1. Introduction
The world population is about to reach 10 billion in the coming era. This causes the global need for food followed by production [
1] pressing the global need to increase food production. However, the reduction of agricultural land caused by rapid urbanization and industrialization and extreme weather accompanied by environmental stressors like climate change and global warming limit agriculture and food production, which may threaten food security [
2]. Production may be slightly enhanced by some of the previously developed technologies that can help in crop improvement. The method used in plant breeding for the purpose of improving the yield of crops is characterized by the use of selective breeding and hybridization. This process is largely dependent on the production of genetic diversity through homologous re-arrangement between chromosome pairs, which has been practiced since ancient times. Plant breeders have also looked into using chemicals or irradiation to broaden the range of natural variation for characteristics. Although traditional and mutational breeding techniques have yielded considerable improvements in various agronomic attributes, such as nutritional needs, yield, quality, and resistance to stresses, there are limitations to labor, time, and exact knowledge of selection. CRISPR/Cas9 can improve crop performance by increasing yields, enhancing nutrition, and fighting biotic and abiotic stresses [
3]. Biotechnological methods have advanced to the point where researchers have been able to incorporate desired characteristics into crops through the incorporation of transgenic genes from other organisms in the plant genome, a process commonly referred to as “transgenesis”. The transgenic techniques have possessed faster, accessibility and also easy producibility. Various techniques have been developed which allow us to modify genetic sequences specifically, summarized in
(Figure 1). Another barrier to de novo domestication through gene editing is the lack of expertise of botanists in wild-plant biology [
4]. New technologies such as, transcription activator-like effector nucleases (TALENs), RNAi (RNAi), zinc finger nucleases (ZFNs) and CRISPR (Cas9) have revolutionized crop engineering by making it easier to create crops with higher yield and other desirable characteristics. Among them, the one of the best state-of-the-art technology is CRISPR/Cas genome editing for developing high-yielding, nutrition-rich crops that are more tolerant of both biotic and abiotic stressors [
5]. CRISPR/Cas9 in offspring can cause unpredictable traits, posing research and commercial application challenges due to ongoing editing activity [
6]. Genetic alteration can be achieved by the method of gene editing, which involves altering or removing an organism’s DNA sequence. Actually, the CRISPR–Cas systems originate from the defence mechanism that bacteria use based on RNA, and they recognize and eliminate foreign DNA from invading plasmids and bacteriophages and are thus considered a type of bacterial “immune system”.
Overall the, CRISPR/Cas9 modifies genes through exact cutting of DNA, followed by the natural mechanisms of DNA repair [
5,
7]. The adoption of genome-edited plants by the general public and regulatory agencies is also impacted by these strategies [
8].
Although gene knockout is the most commonly used use of the CRISPR-Cas9 system, it has also been successfully used for gene regulation and gene knock-in. (Zhao et al., 2019) [
9]. The various applications of CRISPR-cas9 in agricultural crop refinement have been also described earlier [
5,
7,
10,
11]. The application of CRISPR/Cas has been successfully applied in editing the genome of many plant species, including Arabidopsis [
12], tobacco [
13], rice [
14], tomato [
15], maize [
16], wheat [
17], potato [
18,
19], Brassica [
20] and others, Clearly, its application has been proven to provide valuable trait improvement in multiple crop plants [
10].
In this study, we provide an overview of the most recent developments in CRISPR/Cas technology, the functional processes of all identified Cas proteins, and their use in contemporary horticulture and agriculture.
2. History of CRISPR/Cas System
This system refers to the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR), which are CRISPR-associated proteins. It is a revolutionary genome-editing mechanism that allows researchers to modify the DNA of various organisms, including humans, animals, plants, and microorganisms. Since the
E. coli K-12 strain’s IAP isozyme conversion is caused by the analysis of the alkyl phosphatase gene (IAP), CRISPRs have their roots in this process. They discovered a section of the genome on the 3′ ends of the iap gene with 32 nucleotides of distinct sequences surrounded by persistent palindromic repeats. Later, particular parallel sequences have been identified in
Salmonella Enterica and
Shigella dysenteries, two types of enterobacteria, as well as other
E. coli strains. Similar to this, when examining Mycobacterium TB strains, researchers found 36 bp repetitions scattered with unique 35–41 bp spacers. In the ensuing investigations, the CRISPR array was found in the genomes of 40% of archaea and 90% of bacteria, specifically in
Haloferax mediterranei and Streptococcus thermopile. However, this genetic sequence initially demonstrated to reflect the overall form of the CRISPR array. This genetic sequence, however, first proved to represent the CRISPR array’s general shape. The biological purpose of CRISPR was still a mystery due to a lack of required spacer or protospacer genome sequence data. The CRISPR-associated genes were first reported by Yoshizumi Ishino and colleagues in their ground-breaking paper published in 1987. This was followed by the identification of other CRISPR/Cas genes. Spacer sequences are short, single-nucleotide polymorphic (SN polymorphic) DNA sequences found in CRISPR arrays of bacterial or archaeal genomic material. Spacer DNA is also referred to as spacer or simply spacer. Spacer sequences are a vital component of the immune system involved in the CRISPR /Cas [
21].
3. Nomenclature of CRISPR/Cas System
The CRISPR system has been divided into two main categories based on the complexity of the effector proteins involved in the interference stage of the system. In the Class 1 system, multiple effector proteins are required for the RNA-guided target cleavage, while in Class 2, only one RNA-guided endonuclease is required for the DNA sequence cleavage. Three types of CRISPR present in the Class 1 system: I, III, and IV, while three types present in the Class 2 system: II, V, and VI. The Cas3 signature gene, which codes for an immense protein containing a helicase to unravel DNA-DNA and RNA-DNA duplexes, is found in the type I system’s CRISPR/Cas locus. The multidomain Cas9 (CRISPR-associated protein 9) that the type II locus generates cleaves and targets dsDNA. The cas10 signature gene is present in type III CRISPR/Cas, along with a multidomain protein with a palm domain that can target ssDNA. The ribonucleic protein encoded by the CRISPR-associated splicing factor systems, although its precise function is still unknown. The RuvC gene is encoded by the CRISPR from
Prevotella and
Francisella 1 (Cpf1), C2c1 or C2c3 protein, which is carried by the type V locus and carries the Cas12 signature gene (a DNA repair-related
E. coli protein) domain. This domain cleaves either dsDNA or ssDNA. Type VI has a nucleotide-binding domain (HEPN) called Cas13 (C2c2) that is responsible for cleaving ssRNA in higher eukaryotes and prokaryotes
(Table 1).
A novel microbial defence mechanism that guards against infections and mobile inheritable primitives has been found by scientists. The CRISPR/Cas system is one of the defence mechanisms that bacteria and archaea have developed.
(Figure 2). Bacteria and archaea produce cellular memory using DNA sequences that are compatible with those of previous raiders in their genomes. These ways include (i) fitting a short overrunning DNA as a spacer sequence into the CRISPR network (ii) recap of CRISPR precursor RNA (pre-cRNA); It develops into separate cRNAs, each made up of a raider-targeting spacer member and a reprise member. And (iii) crRNA-directed fractionalization of foreign nucleic acids by Cas proteins at spots reciprocal to crRNA spacer sequences. The three CRISPR-Cas systems (I, II, and III) within this broad theme achieve nucleic acid recognition and fractionalization by distinct molecular mechanisms. Motifs bordering crRNA target sequences on overrunning DNA play important places in the adaption and hindrance way in both type I and type II systems. Type I and III systems employ big Cas protein complexes for cRNA targeting. still, in type II systems, only one protein is needed to fete and stick DNA by RNA, which has proven veritably useful for genome engineering operations organism. The targeted genome editing technique of using finaxel nucleases has quickly evolved from a specialised technique to a widely accepted platform utilised by many natural experimenters. The development of CRISPR technology, a novel method for producing RNA-guided nucleases, such as Cas9, has significantly contributed to this widespread renunciation. These CRISPR-Cas9 genome editing systems act as an intermediary in the process of genome editing. Using adaptable and vulnerable CRISPR systems, bacteria can defend themselves against foreign nucleic acids, such as viruses and plasmids. The bacterial host genome’s CRISPR repeat sequences are decoded as arrays, and type II CRISPR systems integrate sequences from overrunning DNA between them. The CRISPR reprise arrays are repeated to form crRNA, each containing a different sequence taken from the DNA it overlaps, called the “protospacer” sequence. This sequence makes up a part of the CRISPR reprise. The trans-activating crRNA (tracrRNA), an alternative RNA that hybridises with each crRNA, forms a complex with the Cas9 nuclease. Only when PAMs are located conterminous to the protospacer-decoded portion of the crRNA will Cas9 be directed to stick reciprocal target-DNA sequences. The Streptococcus pyogenes type II CRISPR system has been adapted for targeted genome editing and the conversion of sequence-specific DSBs. Genome editing can be achieved by incorporating Cas9 nuclease and a companion RNA into cells or an organism, which is analogous to an emulsion of a crRNA and a tracrRNA. These components need to be expressed for the editing process to occur. The use of manipulated nucleases for precise genome editing has rapidly become a widely adopted technique among many researchers, transitioning from a specialized technology to a common approach. The introduction of CRISPR technology, a novel method for producing RNA-guided nucleases (such as Cas9), has significantly contributed to this widespread renunciation. Nucleases have facilitated genome editing, which has been utilized to modify endogenous genes snappily, fluently, and effectively for various biomedical purposes. Several bacteria utilise the CRISPR system, an adaptable and fragile medium, to protect themselves against external nucleic acids like plasmids and contagions in the genome of the host. The CRISPR reprise template repeats are translated into cRNA, each with a segment of the CRISPR reprise plus a variable sequence called the “protospacer” sequence transcribed from the overrunning DNA. The nuclease Cas9 forms a complex with each cRNA when it hybridises with an alternative RNA known as trans-activating cRNA(track). Cas9 is directed to stick reciprocal target DNA sequences only when PAM is located near the protospace garbling portion of the cRNA. The Streptococcus pyogenes type II CRISPR system has been adapted to create sequence-specific DSBs and enable targeted genome editing. This process involves a mixture of Cas9 nuclease and companion RNA, which, when combined with tra cRNA, can be integrated and expressed in cells or organisms to carry out genome editing. Several natural experimenters are utilising targeted genome editing with modifying nucleases, which has quickly moved from a niche to a mainstream practice. This wide relinquishment was greatly eased by the emergence of his CRISPR technology, a new approach for designing RNA-guided nucleases similar to Cas9. These nucleases’ role as intermediates in genome editing allows for the quick, fluid, and effective modification of endogenous genes in a wide range of biologically useful cells and species that are typically sensitive to genetic alterations. It’s been done. The capability of these systems to deliver targeted and largely effective changes in genome sequencing and gene expression will surely transfigure natural exploration and ease the development of new molecular rectifiers for humans [
5].
4. Cas Proteins of the CRISPR System
CRISPR-associated protein systems (CRISPR/Cas) are revolutionizing life wisdom exploration to manipulate, describe, fantasize, and annotate DNA sequences or specific RNAs from a variety of organisms
(Table 2). It offers enormous openings for attachment. The system incorporates foreign DNA fractions (spacers) into CRISPR cassettes, transcribes them onto the CRISPR chip, and processes them to produce companion RNAs (gRNAs). A CRISPR chip is a gene that encodes a Cas protein. Cas proteins give the enzymatic mechanisms necessary for the accession of new spacers that target irruption rudiments. Many Cas proteins, including Cas9, Cas12, Cas13, and Cas14, have been exploited to generate novel equipments for genetic engineering because of their programmable sequence particularity. [
22]
(Table 3).
4.1. Cas 1 and Cas 2 Proteins
The first step in CRISPR-Cas immunization is to obtain a spacer DNA segment distinct from the host’s CRISPR genomic locus. The only conserved proteins in all CRISPR-Cas systems are the nucleases Cas1 and Cas2, yet it is unclear what role these proteins play molecularly during immunisation. To confer immunity, CRISPR RNAs (crRNAs) from the CRISPR8–12 network are used by Cas proteins as molecular guides for base-pairing with complementary sequences of foreign DNA, inducing their degradation during the step of interference.
Mechanism and Application of Cas 1 and Cas 2 Proteins
Cas1 and Cas2 are two proteins found in
E. coli K12, forming a hexameric complex. Cas1 is an asymmetric homodimer with a central ferredoxin fold, while Cas2 is a symmetric homodimer with a central ferredoxin fold [
23]. The complex produces crystals diffracting X-rays with a resolution of 2.3 Å. Both are heterozygous molecules that catalyse spacer integration via trans esterification reactions.
4.2. Cas9 Protein
Cas9, also known as SpyCas9 protein, is a protein connected to the S. pyogenes CRISPR mechanisms of adaptive immunity. In natural and synthetic CRISPR/Cas systems, the SpyCas9 protein functions as a DNA endonuclease and Contains 1368 amino acids in its big multifunctional domain. The genetic editing technique of CRISPR/Cas-9 type II is extensively researched and applied [
24]. The bacteria
Streptococcus pyogenes provided the Cas-9 protein, which was the first Cas protein to be employed in genome editing (SpCas-9). Cas-9 is a large multidomain DNA endonuclease that cleaves target DNA to form duplexes and is known as genome scissors. The nuclease portion of Cos-9 is divided into two sections (NUC). The interaction domains RuvC, HNH, and Protospacer Adjacent Motive (PAM) make up the NUC component, whereas the REC part is composed of the REC1 and REC2 domains, which are in charge of binding guide RNA. Target DNA binding is initiated by the PAM-interacting domain, which confers PAM specificity. Single-stranded DNA is broken by the RuvC and HNH domains. The size of the crRNA is 18–20 bp. It uses its binding to the target sequence to recognise the target DNA, tracrRNA is a lengthy loop that acts as a support structure for the nuclear binding of Cas-9. While guide RNAs are utilised in prokaryotes to target viral DNA, gene editing techniques can target nearly any gene sequence by combining and intentionally modifying rRNA and tRNA.
4.2.1. Mechanism of Cas9 Protein and Application
The Cas9 protein consisting of six domains, is responsible for binding with gRNA and cleaving specific sequences. Its primary function is to cut three base pairs upstream of the PAM sequence in dsDNA. Since the CRISPR/Cas9 system was created by fusing twin tracrRNA: crRNA into a single guide RNA (sgRNA), it can cut particular dsDNA or ssDNA sequences.
(Figure 3). This protein has significant potential for genome engineering applications in science, medicine, and biotechnology. More comprehensive exploration of genome function can be achieved through large-scale genome editing experiments compared to traditional methods like as TALEN and ZFN. The transcription of specific genomic loci can be modified, or the genome can be reorganized using the CRISPR/Cas9 system. This expands the range of possible genome engineering modifications. Researchers have widely adopted this system in various fields, including microorganisms, plants, animals, insects, and human cell lines [
25]
(Figure 4).
4.2.2. Pros and Cons of Cas9 Protein
The mechanism of CRISPR/Cas9 is preferred over ZFN and TALEN systems due to its simple design and higher effectiveness. The ability to create several sequence-specific gRNAs simultaneously using Cas9 allows for multiplex genome editing, another significant advantage of the technology. Despite numerous advances, CRISPR/ Cas9 systems have numerous failings that raise numerous questions about the pitfalls involved in editing. As an example, target organisms exhibiting sophisticated on- and off-target mutations were shown by gRNAs instructing Cas9 to stick dsDNA or ssDNA. Indeed, the stylish CRISPR/ Cas systems with HDR produce unwanted mutations. Still, these off-target goods can be eased by using a modified interpretation of Cas9 called Null-Cas9 (nCas9). This creates a larger gap in single beaches than in DSB. Still, nCas9 cannot reduce target divagation by 100, so we need a streamlined interpretation of Cas9 in the future [
26].
4.3. Cas12 Protein
The CRISPR-Cas system, which was partially repurposed as a programmable genome-editing tool, was identified as an adaptively susceptible mechanism in prokaryotes. [
27]. In programmable genome editing, the Cas9 and Cas12a proteins are widely utilised; their unique characteristics may influence editing issues. The ability of Cas12a to perform double-nuclease exertion under the control of CRISPR RNA (crRNA) (cis exertion) double-stranded target DNA (dsDNA) and destroy non-specifically represents a potentially significant distinction in the biochemical behaviour of Cas9 and Cas12a. Targeted DNA bifurcation (trans-activation) by single-stranded DNA (ssDNA) and cRNA after binding. The trans-ssDNase exertion has been employed in viral diagnostics and CCas12a-interceded nucleic acid finding [
28]. Still, it’s not fully understood how trans nuclease exertion affects the outgrowth of genome editing.
4.3.1. Mechanism and Application of Cas12 Protein
Cas12a, a CRISPR-Cas protein, is responsible for cleaving double-stranded target DNA in a well-defined order with a cis force and anon-specific single beachfront fractionalization with a trans force. It has been shown to stick to beachfront DNA response steps. Mutations in the bridging helix (BH) region may intrude with recap while maintaining target-specific cis force [
29]
(Figure 5). The BH of the bacteria ND2006 Cas12a (LbCas12a) belonging to the family
Lachnospiraceae experiences a loss of trans force and a minor decrease in cis force due to the W890A mutation. Combining salutary mutations with directed elaboration, researchers generated a Cas12a variant with high cis-nuclease exertion and minimum trans-ssDNase exertion [
30]. The utility of genome manipulation and nucleic acid discovery using CRISPR-Cas12a can be improved by modifying its binary exertion, as demonstrated in this study (
Figure 6a,b).
4.2.2. Pros and Cons of Cas12 Protein
Cas12 protein was also once thought to be the only member of the CRISPR family that could change the genome, similar to Cas9. Cas12 protein is often considered superior to Cas9 in many situations because it generates staggered double-stranded breaks, promoting homology-directed repair mechanisms rather than non-homologous end joining and homology-directed repair [
31]. Cas12 framework moreover overcomes drawbacks related to symptomatic methodologies. For instance, utilising quantitative qRT-PCR to diagnose SARS-COV-2 takes longer than expected. However, the CRISPR/Cas12-based DETECTR identifies the SARS-CoV-2 in less than an hour. The CRISPR/Cas12 technology can be used in the future to identify recently found ailments, as demonstrated by these results that demonstrate its utility. The CRISPR-Cas12 genome editing framework’s rapid advancement has shown advancements in the biological sciences. Although this technology has many applications, the CRISPR/Cas12 architecture has some notable flaws. For example, the CRISPR/Cas12 framework is dependent upon the presence of a cell DNA repair mechanism, whether or not a layout is present. [
32]. The feasibility of this method varies depending on the type of cell, despite the fact that it has been effectively utilised to guarantee precise DNA insertion into the specified genomic loci. Since DNA repair by HDR is also linked to dynamic cell division, these methods are ineffective in certain situations, such as neuronal cell division. In any case, noteworthy proceeding investigates points to tailor the Cas12 framework encourage to guarantee exact DNA inclusion into the focused genome [
33]. Separated from this disadvantage, this framework contains a wide run of pertinence, and continuous endeavors are endeavoring to create upgraded CRISPR/Cas12 for vigorous genome building.
4.4. Cas13 Protein
Cas13 proteins are display in at slightest 21 bacterial genomes. They all have two HEPN spaces, but the measure of the full-length protein can change depending on which microscopic organisms they come from. That’s why they are subclassified as Cas13 a, b, c, d, etc. Moreover, since the arrangement of the hairpin-like structure they tie to is distinctive among the distinctive species, the logical title of the microbes they come from is often included within the title; for case, LbaCa13a comes from Lachnospiracea bacterium.
Figure 2 shows how diverse Cas13 proteins compare. Since its disclosure and characterization, this programmable RNA official protein has changed the way we quiet quality expression and how we consider, alter, track, and identify RNA [
34].
4.4.1. Mechanism and Applications of Cas13 Protein
The double Cas13a-crRNA complexes share a common crRNA structure with RcCas13a, which recognizes its ~58 nt long crRNA transcendentally. Base-specific connections with an A and a G are made by LbuCas13a and LseCas13a. [
35]
(Figure 7). According to research, RcCas13a can prepare pre-crRNA without the aid of metal particles and does not require catalytic HEPN-domains for this purpose [
36]. LbaCas13a and RcaCas13a both utilize build-ups within the HEPN2 space for pre-crRNA processing. Cas13 is primarily used for knocking down mRNAs in eukaryotic cells, offering over 90% knock-down productivity and less off-target effects than RNA impedances
(Figure 8).
4.4.2. Pros and Cons of Cas13 Protein
CRISPR /Cas13 is also an efficient, accurate, and flexible RNA targeting framework, similar to that of Cas9 or Cas12, which provides new opportunities for research in several areas. CRISPR/Cas13 offers a number of interesting differences from earlier RNA control frameworks. For illustration, its measured construction provides both a simple and fast plan and noteworthy scalability by permitting the synthesis of entire libraries of distinct direct RNAs. It consists of RNA direct modules and a single protein effector [
37]. The RNA-binding proteins known as programmable Cas13 mutant forms (dCas13, Cas13x) can effectively target specific effectors to specific RNAs in order to elicit specific modifications [
37]. RNAs can be selectively targeted to use Cas13 entirely because of the innate crRNA biosynthesis. In contrast to RNA interference, Genome changes induced by CRISPR/Cas13 are not restricted to focusing on cytoplasmic transcripts. In expansion, Cas13 selectively knocks off cytoplasmic mRNA transcripts, enabling quicker downregulation of quality expression [
37]. Recently, SHERLOCK and SHERLOCKv2, based on CRISPR/Cas13, have been instrumental in developing innovative diagnostic tools to detect various infections, including SARS-CoV-2. CRISPR/Cas13, despite its primary advantages, faces issues related to off-target transformations, which is considered as one of the major drawbacks of this system. Be that as it may, future investigations would offer assistance to overcome this deterrent and facilitate.
4.5. Cas14 Protein
Using a Phylogenetic analysis, it is proposed that DNA-binding, Lesson 2 (Cas) proteins are derived from a genetic quality that has the characteristic RuvC endonuclease space. In a subsequent publication, Doudna, Banfield, and associates exploit this reasoning to identify a smaller, less complex Cas protein, which suggests a glimpse of the Cas developmental process and, surprisingly, provides a few biotechnological focal points in comparison to Cas proteins so far identified. In brief, the researchers identified a group of remarkably unique, potential single-effector Cas proteins by searching across terabase sequencing data sets for characteristics around a CRISPR locus and with a RuvC-like region. These variations, known as Cas14s, are similar to other Sort V Cas proteins, such as the recently discovered CasX variation and the well-characterized Cas12a protein. However, what sets them apart is their petite size, typically ranging from 400–700 amino acids, as opposed to the typical ≥1000 amino acids of Lesson 2 variations.
4.5.1. Mechanism and Application of Cas14 Protein
Cas14, a protein in the CRISPR-Cas framework, has been characterized for its unique biochemical properties. It has a strict protospacer adjoining motif (PAM) DNA authoritative requirement [
38], which ensures that the CRISPR-Cas framework does not target itself. Cas14 is sensitive to jumbles in a 6 bp extent within its target region
(Figure 9). The CRISPR/Cas14 framework has been used to detect single nucleotide polymorphisms (SNPs) in human spit tests, providing an economical approach of identifying pathogenic transformations and mapping candidate traits related to different pathogens. This novel system has potential applications in phylogenetic association, epidemiology, and ordered examination.
4.5.2. Pros and Cons of Cas14 Protein
Different benefits are associated with the Cas14 protein than with the traditional Cas9 protein, which modifies dsDNA, ssDNA, and RNA. For example, the 500 amino acids that make up the Cas14 protein are incredibly small, meaning that it may be delivered to any target more efficiently than the Cas9 protein. In addition, the selectivity of the Cas14 enzyme advances the consistency of single nucleotide polymorphism (SNP) [
37]. At long last, the protein Cas14 has fewer PAM requirements and only uses T-rich sequences [
37]; comparatively, genomic sequence is more focused than cas9 protein. In any case, only many studies have been illustrated, which needs broad ponders to help Cas14 in a few areas of investigation within the future
(Table 4).
6. Application against Abiotic Stress
Plants under salt stress may have negative effects that lower yield and quality. This is due to the plant’s formation of osmotic, ionic, and secondary stress. A rice variety that is resistant to salinity was developed by Alam et al. in 2022 by removing the
OsbHLH024 gene and increasing the expression of the ion transporter genes
OsHKT1-3,
OsHAK7, and
OsSOS1. [
39]. CRISPR/Cas9-induced
OsRR22 gene mutation increased rice’s resistance to salt without affecting other agronomic characteristics [
40]. CRISPR/Cas9 was utilized to successfully alter
OsRAV2, and the resultant mutant exhibited increased survivability under salt stress. Furthermore, it is also possible to significantly increase the resistance of crops to salt stress by deleting the genes for tomato SlARF4, barley HvITPK1 [
43], rice OsDST [
41], OsNAC041 and OsmiR535 [
42], and barley HvITPK1 [
43]. Excellent drought stress resistance was achieved in rice using
OsERA1 mutagenesis driven by CRISPR/Cas9 [
44]. TaDREB2 and TaERF3 CRISPR/Cas editing improved wheat’s ability to withstand drought. Santosh Kumar et al. (2020) used CRISPR/Cas9 to modify the
OsDST gene in order to produce a mutant of the indica mega rice cultivar
MTU1010 with wider leaves, a lower stomatal density, and better leaf water retention capacity under drought stress [
41]. The ospyl9 mutant, which CRISPR/Cas9 produced, boosts rice yield and drought tolerance, according to Usman et al. (2020). To give rice the phenotype of curled leaves and drought tolerance, CRISPR/Cas9 produced mutations in the
SRL1 and
SRL2 genes by altering protein expression patterns and scavenging reactive oxygen species [
45]. Ding et al. 2020 suggest that cold stress, encompassing temperatures lower than 20°C and 0°C, obstructs growth and plant development and significantly restricts plant geographical distribution and agricultural output. [
46]. Low temperatures directly decrease plants’ metabolic response, causing osmotic stress, oxidative stress, and other types of stress. The CRISPR/Cas9-generated ospin5b, gs3, and osmyb30 mutants exhibited enhanced cold tolerance, spike length, and grain size, as reported by Zeng et al. [
47]. The entire crop growth cycle is impacted by high temperatures, particularly in heat-sensitive stages like early establishment, flowering, and gameto phytogenesis. Compared to the natural type, tomato plants with the CRISPR/Cas9-mediated slmapk3 mutant were better able to withstand high temperatures because the production of antioxidant enzymes and HSPs/HSFs genes was controlled. CRISPR/Cas9 editing was used to create rice with pyl1/4/6 triple knockdown. The mutant showed less germination before harvest, a higher yield, and a higher tolerance to temperature than the natural variety. One of the most damaging abiotic stresses is heavy metal poisoning. A list of crops with gene details is included in (
Table 6).
7. Application against Biotic Stress
Powdery mildew is one of the most destructive types of fungus diseases that crops can suffer, as it significantly reduces crop yield. Wheat plants now have a higher resilience to powdery mildew due to removing all three TaMLO alleles using CRISPR/Cas9. Similarly, grapes [
48] and tomatoes [
49] became resistant to powdery mildew due to the CRISPR/Cas9-mediated reduction of SlMLO and VvMOL3. Furthermore, tomato powdery mildew resistance was markedly enhanced by the CRISPR/Cas9-mediated SlPMR4 mutation, although immunity was not entirely restored [
50]. A damaging fungal disease is called rice blast. The OsERF922 and OsSEC3A genes in rice were mutated using CRISPR/Cas9 to increase resistance to rice blast disease [
51,
52]. While dwarfing was brought on by an increase in SA content in ossec3a, other agronomic features of the oserf922 mutant remained unchanged. Although the effect was not as great as that of oserf922, rice Bsr-d1 and Pi21 mutations generated by CRISPR/Cas9 could potentially result in partial resistance to rice blast [
53,
54]. A method enabled by CRISPR/Cas9 technology allows for the mutation of the acetylegenase-encoding genes ACER1a and ACET1b to generate distinct resistant materials with increased resistance to bacterial and fungal diseases. Tomato susceptibility gene SlDMR6-1 mutations produced by CRISPR/Cas9 offer resistance against a variety of diseases, such as bacteria, oomycetes, and fungus [
55]. Broad-spectrum resistance to bacteria and fungi was demonstrated by CRISPR/Cas9-mediated osnramp1 mutants, which showed decreased catalase (CAT) activity but elevated hydrogen peroxide (H
2O
2) content and superoxide dismutase (SOD) activity [
56]. The list of crops detailed with genes is mentioned in
(Table 7).
8. Prime Technology
The CRISPR-Cas technology was first introduced and has since been extensively utilised in the engineering of live cells. Prime editing technology is an advanced form of this system that has been rapidly adopted. A prime editing system is used to develop resistance against biotic stress by successfully integrating a 30-nucleotide cis-regulatory element through knock-in with high efficiency [
57]. The first application of prime technology was to mammalian cells. The initial prime technology system was found to repair up to 89% of the known gene variants associated with human disorders [
58]. The prime technology derivative technologies have not only been developed and used in mammalian cells but have also been rapidly adapted to be used in plants. The low homozygous recombination rate and restricted donor transmission pose obstacles to the advancement of plant genome editing, despite its immense potential [
59]. Microorganisms play an integral role in the study of life sciences, as well as being closely associated with human health, industry, and agricultural production. In the most recent instance, Prime technology has also been employed in the model of the microorganism “
E. coli” [
60]. Monocots and dicots underpin the foundation of modern agriculture. Monocots include critical crops like rice, wheat, and maize, while dicots encompass essential plants like soybeans, peas, and sunflowers. Understanding the unique characteristics and growth patterns of these plant groups is essential for crop breeding, efficient farming practices, and sustainable food production
(Table 8). The methods used against tolerance to biotic stresses include CRISPR-Cas9, TALENs (Transcription Activator-Like Effector Nucleases), and ZFNs (Zinc Finger Nucleases). The stress caused by non-living factors such as extreme temperatures, drought, and salinity also poses a great loss to the farmers. To overcome these stress factors, different gene editing technologies like CRISPR-Cas9, TALENs, and ZFNs have been explored for modifying specific genes to enhance abiotic stress tolerance and increase productivity.
Application of the Prime Technology in Plants
It is difficult to achieve accurate genome editing through Homologous Directed Repairs (HDR) due to the low rate of efficiency of HDR and the challenges associated with providing sufficient plant cell donor repair templates. Prime technology enables precise genome target sequence editing in plant cells without DNA restriction technology (DRT) or DNA double-strand breaks (DSB). This technology can provide technical support for improved crop breeding.
9. Conclusion and Future Perspectives
Nowadays, CRISPR/Cas9 technology is being used to improve the different traits such as abiotic stress tolerance, disease resistance, quality and yield of both the monocots and dicots. Crop genomic sequences can be modified to give higher yields by diversifying the approaches used to characterise the activities of individual genes. These findings imply that creative agricultural methods can support the production of nutrient-dense food in a sustainable manner to satisfy the growing demands of a worldwide population. The Cas proteins has boost the basic, therapeutics and diagnostic research. Because these CRISPR/Cas systems are inexpensive and simple to use, many researchers are using them to investigate the functions of different organisms’ genes. Researchers have been able to identify distinct Cas variations, like the proteins Cas12, Cas13, and Cas14, with greater ease since the discovery of the Cas9 protein, which emphasises its role within the CRISPR system. This research on Cas proteins has led to exciting developments in the construction of genomes, the counting and identification of RNA infections in plants, animals, and humans, as well as the treatment of contaminations. Some research is now being done to find new Cas variants found in nature. Be that as it may, various viewpoints of the instrument of Cas proteins still need adequate getting. Consequently, understanding the atomic tool that underpins Cas proteins, being able to recognise evidence of PAM-less Cas proteins, and placing a precise emphasis on specificity to limit off-target effects would boost the potential of exploiting Cas proteins for precise genome engineering applications. Furthermore, identifying the unexplained evolution of Cas masteries, which persist in a variety of microscopic organisms or archaea, would transform a variety of fields, such as the diagnosis of new illnesses, medicines, agriculture, breeding, and so forth. If these proteins’ advanced genome-editing potential is fully explored, it may ignite an untapped CRISPR “fever” that might lead to the widespread adoption of compelling and revolutionary CRISPR technologies in the near future.
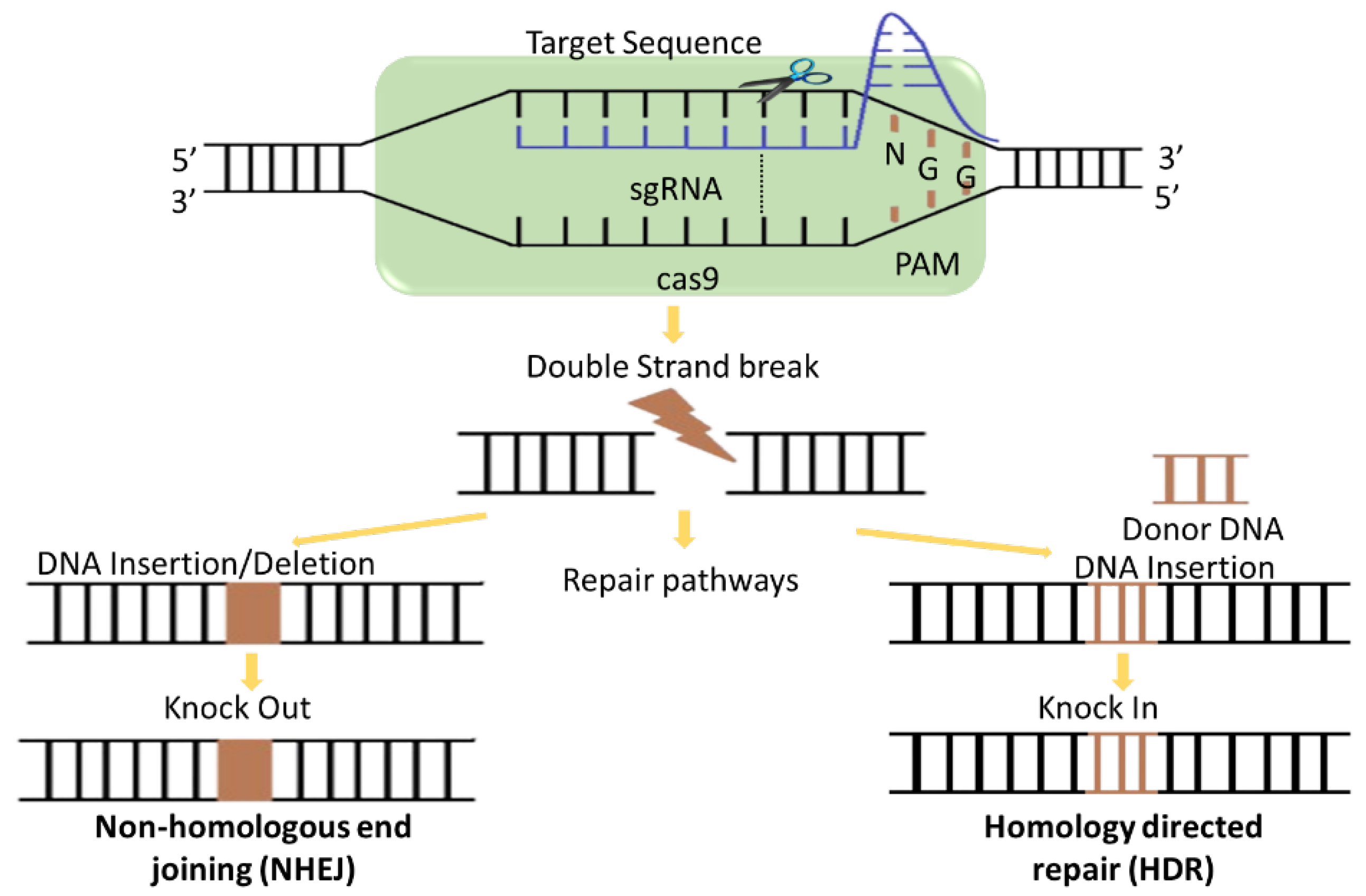
Figure 1.
CRISPER-Cas 9 Genome Editing Mechanism CRISPR stands for spacer guide RNA, and the sgRNA is positioned next to the protospacer adjacent motif (PAM) site (5′NGG-3′). Scissors represent the cleavage sites of the Cas9 protein, and dotted lines represent the blunt ends. The Cas 9 protein induces a double-strand break (DSB) in the DNA. The break is repaired by a non-homologous end (NHEJ) joining, resulting in DNA sequence insertion/deletion, or in a homologous directed repair (HDR) resulting in DNA insertion from homologously donor DNA dotted lines representing the blunt ends.
Figure 1.
CRISPER-Cas 9 Genome Editing Mechanism CRISPR stands for spacer guide RNA, and the sgRNA is positioned next to the protospacer adjacent motif (PAM) site (5′NGG-3′). Scissors represent the cleavage sites of the Cas9 protein, and dotted lines represent the blunt ends. The Cas 9 protein induces a double-strand break (DSB) in the DNA. The break is repaired by a non-homologous end (NHEJ) joining, resulting in DNA sequence insertion/deletion, or in a homologous directed repair (HDR) resulting in DNA insertion from homologously donor DNA dotted lines representing the blunt ends.
Figure 2.
CRISPR/Cas alters the immune system through three steps: adaptation, RNA biogenesis, and interference. During replication, Cas1 and Cas2 cleave invasion sequences, while CRISPR/RNA biogenesis transcribes CRISPR fragments into precursor cRNA genes, which form complexes with Cas proteins. CRISPR interference occurs when foreign gene sequences insert into CRISPR spacers.
Figure 2.
CRISPR/Cas alters the immune system through three steps: adaptation, RNA biogenesis, and interference. During replication, Cas1 and Cas2 cleave invasion sequences, while CRISPR/RNA biogenesis transcribes CRISPR fragments into precursor cRNA genes, which form complexes with Cas proteins. CRISPR interference occurs when foreign gene sequences insert into CRISPR spacers.
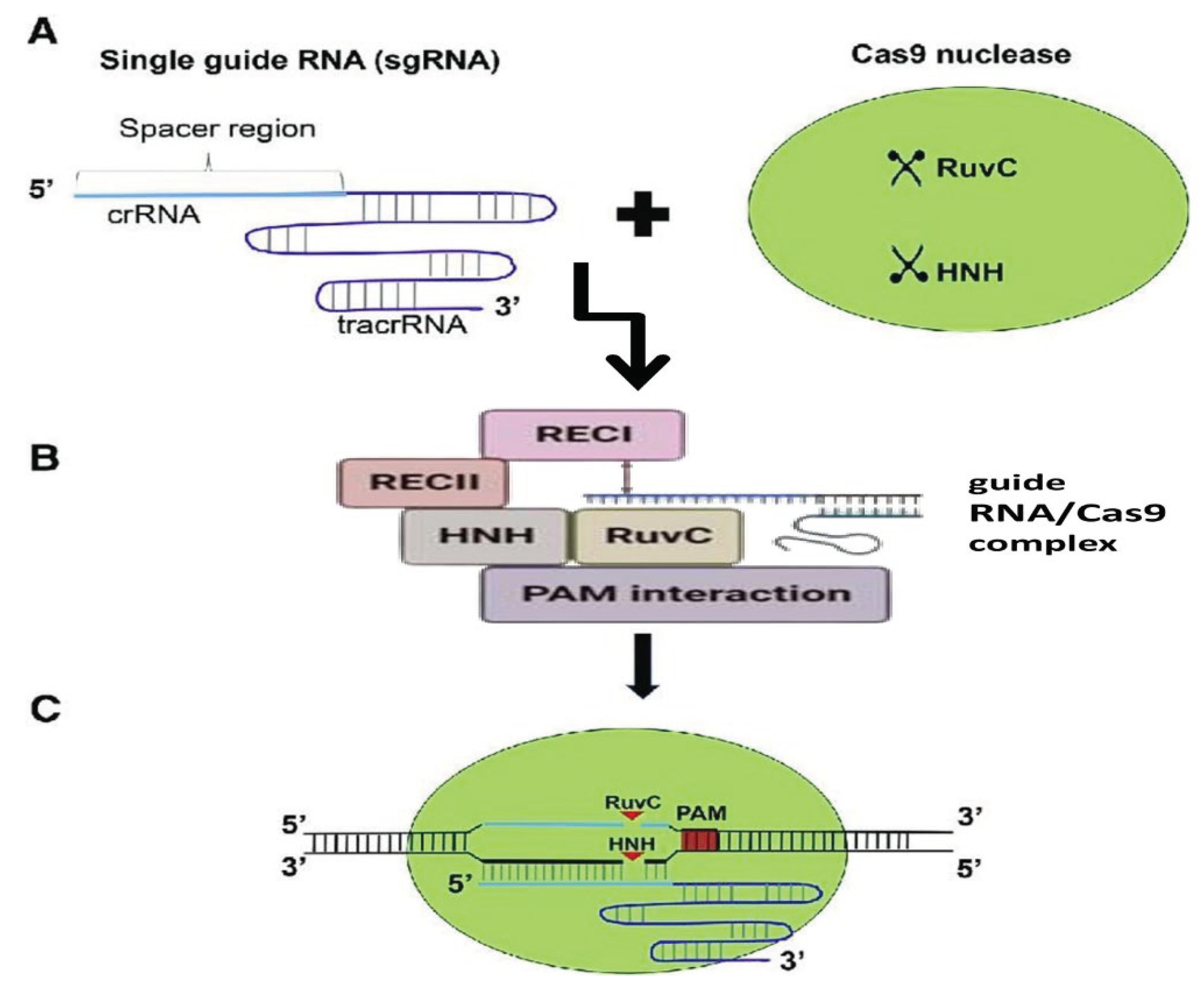
Figure 3.
The CRISPR/Cas9 mechanism involves six domains: recognition flap (REC I), arginine-rich bridging helix, PAM interaction, HNH, and RuvC. REC I is responsible for hRNA binding, while HNH and RuvC initiate cleavage. B-programmed gRNA binds to Cas9, transforming it into an active form. Cas9 generates a 3bp DSP using HNH and RuvC domains.
Figure 3.
The CRISPR/Cas9 mechanism involves six domains: recognition flap (REC I), arginine-rich bridging helix, PAM interaction, HNH, and RuvC. REC I is responsible for hRNA binding, while HNH and RuvC initiate cleavage. B-programmed gRNA binds to Cas9, transforming it into an active form. Cas9 generates a 3bp DSP using HNH and RuvC domains.
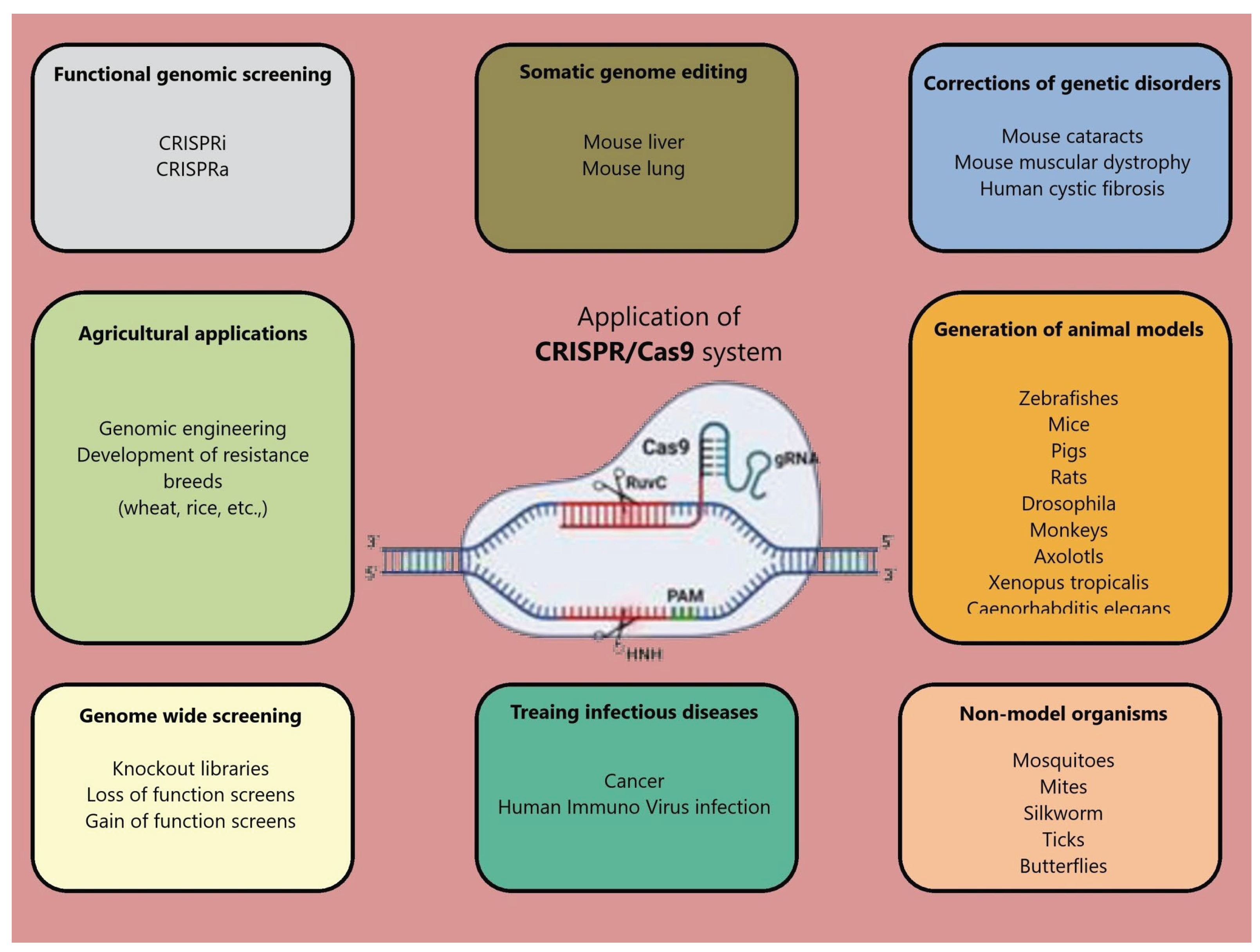
Figure 4.
The CRISPR/Cas9 system revolutionizes genetic engineering with its precision, speed, and cost-effectiveness. It has been used to treat diseases, develop precision medicine, study insect biology, edit body genes, produce biofuel, and study crops. The system uses gRNA to direct Cas9 to cleave dsDNA or ssDNA, reducing unwanted mutations.
Figure 4.
The CRISPR/Cas9 system revolutionizes genetic engineering with its precision, speed, and cost-effectiveness. It has been used to treat diseases, develop precision medicine, study insect biology, edit body genes, produce biofuel, and study crops. The system uses gRNA to direct Cas9 to cleave dsDNA or ssDNA, reducing unwanted mutations.
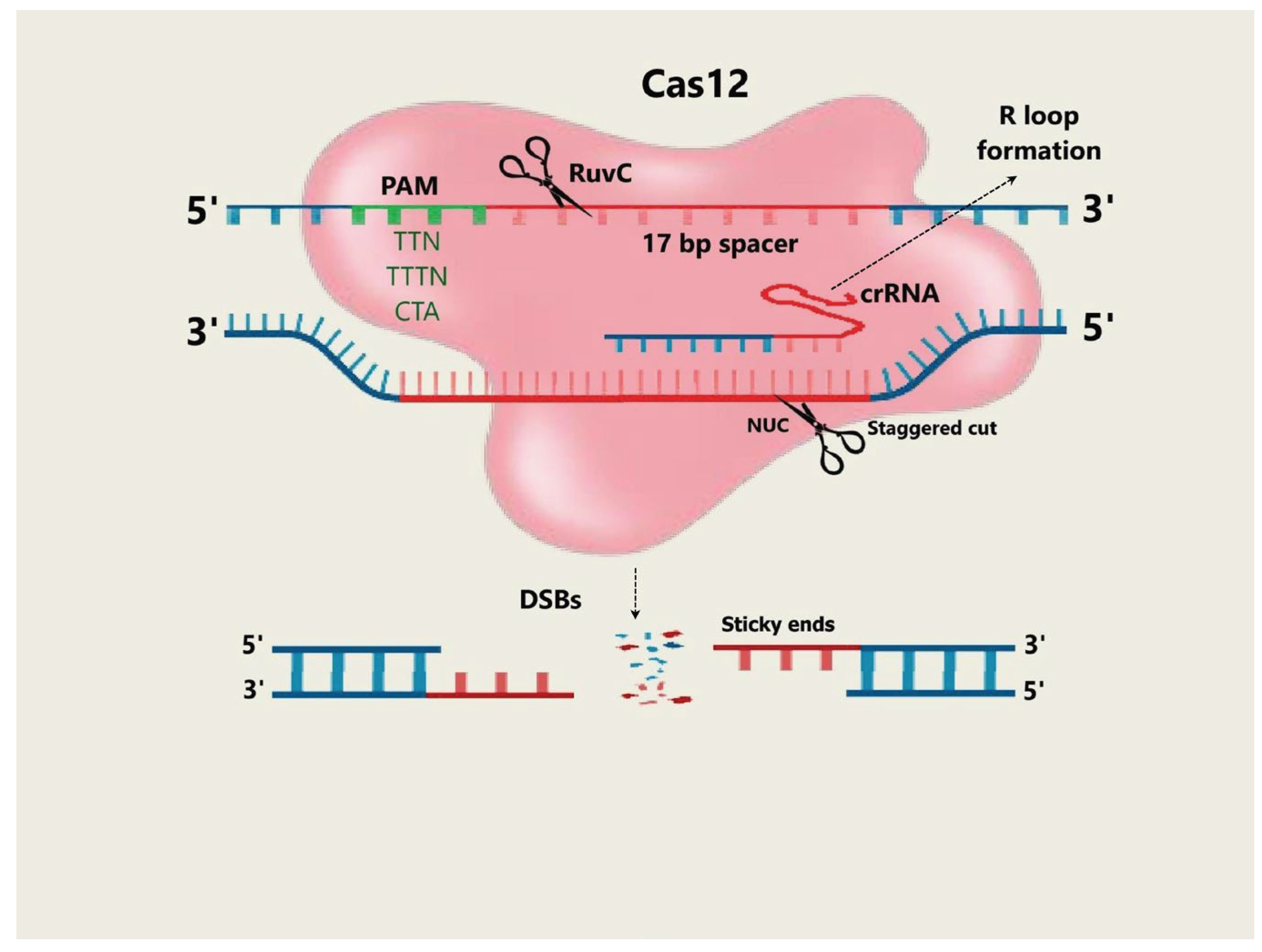
Figure 5.
The CRISPR/Cas12 mechanism involves cleaving the target region near the PAM sequence, creating an R-loop that mixes bases with the target DNA thread, and creating an R cycle, designed using the Cas12 protein’s active domain, RuvC.
Figure 5.
The CRISPR/Cas12 mechanism involves cleaving the target region near the PAM sequence, creating an R-loop that mixes bases with the target DNA thread, and creating an R cycle, designed using the Cas12 protein’s active domain, RuvC.
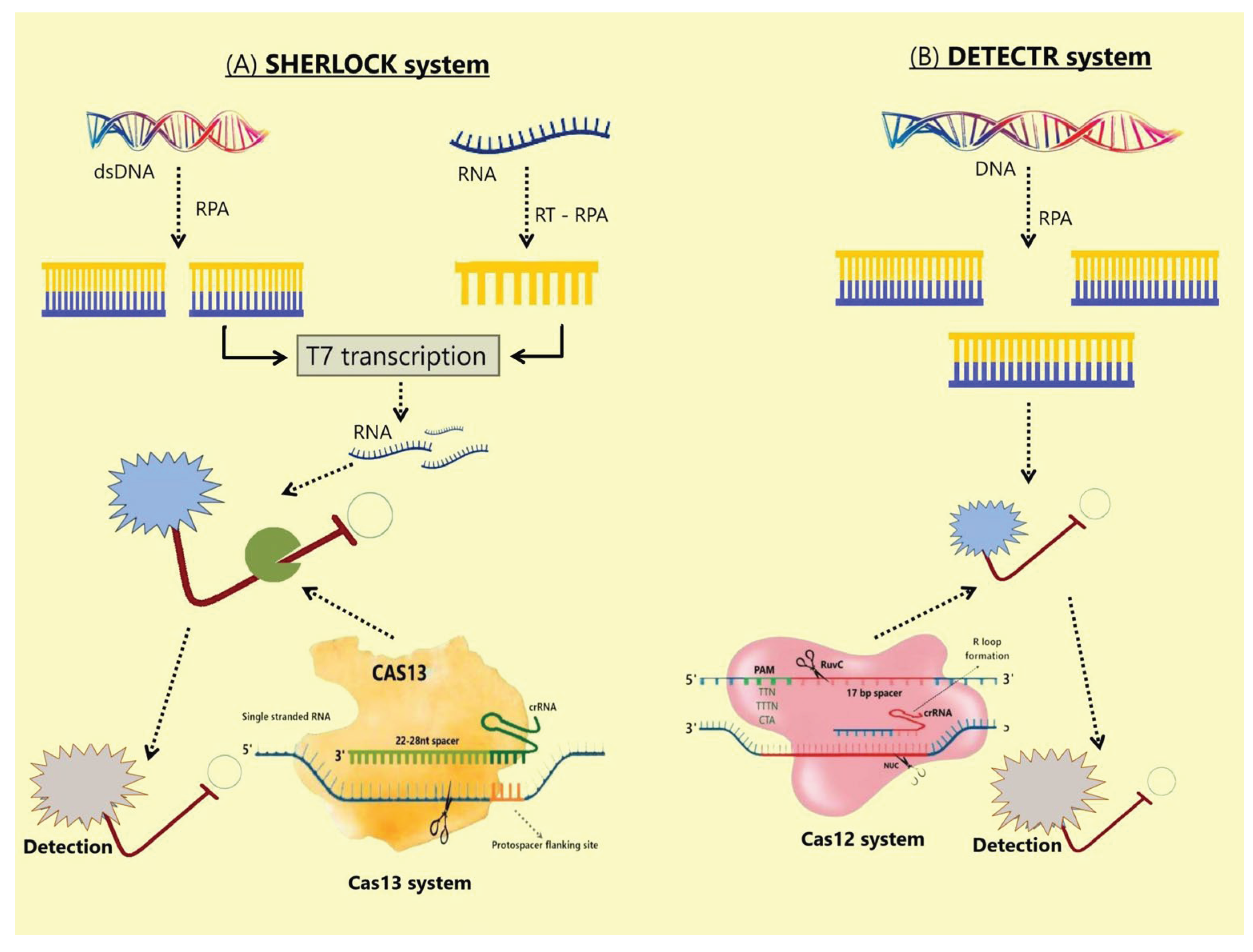
Figure 6.
Sherlock and the detector system are used to amplify target double-stranded DNA or RNA using recombinant polymerase amplification (RPA) or reverse transcription (RT)-RPA. This enables the amplification of enzyme-sensitive reporter expression (SHERLOCK) with a CRISPR endonuclease-targeted trans reporter (DETECTR), allowing detection of target sequences.
Figure 6.
Sherlock and the detector system are used to amplify target double-stranded DNA or RNA using recombinant polymerase amplification (RPA) or reverse transcription (RT)-RPA. This enables the amplification of enzyme-sensitive reporter expression (SHERLOCK) with a CRISPR endonuclease-targeted trans reporter (DETECTR), allowing detection of target sequences.
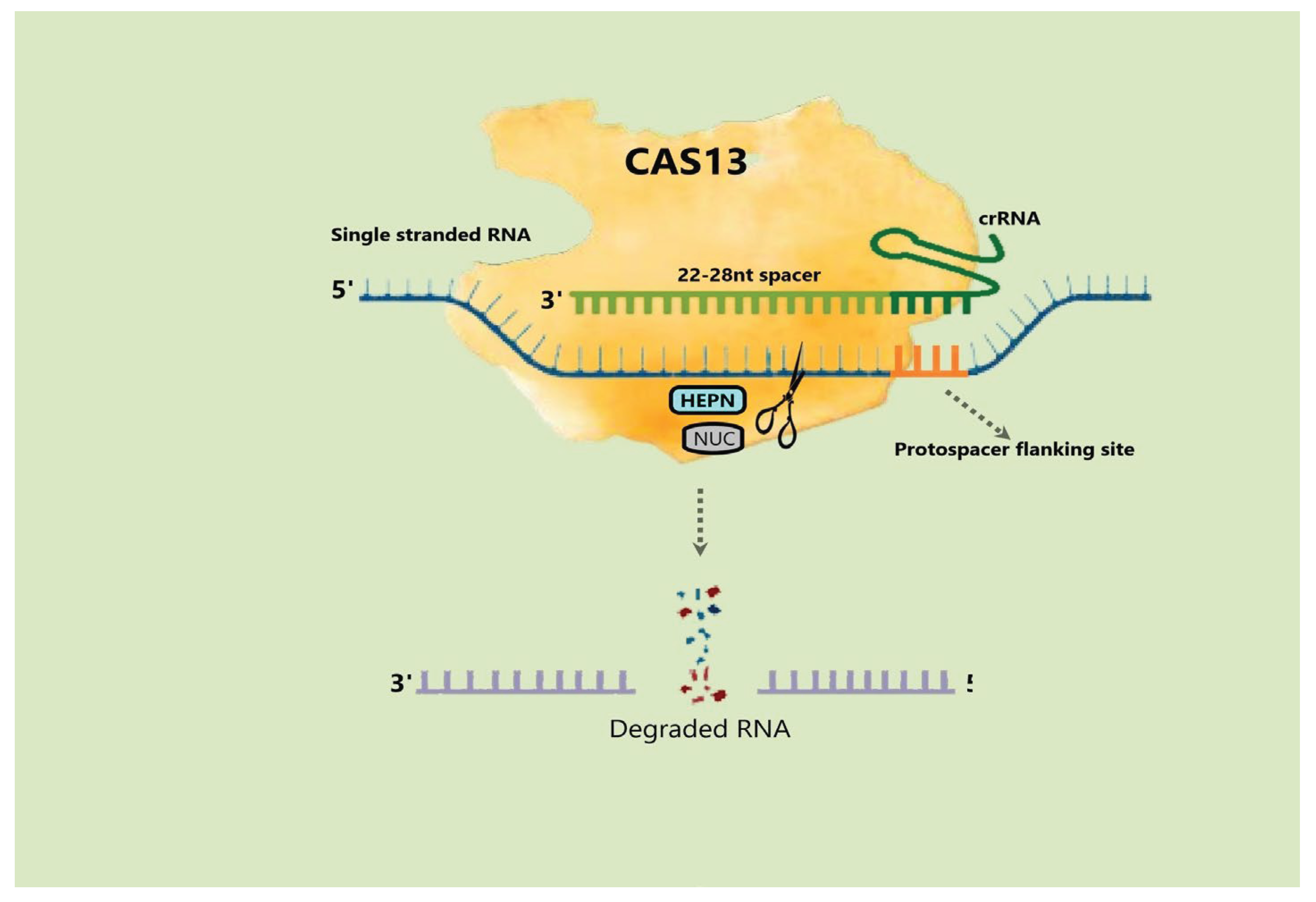
Figure 7.
The CRISPR/Cas13a mechanism involves activating Cas13a protein, which encodes crRNA, NUC particles, and HEPN domains for target RNA. Cas13a cleaves ssRNA complementary to the target sequence, while crRNA binds and cleaves the target region without tracRNA, forming LshCas13a.
Figure 7.
The CRISPR/Cas13a mechanism involves activating Cas13a protein, which encodes crRNA, NUC particles, and HEPN domains for target RNA. Cas13a cleaves ssRNA complementary to the target sequence, while crRNA binds and cleaves the target region without tracRNA, forming LshCas13a.
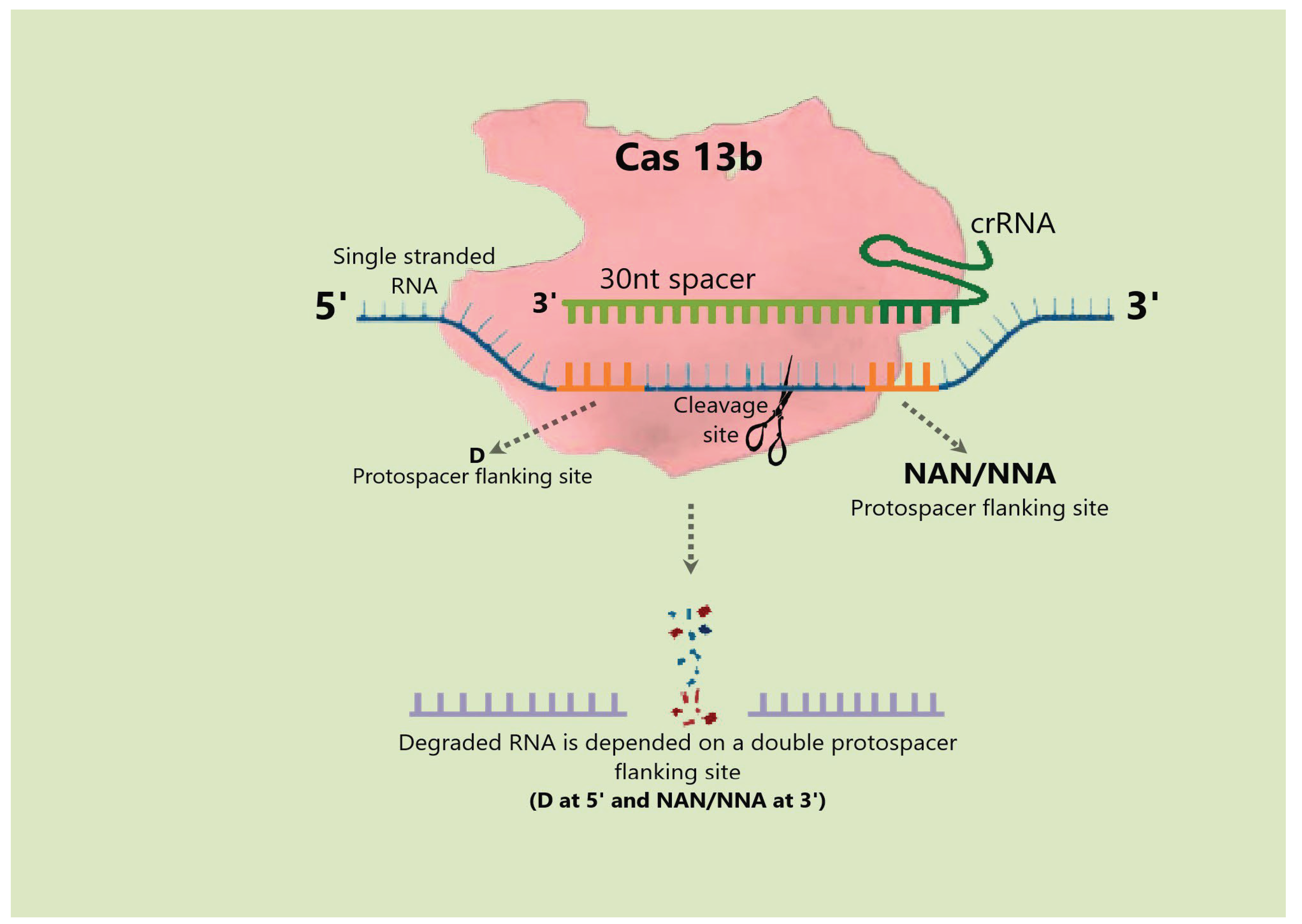
Figure 8.
Schematic representation of the CRISPR/Cas13b mechanism. Cas13b protein is associated with mature crRNA. This ÇRISPR/Cas13b ssRNA complex seeks out the target ssRNA and, with the help of a protospacer flanking site (PFS) at the 5′ end of the target RNA and a PAM sequence (NAN/NNA), makes precise conformational changes in the target ssRNA. Nonspecific cleavage of RNA occurs at the 3′ end.
Figure 8.
Schematic representation of the CRISPR/Cas13b mechanism. Cas13b protein is associated with mature crRNA. This ÇRISPR/Cas13b ssRNA complex seeks out the target ssRNA and, with the help of a protospacer flanking site (PFS) at the 5′ end of the target RNA and a PAM sequence (NAN/NNA), makes precise conformational changes in the target ssRNA. Nonspecific cleavage of RNA occurs at the 3′ end.
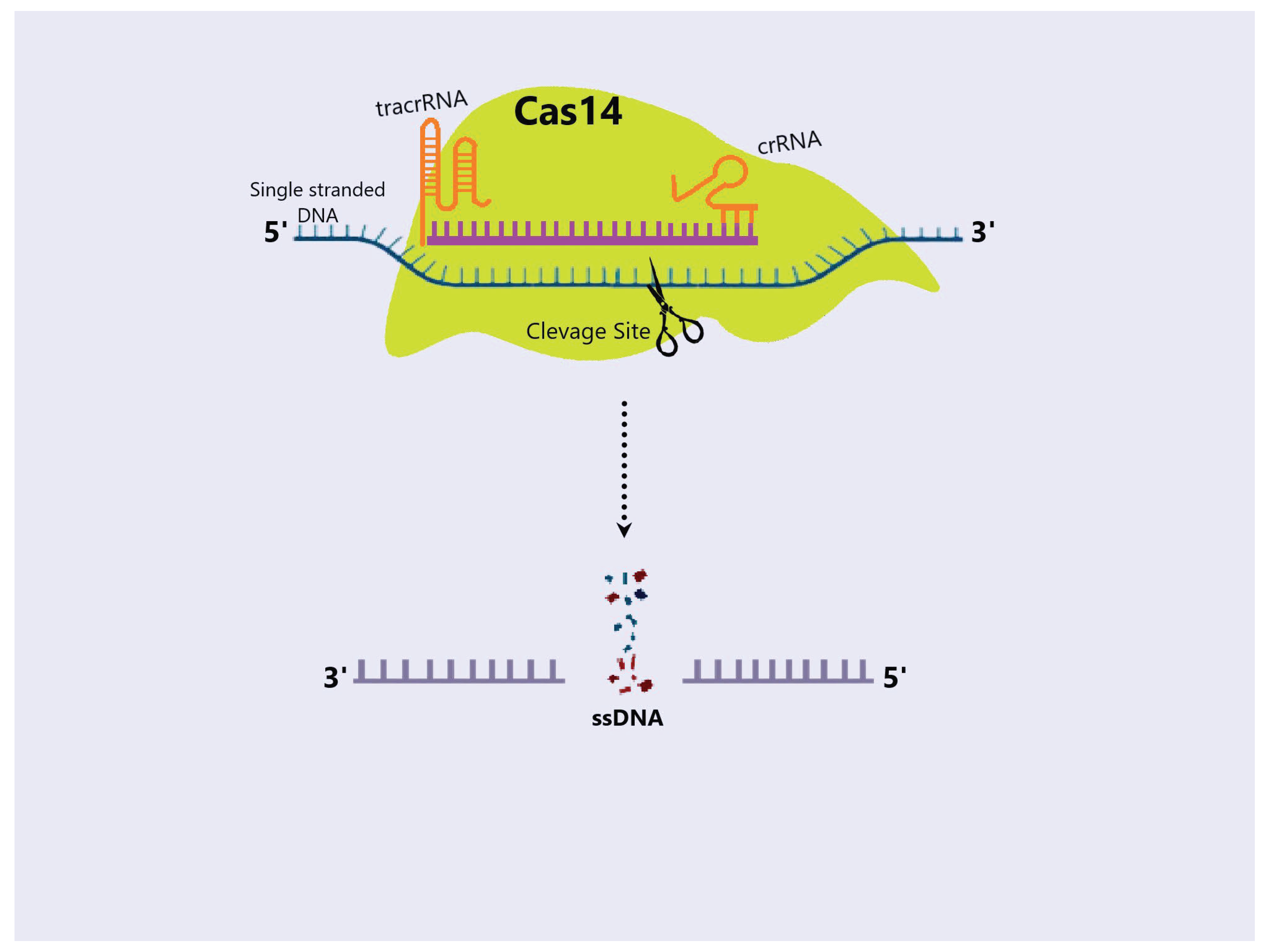
Figure 9.
The CRISPR/Cas14 system consists of a Cas14 protein, TracRNA and crRNA, Case 14, and target proteins of ssDNA. The seed system interacts with ssDNA targets, destroying ssDNA. The PAM domain meets usability and application criteria for future genetic engineering.
Figure 9.
The CRISPR/Cas14 system consists of a Cas14 protein, TracRNA and crRNA, Case 14, and target proteins of ssDNA. The seed system interacts with ssDNA targets, destroying ssDNA. The PAM domain meets usability and application criteria for future genetic engineering.
Figure 10.
CRISPR/Cas9 system in agriculture CRISPR / Cas9 genome editing system in agriculture Wide application Agricultural and crop improvement Improvements in abiotic and biotic stress Flood tolerance crops Vegetable Fruits.
Figure 10.
CRISPR/Cas9 system in agriculture CRISPR / Cas9 genome editing system in agriculture Wide application Agricultural and crop improvement Improvements in abiotic and biotic stress Flood tolerance crops Vegetable Fruits.
Figure 11.
CRISPR/Cas9 Genome Editing in Biological Sciences CRISPR / Cas9 genome editing system is widely used in the fields of biological research and applications, such as: Agriculture Synthetic Biology Viral gene editing Ecological vector control Human gene therapy Drug screening for targeted delivery Programmable RNA targeting.
Figure 11.
CRISPR/Cas9 Genome Editing in Biological Sciences CRISPR / Cas9 genome editing system is widely used in the fields of biological research and applications, such as: Agriculture Synthetic Biology Viral gene editing Ecological vector control Human gene therapy Drug screening for targeted delivery Programmable RNA targeting.
Table 1.
Classification of CRISPR/Cas system with their protein, target molecule, spacer acquisition mechanism, Pre-CRISPR processing, self vs. non-self-discrimination, and their effectors with the name of isolated organisms.
Table 1.
Classification of CRISPR/Cas system with their protein, target molecule, spacer acquisition mechanism, Pre-CRISPR processing, self vs. non-self-discrimination, and their effectors with the name of isolated organisms.
| Class (C)&Type (T) |
Protein(P)
&
Target (T) |
Spacer
acquisition
strategy |
Name of
CRISPR/Cas
system |
Pre-Crispr
Processing |
Self vs. Non-self
Discrimination |
Effectors of
CRISPR
System |
Host
Organism |
Reference |
C – 1
T – III |
P – Cas3
T-ssDNAssDNA |
Cas1/Cas2 |
Cas7, Cas5, Cas8, and Cas3 |
Cas6 |
- |
Cas3, Cascade, and crRNA |
E. coli
|
[61] |
C – 1
T – I |
P – Cas10
T- A |
Cas4Cas1/Cas2 |
Cas7, Cas5, and Cas1 |
Cas6 |
- |
Cmr/Csm, crRNA, and Cas10 |
S.epidermics
|
[61] |
C – 1
T - IV |
P – Csf1
T - - |
- |
Cas7, Cas5, and |
- |
- |
- |
- |
[61] |
C – 2
T – II |
P – Cas9
T – dsDNA |
Cas1/Cas2/ Cas4 |
Csf1Cas9 |
RNase III, and tracr RNA |
- |
Cas9, tracrRNA, and crRNA |
S. thermophilus and S. pyogenes |
[61] |
C – 2
T – V |
P – Cpf1
T – ssDNA & dsDNA |
Cas1/Cas2/ Cas4 |
Cas12 |
Cpf1 |
PAM |
Cpf1, crRNA and tracrRNA |
F.novicida
|
[61] |
C – 2
T - VI |
P – C2c2
T - ssRNA |
Cas1/Cas2 |
Cas13 |
- |
- |
C2c1, and crRNA |
- |
[61] |
Table 2.
Details various CRISPR/Cas proteins with their host organism, sgRNA size, PAM sequence with their target molecule and cut site.
Table 2.
Details various CRISPR/Cas proteins with their host organism, sgRNA size, PAM sequence with their target molecule and cut site.
| ProteinName |
Host organism |
sgRNAsize |
PAM sequence |
Target |
Cut
site |
References |
| Cas9 |
S.pyogenes |
20 |
5ʹ-NGG-3ʹ |
dsDNA |
5ʹofPAM |
[61] |
| Cas9 |
S.pyogenes |
- |
5ʹ-NAC,NTG,NTT,andNCG-3ʹ
|
DNA
|
5ʹofPAM
|
[61] |
| Cas9 |
F.novicida |
20 |
5ʹ-NGG-3ʹ |
DNA |
5ʹofPAM |
[61] |
| Cas9 |
S.aureus |
21 |
5ʹ-NNGRRT-3ʹ |
DNA |
5ʹofPAM |
[61] |
| Cas9 |
Neisseriameningitidis |
24 |
5ʹ-NNNNGATT-3ʹ |
DNA |
5ʹofPAM |
[61] |
| Cas9 |
S. thermophilus |
20 |
5ʹ-NNAGAAW5ʹ |
DNA |
5ʹofPAM |
[61] |
| Cas9 |
S. thermophilus |
20 |
5ʹ-NGGNG-3 |
DNA |
5ʹofPAM |
[61] |
| Cas9 |
Campylobacterjejuna |
22 |
NNNNACAC and NNNRYAC |
DNA |
5ʹofPAM |
[61] |
| C2c1 |
Alicyclobacillusacidoterrestris |
20 |
T-rich PAM |
DNA |
5ʹofPAM |
[61] |
| Cpf1 |
PrevotellaandFrancisella |
20 |
TTTV |
DNA |
5ʹofPAM |
[61] |
| Cpf1 |
Acidaminococcussp. |
24 |
5ʹ-TTTN-3ʹ |
DNA |
3ʹofPAM |
[61] |
| Cas12a |
Acidaminococcussp. |
- |
Thymine-rich PAM |
DNA |
5ʹofPAM |
[61] |
| Cas13 |
Lb |
28 |
Non-Gnucleotideatthe3ʹproto-spacerflankingsite (PFS) |
ssRNA |
NA |
[61] |
| Cas14 |
Uncultivatedarchaea |
NA |
NA |
ssDNA |
NA |
[61] |
Table 3.
Major naturally occurring and genetically modified Cas enzymes used for genome editing.
Table 3.
Major naturally occurring and genetically modified Cas enzymes used for genome editing.
| Name |
Cas |
CRISPR/Cas |
PAM |
PAM location |
Resources |
Reference |
SpRY
SpG
Cas9-NRNH
HypaCas9
evoCas9
Sniper-Cas9
xCas9
SpCas9-NG
eSpCas9
SpCas9-HF
SaCas9-KKH
Modified SpCas9
FnCas9variant
SpCas9
SaCas9
FnCas9
NmCas9
CjCas9
St1Cas9
St1Cas9
FnCas12a
LbCas12a
AsCas12a
LsCas13#
Cas14 |
Cas9
Cas9
Cas9
Cas9
Cas9
Cas9
Cas9
Cas9
Cas9
Cas9
Cas9
Cas9
Cas9
Cas9
Cas9
Cas9
Cas9
Cas9
Cas9
Cas9
Cas12a(cpf1)
Cas12a(cpf1)
Cas12a(cpf1)
Cas13(C2c2)
Cas14 |
Type II
Type II
Type II
Type II
Type II
Type II
Type II
Type II
Type II
Type II
Type II
Type II
Type II
Type II
Type II
Type II
Type II
Type II
Type II
Type II
Type II
Type II
Type II
TypeVI
NA
|
NRN or NYN
NGN
NRNH
NGG
NGG
NGG
NG
NG
NGG
NGG
NNNRRT
NGA or NAG
YG
NGG
NNGRRT
NGG
NNNNGATT
NNNNRYAC
NNAGAAW
NGGNG
TTTN or YTN
TTTV
TTTV
NA
NA |
3′
3′
3′
3′
3′
3′
3′
3′
3′
3′
3′
3′
3′
3′
3′
3′
3′
3′
3′
3′
5′
5′
5′
NA
NA. |
Engineered SpCas9
Engineered SpCas9
Engineered SpCas9
Mutated SpCas9-HF
Mutated SpCas9
Engineered SpCas9
Engineered SpCas9
Engineered SpCas9
Engineered SpCas9
Engineered SpCas9
Engineered SaCas9
Engineered SpCas9
Modified FnCas9
Streptococcus pyogenes
Streptococcus aureus
Francisella Novicida
Neisseria meningitidis
Campylobacter jejuni
Streptococcusthermophilus
Streptococcusthermophilus
Francisella novicida
Lachnospiracea bacterium
Acidaminococcus sp.
Leptotrichia shahii
Archaea |
[31]
[31]
[31]
[31]
[31]
[31]
[31]
[31]
[31]
[31]
[31]
[31]
[31]
[31]
[31]
[31]
[31]
[31]
[31]
[31]
[31]
[31]
[31]
[31]
[31] |
Table 4.
Comparision of Cas9 vs Cas12 vs Cas13 vs Cas14.
Table 4.
Comparision of Cas9 vs Cas12 vs Cas13 vs Cas14.
| Parameter |
Cas9 |
Cas12 |
Cas13 |
Cas14 |
Size of Protein
(Amino Acid) |
~1000-1600 |
~1300 |
~1400 |
~400-700 |
| Target |
DNA |
DNA |
RNA |
DNA |
| RNA |
Two RNA
molecules |
Single RNA
molecules |
Two RNA
molecules |
Single RNA
molecules |
| Nuclease Site |
2 nuclease domains HNH and RuVc |
Single nuclease
RucV-Nuc |
Target RNA
domain HEPN |
DNA binding
domain RuVc |
| Pattern of cut |
Blunt |
Sticky-ended |
Degraded |
NA |
| Spacer Size |
16-20nt |
16-25nt |
25-35nt |
NA |
| Protospacer restriction |
PAM |
PAM |
PFS |
PAM |
|
Single guide molecular size (Nucleotides, nt) |
17-24nt |
42-44nt |
-64nt |
-140nt |
Non-specifically cut nucleic acids (DNA or RNA) |
DNA(SS) |
DNA(SS) |
RNA(SS) |
DNA(SS) |
Table 5.
List of crop improvement using CRISPR-Cas technology.
Table 5.
List of crop improvement using CRISPR-Cas technology.
| Name of the crops |
Types of CRISPR- Cas |
Resistance to stress |
Reference |
| Rice |
Cas 9 |
Drought tolerance, Pest resistance
Salinity tolerance |
[62] |
| Cas 12 |
Salinity Tolerance |
[62] |
| Cas13 |
Enhancing resistance to specific stress conditions |
[62] |
| Wheat |
Cas 9 |
Drought tolerance, Disease resistance
Salinity tolerance, Abiotic Stress |
[62] |
| Cas 12 |
Salt tolerance, Drought tolerance
Pest resistance |
[62] |
| Cas 13 |
It is used to target genes related to specific stress response |
[62] |
| Maize |
Cas 9 |
Drought tolerance, Disease resistance |
[62] |
| Cas 12 |
Pest resistance |
[62] |
| Cas 13 |
To enhance stress resistance. |
[62] |
| Arabidopsis |
Cas 9 |
Drought tolerance, Salt tolerance
Stress-related traits. |
[62] |
| Cas 12 |
Drought tolerance, Disease resistance. |
[62] |
| Cas 13 |
Potential for RNA-based genome editing and may be applied to enhance stress resistance |
[62] |
| Cotton |
Cas 9 |
Pest resistance |
[62] |
| Cas 12 |
Disease Resistance |
[62] |
| Cas 13 |
RNA Virus Resistance |
[62] |
| Soybean |
Cas 9 |
Drought tolerance, Pest resistance
Disease resistance. |
[62] |
| Cas 12 |
Drought tolerance, Disease resistance. |
[62] |
| Cas 13 |
RNA Virus Resistance |
[62] |
| Tomato |
Cas 9 |
Drought tolerance, Pest resistance
Disease resistance. |
[62] |
| Cas 12 |
Drought tolerance |
[62] |
| Cas 13 |
RNA Virus Resistance |
[62] |
| Potato |
Cas 9 |
Drought tolerance,Pest resistance
Disease resistance. |
[62] |
| Cas 12 |
Disease resistance |
[62] |
| Cas 13 |
The potential for RNA-based genome editing, and may be applied to enhance stress resistance |
[62] |
| Citrus |
Cas 9 |
Disease resistance |
[62] |
| Cas 12 |
Disease resistance, Drought tolerance. |
[62] |
| Cas 13 |
It holds potential for RNA-based genome editing, which may be applied to enhance stress resistance |
[62] |
| Grape |
Cas 9 |
Disease resistance |
[62] |
| Cas 12 |
Disease resistance and improving grape quality traits. |
[62] |
| Cas 13 |
Enhance stress resistance and control grapevine diseases. |
[62] |
Table 6.
CRISPR/Cas genes intended to confer tolerance against abiotic stress.
Table 6.
CRISPR/Cas genes intended to confer tolerance against abiotic stress.
| Stress |
Crop |
The name of the target gene |
References |
| Salinity |
Rice (Oryza sativa) |
BASIC HELIX-LOOP-HELIX 024 (OsbHLH024) |
[39] |
| Rice (Oryza sativa) |
RESPONSE REGULAT ORS 22 (OsRR22) |
[40] |
| Rice (Oryza sativa) |
RELATED TO ABI3/VP1 2 (OsRAV2) |
[62] |
| Rice (Oryza sativa) |
DROUGHT AND SALT TOLERANCE (OsDST) |
[41] |
| Rice (Oryza sativa) |
NAM, ATAF and CUC 041 (OsNAC041) |
[51] |
| Rice (Oryza sativa) |
OsmiR535 |
[42] |
| Barley (Hordeum vulgare) |
INOSITOLTRISPHOSPHATE 5/6 KINASES 1 (HvITPK1) |
[43] |
| Tomato (Solanum lycopersicum) |
HYBRID PROLINE-RICH PROTEIN 1 (SlHyPRP1) |
[62] |
| Tomato (Solanum lycopersicum) |
Auxin Response Factor 4 (SlARF4) |
[62] |
| Drought |
Rice (Oryza sativa) |
ENHANCED RESPONSE TO ABA1 (OsERA1) |
[44] |
| Rice (Oryza sativa) |
OsDST |
[41] |
| Rice (Oryza sativa) |
PYRABACTIN RESISTANCE-LIKE 9 (OsPYL9) |
[45] |
| Rice (Oryza sativa) |
SEMI-ROLLED LEAF 1 (SRL1) and SEMI-ROLLED LEAF 2 (SRL2) |
[47] |
| Maize (Zea mays) |
AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE 8 (ZmARGOS8) |
[62] |
| Wheat (Triticum aestivum) |
DEHYDRATION RESPONSIVE ELEMENT BINDING PROTEIN 2 (TaDREB2) |
[23] |
| Wheat (Triticum aestivum) |
ETHYLENE-RESPONSE FACTOR 3 (TaERF3) |
[23] |
| Tomato (Solanum lycopersicum) |
GA-INSENSITIVE DWARF1 1 (SlGID1) |
[62] |
| Tomato (Solanum lycopersicum) |
LATERAL ORGAN BOUNDARIES DOMAIN 40 (SlLBD40) |
[24] |
| Arsenic Caesium |
Rice (Oryza sativa) |
HIGH-AFFINITY POTASSIUM TRANSPORTER 1 (OSHAK1) |
[62] |
| Rice (Oryza sativa) |
ARSENITE-RESPONSIVE MYB1 (OsARM1) |
[62] |
| Low temperature |
Rice (Oryza sativa) |
PIN-FORMED 5b (OsPIN5b) |
[62] |
| Rice (Oryza sativa) |
GRAIN SIZE (GS3) |
[62] |
| Rice (Oryza sativa) |
V-MYB AVIAN MYELOBLASTOSIS VIRAL ONCOGENE HOMOLOG 30(OsMYB30) |
[62] |
| High temperature |
Rice (Oryza sativa) |
PYRABACTIN RESISTANCE-LIKE 1/4/6 (OsPYL1/4/6) |
[62] |
| Tomato (Solanum lycopersicum) |
MITOGEN-ACTIVATED PROTEIN KINASES 3 (SlMAPK3) |
[62] |
| Cadmium |
Rice (Oryza sativa) |
NATURAL RESISTANCE-ASSOCIATED MACROPHAGE PROTEIN 5(OsNRAMP5) |
[56] |
| Rice (Oryza sativa) |
LOW-AFFINITY CATION TRANSPORTER 1 (OsLCT1) |
[62] |
| Rice (Oryza sativa) |
NATURAL RESISTANCE-ASSOCIATED MACROPHAGE PROTEIN 1(OsNRAMP1) |
[56] |