1. Introduction
Carboxylic acid is among the most abundant functional groups in biologically important small molecules, particularly relevant in central carbon energy metabolism such as tricarboxylic acid (TCA) cycle and, fatty acid and cholesterol metabolism etc. Of ∼5000 endogenous human metabolites about 65% contain at least one carboxylic acid group [
1] and their abundance can vary in abnormal physiological states. Hence, deeper insights into the carboxyl-containing metabolites (CCMs) in biofluids would be helpful in furthering our understanding of key biological events, as well as the onset and progression of a disease. For example, glycolytic carbon flux results in increased intracellular levels of lactic acid in cancer [
2,
3,
4,
5] while fumaric and succinic acids have been reported as potential biomarkers of renal carcinoma [
6]. Methyl malonic acid has been reported as potential biomarker of vitamin B12 deficiency while α-ketoglutaric and pyruvic acid levels in urine are considered to be potential biomarkers for type II diabetes and liver cancer [
7,
8].
Mass spectrometry based analytical methods for profiling various biofluids are robust and reproducible and offer several advantages including great specificity and excellent sensitivity. However, for CCMs, those are limited by metabolite instability, poor chromatographic resolution, and retention, resulting in compromised sensitivity and/or specificity. Although, UPLC coupled multiple-reaction monitoring (MRM)-based mass spectrometry (MS) methods are benefitted from increased sensitivity and specificity, however, poor ionization efficiency for CCMs combined with other challenges such as high structural diversity led polarity differences, and within class structural isomerism imposes limits on their accurate and reliable quantification. Poor retention using traditional reverse phase LC methods and ion suppression remain other methodological challenges with LC-MRM based MS analyses of CCMs [
9]. Also, spontaneous decarboxylation caused by changes in temperature and pH during sample processing, could lead to lower measured levels of CCMS, ultimately, compromising the sensitivity of the assay [
9].
Currently, there is a glaring lack of methodologies that could accurately analyze and precisely quantify CCMs in the physiological range [
10]. Some of the current approaches used to improve LC–MS analysis of such metabolites include the use of ion pairing agents [
11,
12,
13,
14] or hydrophilic interaction chromatography-based column separations [
15,
16,
17,
18,
19]. Structurally, the lack of functional group and/or atom that could help in ionization in ESI-MS, is the main cause of low detection potential/sensitivity for this class of metabolites. Hence, chemical labelling/derivatization with small molecular weight compounds to improve CCMs ease of ionization could yield desired sensitivity for their quantification, especially for matrices where samples volumes are limited. Chemical derivatization has been used in analytical chemistry and instrumental analysis for different classes of analytes to improve their ionization efficiency, chromatographic separation, and chemical stability. Several chemical derivatization methods have been reported for analyzing carboxylic acid using different MS platforms [
20,
21,
22,
23,
24,
25,
26]. Most of the reactions for carboxylic acid derivatization are based on condensation reaction with amines [
27,
28,
29,
30,
31,
32,
33] or esterification reaction [
21,
34,
35]. However, most chemical derivatizations suffer from multiple limitations such as lack of selective MRM transitions for isomeric species or non-specificity for COOH group, chemical derivatizations degradation of derivatives during sample processing, or harsh reaction conditions that are not favorable for biological samples. In summary, there is a critical need to develop a high throughput method for the accurate and reliable quantification of such an important class of metabolites, in biological matrices, which may tolerate aqueous media as present in most of the biological matrices (without sacrificing sensitivity of the method) to augment clinical chemistry-based applications. This work presents a phenylenediamine-based chemical derivatization method that has been applied to the quantitative LC–MS/MS analyses of carboxylic acid sub-metabolome in a variety of biological matrices. Moreover, we observed wide linear dynamic range and excellent sensitivity. Benzimidazole-based ionizable groups were purposely introduced to the molecules of interest wherein the presence of nitrogen atoms helps to improve ionization efficiency, and subsequently, the detection sensitivity of the derivatized CCMs. More importantly, the enhanced hydrophobicity of polar CCMs including amino acids, TCA cycle intermediates, and fatty acids resulted in remarkable separation efficiency. The method was validated in commonly employed biological matrices such as urine, plasma, serum, cells, tissue, and saliva, with very low sample volumes.
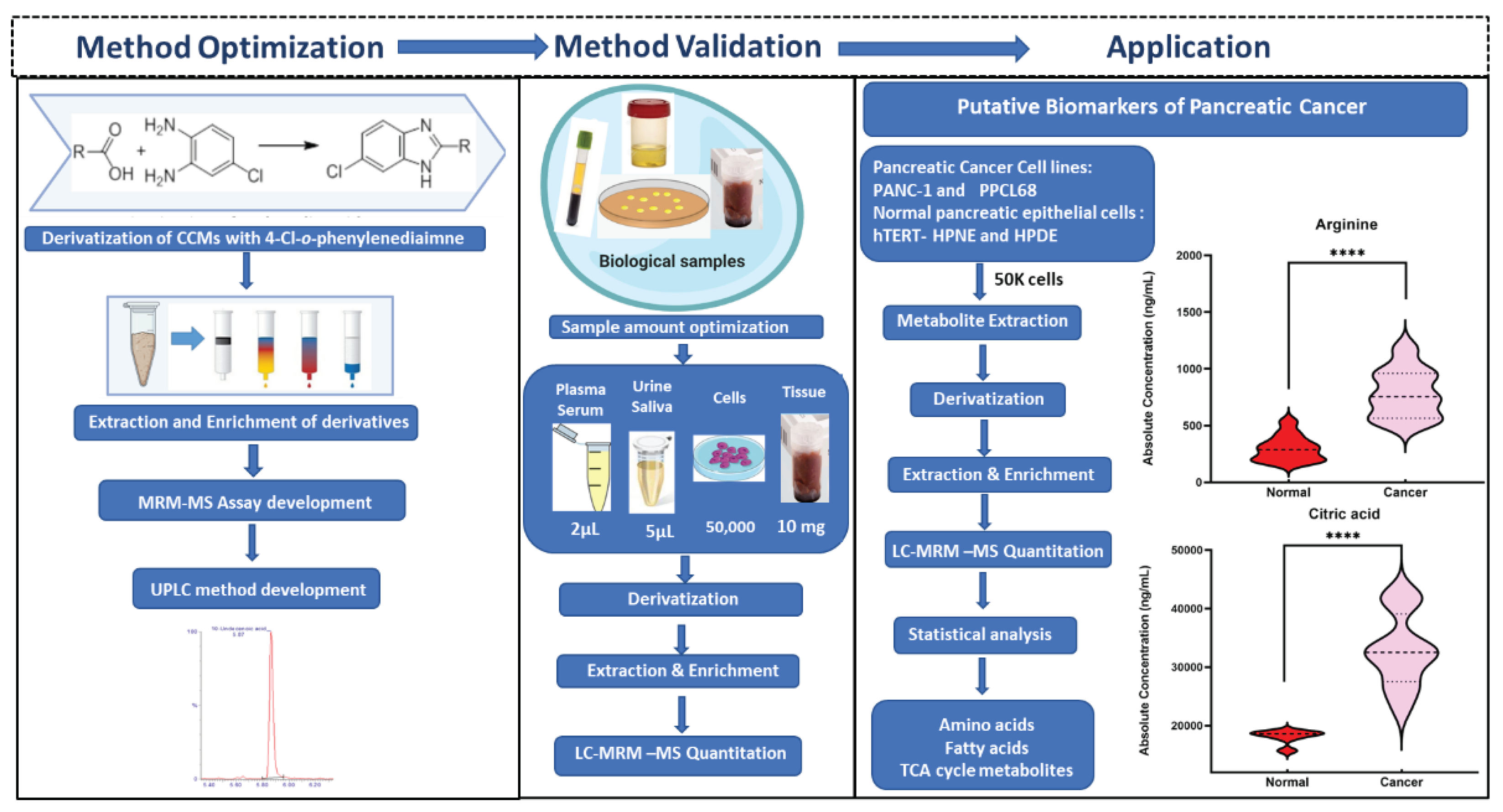
To our knowledge, this is the first report that provides a highly accurate, sensitive, and reliable assay for the analyses of small, medium, and long chain CCMs belonging to different classes of compounds that are integral to endogenous metabolism and important as potential clinical biomarkers. The workflow for the derivatization protocol for the quantitation of CCMs is illustrated in
Figure 1. The developed analytical method was thereafter validated with biological samples using pancreatic cancer and normal pancreatic epithelial cell samples as an example of a complex metabolomic system. Pancreatic ductal adenocarcinoma (PDAC) is a lethal malignancy that aggressively invades the adjacent vasculatures and quickly spreads to far-flung areas by the time it is clinically detected. Although complete surgical resection remains the only curative treatment, fewer than 20% of patients are candidates for surgery at the time of presentation. Hence, there is a critical need to identify diagnostic biomarkers with potential clinical utility in this pathology. In this context, metabolomics could be a powerful tool to search for new robust biomarkers. Comparative metabolomic profiling for the assessment of differential CCMs abundance was performed from two pancreatic cancer cell lines (PANC-1, PPCL68, n = biological replicates each) and two normal pancreatic epithelial cells (HPDE and hTERT-HPNE, control, n=6 biological replicates each) cell lines. Several CCMs were observed to exhibit altered levels in cancer cells, including TCA cycle intermediates, amino acids, fatty acids etc. The dysregulation corresponds to pathways such as energy and protein metabolism, and fatty acid biosynthesis and are hallmarks of pancreatic and other cancer types. These findings offer an information-rich matrix for discovering novel candidate biomarkers with diagnostic or prognostic potential in PDAC and are well supported by several literature reports.
2. Results
A total of 76 different CCMs including TCA metabolites, short chain organic acids (e.g. pyruvic acid), short chain fatty acids (e.g. propionic acid) and hydroxyl fatty acids (e.g. hydroxybutyric acid), heterocyclic amino acids (e.g. 4-OH proline), long chain mono (palmitic acid) and di-carboxylic acids (octadecane dioic acid) were tested for quantitative analysis using current protocol in six different biological matrices (
Supplementary Table S2). Chromatograms and calibration curves for all the quantified metabolites are provided in
Supplementary Figure S1 and S2, respectively. All derivatized CCMs showed symmetrical peak shapes, and baseline separations were obtained for most of the derivatives. Peak separation was achieved within a short time of 4 to 10 min, making the method ideal for high throughput analysis. UPLC-MRM-MS profiles of standard mixture of derivatized CCMs is shown in
Figure 2.
2.1. MRM Protocol Optimization
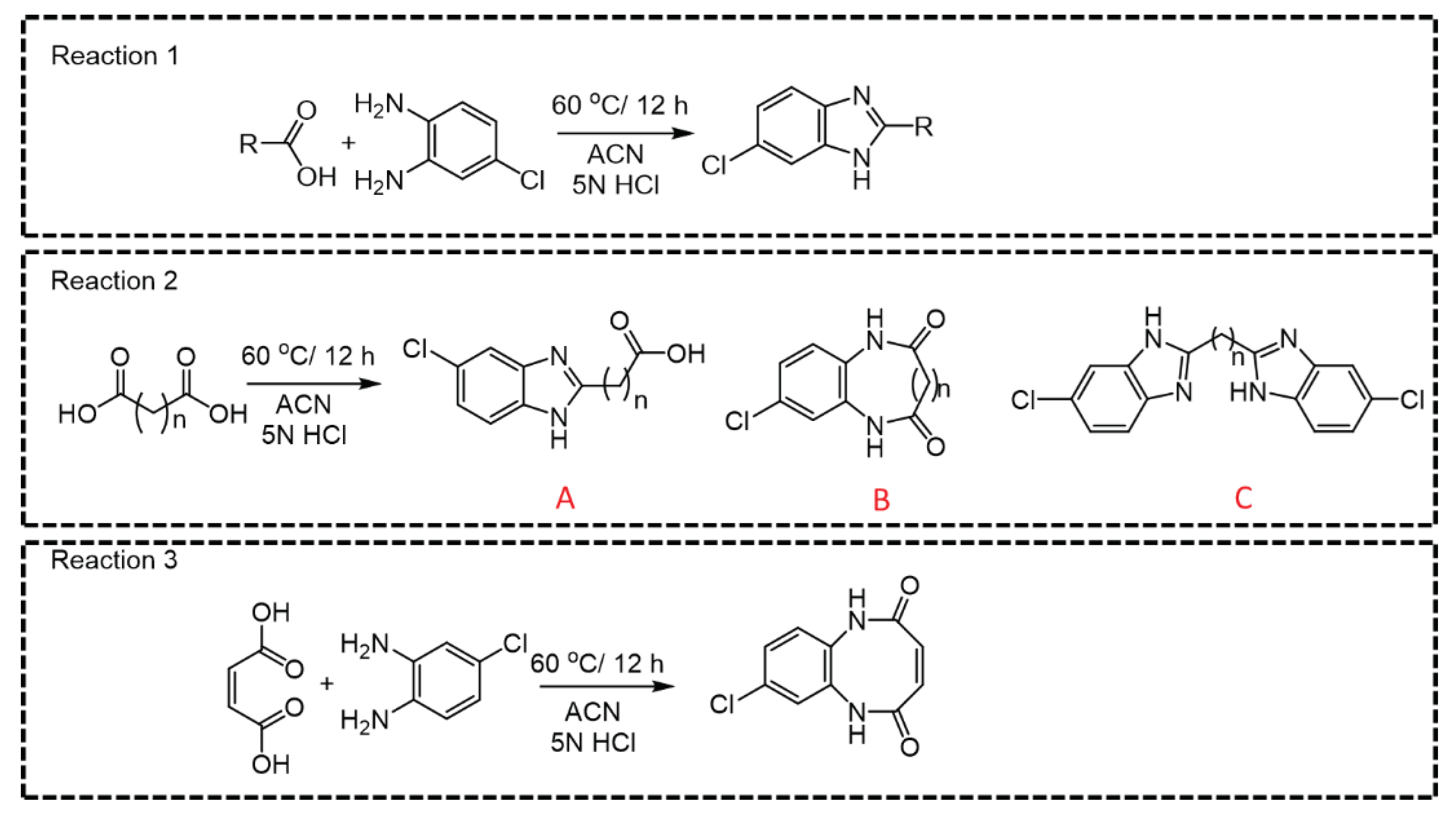
Among all CCMs studied, for the parent ions where linear derivative was formed with mono or di-carboxylic acids, m/z 166.06 was observed as the most intense daughter ion (Q3) (
Figure 3, Reaction 1, 2A and 2C). In the case of dicarboxylic acids like fumaric acid, oxalic acid, malic acid, succinic acid, malonic acid, methylmalonic acid, glutaric acid, adipic acid, 2-hydroxy glutaric acid, 2,2-dimethylglutaric acid, 3-methyl adipic acid and dodecanedioic acid, two kinds of products; linear- (A or C) and cyclic derivatives (B) are possible (
Figure 3, Reaction 2) depending upon the number of carboxylic acid groups participating in the derivatization reaction. For example, linear derivative A (Q1>Q3; 225.195>166.01) was observed as the major product for succinic acid. However, linear derivative C (Q1>Q3; 353.14>180.01) was the preferred product with methylmalonic acid. Due to enhanced lipophilicity, the derivatized product of methyl malonic acid eluted at relatively higher retention time (5.24 min) compared to that of succinic acid (4.05 min). Previous reports on methylmalonic acid quantification have cited issues either with the identification of Q1>Q3 ions, interference of succinic acid in chromatographic resolution or sensitivity.
For small chain acids, such as maleic acid and oxalic acid, cyclic derivatives (
Figure 3, Reaction 3) were observed as the major product which is possible with flat planer structure. Moreover, it was also possible to distinguish between maleic acid and fumaric based upon the difference in their major derivative formed (
Supplementary Table S1, entry 10 and 11) that resulted in unique Q1>Q3 pairs. For malic acid, along with the linear major product (m/z 240.6790 [M+H]+
Figure 3, Reaction 2A), we also detected another two major peaks (m/z 245.1729 and 351.1878). Perhaps malic acid dehydrate immediately to maleic acid and yields the cyclic derivative (m/z 245.1729 [M+Na]+) while m/z 351.2517 [M+Na]+ could be the parent ion where both carboxylic acid groups react during derivatization reaction. The observed m/z (Q1; 301.05) for the derivatized ascorbic acid is suggestive of the lactone-ring hydrolysis under the optimized reaction conditions (
Supplementary Table S1, entry 25) [
36]. The derivatized product of citric acid was observed as unstable and immediately converted to a metastable parent ion. Observance of [M+H]+ ion peak (511.021) indicates the derivatization reaction occurring at all 3 carboxylic acid groups, that quickly converted to 493.074 [M+H-H2O]+ indicative of the respective product for cis-aconitic acid. For quantification purpose, we developed specific MRM pair as (Q1>Q3; 281.288>166.06) for cis-aconitic acid (
Supplementary Table S1, entry 39). The parent ion at 237.259 was reflective of a linear derivative from one carboxylic acid group, that may be indicative of citric acid degradation to citraconic acid/itaconic acid (
Supplementary Table S1, entry 39 and 40) [
37]. While we also observed 511.021>248.9438 and 493.074>338.9858 MRM pairs, the transition 237.259>166.115 was selected for quantification of citric acid and the quantification results are the sum of citric, iso-citric, itaconic and citraconic acids.
2.2. Assay Development
The developed LC-MRM methods were evaluated for various quantification parameters such as sensitivity (LOD, S/N >3), lower limits of quantitation (LLOQ, S/N > 10), linearity range, S/N ratios, linear regression coefficients (r2) from the calibrators of each derivatized CCM etc., as listed in
Supplementary Table S3. The linearity of standard curve ranges from 3x103 to 3x104 fold of LOD and linear regression coefficients (r2) were 0.9990, in most cases. The calibration curves and related details for each quantified metabolite are provided in
Supplementary Figure S2. To our knowledge, these are the lowest reported LLOQs for all the CCMs including 0.01 ng/mL for lactic acid, 2-hydroxy glutaric acid, 3-hydroxybutyric acid, and 1.0 ng/mL for pyruvic acid and α-ketoglutaric acid, with improved S/N ratios. Quantitative results of all the metabolites from 6 different biological matrices studied herein are compiled in (
Supplementary Table S2). For assessing intra and inter-day variability, the samples (n=6) including the standard solutions were analyzed for consecutive six days. While we observed consistent peak areas for all the CCMs, the intra- and inter-day precision (RSDs <0.5% and <2%), were well indicative of metabolite stability and assay reproducibility. To study the matrix effects and have estimation of percent metabolite recovery for each matrix, known amounts of derivatized standards were spiked in all six different matrices studied herein at three different concentrations. Thereafter, the same protocol as defined for biological samples was followed except that 50 µL of methanol (solvent alone) was added instead of derivatizing solution. The percent recovery calculated as [(mean observed concentration/ spiked concentration) × 100%] were observed to be ranging from 90% to 105%.
2.3. Method Validation
Once validated across six different biological matrices, we investigated the utility of this methodology to search the potential biomarkers of pancreatic cancer. The metabolomic investigation of cellular content is of fundamental biological interest to unravel metabolic processes [
38,
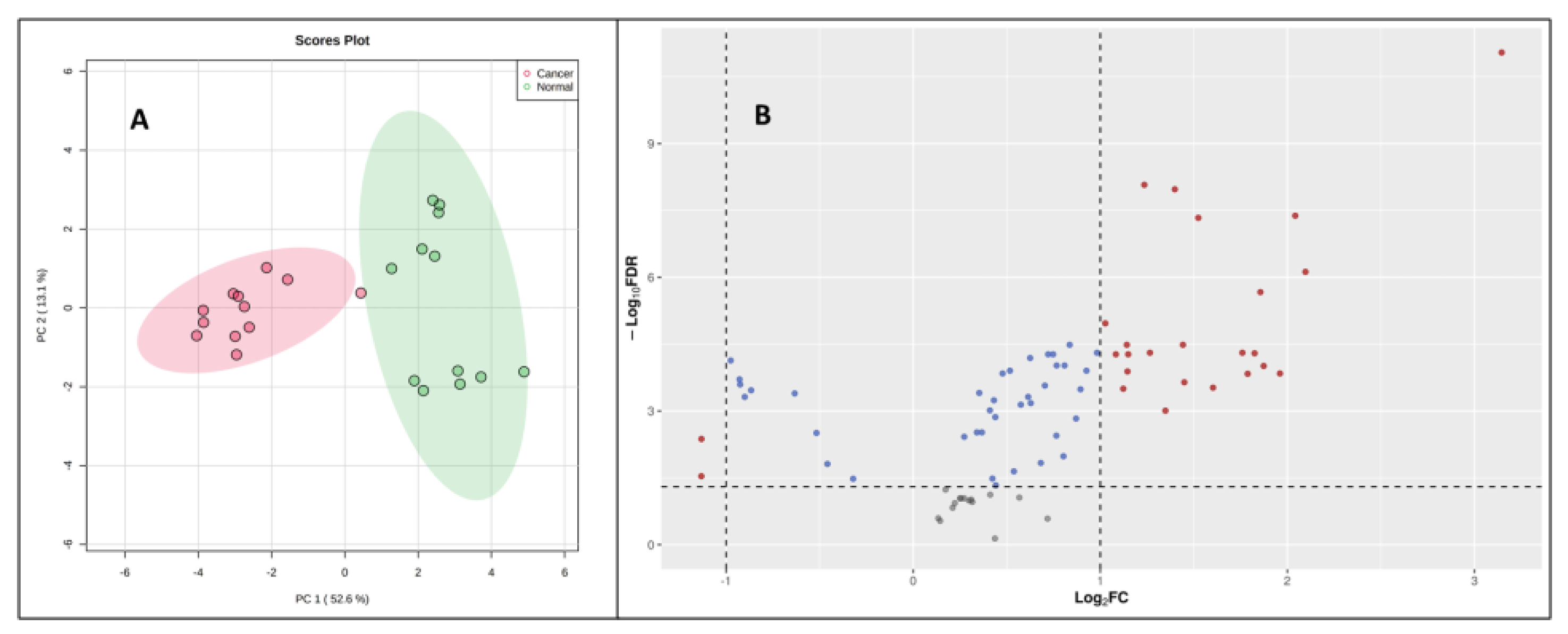
39]. For this, two pancreatic cancer cell lines (PANC-1, PPCL68) and two normal pancreatic epithelial cells (HPDE and HPNE) were grown to a density of 50,000 cells/ml in twelve-well costar plates as explained previously. The cell samples (24) for all four cell lines (n=6) were processed with the derivatization reaction and followed by LC-MS data acquisition. Principal component analysis (PCA) demonstrated distinctly CCMs profiles of cancerous cells when compared to normal pancreatic epithelial cells (
Figure 4A). Volcano plot further helped visualize the significant dysregulations for CCMs in cancerous cells (
Figure 4B). Univariate analysis was performed to compare the CCM profiles of cancerous cells to normal cells revealed altered levels of several CCMs in cancer cells. Overall, 25 CCMs which were identified as significantly altered (0.5>FC>2, FDR adjusted p-value < 0.05) across the two cancer cell lines when compared to normal pancreatic epithelial cells (
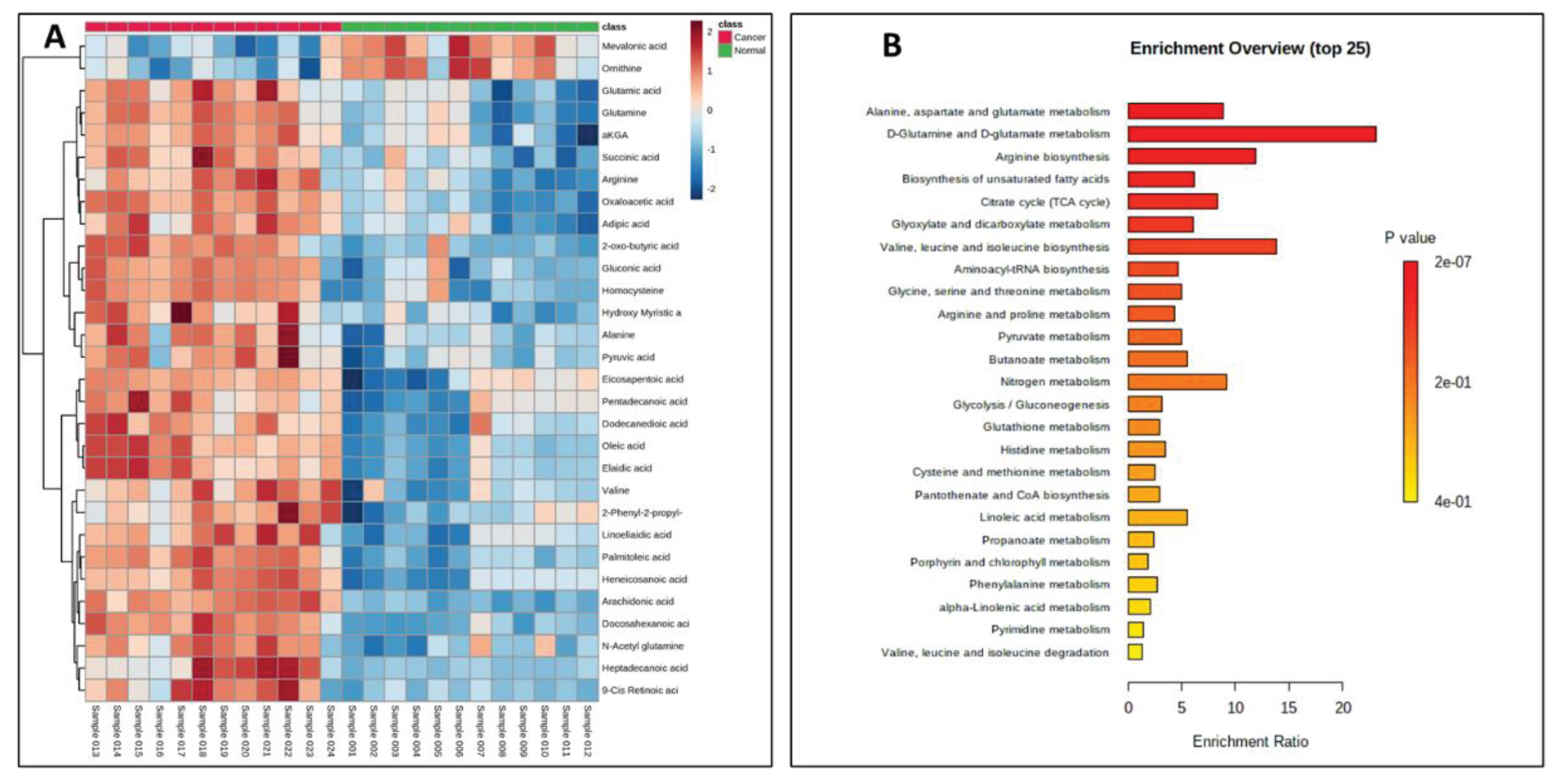
Supplementary Table S4). In addition to the majorly impacted energy and protein metabolism, and fatty acid biosynthesis, pathway analyses reveled perturbations in the glyoxylate and dicarboxylate metabolism, aminoacyl-tRNA biosynthesis, pyruvate metabolism, butanoate metabolism etc., in pancreatic cancer cells (
Figure 5). The media isolated from cancerous and normal pancreatic epithelial cells were also collected and their secretory CCMs profiles (
Supplementary Table S5) were compared with control media. The upregulation of γ-amino butyric acid and pyruvic acid were common for both cancerous and normal pancreatic epithelial cell media, compared to control. On the other hand, media from normal pancreatic epithelial cell lines showed elevated levels of some amino acids, organic acids, and fatty acids, compared to control media. These set of metabolites discussed herein could potentially be a biomarker group for pancreatic cancer to distinguish between pancreatic cancer and healthy individuals and increase the likelihood of providing a reliable diagnosis of PDAC.
3. Discussion
Spontaneous degradation/decarboxylation during ionization, poor chromatographic resolution, and retention on a reverse-phase column, are several factors that limit the sensitivity, specificity, and reproducibility during the down-stream LC-MS analysis of CCMs in biological matrices. There is an urgent and unmet need to develop a reliable method for quantitative analysis of CCMs from biological samples, that would provide adequate sensitivity for quantitation in the physiological range. The discovery of a universal derivatizing reagent to label the carboxylic acid sub-metabolome that could impart stability to the CCMs from any degradation could be the way forward. For the reliable quantification of CCMs, we used a phenylenediamine-based (4-Cl-o-PD) derivatization reaction that would generate benzimidazoles (N-containing/basic heterocyclic moiety) which ionize/protonate readily in ESI positive (+) mode, in contrast to free CCMs and thus can be detected with ultra-high sensitivity in conjunction with reverse-phase chromatography [
40,
41]. The benzimidazole derivatives with enhanced lipophilicity (higher log P values calculated with ChemDraw 18.1, PerkinElmer) as imparted by phenylenediamine-based derivatization of CCMs showed enhanced resolution and optimal retention on reverse-phase column, especially in complex biological matrices like plasma or cell extracts. We describe an optimized method for the analysis of this functionally important class of compounds from an array of biological matrices, hitherto unreported.
Central carbon metabolism is the most significantly perturbed pathway in cancer, in addition to fatty-acid biosynthesis and amino acid metabolism which include CCMs [
42,
43]. Some of the CCMs including tricarboxylic acid (TCA) cycle intermediates, amino acids (AAs), fatty acids/oxylipins, short-chain organic acids etc., have been reported to be dysregulated in pancreatic and other cancer types [
44,
45,
46]. We observed upregulation for several amino acids (AAs) which is in accordance with our previous cancer metabolomics studies in pancreatic cancer [
45,
47,
48,
49]. For example, arginine and glutamate metabolism were found to be upregulated in pancreatic cancer cells. Arginine and its metabolites regulate the proliferation, growth, autophagy, apoptosis, and metastasis of cancer cells [
50]. The pathogenesis of pancreatic cancer is reported to cause perturbations in the Gln-Glu pathway [
46,
51]. Glutamate has been shown to promote pancreatic cancer cell growth, invasion, and migration [
52]. In cancer cells, glutamine is the major amino acid that replenish and drives the tricarboxylic acid (TCA) cycle for continuous energy production in mitochondria [
53]. Elevated homocysteine levels could be due to lower betaine abundance. Betaine as a methyl donor supports methionine homeostasis and thus, maintains proper pancreatic function and cellular replication [
54]. Elevated -Aminobutyric acid (GABA) could play important roles in PDAC development and progression growth through overexpressing GABAA receptor π-subunit [
55]. Histamine, another upregulated metabolite, is an important mediator of cell proliferation in different types of cancers and thus, is critical for tumor development and progression [
56]. Observations in our study, well supported by several literature reports, point to the potential use of AAs as biomarkers for PDAC detection.
Free fatty acids (FFAs) are vital endogenous molecules for cellular energy metabolism and their altered levels could be attributed to disorders associated with fatty-acid beta oxidation and perturbed carnitine shuttle, indicative of disrupted mitochondrial function. Polyunsaturated fatty acids (PUFAs) such as arachidonic acid, EPA, and DHA form precursors to various PGLs, TXs and LTs which are pro-inflammatory in nature, cause immunosuppression and promote tumor cell proliferation [
57,
58,
59].
In summary, we demonstrate the applicability of a novel analytical method that can be used for the quantification of a wide range of clinically important CCMs including amino acids, short to medium chain organic/fatty acids, keto acids, hydroxyl acids, di/tri-carboxylic acids. This method enables effective quantification with small sample size and allows distinctive quantification of structural isomers such as methylmalonic acid and succinic acid. To our knowledge, this is the first report, wherein 4-Cl-o-PD was used for chemical derivatization of CCMs followed by their quantification by LC-MRM based mass spectrometry. This method was optimized for targeted analysis of human plasma, serum, urine, saliva, tissue, and cell extracts. Since these matrices are widely used in clinical chemistry-based biomedical research, we anticipate broad applicability of this high throughput methodology for bio-medical researchers. This method may be extended to other CCMs like bile acids, prostaglandins etc. Clinical metabolomics, though, has made quick progress to identify potential metabolic biomarkers for PDAC, however, there are still important informative gaps to be filled to increase the robustness and the reliability of these outcomes. We believe that phenylenediamine derivatization-based LC-MS approach would be imperative in future studies on any biological sample for comprehensive metabolome profiling to investigate the subtle physiological changes in various diseased states and biomarker discovery.
4. Materials and Methods
4.1. Chemicals and Reagents
LC-MS grade acetonitrile, methanol, water and other organic solvents, and formic acid were purchased from Optima, Fisher Scientific. HPLC grade ethyl acetate was purchased from Thermo Fisher Scientific. 4-Chloro-o-phenylenediamine (4-Cl-o-PD) and HCl were obtained from Sigma-Aldrich. All the standards were purchased from Sigma-Aldrich or CDN Isotopes (Quebec, Canada). The reagents were stored at -20 °C and vacuum-dried in a desiccator over anhydrous phosphorus pentoxide under a vacuum of 30 mmHg, at 4 °C for 12 h, before use. NIST plasma was purchased as Standard Reference Materials (SRM) from National Institute of Standards and Technology (NIST), a physical sciences laboratory, MD, USA. All the biological reagents for cell culture were purchased from Gibco Fisher Scientific.
Detailed information on the collection and handling of biospecimens, cell culture, metabolite extraction from cells, metabolite extraction from growth media has been provided in the supplementary information.
4.2. Derivatization Protocol for Standards, Internal Standards, and Biological Samples
Literature search revealed that o-phenylenediamines (o-PD) react readily with most CCMs to produce 2-substituted benzimidazoles, usually in high yields [
60]. This involves the condensation of o-PD with carboxylic acid group in the presence of concentrated hydrochloric acid. However, most of the currently reported phenylenediamine-based derivatization methods need non-aqueous/moisture free conditions which are incompatible with down-stream LC-MS analysis of biological matrices [
61]. The derivatization methodology reported here can be performed in almost all polar protic (including water) and aprotic solvents, commonly used in research laboratories. The amount of reagent was selected based on a preliminary experiment. Standard compounds and internal standards (ISs) (1 mmol each) were subjected to derivatization with 4-Cl-o-PD (1.1 mmol for mono-carboxylic acids and 2.1 mmol for di-carboxylic acids). For example, 0.9 mg of lactic acid (mono-carboxylic acid) would need 1.50 mg of 4-Cl-o-PD while for 1.19 mg of succinic acid (di-carboxylic acid) would need 3.0 mg of 4-Cl-o-PD. For all biological matrices and cell samples, 50 µg of 4-Cl-o-PD was found as enough for the optimized sample amounts. Addition of a 10 µL of 5N HCl drastically improved the product yield at 60 °C for 12 h in methanol as general reaction conditions for the derivatization method. Sample processing, extraction and derivatization steps were performed simultaneously to avoid loss of analyte content due to spontaneous decarboxylation of CCMs under normal laboratory conditions. We used 2 µL of plasma, serum and NIST plasma, and 5 µL of urine and saliva, 10 mg of tissue and 50,000 cells for the developed derivatization assay. Each biological sample (matrix) was analyzed in duplicate over a 6-day period. Without sacrificing sensitivity, we observed excellent S/N ratios for derivatized CCMs for the optimized sample amounts taken, based upon preliminary experiments. The derivation reaction for standards, ISs and biological samples were done as per the standard operating procedure (SOP) as mentioned in Supplemental Information.
4.3. MRM Development and LC-MS Conditions
The derivatized standards (1 µg/mL in water: MeOH (1:1)) as enriched by cartridge fractionation were infused onto Xevo-TQS (Waters Corporation, USA) and the sample cone voltage and collision energies were optimized for the analyte to obtain maximum ion intensity for parent and daughter ions using “IntelliStart” feature of MassLynx V4.2 software (Waters Corporation, USA). The most intense precursor and fragment ion pair (Q1>Q3) of each analyte was selected for MRM-based quantitation. A complete list of CCMs, the structures of parent molecules and their corresponding benzimidazole derivatives along with MRM transitions (Q1>Q3) and other parameters are provided in
Supplementary Table S1.
The derivatized CCMs were resolved on BEH C-18, 1.7 µM, 2.1 mm, 100 mm column (35 °C) using an Acquity UPLC system coupled with a Xevo-triple quadrupole mass spectrometer (Xevo-TQS, Waters Corporation, MA, USA) operating in the ESI+ MRM mode. Further details on the LC conditions, data acquisition and data processing have been provided in the supplementary information.
4.4. Statistical Analysis
Binary comparisons were done using unpaired t-tests and fold changes were calculated to identify dysregulations in CCMs levels for pancreatic cancer cells, compared to normal pancreatic epithelial cells. Similarly, we also compared the CCMs profiles of media collected during cell harvest to that of control media. All analyses were performed using MetaboAnalyst 5.0. Figures were created using R, ChemDraw 18.1, and BioRender (
www.BioRender.com).
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Author Contributions
Conceptualization, Shreyans K. Jain, Shivani Bansal, Amrita Cheema; methodology, Shreyans K. Jain, Shivani Bansal, Sunil Bansal; software, Shreyans K. Jain, Shivani Bansal, Sunil Bansal, Amrita Cheema; validation, Shreyans K. Jain, Shivani Bansal, Sunil Bansal, Baldev Singh; formal analysis, Shreyans K. Jain, Shivani Bansal, Sunil Bansal, Baldev Singh; investigation, Amrita Cheema; resources, Amrita Cheema; data curation, Shreyans K. Jain, Shivani Bansal, Sunil Bansal, Baldev Singh, Khyati Y. Mehta; writing—review and editing, Shreyans K. Jain, Shivani Bansal, Sunil Bansal; writing—review and editing, Shreyans K. Jain, Shivani Bansal, Sunil Bansal, William Klotzbier, Amrita Cheema; visualization, Shreyans K. Jain, Shivani Bansal, Sunil Bansal; supervision, Amrita Cheema; project administration, Amrita Cheema, funding acquisition: Amrita Cheema.
Funding
The authors would like to acknowledge the Metabolomics Shared Resource in Georgetown University (Washington DC, USA) partially supported by NIH/NCI/CCSG grant P30-CA051008.
Acknowledgments
The authors would also like to acknowledge the Metabolomics Shared Resource in Georgetown University (Washington DC, USA) partially supported by NIH/NCI/CCSG grant P30-CA051008.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wishart, D. S.; Knox, C.; Guo, A. C.; Eisner, R.; Young, N.; Gautam, B.; Hau, D. D.; Psychogios, N.; Dong, E.; Bouatra, S.; Mandal, R.; Sinelnikov, I.; Xia, J.; Jia, L.; Cruz, J. A.; Lim, E.; Sobsey, C. A.; Shrivastava, S.; Huang, P.; Liu, P.; Fang, L.; Peng, J.; Fradette, R.; Cheng, D.; Tzur, D.; Clements, M.; Lewis, A.; De Souza, A.; Zuniga, A.; Dawe, M.; Xiong, Y.; Clive, D.; Greiner, R.; Nazyrova, A.; Shaykhutdinov, R.; Li, L.; Vogel, H. J.; Forsythe, I., HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res 2009, 37 (Database issue), D603-10. [CrossRef]
- Holm, E.; Hagmüller, E.; Staedt, U.; Schlickeiser, G.; Günther, H. J.; Leweling, H.; Tokus, M.; Kollmar, H. B., Substrate balances across colonic carcinomas in humans. Cancer Res 1995, 55 (6), 1373-8.
- Shim, H.; Dolde, C.; Lewis, B. C.; Wu, C. S.; Dang, G.; Jungmann, R. A.; Dalla-Favera, R.; Dang, C. V., c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci U S A 1997, 94 (13), 6658-63. [CrossRef]
- Doherty, J. R.; Cleveland, J. L., Targeting lactate metabolism for cancer therapeutics. J Clin Invest 2013, 123 (9), 3685-92. [CrossRef]
- Warburg, O., On the origin of cancer cells. Science 1956, 123 (3191), 309-14. [CrossRef]
- Linehan, W. M.; Rouault, T. A., Molecular pathways: Fumarate hydratase-deficient kidney cancer--targeting the Warburg effect in cancer. Clin Cancer Res 2013, 19 (13), 3345-52. [CrossRef]
- Chatthanawaree, W., Biomarkers of cobalamin (vitamin B12) deficiency and its application. J Nutr Health Aging 2011, 15 (3), 227-31. [CrossRef]
- Makahleh, A.; Ben-Hander, G. M.; Saad, B., Determination of alpha-ketoglutaric and pyruvic acids in urine as potential biomarkers for diabetic II and liver cancer. Bioanalysis 2015, 7 (6), 713-23. [CrossRef]
- Johnson, D. W., Contemporary clinical usage of LC/MS: analysis of biologically important carboxylic acids. Clin Biochem 2005, 38 (4), 351-61. [CrossRef]
- Halket, J. M.; Waterman, D.; Przyborowska, A. M.; Patel, R. K.; Fraser, P. D.; Bramley, P. M., Chemical derivatization and mass spectral libraries in metabolic profiling by GC/MS and LC/MS/MS. J Exp Bot 2005, 56 (410), 219-43. [CrossRef]
- Kiefer, P.; Delmotte, N.; Vorholt, J. A., Nanoscale ion-pair reversed-phase HPLC-MS for sensitive metabolome analysis. Anal Chem 2011, 83 (3), 850-5. [CrossRef]
- Luo, B.; Groenke, K.; Takors, R.; Wandrey, C.; Oldiges, M., Simultaneous determination of multiple intracellular metabolites in glycolysis, pentose phosphate pathway and tricarboxylic acid cycle by liquid chromatography-mass spectrometry. J Chromatogr A 2007, 1147 (2), 153-64. [CrossRef]
- Buescher, J. M.; Moco, S.; Sauer, U.; Zamboni, N., Ultrahigh performance liquid chromatography-tandem mass spectrometry method for fast and robust quantification of anionic and aromatic metabolites. Anal Chem 2010, 82 (11), 4403-12. [CrossRef]
- Lu, W.; Clasquin, M. F.; Melamud, E.; Amador-Noguez, D.; Caudy, A. A.; Rabinowitz, J. D., Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone orbitrap mass spectrometer. Anal Chem 2010, 82 (8), 3212-21. [CrossRef]
- Li, Z.; Han, J.; Sun, S. A.; Chen, K.; Tang, D. Q., Hydrophilic Interaction Liquid Chromatography/Mass Spectrometry: An Attractive and Prospective Method for the Quantitative Bioanalysis in Drug Metabolism. Curr Drug Metab 2016, 17 (4), 386-400. [CrossRef]
- Buszewski, B.; Noga, S., Hydrophilic interaction liquid chromatography (HILIC)--a powerful separation technique. Anal Bioanal Chem 2012, 402 (1), 231-47. [CrossRef]
- Schiesel, S.; Lammerhofer, M.; Lindner, W., Multitarget quantitative metabolic profiling of hydrophilic metabolites in fermentation broths of beta-lactam antibiotics production by HILIC-ESI-MS/MS. Anal Bioanal Chem 2010, 396 (5), 1655-79. [CrossRef]
- Bajad, S. U.; Lu, W.; Kimball, E. H.; Yuan, J.; Peterson, C.; Rabinowitz, J. D., Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J Chromatogr A 2006, 1125 (1), 76-88. [CrossRef]
- Yuan, M.; Breitkopf, S. B.; Yang, X.; Asara, J. M., A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc 2012, 7 (5), 872-81. [CrossRef]
- Yang, W. C.; Sedlak, M.; Regnier, F. E.; Mosier, N.; Ho, N.; Adamec, J., Simultaneous quantification of metabolites involved in central carbon and energy metabolism using reversed-phase liquid chromatography-mass spectrometry and in vitro 13C labeling. Anal Chem 2008, 80 (24), 9508-16. [CrossRef]
- Bollinger, J. G.; Thompson, W.; Lai, Y.; Oslund, R. C.; Hallstrand, T. S.; Sadilek, M.; Turecek, F.; Gelb, M. H., Improved sensitivity mass spectrometric detection of eicosanoids by charge reversal derivatization. Anal Chem 2010, 82 (16), 6790-6. [CrossRef]
- Yang, W. C.; Regnier, F. E.; Adamec, J., Comparative metabolite profiling of carboxylic acids in rat urine by CE-ESI MS/MS through positively pre-charged and (2)H-coded derivatization. Electrophoresis 2008, 29 (22), 4549-60. [CrossRef]
- Yang, W. C.; Adamec, J.; Regnier, F. E., Enhancement of the LC/MS analysis of fatty acids through derivatization and stable isotope coding. Anal Chem 2007, 79 (14), 5150-7. [CrossRef]
- Kloos, D.; Derks, R. J.; Wijtmans, M.; Lingeman, H.; Mayboroda, O. A.; Deelder, A. M.; Niessen, W. M.; Giera, M., Derivatization of the tricarboxylic acid cycle intermediates and analysis by online solid-phase extraction-liquid chromatography-mass spectrometry with positive-ion electrospray ionization. J Chromatogr A 2012, 1232, 19-26. [CrossRef]
- Koulman, A.; Petras, D.; Narayana, V. K.; Wang, L.; Volmer, D. A., Comparative high-speed profiling of carboxylic acid metabolite levels by differential isotope-coded MALDI mass spectrometry. Anal Chem 2009, 81 (18), 7544-51. [CrossRef]
- Williams, J.; Pandarinathan, L.; Wood, J.; Vouros, P.; Makriyannis, A., Endocannabinoid metabolomics: a novel liquid chromatography-mass spectrometry reagent for fatty acid analysis. Aaps j 2006, 8 (4), E655-60. [CrossRef]
- Bartók, T.; Komoróczy, R.; Börcsök, G.; Sági, F.; Hegyes, P.; Bartók, M., A New Derivatization Reaction for the Identification and Determination of Organic Acids by Positive-ion Electrospray Mass Spectrometry. Rapid Communications in Mass Spectrometry 1997, 11 (1), 133-135.
- Zhu, Q. F.; Zhang, Z.; Liu, P.; Zheng, S. J.; Peng, K.; Deng, Q. Y.; Zheng, F.; Yuan, B. F.; Feng, Y. Q., Analysis of liposoluble carboxylic acids metabolome in human serum by stable isotope labeling coupled with liquid chromatography-mass spectrometry. J Chromatogr A 2016, 1460, 100-9. [CrossRef]
- Yuan, B. F.; Zhu, Q. F.; Guo, N.; Zheng, S. J.; Wang, Y. L.; Wang, J.; Xu, J.; Liu, S. J.; He, K.; Hu, T.; Zheng, Y. W.; Xu, F. Q.; Feng, Y. Q., Comprehensive Profiling of Fecal Metabolome of Mice by Integrated Chemical Isotope Labeling-Mass Spectrometry Analysis. Anal Chem 2018, 90 (5), 3512-3520. [CrossRef]
- Marquis, B. J.; Louks, H. P.; Bose, C.; Wolfe, R. R.; Singh, S. P., A New Derivatization Reagent for HPLC-MS Analysis of Biological Organic Acids. Chromatographia 2017, 80 (12), 1723-1732. [CrossRef]
- Jiang, R.; Jiao, Y.; Zhang, P.; Liu, Y.; Wang, X.; Huang, Y.; Zhang, Z.; Xu, F., Twin Derivatization Strategy for High-Coverage Quantification of Free Fatty Acids by Liquid Chromatography-Tandem Mass Spectrometry. Anal Chem 2017, 89 (22), 12223-12230. [CrossRef]
- Han, J.; Gagnon, S.; Eckle, T.; Borchers, C. H., Metabolomic analysis of key central carbon metabolism carboxylic acids as their 3-nitrophenylhydrazones by UPLC/ESI-MS. Electrophoresis 2013, 34 (19), 2891-900. [CrossRef]
- Han, J.; Lin, K.; Sequeira, C.; Borchers, C. H., An isotope-labeled chemical derivatization method for the quantitation of short-chain fatty acids in human feces by liquid chromatography-tandem mass spectrometry. Anal Chim Acta 2015, 854, 86-94. [CrossRef]
- Harder, U.; Koletzko, B.; Peissner, W., Quantification of 22 plasma amino acids combining derivatization and ion-pair LC-MS/MS. J Chromatogr B Analyt Technol Biomed Life Sci 2011, 879 (7-8), 495-504. [CrossRef]
- O’Maille, G. G., E. P.; Hoang, L.; Want, E. J.; Smith, C.; O’Maille, P.; Nordstrom, A.; Morita, H.; Qin, C.; Uritboonthai, W.; Apon, J.; Moore, R.; Garrett, J.; Siuzdak, G. , Metabolomics relative quantitation with mass spectrometry using chemical derivatization and isotope labeling. Spectroscopy 2008, 22, 327-343. [CrossRef]
- Phillips, M. A., CCCXVII.—The formation of 2-substituted benziminazoles. J. Chem. Soc. 1928, 0 (0), 2393-2399.
- Fujiwara, T.; Inoue, R.; Ohtawa, T.; Tsunoda, M., Liquid-Chromatographic Methods for Carboxylic Acids in Biological Samples. Molecules (Basel, Switzerland) 2020, 25 (21). [CrossRef]
- Kurata, T.; Sakurai, Y., Degradation of L-Ascorbic Acid and Mechanism of Nonenzymic Browning Reaction. Agricultural and Biological Chemistry 2014, 31 (2), 170-184. [CrossRef]
- Umbdenstock, R. R.; Bruins, P. F., Aconitic acid from citric acid by catalytic dehydration. Ind. Eng. Chem. 1945, 37 (10), 963-967. [CrossRef]
- Ni, Q.; Reid, K. R.; Burant, C. F.; Kennedy, R. T., Capillary LC-MS for high sensitivity metabolomic analysis of single islets of Langerhans. Anal Chem 2008, 80 (10), 3539-46. [CrossRef]
- Keefer, J. F.; Schuster, S. M., Separation of citric acid cycle intermediates by high-performance liquid chromatography with ion pairing. Journal of Chromatography B: Biomedical Sciences and Applications 1986, 383, 297-305. [CrossRef]
- Yang, W.-C.; Adamec, J.; Regnier, F. E., Enhancement of the LC/MS analysis of fatty acids through derivatization and stable isotope coding. Anal. Chem. 2007, 79 (14), 5150-5157. [CrossRef]
- Ehrmann, B. M.; Henriksen, T.; Cech, N. B., Relative importance of basicity in the gas phase and in solution for determining selectivity in electrospray ionization mass spectrometry. J Am Soc Mass Spectrom 2008, 19 (5), 719-28. [CrossRef]
- Chan, A. K.; Bruce, J. I.; Siriwardena, A. K., Glucose metabolic phenotype of pancreatic cancer. World J Gastroenterol 2016, 22 (12), 3471-85. [CrossRef]
- Hui, S.; Ghergurovich, J. M.; Morscher, R. J.; Jang, C.; Teng, X.; Lu, W.; Esparza, L. A.; Reya, T.; Le, Z.; Yanxiang Guo, J.; White, E.; Rabinowitz, J. D., Glucose feeds the TCA cycle via circulating lactate. Nature 2017, 551 (7678), 115-118. [CrossRef]
- Patel, S.; Ahmed, S., Emerging field of metabolomics: big promise for cancer biomarker identification and drug discovery. J Pharm Biomed Anal 2015, 107, 63-74. [CrossRef]
- Nishiumi, S.; Shinohara, M.; Ikeda, A.; Yoshie, T.; Hatano, N.; Kakuyama, S.; Mizuno, S.; Sanuki, T.; Kutsumi, H.; Fukusaki, E.; Azuma, T.; Takenawa, T.; Yoshida, M., Serum metabolomics as a novel diagnostic approach for pancreatic cancer. Metabolomics 2010, 6 (4), 518-528. [CrossRef]
- Long, N. P.; Yoon, S. J.; Anh, N. H.; Nghi, T. D.; Lim, D. K.; Hong, Y. J.; Hong, S. S.; Kwon, S. W., A systematic review on metabolomics-based diagnostic biomarker discovery and validation in pancreatic cancer. Metabolomics 2018, 14 (8), 109. [CrossRef]
- Rajagopal, M. U.; Bansal, S.; Kaur, P.; Jain, S. K.; Altadil, T.; Hinzman, C. P.; Li, Y.; Moulton, J.; Singh, B.; Bansal, S.; Chauthe, S. K.; Singh, R.; Banerjee, P. P.; Mapstone, M.; Fiandaca, M. S.; Federoff, H. J.; Unger, K.; Smith, J. P.; Cheema, A. K., TGFβ Drives Metabolic Perturbations during Epithelial Mesenchymal Transition in Pancreatic Cancer: TGFβ Induced EMT in PDAC. Cancers (Basel) 2021, 13 (24). [CrossRef]
- Roth, H. E.; Powers, R., Meta-Analysis Reveals Both the Promises and the Challenges of Clinical Metabolomics. Cancers (Basel) 2022, 14 (16). [CrossRef]
- Tumas, J.; Kvederaviciute, K.; Petrulionis, M.; Kurlinkus, B.; Rimkus, A.; Sakalauskaite, G.; Cicenas, J.; Sileikis, A., Metabolomics in pancreatic cancer biomarkers research. Med Oncol 2016, 33 (12), 133. [CrossRef]
- Yang, J. S.; Wang, C. C.; Qiu, J. D.; Ren, B.; You, L., Arginine metabolism: a potential target in pancreatic cancer therapy. Chin Med J (Engl) 2020, 134 (1), 28-37. [CrossRef]
- Xiong, Y.; Shi, C.; Zhong, F.; Liu, X.; Yang, P., LC-MS/MS and SWATH based serum metabolomics enables biomarker discovery in pancreatic cancer. Clin Chim Acta 2020, 506, 214-221. [CrossRef]
- Herner, A.; Sauliunaite, D.; Michalski, C. W.; Erkan, M.; De Oliveira, T.; Abiatari, I.; Kong, B.; Esposito, I.; Friess, H.; Kleeff, J., Glutamate increases pancreatic cancer cell invasion and migration via AMPA receptor activation and Kras-MAPK signaling. Int J Cancer 2011, 129 (10), 2349-59. [CrossRef]
- Bott, A. J.; Shen, J.; Tonelli, C.; Zhan, L.; Sivaram, N.; Jiang, Y. P.; Yu, X.; Bhatt, V.; Chiles, E.; Zhong, H.; Maimouni, S.; Dai, W.; Velasquez, S.; Pan, J. A.; Muthalagu, N.; Morton, J.; Anthony, T. G.; Feng, H.; Lamers, W. H.; Murphy, D. J.; Guo, J. Y.; Jin, J.; Crawford, H. C.; Zhang, L.; White, E.; Lin, R. Z.; Su, X.; Tuveson, D. A.; Zong, W. X., Glutamine Anabolism Plays a Critical Role in Pancreatic Cancer by Coupling Carbon and Nitrogen Metabolism. Cell Rep 2019, 29 (5), 1287-1298 e6. [CrossRef]
- Xie, G.; Lu, L.; Qiu, Y.; Ni, Q.; Zhang, W.; Gao, Y. T.; Risch, H. A.; Yu, H.; Jia, W., Plasma metabolite biomarkers for the detection of pancreatic cancer. J Proteome Res 2015, 14 (2), 1195-202. [CrossRef]
- Takehara, A.; Hosokawa, M.; Eguchi, H.; Ohigashi, H.; Ishikawa, O.; Nakamura, Y.; Nakagawa, H., Gamma-aminobutyric acid (GABA) stimulates pancreatic cancer growth through overexpressing GABAA receptor pi subunit. Cancer Res 2007, 67 (20), 9704-12. [CrossRef]
- Medina, V. A.; Rivera, E. S., Histamine receptors and cancer pharmacology. Br J Pharmacol 2010, 161 (4), 755-67. [CrossRef]
- Das, U. N., Essential fatty acids: biochemistry, physiology and pathology. Biotechnol J 2006, 1 (4), 420-39. [CrossRef]
- Serhan, C. N.; Clish, C. B.; Brannon, J.; Colgan, S. P.; Chiang, N.; Gronert, K., Novel Functional Sets of Lipid-Derived Mediators with Antiinflammatory Actions Generated from Omega-3 Fatty Acids via Cyclooxygenase 2–Nonsteroidal Antiinflammatory Drugs and Transcellular Processing. Journal of Experimental Medicine 2000, 192 (8), 1197-1204. [CrossRef]
- Das, U. N., Essential Fatty Acids and Their Metabolites in the Pathobiology of Inflammation and Its Resolution. Biomolecules 2021, 11 (12). [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).