Submitted:
10 April 2024
Posted:
10 April 2024
You are already at the latest version
Abstract
Keywords:
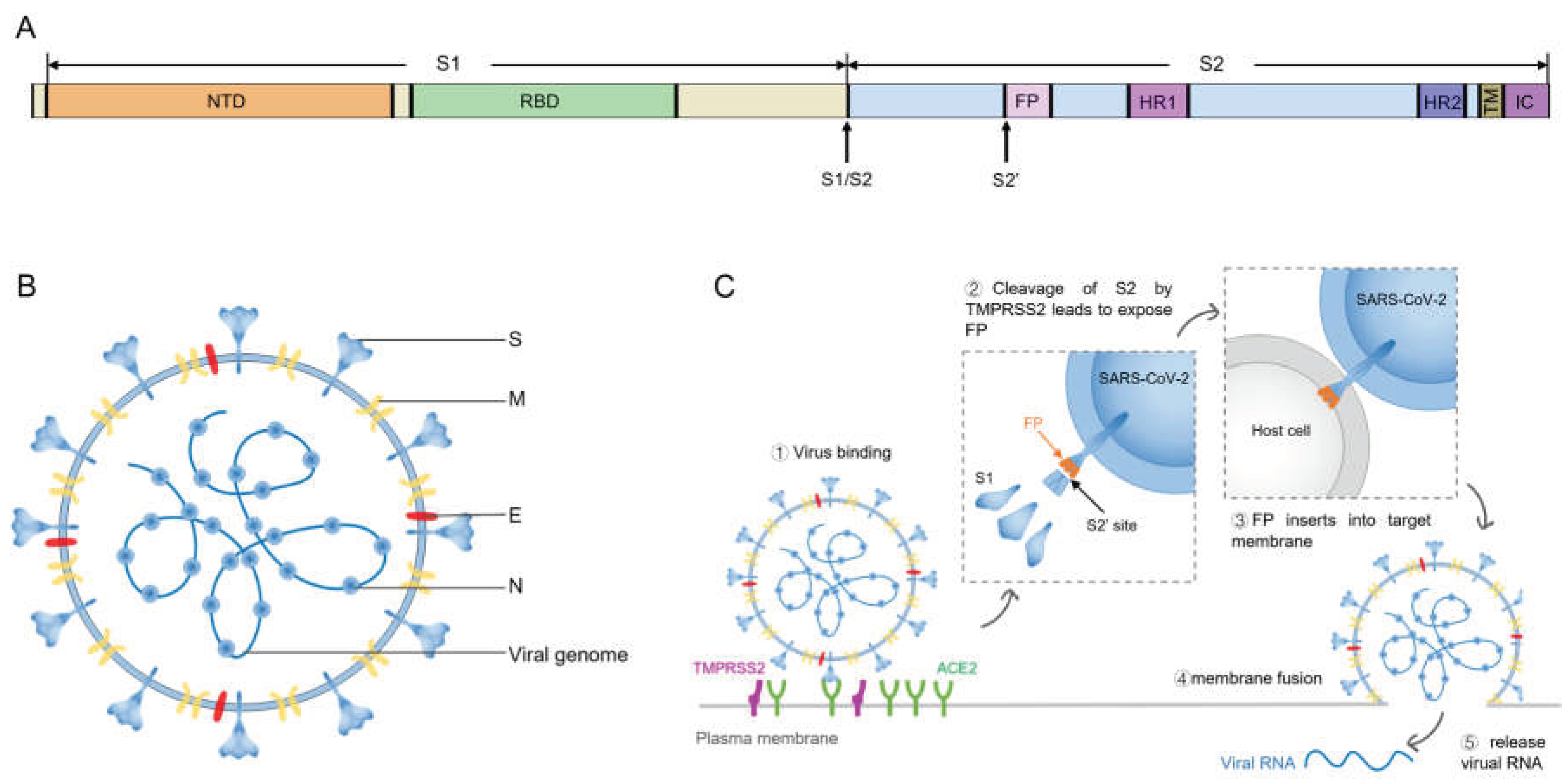
1. Introduction
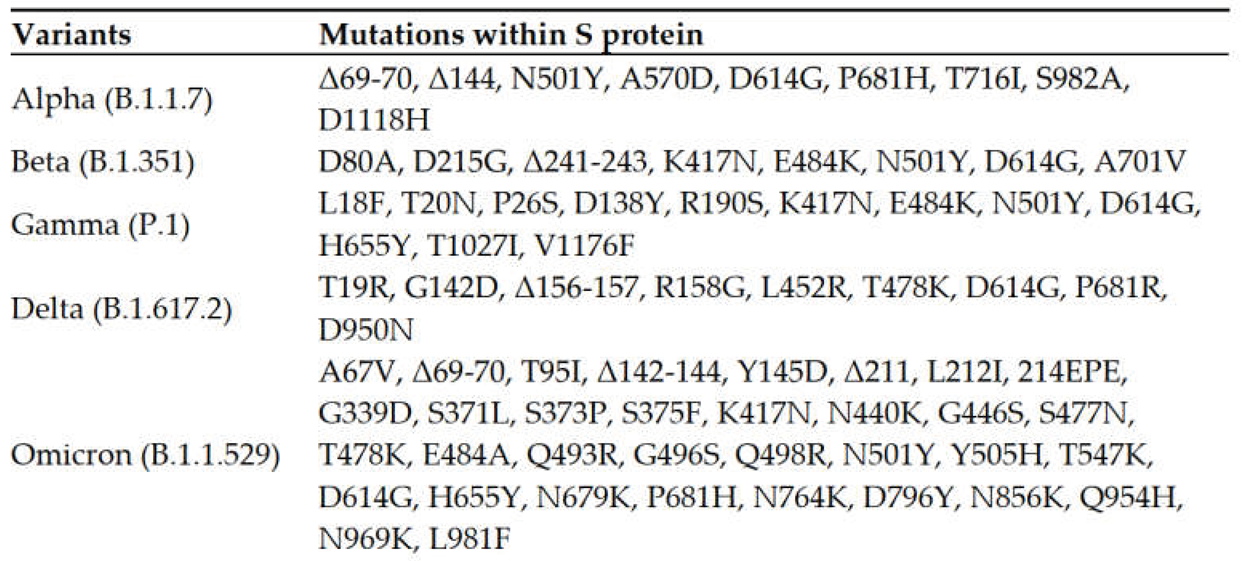
2. Cumulative Mutations in SARS-CoV-2 Spike
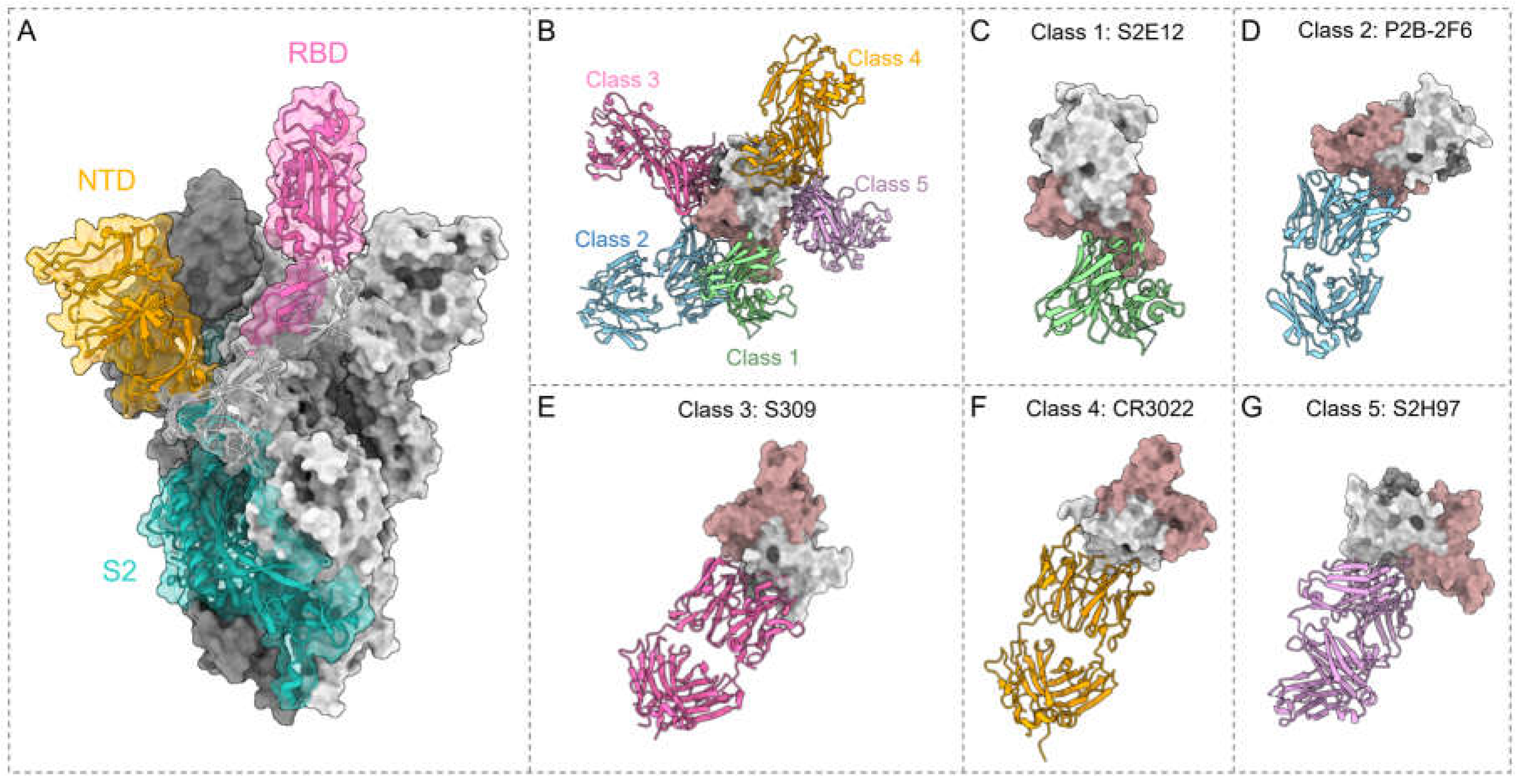
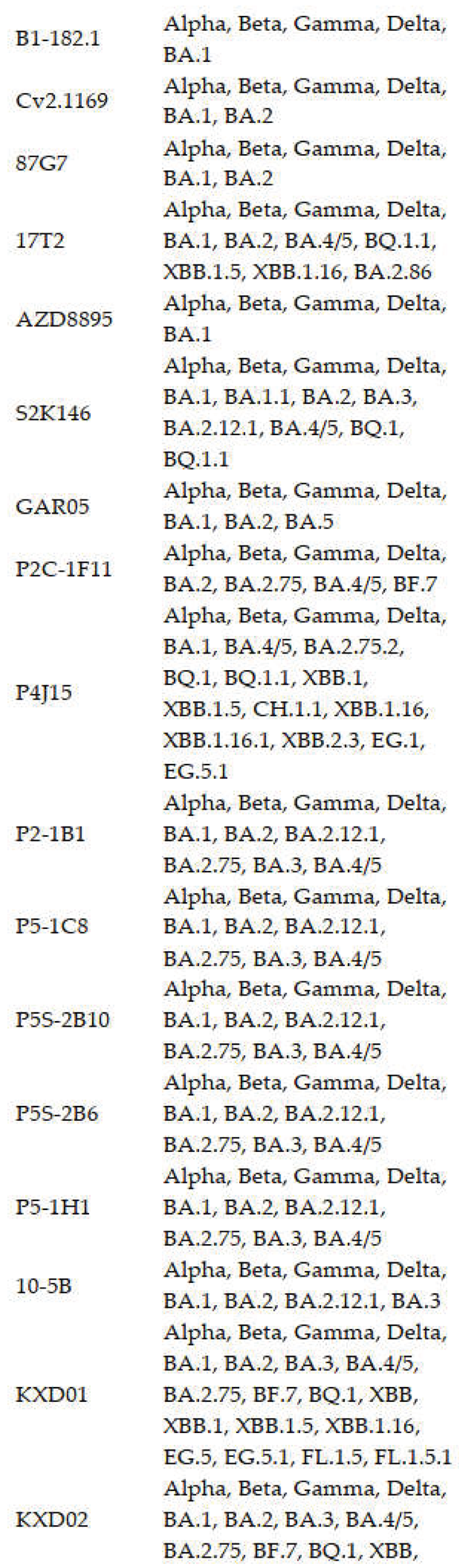
3. Potent bnAbs against SARS-CoV-2
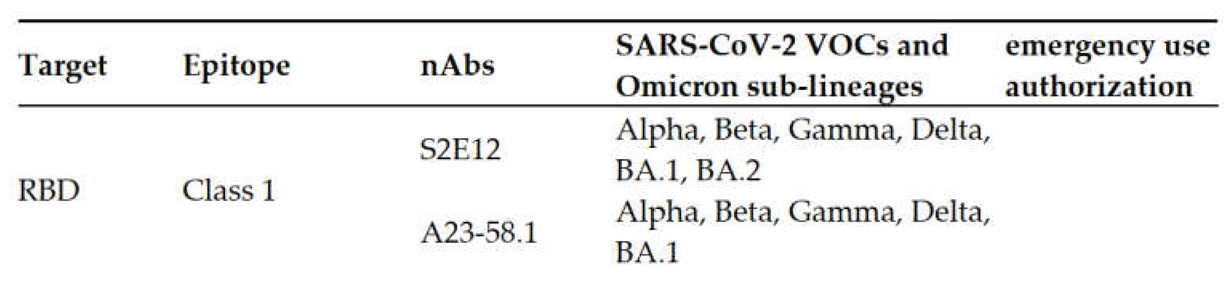
3.1. RBD Antibodies
3.1.1. Class 1 Antibodies
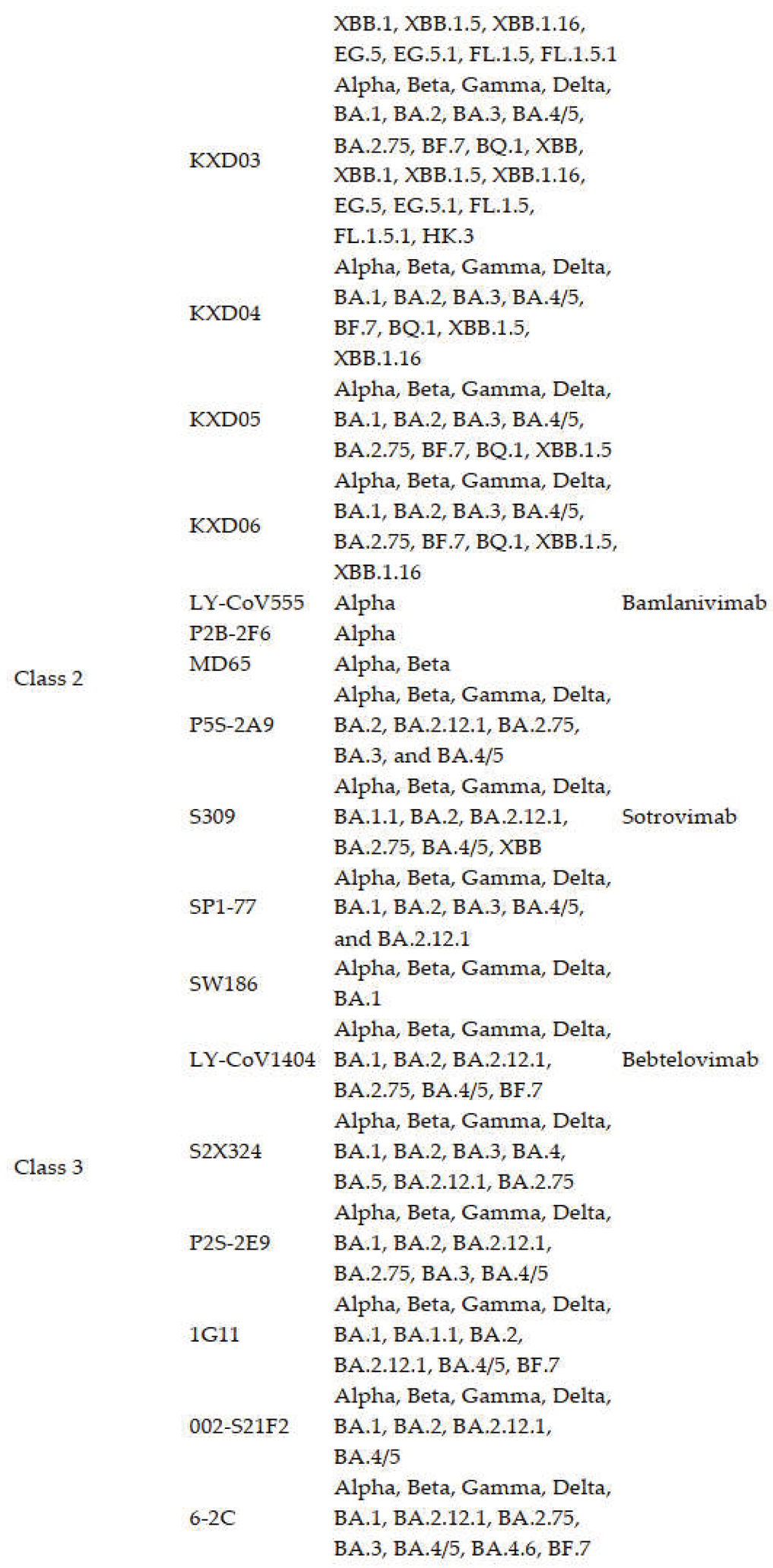
3.1.2. Class 2 Antibodies
3.1.3. Class 3 Antibodies
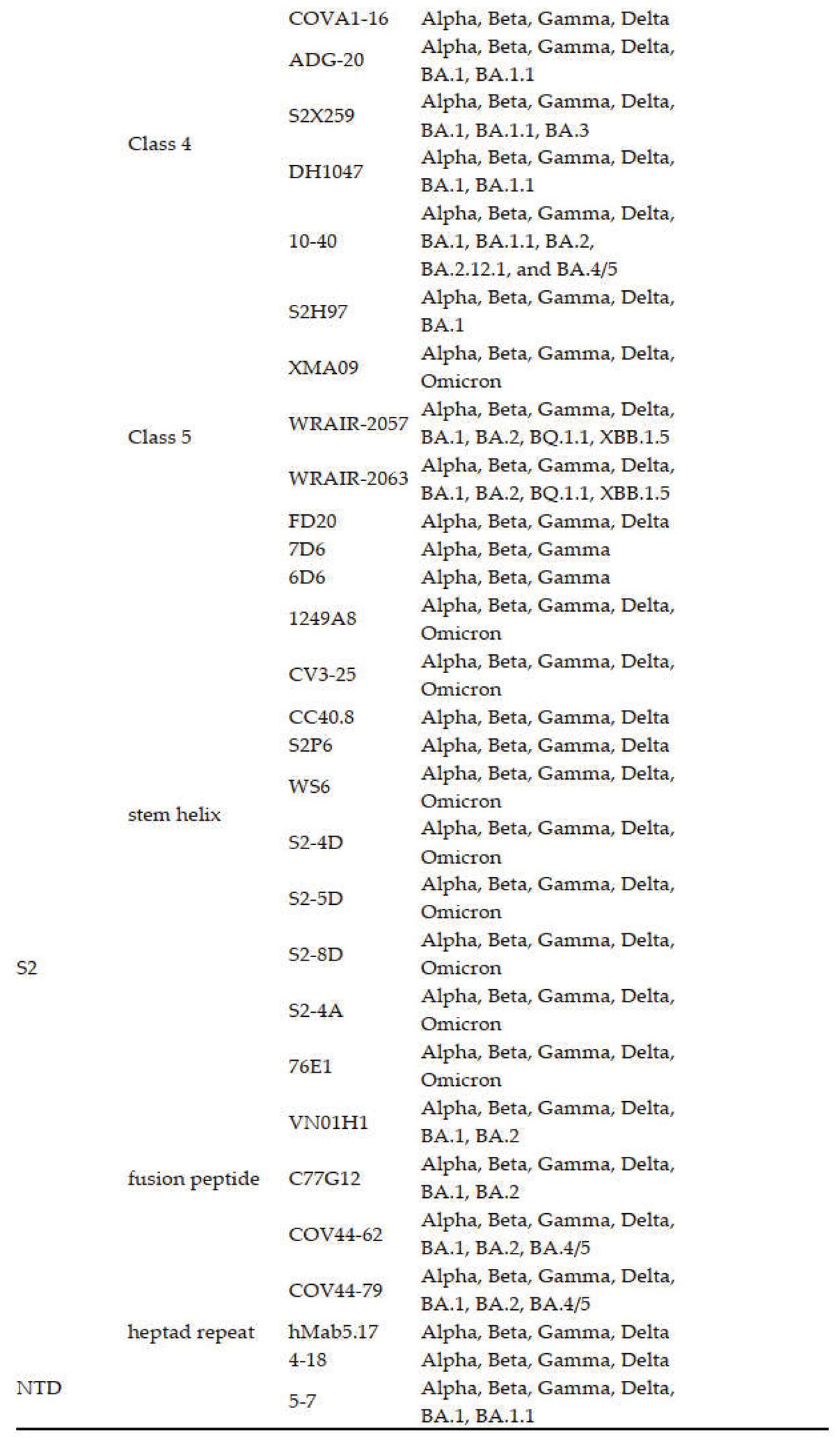
3.1.4. Class 4 Antibodies
3.1.5. Class 5 Antibodies
3.2. NTD Antibodies
3.3. S2 Antibodies
4. Conclusions
Author Contributions
Data Availability Statement
Acknowledgments
Conflicts of Interests
References
- Hu, B.; et al. Characteristics of SARS-CoV-2 and COVID-19. Nature Reviews Microbiology 2021, 19, 141–154. [Google Scholar] [CrossRef]
- Wang, M.Y.; et al. SARS-CoV-2: Structure, Biology, and Structure-Based Therapeutics Development. Front Cell Infect Microbiol 2020, 10, 587269. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef]
- Lu, R.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Bai, C.; Zhong, Q.; Gao, G.F. . Overview of SARS-CoV-2 genome-encoded proteins. Sci China Life Sci 2022, 65, 280–294. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.B.; et al. Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol 2022, 23, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Schoeman, D.; Fielding, B.C. Coronavirus envelope protein: current knowledge. Virology Journal 2019, 16, 69. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ye, Q. . The Key Site Variation and Immune Challenges in SARS-CoV-2 Evolution. Vaccines (Basel) 2023, 11. [Google Scholar] [CrossRef]
- Masters, P.S. The Molecular Biology of Coronaviruses, in Advances in Virus Research; Academic Press, 2006; pp. 193–292. [Google Scholar]
- Chang, C.-k.; et al. The SARS coronavirus nucleocapsid protein – Forms and functions. Antiviral Research 2014, 103, 39–50. [Google Scholar] [CrossRef]
- Yadav, R.; et al. Role of Structural and Non-Structural Proteins and Therapeutic Targets of SARS-CoV-2 for COVID-19. Cells 2021, 10. [Google Scholar] [CrossRef]
- Ke, Z.; et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 2020, 588, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- V'Kovski, P.; et al. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol 2021, 19, 155–170. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Y.; Zhou, H. Potent antibodies against immune invasive SARS-CoV-2 Omicron subvariants. Int J Biol Macromol 2023, 249, 125997. [Google Scholar] [CrossRef]
- Wang, L.; et al. Immune evasion of neutralizing antibodies by SARS-CoV-2 Omicron. Cytokine Growth Factor Rev 2023, 70, 13–25. [Google Scholar] [CrossRef]
- Markov, P.V.; et al. The evolution of SARS-CoV-2. Nature Reviews Microbiology 2023, 21, 361–379. [Google Scholar] [CrossRef]
- Viana, R.; et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature 2022, 603, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature 2022, 608, 603–608. [Google Scholar] [CrossRef] [PubMed]
- Challen, R.; et al. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study. Bmj 2021, 372, n579. [Google Scholar] [CrossRef]
- Tegally, H.; et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature, 2021, 592, 438–443. [Google Scholar] [CrossRef]
- Iketani, S.; et al. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature 2022, 604, 553–556. [Google Scholar] [CrossRef] [PubMed]
- Planas, D.; et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 2022, 602, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; et al. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell 2023, 186, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Omotuyi, I.O.; et al. Atomistic simulation reveals structural mechanisms underlying D614G spike glycoprotein-enhanced fitness in SARS-COV-2. J Comput Chem 2020, 41, 2158–2161. [Google Scholar] [CrossRef]
- Yurkovetskiy, L.; et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell 2020, 183, 739–751. [Google Scholar] [CrossRef]
- Meng, B.; et al. Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7. Cell Rep 2021, 35, 109292. [Google Scholar] [CrossRef] [PubMed]
- McCallum, M.; et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell 2021, 184, 2332–2347. [Google Scholar] [CrossRef]
- Liu, H.; et al. The basis of a more contagious 501Y.V1 variant of SARS-CoV-2. Cell Res 2021, 31, 720–722. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; et al. SARS-CoV-2 and Emerging Variants: Unmasking Structure, Function, Infection, and Immune Escape Mechanisms. Front Cell Infect Microbiol 2022, 12, 869832. [Google Scholar] [CrossRef]
- Starr, T.N.; et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 2020, 182, 1295–1310. [Google Scholar] [CrossRef]
- Han, P.; et al. Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS-CoV-2. Cell 2022, 185, 630–640. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; et al. Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: An insight from structural data. J Cell Physiol 2021, 236, 7045–7057. [Google Scholar] [CrossRef] [PubMed]
- Motozono, C.; et al. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe 2021, 29, 1124–1136. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; et al. Transmission, infectivity, and neutralization of a spike L452R SARS-CoV-2 variant. Cell 2021, 184, 3426–3437. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 2022, 602, 657–663. [Google Scholar] [CrossRef]
- West, A.P., Jr.; et al. Detection and characterization of the SARS-CoV-2 lineage B.1.526 in New York. Nat Commun 2021, 12, 4886. [Google Scholar] [CrossRef] [PubMed]
- Barnes, C.O.; et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 2020, 588, 682–687. [Google Scholar] [CrossRef] [PubMed]
- Lubinski, B.; et al. Functional evaluation of the P681H mutation on the proteolytic activation of the SARS-CoV-2 variant B.1.1.7 (Alpha) spike. iScience 2022, 25, 103589. [Google Scholar] [CrossRef] [PubMed]
- Saito, A.; et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature 2022, 602, 300–306. [Google Scholar] [CrossRef]
- Shah, M.; Woo, H.G. . Omicron: A Heavily Mutated SARS-CoV-2 Variant Exhibits Stronger Binding to ACE2 and Potently Escapes Approved COVID-19 Therapeutic Antibodies. Front Immunol 2021, 12, 830527. [Google Scholar] [CrossRef]
- Miller, N.L.; et al. Insights on the mutational landscape of the SARS-CoV-2 Omicron variant receptor-binding domain. Cell Rep Med 2022, 3, 100527. [Google Scholar] [CrossRef] [PubMed]
- Tortorici, M.A.; et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science 2020, 370, 950–957. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; et al. Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529. Science 2022, 376, eabn8897. [Google Scholar] [CrossRef] [PubMed]
- de Campos-Mata, L.; et al. A monoclonal antibody targeting a large surface of the receptor binding motif shows pan-neutralizing SARS-CoV-2 activity. Nat Commun 2024, 15, 1051. [Google Scholar] [CrossRef] [PubMed]
- Planchais, C.; et al. Potent human broadly SARS-CoV-2-neutralizing IgA and IgG antibodies effective against Omicron BA.1 and BA.2. J Exp Med 2022, 219. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; et al. Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants. Science 2021, 373, eabh1766. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; et al. An ACE2-blocking antibody confers broad neutralization and protection against Omicron and other SARS-CoV-2 variants of concern. Sci Immunol 2022, 7, eabp9312. [Google Scholar] [CrossRef]
- Dong, J.; et al. Genetic and structural basis for SARS-CoV-2 variant neutralization by a two-antibody cocktail. Nat Microbiol 2021, 6, 1233–1244. [Google Scholar] [CrossRef] [PubMed]
- Zost, S.J.; et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 2020, 584, 443–449. [Google Scholar] [CrossRef]
- Starr, T.N.; et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature 2021, 597, 97–102. [Google Scholar] [CrossRef]
- Qu, P.; et al. Enhanced neutralization resistance of SARS-CoV-2 Omicron subvariants BQ.1, BQ.1.1, BA.4.6, BF.7, and BA.2.75.2. Cell Host Microbe 2023, 31, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.J.; et al. Antibody-mediated broad sarbecovirus neutralization through ACE2 molecular mimicry. Science 2022, 375, 449–454. [Google Scholar] [CrossRef] [PubMed]
- Cameroni, E.; et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature 2022, 602, 664–670. [Google Scholar] [CrossRef]
- Rouet, R.; et al. Broadly neutralizing SARS-CoV-2 antibodies through epitope-based selection from convalescent patients. Nat Commun 2023, 14, 687. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.; et al. Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry. Nat Commun 2021, 12, 250. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; et al. Breakthrough infection elicits hypermutated IGHV3-53/3-66 public antibodies with broad and potent neutralizing activity against SARS-CoV-2 variants including the emerging EG.5 lineages. PLOS PATHOGENS 2023, 19. [Google Scholar] [CrossRef]
- Fenwick, C.; et al. Broadly potent anti-SARS-CoV-2 antibody shares 93% of epitope with ACE2 and provides full protection in monkeys. Journal of Infection 2023, 87, 524–537. [Google Scholar] [CrossRef]
- Ju, B.; et al. Infection with wild-type SARS-CoV-2 elicits broadly neutralizing and protective antibodies against omicron subvariants. Nat Immunol 2023, 24, 690–699. [Google Scholar] [CrossRef]
- Liu, Y.; et al. Inactivated vaccine-elicited potent antibodies can broadly neutralize SARS-CoV-2 circulating variants. Nat Commun 2023, 14, 2179. [Google Scholar] [CrossRef]
- Yuan, M.; et al. Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants. Science 2021, 373, 818–823. [Google Scholar] [CrossRef]
- Jones, B.E.; et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci Transl Med 2021, 13. [Google Scholar] [CrossRef]
- Chen, J.; et al. Rational optimization of a human neutralizing antibody of SARS-CoV-2. Comput Biol Med 2021, 135, 104550. [Google Scholar] [CrossRef] [PubMed]
- Ju, B.; et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 2020, 584, 115–119. [Google Scholar] [CrossRef]
- Patrick, C.; et al. Biophysical Fitness Landscape of the SARS-CoV-2 Delta Variant Receptor Binding Domain. J Mol Biol 2022, 434, 167622. [Google Scholar] [CrossRef]
- Makdasi, E.; et al. The neutralization potency of anti-SARS-CoV-2 therapeutic human monoclonal antibodies is retained against viral variants. Cell Rep 2021, 36, 109679. [Google Scholar] [CrossRef]
- Sun, H.; et al. Structural basis for broad neutralization of human antibody against Omicron sublineages and evasion by XBB variant. J Virol 2023, 97, e0113723. [Google Scholar] [CrossRef] [PubMed]
- Yue, C.; et al. ACE2 binding and antibody evasion in enhanced transmissibility of XBB.1.5. Lancet Infect Dis 2023, 23, 278–280. [Google Scholar] [CrossRef] [PubMed]
- Pinto, D.; et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 2020, 583, 290–295. [Google Scholar] [CrossRef]
- Gupta, A.; et al. Early Treatment for Covid-19 with SARS-CoV-2 Neutralizing Antibody Sotrovimab. New England Journal of Medicine 2021, 385, 1941–1950. [Google Scholar] [CrossRef]
- Magnus, C.L.; et al. Targeted escape of SARS-CoV-2 in vitro from monoclonal antibody S309, the precursor of sotrovimab. Frontiers in Immunology 2022, 13. [Google Scholar] [CrossRef]
- Luo, S.; et al. An antibody from single human VH-rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion. Science Immunology 2022, 7, eadd5446. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; et al. An antibody that neutralizes SARS-CoV-1 and SARS-CoV-2 by binding to a conserved spike epitope outside the receptor binding motif. Science Immunology 2022, 7, eabp9962. [Google Scholar] [CrossRef] [PubMed]
- Westendorf, K.; et al. LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants. Cell Rep 2022, 39, 110812. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.-J.; et al. Imprinted antibody responses against SARS-CoV-2 Omicron sublineages. Science 2022, 378, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; et al. Structural insights for neutralization of Omicron variants BA.1, BA.2, BA.4, and BA.5 by a broadly neutralizing SARS-CoV-2 antibody. Science Advances 2022, 8, eadd2032. [Google Scholar] [CrossRef]
- Yuan, M.; et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science 2020, 368, 630–633. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; et al. A broad and potent neutralization epitope in SARS-related coronaviruses. Proc Natl Acad Sci U S A 2022, 119, e2205784119. [Google Scholar] [CrossRef]
- Tortorici, M.A.; et al. Broad sarbecovirus neutralization by a human monoclonal antibody. Nature 2021, 597, 103–108. [Google Scholar] [CrossRef]
- Huo, J.; et al. Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike. Cell Host & Microbe 2020, 28, 497. [Google Scholar]
- Sankhala, R.S.; et al. Antibody targeting of conserved sites of vulnerability on the SARS-CoV-2 spike receptor-binding domain. Structure 2024, 32, 131–147. [Google Scholar] [CrossRef]
- Liu, H.; et al. Cross-Neutralization of a SARS-CoV-2 Antibody to a Functionally Conserved Site Is Mediated by Avidity. Immunity 2020, 53, 1272–1280. [Google Scholar] [CrossRef]
- Li, D.; et al. In vitro and in vivo functions of SARS-CoV-2 infection-enhancing and neutralizing antibodies. Cell 2021, 184, 4203–4219. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; et al. An antibody class with a common CDRH3 motif broadly neutralizes sarbecoviruses. Science Translational Medicine 2022, 14, eabn6859. [Google Scholar] [CrossRef] [PubMed]
- Jensen, J.L.; et al. Targeting the Spike Receptor Binding Domain Class V Cryptic Epitope by an Antibody with Pan-Sarbecovirus Activity. J Virol 2023, 97, e0159622. [Google Scholar] [CrossRef]
- Wang, S.; et al. Three SARS-CoV-2 antibodies provide broad and synergistic neutralization against variants of concern, including Omicron. Cell Rep 2022, 39, 110862. [Google Scholar] [CrossRef] [PubMed]
- Bullen, G.; et al. Cross-Reactive SARS-CoV-2 Neutralizing Antibodies From Deep Mining of Early Patient Responses. Front Immunol 2021, 12, 678570. [Google Scholar] [CrossRef]
- Dussupt, V.; et al. Low-dose in vivo protection and neutralization across SARS-CoV-2 variants by monoclonal antibody combinations. Nature Immunology 2021, 22, 1503–1514. [Google Scholar] [CrossRef]
- Li, T.; et al. Uncovering a conserved vulnerability site in SARS-CoV-2 by a human antibody. EMBO Molecular Medicine 2021, 13, e14544. [Google Scholar] [CrossRef]
- Li, T.; et al. Cross-neutralizing antibodies bind a SARS-CoV-2 cryptic site and resist circulating variants. Nat Commun 2021, 12, 5652. [Google Scholar] [CrossRef]
- Olukitibi, T.A.; et al. Significance of Conserved Regions in Coronavirus Spike Protein for Developing a Novel Vaccine against SARS-CoV-2 Infection. Vaccines 2023, 11. [Google Scholar] [CrossRef]
- Shrock, E.; et al. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science 2020, 370, eabd4250. [Google Scholar] [CrossRef] [PubMed]
- Ladner, J.T.; et al. Epitope-resolved profiling of the SARS-CoV-2 antibody response identifies cross-reactivity with endemic human coronaviruses. Cell Reports Medicine 2021, 2, 100189. [Google Scholar] [CrossRef] [PubMed]
- Crowley, A.R.; et al. Boosting of Cross-Reactive Antibodies to Endemic Coronaviruses by SARS-CoV-2 Infection but not Vaccination with Stabilized Spike. medRxiv 2021. [Google Scholar]
- Pannus, P.; et al. Poor Antibody Response to BioNTech/Pfizer Coronavirus Disease 2019 Vaccination in Severe Acute Respiratory Syndrome Coronavirus 2-Naive Residents of Nursing Homes. Clin Infect Dis 2022, 75, e695–e704. [Google Scholar] [CrossRef] [PubMed]
- Piepenbrink, M.S.; et al. Potent universal beta-coronavirus therapeutic activity mediated by direct respiratory administration of a Spike S2 domain-specific human neutralizing monoclonal antibody. PLoS Pathog 2022, 18, e1010691. [Google Scholar] [CrossRef] [PubMed]
- Li, C.J.; Chang, S.C. . SARS-CoV-2 spike S2-specific neutralizing antibodies. Emerg Microbes Infect 2023, 12, 2220582. [Google Scholar] [CrossRef] [PubMed]
- Pinto, D.; et al. Broad betacoronavirus neutralization by a stem helix–specific human antibody. Science 2021, 373, 1109–1116. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; et al. Vaccine-elicited murine antibody WS6 neutralizes diverse beta-coronaviruses by recognizing a helical stem supersite of vulnerability. Structure 2022, 30, 1233–1244. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; et al. A human antibody reveals a conserved site on beta-coronavirus spike proteins and confers protection against SARS-CoV-2 infection. Science Translational Medicine 2022, 14, eabi9215. [Google Scholar] [CrossRef]
- Hurlburt, N.K.; et al. Structural definition of a pan-sarbecovirus neutralizing epitope on the spike S2 subunit. Communications Biology 2022, 5, 342. [Google Scholar] [CrossRef]
- Jennewein, M.F.; et al. Isolation and characterization of cross-neutralizing coronavirus antibodies from COVID-19+ subjects. Cell Reports 2021, 36, 109353. [Google Scholar] [CrossRef] [PubMed]
- Li, C.J.; et al. Neutralizing Monoclonal Antibodies Inhibit SARS-CoV-2 Infection through Blocking Membrane Fusion. Microbiol Spectr 2022, 10, e0181421. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; et al. Neutralization mechanism of a human antibody with pan-coronavirus reactivity including SARS-CoV-2. Nature Microbiology 2022, 7, 1063–1074. [Google Scholar] [CrossRef] [PubMed]
- Low, J.S.; et al. ACE2-binding exposes the SARS-CoV-2 fusion peptide to broadly neutralizing coronavirus antibodies. Science 2022, 377, 735–742. [Google Scholar] [CrossRef] [PubMed]
- Dacon, C.; et al. Broadly neutralizing antibodies target the coronavirus fusion peptide. Science 2022, 377, 728–735. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.L.; et al. Monoclonal antibody targeting the conserved region of the SARS-CoV-2 spike protein to overcome viral variants. JCI Insight 2022, 7. [Google Scholar] [CrossRef] [PubMed]
- Piccoli, L.; et al. Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell 2020, 183, 1024–1042. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; et al. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292. [Google Scholar] [CrossRef]
- Chi, X.; et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 2020, 369, 650–655. [Google Scholar] [CrossRef]
- Cerutti, G.; et al. Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host & Microbe 2021, 29, 819–833. [Google Scholar]
- Souza, P.F.N.; et al. The spike glycoprotein of SARS-CoV-2: A review of how mutations of spike glycoproteins have driven the emergence of variants with high transmissibility and immune escape. Int J Biol Macromol 2022, 208, 105–125. [Google Scholar] [CrossRef]
- Lok, S.-M. An NTD supersite of attack. Cell Host & Microbe 2021, 29, 744–746. [Google Scholar]
- Liu, L.; et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 2020, 584, 450–456. [Google Scholar] [CrossRef] [PubMed]
- Cerutti, G.; et al. Neutralizing antibody 5-7 defines a distinct site of vulnerability in SARS-CoV-2 spike N-terminal domain. Cell Reports 2021, 37, 109928. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; et al. SARS-CoV-2 Neutralizing Antibodies for COVID-19 Prevention and Treatment. Annual Review of Medicine 2022, 73, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; et al. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature 2023, 614, 521–529. [Google Scholar] [CrossRef]
- Casadevall, A.; Focosi, D. SARS-CoV-2 variants resistant to monoclonal antibodies in immunocompromised patients constitute a public health concern. J Clin Invest 2023, 133. [Google Scholar] [CrossRef]
- Levin, M.J.; et al. LB5. PROVENT: Phase 3 Study of Efficacy and Safety of AZD7442 (Tixagevimab/Cilgavimab) for Pre-exposure Prophylaxis of COVID-19 in Adults. Open Forum Infectious Diseases 2021, 8 (Supplement_1), S810–S810. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





