.; Haddad, F.; Santos-Serejo, J.A.D.; Amorim, E.P. Improvements in the Resistance of the Banana Species to Fusarium Wilt: A Systematic Review of Methods and Perspectives. J. Fungi 2022, 7, 249. [Google Scholar]
- Madigan, M.T.; Martinko, J.M.; Bender, K.S.; Buckley, D.H.; Stahl, D.A. Microbiologia de Brock. 14.ed. Porto Alegre: Artmed Editora, 2016.
- Dita, M.A.; Pérez, V.L.; Martinez De La Parte, E. Inoculation of Fusarium oxysporum f. sp. cubense causal agent of Fusarium wilt in banana. In Technical Manual: Prevention and Diagnostic of Fusarium Wilt (Panama Disease) of Banana Caused by Fusarium oysporum f. sp. cubense Tropical Race 4 (TR4); Vicente, L.P., Dita, M.A.R., Martínez, E., Eds.; United Nations: FAO: Rome, Italy. 2014; pp. 55–58. [Google Scholar]
- Mckinney, H.H. Influence of soil, temperature and moisture on infection of wheat seedlings by Helminthosporium sativum. Journal of Agricultural Research. 1923, 26, 195–217. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; The R Foundation for Statistical Computing: Vienna, Austria, 2020; Volume 1. [Google Scholar]
- Phillips, J.M.; Hayman, DS. Improved procedures for clearing roots and staining parasitic and vesicular-arbuscular mycorrhizal fungi for rapid assessment of infection. Trans. Br. Mycol. Soc. 1970, 55, 158–161. [Google Scholar]
- Karnovsk, M.J. A formaldehydeglutaraldehyde fixative of high osmolality for use in electron microscopy. J. Cell Biol.. 1965, 27, 137. [Google Scholar]
- Foster, AS. Practical plant anatomy. 2. ed. Torronto. Van Nostrand. 1949, 228 p.
- Rocha, A.D.J.; Soares, J.M.D.S.; Nascimento, F.D.S.; Rocha; Amorim, V. B.O. D.; Ramos, E.S.T.E.; Ferreira, C.F.; Haddad, F.; Amorim, E.P. Molecular, histological and histochemical responses of banana cultivars challenged with Fusarium oxysporum f. sp. cubense with different levels of virulence. Plants. 2022, 11, 2339. [Google Scholar]
- Ndayihanzamaso, P.; Mostert, D.; Matthews, M.C.; Mahuku, G.; Jomanga, K.; Mpanda, H.J.; Mduma, H.; Brown, A.; Uwimana, B.; Swennen, R.; et al. Evaluation of Mchare and Matooke Bananas for Resistance to Fusarium oxysporum f. sp. cubense Race 1. Plants. 2020, 9, 1082. [Google Scholar]
- Ribeiro, L.R. ; Oliveira, S de O.; Oliveira, S.S.A. S de.; Amorim, E.P.; Serejo, JAS.; Haddad. F. SOURCES OF RESISTANCE TO Fusarium oxysporum f. sp.cubense IN BANANA GERMPLASM. Rev. Bras. Frutic 2018. [Google Scholar]
- Li, W.M.; Dita, M.; Rouard, M.; Wu, W.; Roux, N.; Xie, J.H.; Ge, X.J. Deep RNA-seq analysis reveals key responding aspects of wild banana relative resistance to Fusarium oxysporum f. sp. cubense tropical race 4. Funct. Integr. Genom. 2020, 20, 551–562. [Google Scholar]
- Rebouças, T.A.; Haddad, F.; Ferreira, C.F.; Oliveira, S.A.S. de.; Ledo, C.A.D.S.; Amorim, E.P. Identification of banana genotypes resistant to Fusarium wilt race 1 under field and greenhouse conditions. Scientia Horticulturae. 2018, 239, 308–313. [Google Scholar]
- Nakata, P.A. Advances in our understanding of the formation and function of plasma calcium oxalate crystals. Protoplasma 2003. [Google Scholar]
- Ceita, G.D.O.; Macedo, J.N.A.; Santos, T.B.; Alemanno, L.; Gesteira, A.D.; Micheli, F.; Mariano, A.C.; Gramacho, K.P.; Silva, D.D.C.; Meinhardt, L.; et al. Involvement of calcium oxalate degradation during programmed cell death in Theobroma cacao tissues triggered by the hemibiotrophic fungus Moniliophthora pemiciosa. Plant Sci. 2007, 173, 106–117. [Google Scholar]
- Zavaliev, R.; Ueki, S.; Epel, BL.; Citovsky, V. Biologia do turnover de callose (β-1,3-glucano) em plasmodes mata. 2010.
- Dong, H.; Ye, Y.; Guo, Y.; Li, H. Comparison transcriptome analysis revealed resistance differences of Cavendish bananas to Fusarium oxysporum f. sp. cubense race1 and race 4. BMC Genom. 2020, 21, 122. [Google Scholar]
- García-Velasco,R.; Portal-González, N.; Santos-Bermúdez,R.; Rodríguez-García,A.; Companioni-González, B. Melhoramento genético para resistência à murcha de Fusarium em banana. Revista Mexicana de Fitopatologia, [S.l.] 2020, 39.
- Peraza-Echeverria,s. Molecular clonando and characterisation of potencial Fusarium resistance genes in bananas (Musa acuminata ssp.malaccensis). 2007.200p. Thesis (Plant Biotechnology Programa) Queensland University of Technology, Queensland, 2007.
- Dale, J.; James, A.; Paul, JY. Transgenic Cavendish bananas with resistance to Fusarium wilt tropical race 4. Nat Commun 2017, 8, 1496. [Google Scholar]
- D’Hont, A.; Denoeud, F.; Aury, J.M.; Baurens, F.C.; Carreel, F.; Garsmeur, O.; Noel, B.; Bocs, S.; et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 2012, 488, 213–217. [Google Scholar]
- Zheng, S.J.; García-Batisdas, F.A.; Li, X. D.; Zeng, L.; Bai, T.T.; Xu, S.T.; Yin, K.S.; Li, H.X.X.; Fu, G.; Yu, Y.C.; Yang, L.; Nguyen, H.C.; Douangboupha, B.; Khaing, A.A.; Drenth, A.; Seidl, M.F.; Meijer, H.G.J.; Kema, G.H.J. New incursions of Fusarium oxysporum f. sp. cubense tropical Race 4 across the Greater Mekong Subregion. Frontier in Plant Science 2018, 4, 208–218. [Google Scholar]
- Li, W.M.; Dita, M.; Rouard, M. Deep RNA-seq analysis reveals key responding aspects of wild banana relative resistance to Fusarium oxysporum f. sp. cubense tropical race 4. Functional and Integrative Genomics 2020, 20, 551–562. [Google Scholar]
- Better Bananas. Panama TR4 Variety Screening Trial (December 2018) Sub-Trial Results (Plant and First Ratoon). Available online: https://betterbananas.com.au/2022/03/04/panama-tr4-variety-screening-trial-december-2018-sub-trial-results-plant-and-first-ratoon/ (acesso em 3 janeiro 2023).
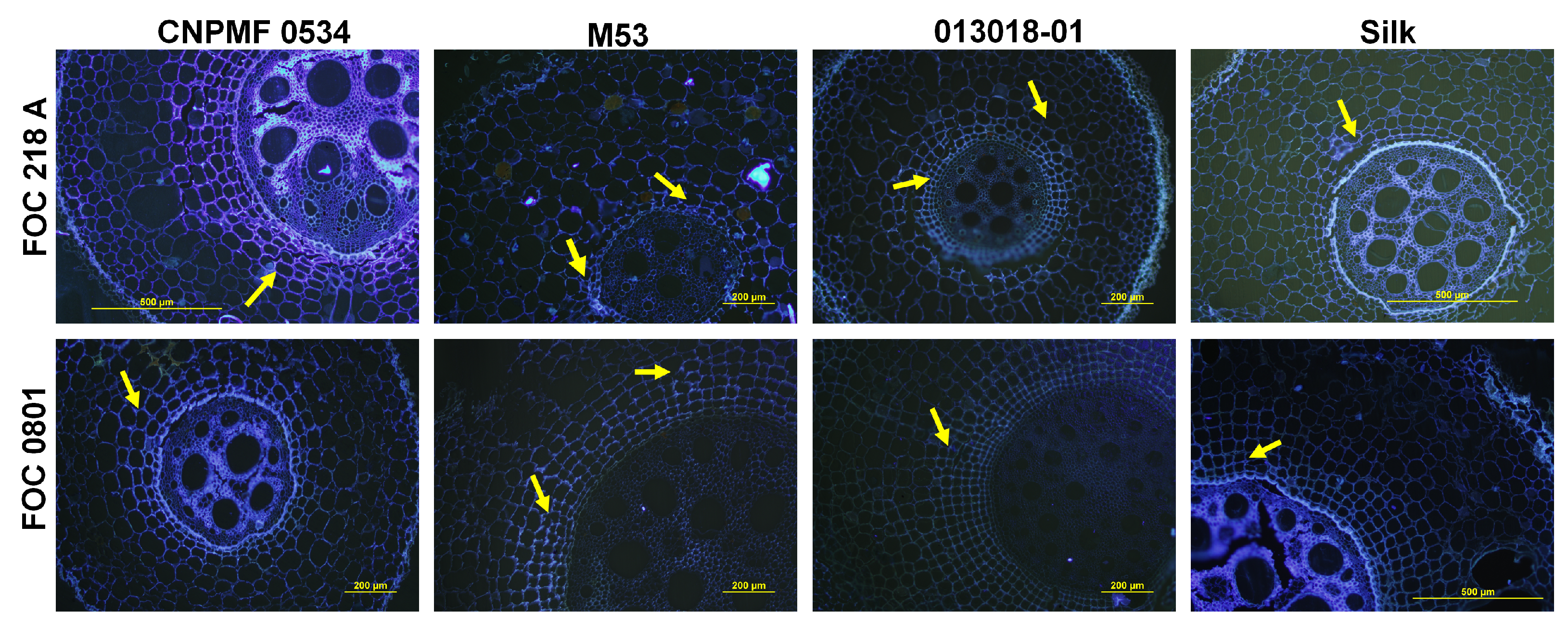
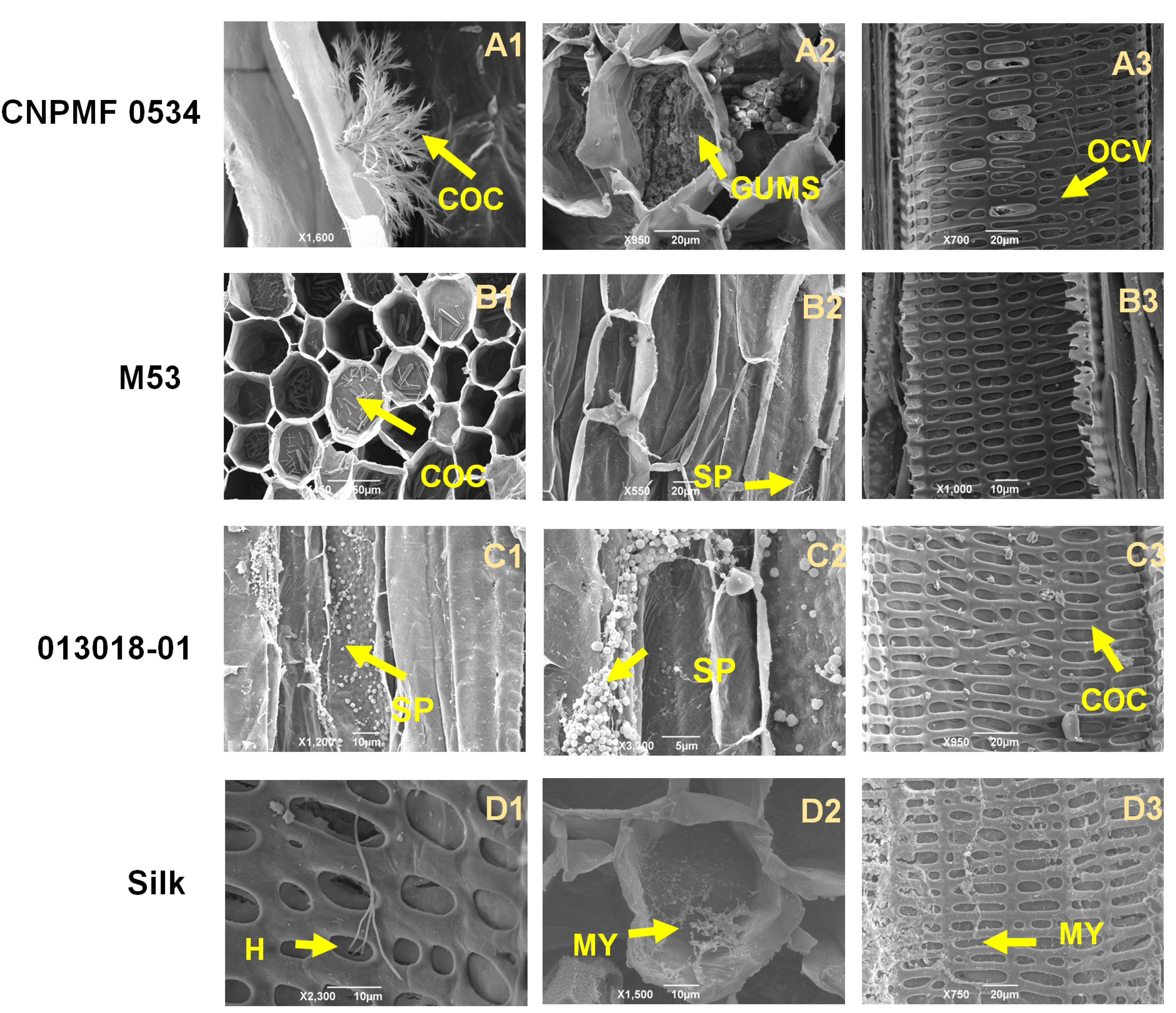
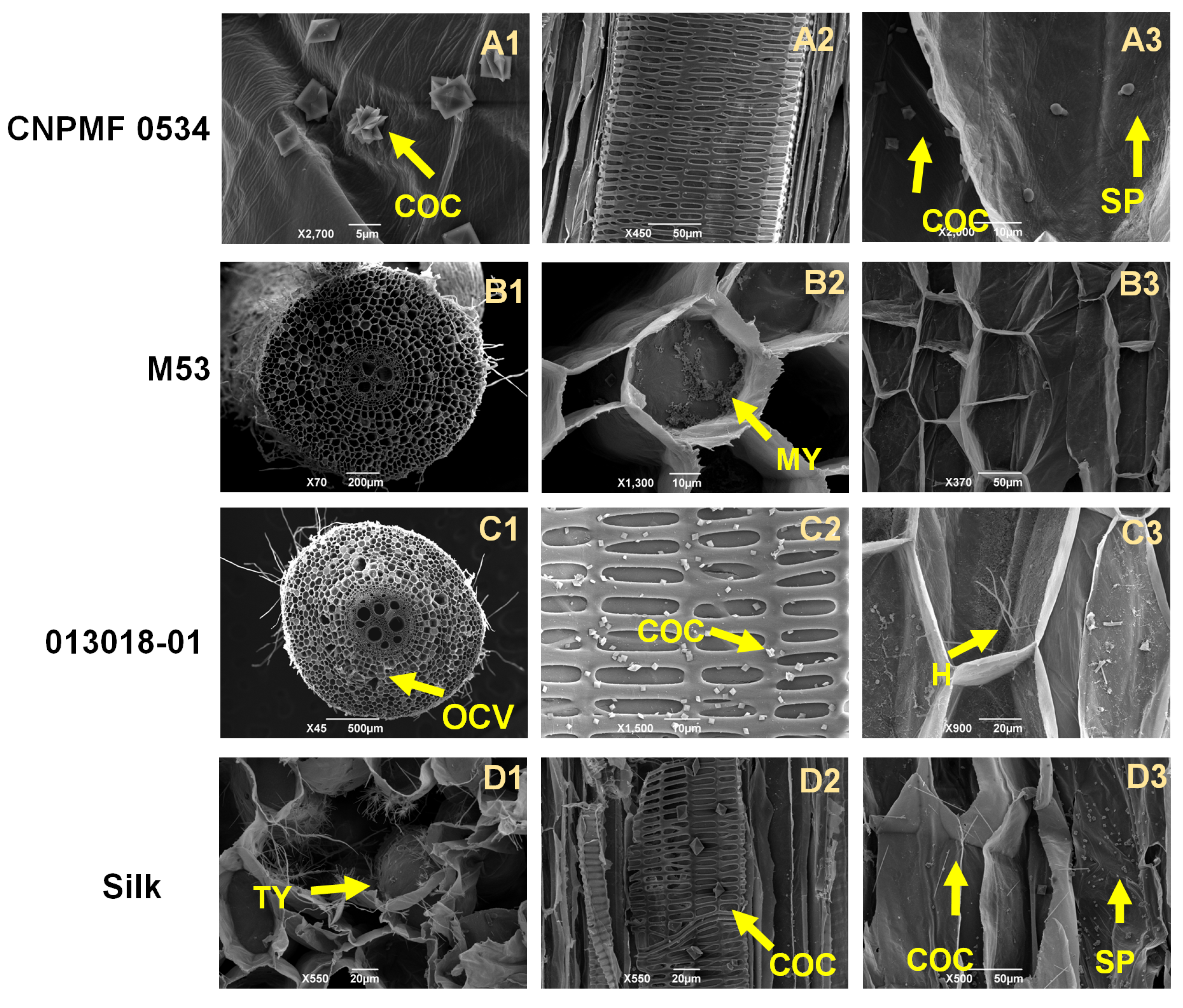
| Genotype | Genealogy |
| M53 | [(Malaccensis – Kedah x Banksii- Samoa)] x [(Paka x Banksii - Samoa)] |
| 001016-01 | Borneo x Guyod |
| 013004-04 | Malaccensis x Madang |
| 013018-01 | Malaccensis x Sinwobogi |
| 013018-02 | Malaccensis x Sinwobogi |
| 042085-02 | M53 x [(Madu x Calcutta 4)] |
| 050012-02 | M61 x Lidi |
| 058054-03 | [(Calcutta 4 x Pahang)] x [(Borneo x Madang)] |
| 086094-20 | [(Calcutta 4 x Galeo)] x SH3263 |
| SH3263 | - |
| SH3362 | - |
| 013019-01 | Malaccensis x Tjau Lagada |
| CNPMF 0557 | [(M61 x Pisang Lilin)] x [(Malaccensis x Tjau Lagada)] |
| CNPMF 0496 | [(M61 x Pisang Lilin)] x [(Terrinha x Calcutta 4)] |
| CNPMF 0536 | [(Calcutta 4 x Madang)] x [(Borneo x Guyod)] |
| CNPMF 0542 | [(SH3263)] x [(Malaccensis x Sinwobogi)] |
| CNPMF 0612 | [(M53 x Madu) x Madu)] x SH3263 |
| CNPMF 0731 | [(Malaccensis x Madang)] x [(Tuu Gia x Calcutta 4)] |
| CNPMF 0998 | [(Borneo x Guyod)] x [(Borneo x Guyod) x SH3263] |
| CNPMF 1323 | [(Malaccensis x Sinwobogi)] x [(Calcutta 4 x Heva)] |
| CNPMF 0513 | [(M61 x Pisang Lilin)] x [(M53 x Kumburgh) |
| CNPMF 0519 | Self-fertilization (wild diploid Tambi) |
| CNPMF 0534 | [(Calcutta 4 x Madang)] x [(Borneo x Guyod)] |
| CNPMF 0993 | [(Borneo x Guyod) x (Tuu Gia x Calcutta 4)] x [(Khai x (Calcutta 4 x Madang)] |
| Silk/Maçã | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





