Submitted:
03 April 2024
Posted:
04 April 2024
You are already at the latest version
Abstract
Keywords:
Introduction
Role of miRNAs in neurodegenerative diseases.
Alzheimer's Disease (AD)
Parkinson's Disease (PD)
Multiple Sclerosis (MS)
ALS
miRNAs as Therapy
Conclusion and Future Perspective
Abbreviations---111
Author's contribution
Acknowledgements
Consent to participate
Ethical Approval
Consent to Publish
Competing Interests
Data availability
Funding
References
- Gitler, A.D.; Dhillon, P.; Shorter, J. Neurodegenerative disease: models, mechanisms, and a new hope. Dis. Model. Mech. 2017, 10, 499–502. [Google Scholar] [CrossRef] [PubMed]
- De Felice, B.; Mondola, P.; Sasso, A.; Orefice, G.; Bresciamorra, V.; Vacca, G.; Biffali, E.; Borra, M.; Pannone, R. Small non-coding RNA signature in multiple sclerosis patients after treatment with interferon-β. BMC Med Genom. 2014, 7, 1–9. [Google Scholar] [CrossRef]
- Li, S.; Lei, Z.; Sun, T. The role of microRNAs in neurodegenerative diseases: a review. Cell Biol. Toxicol. 2022, 39, 53–83. [Google Scholar] [CrossRef]
- Heemels M-T Neurodegenerative diseases. Nature 2016, 539, 179–180. [CrossRef] [PubMed]
- Ettle, B.; Schlachetzki, J.C.M.; Winkler, J. Oligodendroglia and myelin in neurodegenerative diseases: more than just bystanders? Mol Neurobiol 2016, 53, 3046–3062. [Google Scholar] [CrossRef]
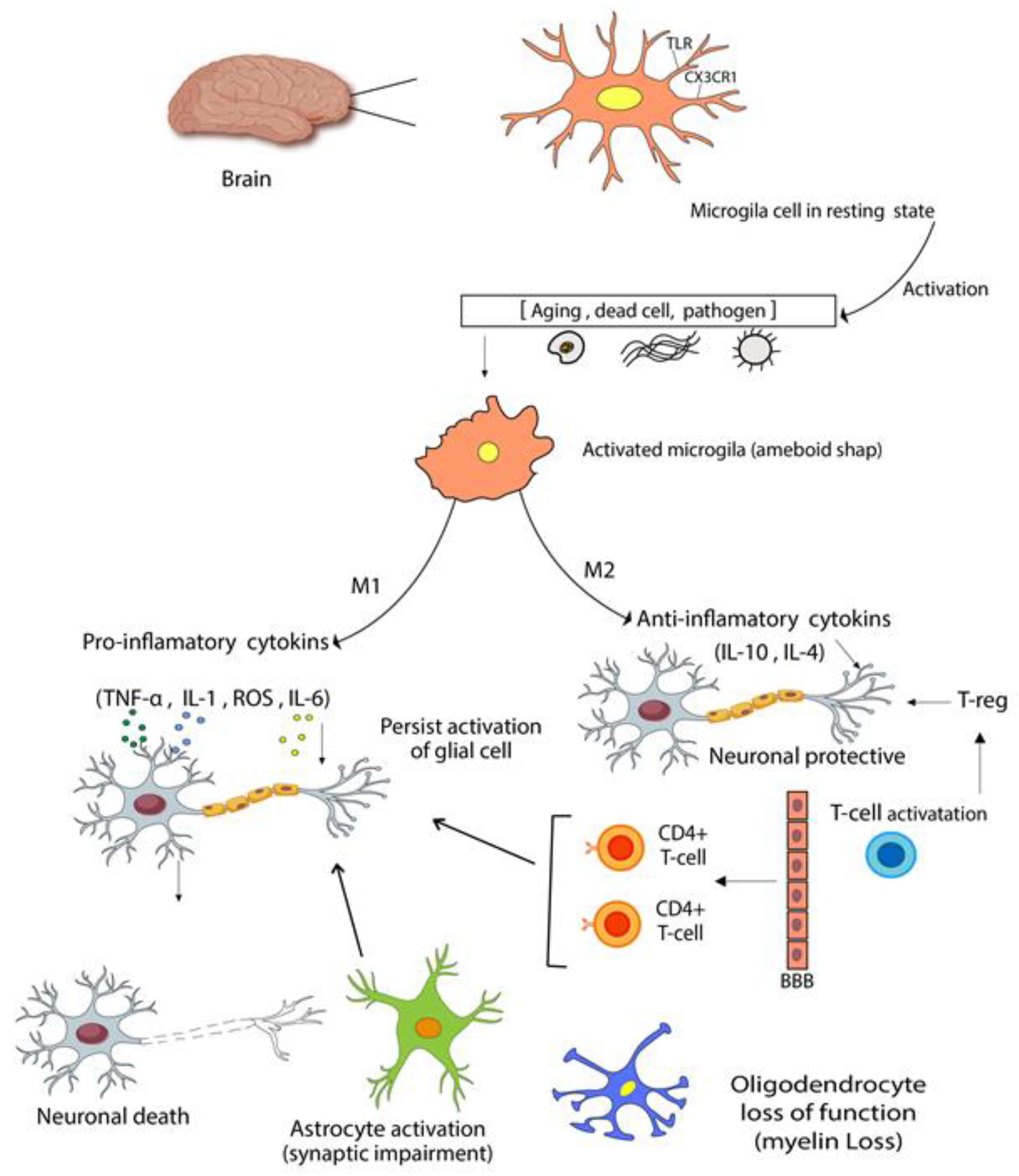
- Heneka, M.T.; Kummer, M.P.; Latz, E. Innate immune activation in neurodegenerative disease. Nat. Rev. Immunol. 2014, 14, 463–477. [Google Scholar] [CrossRef]
- Marogianni, C.; Sokratous, M.; Dardiotis, E.; Hadjigeorgiou, G.M.; Bogdanos, D.; Xiromerisiou, G. Neurodegeneration and Inflammation—An Interesting Interplay in Parkinson’s Disease. Int. J. Mol. Sci. 2020, 21, 8421. [Google Scholar] [CrossRef]
- Tang, Y.; Le, W. Differential Roles of M1 and M2 Microglia in Neurodegenerative Diseases. Mol. Neurobiol. 2016, 53, 1181–1194. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Dong, X.; Zheng, D.; Nao, J. MiR-124 and the Underlying Therapeutic Promise of Neurodegenerative Disorders. Front. Pharmacol. 2020, 10, 1555. [Google Scholar] [CrossRef]
- Essandoh, K.; Li, Y.; Huo, J. ; Fan G-C MiRNA-mediated macrophage polarization and its potential role in the regulation of inflammatory response. Shock 2016, 46, 122. [Google Scholar] [CrossRef]
- Colonna, M.; Butovsky, O. Microglia Function in the Central Nervous System During Health and Neurodegeneration. Annu. Rev. Immunol. 2017, 35, 441–468. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y. ; Holtzman DM Interplay between innate immunity and Alzheimer disease: APOE and TREM2 in the spotlight. Nat Rev Immunol 2018, 18, 759–772. [Google Scholar] [CrossRef] [PubMed]
- Chamera, K.; Trojan, E.; Szuster-Głuszczak, M.; Basta-Kaim, A. The Potential Role of Dysfunctions in Neuron-Microglia Communication in the Pathogenesis of Brain Disorders. Curr. Neuropharmacol. 2020, 18, 408–430. [Google Scholar] [CrossRef] [PubMed]
- Poniatowski. A.; Wojdasiewicz, P.; Krawczyk, M.; Szukiewicz, D.; Gasik, R.; Kubaszewski,.; Kurkowska-Jastrzębska, I. Analysis of the Role of CX3CL1 (Fractalkine) and Its Receptor CX3CR1 in Traumatic Brain and Spinal Cord Injury: Insight into Recent Advances in Actions of Neurochemokine Agents. Mol. Neurobiol. 2016, 54, 2167–2188. [Google Scholar] [CrossRef]
- Tarozzo, G.; Bortolazzi, S.; Crochemore, C.; Chen, S.; Lira, A.; Abrams, J.; Beltramo, M. Fractalkine protein localization and gene expression in mouse brain. J. Neurosci. Res. 2003, 73, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Yuan, Z.; Yang, S.; Zhu, Y.; Xue, M.; Zhang, J.; Leng, L. Brain Energy Metabolism: Astrocytes in Neurodegenerative Diseases. CNS Neurosci. Ther. 2022, 29, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Tcw, J.; Qian, L.; Pipalia, N.H.; Chao, M.J.; Liang, S.A.; Shi, Y.; Jain, B.R.; Bertelsen, S.E.; Kapoor, M.; Marcora, E.; et al. Cholesterol and matrisome pathways dysregulated in astrocytes and microglia. Cell 2022, 185, 2213–2233. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, F.; Huang, N.; et al. Oligodendrocytes and myelin: Active players in neurodegenerative brains? Dev Neurobiol 2022, 82, 160–174. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-H.; Rezzonico, M.G.; Friedman, B.A.; Huntley, M.H.; Meilandt, W.J.; Pandey, S.; Chen, Y.-J.J.; Easton, A.; Modrusan, Z.; Hansen, D.V.; et al. TREM2-independent oligodendrocyte, astrocyte, and T cell responses to tau and amyloid pathology in mouse models of Alzheimer disease. Cell Rep. 2021, 37, 110158. [Google Scholar] [CrossRef]
- Correale, J.; Marrodan, M. ; Ysrraelit MC Mechanisms of neurodegeneration and axonal dysfunction in progressive multiple sclerosis. Biomedicines 2019, 7, 14. [Google Scholar] [CrossRef]
- V. V.; I.T.; Vasu, M.M.; A Poovathinal, S.; Anitha, A. miRNAs as biomarkers of neurodegenerative disorders. Biomarkers Med. 2017, 11, 151–167. [Google Scholar] [CrossRef]
- Szelągowski, A.; Kozakiewicz, M. A Glance at Biogenesis and Functionality of MicroRNAs and Their Role in the Neuropathogenesis of Parkinson’s Disease. Oxidative Med. Cell. Longev. 2023, 2023, 1–18. [Google Scholar] [CrossRef]
- Rege, S.D.; Geetha, T.; Pondugula, S.R.; Zizza, C.A.; Wernette, C.M.; Babu, J.R. Noncoding RNAs in Neurodegenerative Diseases. ISRN Neurol. 2013, 2013, 1–5. [Google Scholar] [CrossRef]
- Cabezas, R.; Baez-Jurado, E.; Hidalgo-Lanussa, O.; Echeverria, V.; Ashrad, G.M.; Sahebkar, A.; Barreto, G.E. Growth Factors and Neuroglobin in Astrocyte Protection Against Neurodegeneration and Oxidative Stress. Mol. Neurobiol. 2019, 56, 2339–2351. [Google Scholar] [CrossRef]
- Jovanovic, M. ; Hengartner MO miRNAs and apoptosis: RNAs to die for. Oncogene 2006, 25, 6176–6187. [Google Scholar] [CrossRef]
- Sharma, S. ; Lu H-C microRNAs in neurodegeneration: current findings and potential impacts. 2018. [Google Scholar]
- Cho, K.H.T.; Xu, B.; Blenkiron, C.; Fraser, M. Emerging Roles of miRNAs in Brain Development and Perinatal Brain Injury. Front. Physiol. 2019, 10, 227. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.-H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Gregory, R.I.; Yan, K.-P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240. [Google Scholar] [CrossRef]
- Okada, C.; Yamashita, E.; Lee, S.J.; et al. A high-resolution structure of the pre-microRNA nuclear export machinery. Science 2009, 326, 1275–1279. [Google Scholar] [CrossRef]
- Zeng, Y. ; Cullen BR Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res 2004, 32, 4776–4785. [Google Scholar] [CrossRef]
- Roy, B.; Lee, E.; Li, T.; Rampersaud, M. Role of miRNAs in Neurodegeneration: From Disease Cause to Tools of Biomarker Discovery and Therapeutics. Genes 2022, 13, 425. [Google Scholar] [CrossRef]
- Weng, Y.-T.; Chang, Y.-M.; Chern, Y. The Impact of Dysregulated microRNA Biogenesis Machinery and microRNA Sorting on Neurodegenerative Diseases. Int. J. Mol. Sci. 2023, 24, 3443. [Google Scholar] [CrossRef]
- Junn, E. ; Mouradian MM MicroRNAs in neurodegenerative diseases and their therapeutic potential. Pharmacol Ther 2012, 133, 142–150. [Google Scholar] [CrossRef]
- Song M-S, Rossi JJ Molecular mechanisms of Dicer: endonuclease and enzymatic activity. Biochem J 2017, 474, 1603–1618. [CrossRef]
- Chmielarz, P.; Konovalova, J.; Najam, S.S.; Alter, H.; Piepponen, P.; Erfle, H.; Sonntag, K.-C.; Schütz, G.; Vinnikov, I.; Domanskyi, A. Dicer and microRNAs protect adult dopamine neurons. Cell Death Dis. 2017, 8, e2813–e2813. [Google Scholar] [CrossRef]
- Saeidi, N.; Goudarzvand, H.; Mohammadi, H.; Mardi, A.; Ghoreishizadeh, S.; Shomali, N.; Goudarzvand, M. Dysregulation of miR-193a serves as a potential contributor to MS pathogenesis via affecting RhoA and Rock1. Mult. Scler. Relat. Disord. 2022, 69, 104468. [Google Scholar] [CrossRef]
- Martín, M.M.-S.; Gómez, I.; Quiroga-Varela, A.; Río, M.G.-D.; Cedeño, R.R.; Álvarez, G.; Buxó, M.; Miguela, A.; Villar, L.M.; Castillo-Villalba, J.; et al. miRNA Signature in CSF From Patients With Primary Progressive Multiple Sclerosis. Neurol. - Neuroimmunol. Neuroinflammation 2023, 10. [Google Scholar] [CrossRef]
- Saeidi, N.N.; Dabiri, A.; Mansouri, R.; Moomivand, A.; Goudarzvand, M. miRNAs as Valuable Diagnostic Biomarkers in Patients with Multiple Sclerosis. J. Biomed. Res. Environ. Sci. 2023, 4, 773–778. [Google Scholar] [CrossRef]
- Abolghasemi, M.; Ashrafi, S.A.; Asadi, M.; et al. 2022; -10.
- Unlu, H.T.; Saridas, F.; Taskapilioglu, O.; Cecener, G.; Egeli, U.; Turan, O.F.; Tunca, B.; Zarifoglu, M. Investigation of miR-146a Expression Profiles in Fecal Samples of Patients With Multiple Sclerosis for Early Diagnosis and Treatment. Neurol. Sci. Neurophysiol. 2023, 40, 81–87. [Google Scholar] [CrossRef]
- Citterio, L.A.; Mancuso, R.; Agostini, S.; Meloni, M.; Clerici, M. Serum and Exosomal miR-7-1-5p and miR-223-3p as Possible Biomarkers for Parkinson’s Disease. Biomolecules 2023, 13, 865. [Google Scholar] [CrossRef]
- Cao, X.-Y.; Lu, J.-M.; Zhao, Z.-Q.; Li, M.-C.; Lu, T.; An, X.-S.; Xue, L.-J. MicroRNA biomarkers of Parkinson’s disease in serum exosome-like microvesicles. Neurosci. Lett. 2017, 644, 94–99. [Google Scholar] [CrossRef]
- Dos Santos, M.C.T.; Barreto-Sanz, M.A.; Correia, B.R.S.; et al. miRNA-based signatures in cerebrospinal fluid as potential diagnostic tools for early stage Parkinson’s disease. Oncotarget 2018, 9, 17455. [Google Scholar] [CrossRef]
- Zago, E.; Dal Molin, A.; Dimitri, G.M.; et al. Early downregulation of hsa-miR-144-3p in serum from drug-naïve Parkinson’s disease patients. Sci Rep 2022, 12, 1330. [Google Scholar] [CrossRef]
- Chen, L.; Yang, J.; Lü, J.; et al. Identification of aberrant circulating mi RNA s in Parkinson’s disease plasma samples. Brain Behav 2018, 8, e00941. [Google Scholar] [CrossRef]
- Kim, J.; Fiesel, F.C.; Belmonte, K.C.; Hudec, R.; Wang, W.-X.; Kim, C.; Nelson, P.T.; Springer, W.; Kim, J. miR-27a and miR-27b regulate autophagic clearance of damaged mitochondria by targeting PTEN-induced putative kinase 1 (PINK1). Mol. Neurodegener. 2016, 11, 1–16. [Google Scholar] [CrossRef]
- Yang, Z.; Li, T.; Li, S.; Wei, M.; Qi, H.; Shen, B.; Chang, R.C.-C.; Le, W.; Piao, F. Altered Expression Levels of MicroRNA-132 and Nurr1 in Peripheral Blood of Parkinson’s Disease: Potential Disease Biomarkers. ACS Chem. Neurosci. 2019, 10, 2243–2249. [Google Scholar] [CrossRef]
- Baumert, B.; Sobuś, A.; Gołąb-Janowska, M.; Ulańczyk, Z.; Paczkowska, E.; Łuczkowska, K.; Zawiślak, A.; Milczarek, S.; Osękowska, B.; Meller, A.; et al. Local and Systemic Humoral Response to Autologous Lineage-Negative Cells Intrathecal Administration in ALS Patients. Int. J. Mol. Sci. 2020, 21, 1070. [Google Scholar] [CrossRef]
- Russell, A.P.; Wada, S.; Vergani, L.; Hock, M.B.; Lamon, S.; Léger, B.; Ushida, T.; Cartoni, R.; Wadley, G.D.; Hespel, P.; et al. Disruption of skeletal muscle mitochondrial network genes and miRNAs in amyotrophic lateral sclerosis. Neurobiol. Dis. 2013, 49, 107–117. [Google Scholar] [CrossRef]
- Kong, Y.; Li, S.; Zhang, M.; Xu, W.; Chen, Q.; Zheng, L.; Liu, P.; Zou, W. Acupuncture Ameliorates Neuronal Cell Death, Inflammation, and Ferroptosis and Downregulated miR-23a-3p After Intracerebral Hemorrhage in Rats. J. Mol. Neurosci. 2021, 71, 1863–1875. [Google Scholar] [CrossRef]
- Takahashi, I.; Hama, Y.; Matsushima, M.; Hirotani, M.; Kano, T.; Hohzen, H.; Yabe, I.; Utsumi, J.; Sasaki, H. Identification of plasma microRNAs as a biomarker of sporadic Amyotrophic Lateral Sclerosis. Mol. Brain 2015, 8, 1–9. [Google Scholar] [CrossRef]
- Alexandrov, P.N.; Dua, P.; Hill, J.M.; Bhattacharjee, S.; Zhao, Y.; Lukiw, W.J. microRNA (miRNA) speciation in Alzheimer’s disease (AD) cerebrospinal fluid (CSF) and extracellular fluid (ECF). Int. J. Biochem. Mol. Biol. 2012, 3, 365–373. [Google Scholar]
- Lukiw, W.J. ; Pogue AI Vesicular transport of encapsulated microRNA between glial and neuronal cells. Int J Mol Sci 2020, 21, 5078. [Google Scholar] [CrossRef]
- Kumar, S.; Reddy, A.P.; Yin, X. ; Reddy PH Novel MicroRNA-455-3p and its protective effects against abnormal APP processing and amyloid beta toxicity in Alzheimer’s disease. Biochim Biophys Acta (BBA)-Molecular Basis Dis 2019, 1865, 2428–2440. [Google Scholar] [CrossRef]
- Kumar, S. ; Reddy PH A new discovery of MicroRNA-455-3p in Alzheimer’s disease. J Alzheimer’s Dis 2019, 72, S117–S130. [Google Scholar] [CrossRef]
- Zhao, Y.; Pogue, A.I. ; Lukiw WJ MicroRNA (miRNA) signaling in the human CNS in sporadic Alzheimer’s disease (AD)-novel and unique pathological features. Int J Mol Sci 2015, 16, 30105–30116. [Google Scholar] [CrossRef]
- Fan, W.; Liang, C.; Ou, M.; Zou, T.; Sun, F.; Zhou, H.; Cui, L. MicroRNA-146a Is a Wide-Reaching Neuroinflammatory Regulator and Potential Treatment Target in Neurological Diseases. Front. Mol. Neurosci 2020, 13, 90. [Google Scholar] [CrossRef]
- Zhao, Y.; Bhattacharjee, S.; Jones, B.M.; et al. Regulation of neurotropic signaling by the inducible, NF-kB-sensitive miRNA-125b in Alzheimer’s disease (AD) and in primary human neuronal-glial (HNG) cells. Mol Neurobiol 2014, 50, 97–106. [Google Scholar] [CrossRef]
- Andersen, E.; Casteigne, B.; Chapman, W.D.; et al. Diagnostic biomarkers in Alzheimer’s disease. Biomarkers in Neuropsychiatry 2021, 5, 100041. [Google Scholar] [CrossRef]
- McKhann, G.; Drachman, D.; Folstein, M.; Katzman, R.; Price, D.; Stadlan, E.M. Clinical diagnosis of Alzheimer's disease: Report of the NINCDS—ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology 2011, 77, 333–333. [Google Scholar] [CrossRef]
- Mayeux, R.; Stern, Y. Epidemiology of Alzheimer disease. Cold Spring Harb Perspect Med 2012, 2, a006239. [Google Scholar] [CrossRef]
- Hyman, B.T.; Phelps, C.H.; Beach, T.G.; Bigio, E.H.; Cairns, N.J.; Carrillo, M.C.; Dickson, D.W.; Duyckaerts, C.; Frosch, M.P.; Masliah, E.; et al. National Institute on Aging–Alzheimer's Association guidelines for the neuropathologic assessment of Alzheimer's disease. Alzheimer's Dement. 2012, 8, 1–13. [Google Scholar] [CrossRef]
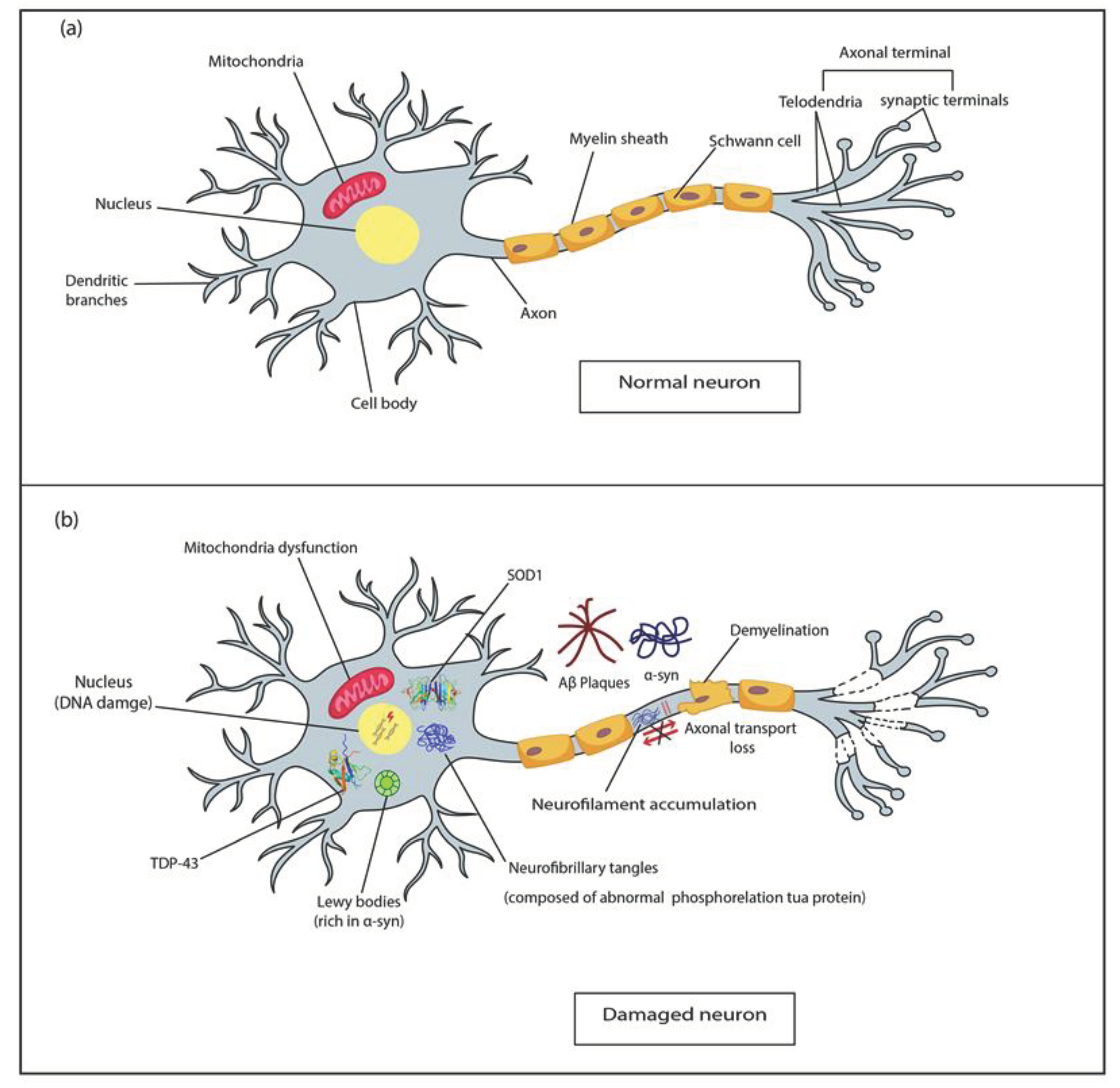
- Masliah, E.; Crews, L.; Hansen, L. Synaptic remodeling during aging and in Alzheimer's disease. J. Alzheimer's Dis. 2006, 9, 91–99. [Google Scholar] [CrossRef]
- Pennanen, C.; Kivipelto, M.; Tuomainen, S.; Hartikainen, P.; Hänninen, T.; Laakso, M.P.; Hallikainen, M.; Vanhanen, M.; Nissinen, A.; Helkala, E.-L.; et al. Hippocampus and entorhinal cortex in mild cognitive impairment and early AD. Neurobiol. Aging 2004, 25, 303–310. [Google Scholar] [CrossRef]
- DeKosky, S.T.; Scheff, S.W. ; Styren SD Structural correlates of cognition in dementia: quantification and assessment of synapse change. Neurodegeneration 1996, 5, 417–421. [Google Scholar] [CrossRef]
- Katsumata, Y.; Fardo, D.W.; Kukull, W.A. ; Nelson PT Dichotomous scoring of TDP-43 proteinopathy from specific brain regions in 27 academic research centers: associations with Alzheimer’s disease and cerebrovascular disease pathologies. Acta Neuropathol Commun 2018, 6, 1–11. [Google Scholar] [CrossRef]
- Berson, A.; Barbash, S.; Shaltiel, G.; Goll, Y.; Hanin, G.; Greenberg, D.S.; Ketzef, M.; Becker, A.J.; Friedman, A.; Soreq, H. Cholinergic-associated loss of hnRNP-A/B in Alzheimer's disease impairs cortical splicing and cognitive function in mice. EMBO Mol. Med. 2012, 4, 730–742. [Google Scholar] [CrossRef]
- Yang, Q.; Zhao, Q.; Yin, Y. miR-133b is a potential diagnostic biomarker for Alzheimer's disease and has a neuroprotective role. Exp. Ther. Med. 2019, 18, 2711–2718. [Google Scholar] [CrossRef]
- Dorval, V.; Nelson, P.T. ; Hébert SS Circulating microRNAs in Alzheimer’s disease: the search for novel biomarkers. Front Mol Neurosci 2013, 6, 24. [Google Scholar]
- Kumar, S.; Vijayan, M. ; Reddy PH MicroRNA-455-3p as a potential peripheral biomarker for Alzheimer’s disease. Hum Mol Genet 2017, 26, 3808–3822. [Google Scholar] [CrossRef]
- Banzhaf-Strathmann, J.; Benito, E.; May, S.; Arzberger, T.; Tahirovic, S.; Kretzschmar, H.; Fischer, A.; Edbauer, D. MicroRNA-125b induces tau hyperphosphorylation and cognitive deficits in Alzheimer's disease. EMBO J. 2014, 33, 1667–1680. [Google Scholar] [CrossRef]
- Hong, H.; Li, Y.; Su, B. Identification of Circulating miR-125b as a Potential Biomarker of Alzheimer’s Disease in APP/PS1 Transgenic Mouse. J. Alzheimer's Dis. 2017, 59, 1449–1458. [Google Scholar] [CrossRef]
- Pan, Y.; Liu, R.; Terpstra, E.; Wang, Y.; Qiao, F.; Wang, J.; Tong, Y.; Pan, B. Dysregulation and Diagnostic Potential of microRNA in Alzheimer’s Disease. J. Alzheimer's Dis. 2015, 49, 1–12. [Google Scholar] [CrossRef]
- Callens, M.; Kraskovskaya, N.; Derevtsova, K.; Annaert, W.; Bultynck, G.; Bezprozvanny, I.; Vervliet, T. The role of Bcl-2 proteins in modulating neuronal Ca2+ signaling in health and in Alzheimer's disease. Biochim. et Biophys. Acta (BBA) - Mol. Cell Res. 2021, 1868, 118997–118997. [Google Scholar] [CrossRef]
- Rohn, T.T.; Vyas, V.; Hernandez-Estrada, T.; Nichol, K.E.; Christie, L.-A.; Head, E. Lack of Pathology in a Triple Transgenic Mouse Model of Alzheimer's Disease after Overexpression of the Anti-Apoptotic Protein Bcl-2. J. Neurosci. 2008, 28, 3051–3059. [Google Scholar] [CrossRef]
- Liu, W.; Zhao, J.; Lu, G. miR-106b inhibits tau phosphorylation at Tyr18 by targeting Fyn in a model of Alzheimer's disease. Biochem. Biophys. Res. Commun. 2016, 478, 852–857. [Google Scholar] [CrossRef]
- Di Meco, A.; Pratico, D. MicroRNAs as therapeutic targets for Alzheimer’s disease. J Alzheimer’s Dis 2016, 53, 367–372. [Google Scholar] [CrossRef]
- Vassar, R. Bace 1: The β-secretase enzyme in alzheimer’s disease. J Mol Neurosci 2004, 23, 105–113. [Google Scholar] [CrossRef]
- Du, W.; Lei, C.; Dong, Y. MicroRNA-149 is downregulated in Alzheimer’s disease and inhibits β-amyloid accumulation and ameliorates neuronal viability through targeting BACE1. Genet. Mol. Biol. 2021, 44, e20200064. [Google Scholar] [CrossRef]
- Kouli, A.; Torsney, K.M. ; Kuan W-L Parkinson’s disease: etiology, neuropathology, and pathogenesis. Exon Publ 2018, 3–26. [Google Scholar]
- Jankovic, J. Parkinson's disease: clinical features and diagnosis. J. Neurol. Neurosurg. Psychiatry 2008, 79, 368–376. [Google Scholar] [CrossRef]
- Narhi, L.; Wood, S.J.; Steavenson, S.; Jiang, Y.; Wu, G.M.; Anafi, D.; Kaufman, S.A.; Martin, F.; Sitney, K.; Denis, P.; et al. Both Familial Parkinson's Disease Mutations Accelerate α-Synuclein Aggregation. J. Biol. Chem. 1999, 274, 9843–9846. [Google Scholar] [CrossRef]
- Alcalay, R.N.; Caccappolo, E.; Mejia-Santana, H.; et al. Frequency of known mutations in early-onset Parkinson disease: implication for genetic counseling: the consortium on risk for early onset Parkinson disease study. Arch Neurol 2010, 67, 1116–1122. [Google Scholar] [CrossRef]
- Chaudhuri, K.R.; Healy, D.G. ; Schapira AH V Non-motor symptoms of Parkinson’s disease: diagnosis and management. Lancet Neurol 2006, 5, 235–245. [Google Scholar] [CrossRef]
- Goh, S.Y.; Chao, Y.X.; Dheen, S.T.; Tan, E.-K.; Tay, S.S.-W. Role of MicroRNAs in Parkinson’s Disease. Int. J. Mol. Sci. 2019, 20, 5649. [Google Scholar] [CrossRef]
- Cressatti, M.; Juwara, L.; Galindez, J.M.; et al. Salivary microR-153 and microR-223 levels as potential diagnostic biomarkers of idiopathic Parkinson’s disease. Mov Disord 2020, 35, 468–477. [Google Scholar] [CrossRef]
- Gui, Y.; Liu, H.; Zhang, L.; Lv, W.; Hu, X. Altered microRNA profiles in cerebrospinal fluid exosome in Parkinson disease and Alzheimer disease. Oncotarget 2015, 6, 37043–37053. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, S.; Qi, W.; et al. Overexpression of miR-153 promotes oxidative stress in MPP+-induced PD model by negatively regulating the Nrf2/HO-1 signaling pathway. Int J Clin Exp Pathol 2018, 11, 4179. [Google Scholar]
- Bellezza, I.; Giambanco, I.; Minelli, A.; Donato, R. Nrf2-Keap1 signaling in oxidative and reductive stress. Biochim. et Biophys. Acta (BBA) - Mol. Cell Res. 2018, 1865, 721–733. [Google Scholar] [CrossRef]
- Bai, X.; Tang, Y.; Yu, M.; Wu, L.; Liu, F.; Ni, J.; Wang, Z.; Wang, J.; Fei, J.; Wang, W.; et al. Downregulation of blood serum microRNA 29 family in patients with Parkinson’s disease. Sci. Rep. 2017, 7, 1–7. [Google Scholar] [CrossRef]
- Bhinge, A.; Namboori, S.C.; Bithell, A.; Soldati, C.; Buckley, N.J.; Stanton, L.W. MiR-375 is Essential for Human Spinal Motor Neuron Development and May Be Involved in Motor Neuron Degeneration. STEM CELLS 2015, 34, 124–134. [Google Scholar] [CrossRef]
- Cai L-J, Tu, L. ; Li, T.; et al. Up-regulation of microRNA-375 ameliorates the damage of dopaminergic neurons, reduces oxidative stress and inflammation in Parkinson’s disease by inhibiting SP1. Aging 2020, 12, 672.
- Citron, B.A.; Dennis, J.S.; Zeitlin, R.S.; Echeverria, V. Transcription factor Sp1 dysregulation in Alzheimer's disease. J. Neurosci. Res. 2008, 86, 2499–2504. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, M.; Wei, M.; Wang, A.; Deng, Y.; Cao, H. MicroRNA-216a inhibits neuronal apoptosis in a cellular Parkinson’s disease model by targeting Bax. Metab. Brain Dis. 2020, 35, 627–635. [Google Scholar] [CrossRef]
- Reed JC Proapoptotic multidomain Bcl-2/Bax-family proteins: mechanisms, physiological roles, and therapeutic opportunities. Cell death Differ 2006, 13, 1378–1386. [CrossRef]
- De Falco, M.; De Luca, L.; Acanfora, F.; Cavallotti, I.; Cottone, G.; Laforgia, V.; De Luca, B.; Baldi, A.; De Luca, A. Alteration of the Bcl-2:Bax ratio in the placenta as pregnancy proceeds. Histochem. J. 2001, 33, 421–425. [Google Scholar] [CrossRef]
- Cantoni, C.; Ghezzi, L.; Choi, J.; Cross, A.H.; Piccio, L. Targeting miR-223 enhances myeloid-derived suppressor cell suppressive activities in multiple sclerosis patients. Mult. Scler. Relat. Disord. 2023, 76, 104839–104839. [Google Scholar] [CrossRef]
- Orton, S.-M.; Herrera, B.M.; Yee, I.M.; Valdar, W.; Ramagopalan, S.V.; Sadovnick, A.D.; Ebers, G.C.; Canadian Collaborative Study Group. Sex ratio of multiple sclerosis in Canada: a longitudinal study. Lancet Neurol. 2006, 5, 932–936. [Google Scholar] [CrossRef]
- Maciak, K.; Dziedzic, A.; Saluk, J. Remyelination in multiple sclerosis from the miRNA perspective. Front. Mol. Neurosci. 2023, 16, 1199313. [Google Scholar] [CrossRef]
- Engelhardt, B. ; Ransohoff RM The ins and outs of T-lymphocyte trafficking to the CNS: anatomical sites and molecular mechanisms. Trends Immunol 2005, 26, 485–495. [Google Scholar] [CrossRef]
- Ahlbrecht, J.; Martino, F.; Pul, R.; Skripuletz, T.; Sühs, K.-W.; Schauerte, C.; Yildiz. ; Trebst, C.; Tasto, L.; Thum, S.; et al. Deregulation of microRNA-181c in cerebrospinal fluid of patients with clinically isolated syndrome is associated with early conversion to relapsing–remitting multiple sclerosis. Mult. Scler. J. 2016, 22, 1202–1214. [Google Scholar] [CrossRef]
- Harris, V.K.; Tuddenham, J.F. ; Sadiq SA Biomarkers of multiple sclerosis: current findings. Degener Neurol Neuromuscul Dis 2017, 19–29. [Google Scholar]
- Ma, Q.; Zhao, H.; Tao, Z.; Wang, R.; Liu, P.; Han, Z.; Ma, S.; Luo, Y.; Jia, J. MicroRNA-181c Exacerbates Brain Injury in Acute Ischemic Stroke. Aging Dis. 2016, 7, 705–714. [Google Scholar] [CrossRef]
- Zhang, L. ; Dong L-Y, Li Y-J, et al. The microRNA miR-181c controls microglia-mediated neuronal apoptosis by suppressing tumor necrosis factor. J Neuroinflammation 2012, 9, 1–12. [Google Scholar]
- Kramer, S.; Haghikia, A.; Bang, C.; et al. Elevated levels of miR-181c and miR-633 in the CSF of patients with MS: A validation study. e: Neurol Neuroinflammation 2019, 6, 2019. [Google Scholar]
- Bergman, P.; Piket, E.; Khademi, M.; James, T.; Brundin, L.; Olsson, T.; Piehl, F.; Jagodic, M. Circulating miR-150 in CSF is a novel candidate biomarker for multiple sclerosis. Neurol. - Neuroimmunol. Neuroinflammation 2016, 3, e219–e219. [Google Scholar] [CrossRef]
- Moore, C.S.; Rao, V.T.S.; Durafourt, B.A.; et al. miR-155 as a multiple sclerosis–relevant regulator of myeloid cell polarization. Ann Neurol 2013, 74, 709–720. [Google Scholar] [CrossRef]
- Leng, R.-X.; Pan, H.-F.; Qin, W.-Z.; Chen, G.-M.; Ye, D.-Q. Role of microRNA-155 in autoimmunity. Cytokine Growth Factor Rev. 2011, 22, 141–147. [Google Scholar] [CrossRef]
- Maciak, K.; Dziedzic, A.; Miller, E.; Saluk-Bijak, J. miR-155 as an Important Regulator of Multiple Sclerosis Pathogenesis. A Review. Int. J. Mol. Sci. 2021, 22, 4332. [Google Scholar] [CrossRef]
- Martín, M.M.-S.; Reverter, G.; Robles-Cedeño, R.; Buxò, M.; Ortega, F.J.; Gómez, I.; Tomàs-Roig, J.; Celarain, N.; Villar, L.M.; Perkal, H.; et al. Analysis of miRNA signatures in CSF identifies upregulation of miR-21 and miR-146a/b in patients with multiple sclerosis and active lesions. J. Neuroinflammation 2019, 16, 1–10. [Google Scholar] [CrossRef]
- Galloway, D.A.; Blandford, S.N.; Berry, T.; Williams, J.B.; Stefanelli, M.; Ploughman, M.; Moore, C.S. miR-223 promotes regenerative myeloid cell phenotype and function in the demyelinated central nervous system. Glia 2018, 67, 857–869. [Google Scholar] [CrossRef]
- Marangon, D.; Boda, E.; Parolisi, R.; Negri, C.; Giorgi, C.; Montarolo, F.; Perga, S.; Bertolotto, A.; Buffo, A.; Abbracchio, M.P.; et al. In vivo silencing of miR-125a-3p promotes myelin repair in models of white matter demyelination. Glia 2020, 68, 2001–2014. [Google Scholar] [CrossRef]
- Lecca, D.; Marangon, D.; Coppolino, G.T.; Méndez, A.M.; Finardi, A.; Costa, G.D.; Martinelli, V.; Furlan, R.; Abbracchio, M.P. MiR-125a-3p timely inhibits oligodendroglial maturation and is pathologically up-regulated in human multiple sclerosis. Sci. Rep. 2016, 6, srep34503. [Google Scholar] [CrossRef]
- Brusati, A.; Ratti, A.; Pensato, V.; Peverelli, S.; Gentilini, D.; Bella, E.D.; Sorce, M.N.; Meneri, M.; Gagliardi, D.; Corti, S.; et al. Analysis of miRNA rare variants in amyotrophic lateral sclerosis and in silico prediction of their biological effects. Front. Genet. 2022, 13, 1055313. [Google Scholar] [CrossRef]
- Hardiman, O.; Al-Chalabi, A.; Chio, A.; et al. Amyotrophic lateral sclerosis. Nat Rev Dis Prim 2017, 3, 1–19. [Google Scholar] [CrossRef]
- Goutman, A.S.; Hardiman, O.; Al-Chalabi, A.; Chió, A.; Savelieff, M.G.; Kiernan, M.C.; Feldman, E.L. Emerging insights into the complex genetics and pathophysiology of amyotrophic lateral sclerosis. Lancet Neurol. 2022, 21, 465–479. [Google Scholar] [CrossRef]
- Joilin, G.; Leigh, P.N.; Newbury, S.F.; Hafezparast, M. An Overview of MicroRNAs as Biomarkers of, A.L.S. Front. Neurol.
- Rinchetti 2019, P.; Rizzuti, M.; Faravelli, I.; Corti, S. MicroRNA metabolism and dysregulation in amyotrophic lateral sclerosis. Mol Neurobiol 2018, 55, 2617–2630. [Google Scholar] [CrossRef]
- Alvia, M.; Aytan, N.; Spencer, K.R.; Foster, Z.W.; Rauf, N.A.; Guilderson, L.; Robey, I.; Averill, J.G.; Walker, S.E.; Alvarez, V.E.; et al. MicroRNA Alterations in Chronic Traumatic Encephalopathy and Amyotrophic Lateral Sclerosis. Front. Neurosci. 2022, 16, 855096. [Google Scholar] [CrossRef]
- Toivonen, J.M.; Manzano, R.; Oliván, S.; Zaragoza, P.; García-Redondo, A.; Osta, R. MicroRNA-206: A Potential Circulating Biomarker Candidate for Amyotrophic Lateral Sclerosis. PLOS ONE 2014, 9, e89065. [Google Scholar] [CrossRef]
- Vaz, A.R.; Vizinha, D.; Morais, H.; Colaço, A.R.; Loch-Neckel, G.; Barbosa, M.; Brites, D. Overexpression of miR-124 in Motor Neurons Plays a Key Role in ALS Pathological Processes. Int. J. Mol. Sci. 2021, 22, 6128. [Google Scholar] [CrossRef]
- Magen, I.; Yacovzada, N.S.; Yanowski, E.; Coenen-Stass, A.; Grosskreutz, J.; Lu, C.-H.; Greensmith, L.; Malaspina, A.; Fratta, P.; Hornstein, E. Circulating miR-181 is a prognostic biomarker for amyotrophic lateral sclerosis. Nat. Neurosci. 2021, 24, 1534–1541. [Google Scholar] [CrossRef]
- Cunha, C.; Santos, C.; Gomes, C.; Fernandes, A.; Correia, A.M.; Sebastião, A.M.; Vaz, A.R.; Brites, D. Downregulated Glia Interplay and Increased miRNA-155 as Promising Markers to Track ALS at an Early Stage. Mol. Neurobiol. 2017, 55, 4207–4224. [Google Scholar] [CrossRef]
- Xu, Q.; Zhao, Y.; Zhou, X.; Luan, J.; Cui, Y.; Han, J. Comparison of the extraction and determination of serum exosome and miRNA in serum and the detection of miR-27a-3p in serum exosome of ALS patients. Intractable Rare Dis. Res. 2018, 7, 13–18. [Google Scholar] [CrossRef]
- Reichenstein, I.; Eitan, C.; Diaz-Garcia, S.; Haim, G.; Magen, I.; Siany, A.; Hoye, M.L.; Rivkin, N.; Olender, T.; Toth, B.; et al. Human genetics and neuropathology suggest a link between miR-218 and amyotrophic lateral sclerosis pathophysiology. Sci. Transl. Med. 2019, 11. [Google Scholar] [CrossRef]
- Yang, L.; Embree, L.J. ; Hickstein DD TLS-ERG leukemia fusion protein inhibits RNA splicing mediated by serine-arginine proteins. Mol Cell Biol 2000, 20, 3345–3354. [Google Scholar] [CrossRef]
- Kawahara, Y.; Mieda-Sato, A. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc. Natl. Acad. Sci. 2012, 109, 3347–3352. [Google Scholar] [CrossRef]
- Lagier-Tourenne, C.; Polymenidou, M. ; Cleveland DW TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet 2010, 19, R46–R64. [Google Scholar] [CrossRef]
- Ling S-C, Polymenidou, M. ; Cleveland DW Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 2013, 79, 416–438.
- Koike, Y.; Onodera, O. Implications of miRNAs dysregulation in amyotrophic lateral sclerosis: Challenging for clinical applications. Front. Neurosci. 2023, 17, 1131758. [Google Scholar] [CrossRef]
- Liu, H.; Lan, S.; Shi, X.-J.; Fan, F.-C.; Liu, Q.-S.; Cong, L.; Cheng, Y. Systematic review and meta-analysis on microRNAs in amyotrophic lateral sclerosis. Brain Res. Bull. 2023, 194, 82–89. [Google Scholar] [CrossRef]
- Di Pietro, L.; Lattanzi, W.; Bernardini, C. Skeletal Muscle MicroRNAs as Key Players in the Pathogenesis of Amyotrophic Lateral Sclerosis. Int. J. Mol. Sci. 2018, 19, 1534. [Google Scholar] [CrossRef]
- Pegoraro, V.; Marozzo, R.; Angelini, C. MicroRNAs and HDAC4 protein expression in the skeletal muscle of ALS patients. Clin. Neuropathol. 2020, 39, 105–114. [Google Scholar] [CrossRef]
- Nie M, Deng Z-L, Liu J, Wang D-Z (2015) Noncoding RNAs, emerging regulators of skeletal muscle development and diseases. Biomed Res Int 2015.
- Williams, A.H.; Valdez, G.; Moresi, V.; Qi, X.; McAnally, J.; Elliott, J.L.; Bassel-Duby, R.; Sanes, J.R.; Olson, E.N. MicroRNA-206 Delays ALS Progression and Promotes Regeneration of Neuromuscular Synapses in Mice. Science 2009, 326, 1549–1554. [Google Scholar] [CrossRef]
- Butovsky, O.; Jedrychowski, M.P.; Cialic, R.; Krasemann, S.; Murugaiyan, G.; Fanek, Z.; Greco, D.J.; Wu, P.M.; Doykan, C.E.; Kiner, O.; et al. Targeting miR-155 restores abnormal microglia and attenuates disease in SOD1 mice. Ann. Neurol. 2014, 77, 75–99. [Google Scholar] [CrossRef]
- Gomes, C.; Sequeira, C.; Likhite, S.; Dennys, C.N.; Kolb, S.J.; Shaw, P.J.; Vaz, A.R.; Kaspar, B.K.; Meyer, K.; Brites, D. Neurotoxic Astrocytes Directly Converted from Sporadic and Familial ALS Patient Fibroblasts Reveal Signature Diversities and miR-146a Theragnostic Potential in Specific Subtypes. Cells 2022, 11, 1186. [Google Scholar] [CrossRef]
- Campos-Melo, D.; Droppelmann, C.A.; He, Z.; Volkening, K.; Strong, M.J. Altered microRNA expression profile in amyotrophic lateral sclerosis: a role in the regulation of NFL mRNA levels. Mol. Brain 2013, 6, 26. [Google Scholar] [CrossRef]
- He, M.; Zhang, H.-N.; Tang, Z.-C.; Gao, S.-G. Diagnostic and Therapeutic Potential of Exosomal MicroRNAs for Neurodegenerative Diseases. Neural Plast. 2021, 2021, 1–13. [Google Scholar] [CrossRef]
- Akbari, A.; Jabbari, N.; Sharifi, R.; Ahmadi, M.; Vahhabi, A.; Seyedzadeh, S.J.; Nawaz, M.; Szafert, S.; Mahmoodi, M.; Jabbari, E.; et al. Free and hydrogel encapsulated exosome-based therapies in regenerative medicine. Life Sci. 2020, 249, 117447. [Google Scholar] [CrossRef]
- Liu, X.; Yang, Y.; Li, Y.; Niu, X.; Zhao, B.; Wang, Y.; Bao, C.; Xie, Z.; Lin, Q.; Zhu, L. Integration of stem cell-derived exosomes with in situ hydrogel glue as a promising tissue patch for articular cartilage regeneration. Nanoscale 2017, 9, 4430–4438. [Google Scholar] [CrossRef]
- Kim, H.J.; Seo, S.W.; Chang, J.W.; Lee, J.I.; Kim, C.H.; Chin, J.; Choi, S.J.; Kwon, H.; Yun, H.J.; Lee, J.M.; et al. Stereotactic brain injection of human umbilical cord blood mesenchymal stem cells in patients with Alzheimer's disease dementia: A phase 1 clinical trial. Alzheimer's Dementia: Transl. Res. Clin. Interv. 2015, 1, 95–102. [Google Scholar] [CrossRef]
- Erdő, F.; Bors, L.A.; Farkas, D.; Bajza, Á.; Gizurarson, S. Evaluation of intranasal delivery route of drug administration for brain targeting. Brain Res. Bull. 2018, 143, 155–170. [Google Scholar] [CrossRef]
- Beutler, A.S.; Reinhardt, M. AAV for pain: steps towards clinical translation. Gene Ther. 2009, 16, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Kaplitt, M.G.; Feigin, A.; Tang, C.; Fitzsimons, H.L.; Mattis, P.; A Lawlor, P.; Bland, R.J.; Young, D.; Strybing, K.; Eidelberg, D.; et al. Safety and tolerability of gene therapy with an adeno-associated virus (AAV) borne GAD gene for Parkinson's disease: an open label, phase I trial. Lancet 2007, 369, 2097–2105. [Google Scholar] [CrossRef] [PubMed]
- Ekin, A.; Karatas, O.F.; Culha, M.; Ozen, M. Designing a gold nanoparticle-based nanocarrier for microRNA transfection into the prostate and breast cancer cells. J. Gene Med. 2014, 16, 331–335. [Google Scholar] [CrossRef]
- Conde, J.; Oliva, N.; Atilano, M.; Song, H.S.; Artzi, N. Self-assembled RNA-triple-helix hydrogel scaffold for microRNA modulation in the tumour microenvironment. Nat. Mater. 2015, 15, 353–363. [Google Scholar] [CrossRef]
- Zhao, J.; Lai, H.; Lu, H.; Barner-Kowollik, C.; Stenzel, M.H.; Xiao, P. Fructose-Coated Nanodiamonds: Promising Platforms for Treatment of Human Breast Cancer. Biomacromolecules 2016, 17, 2946–2955. [Google Scholar] [CrossRef]
- Yu, C.; Qian, L.; Uttamchandani, M.; Li, L.; Yao, S.Q. Single-Vehicular Delivery of Antagomir and Small Molecules to Inhibit miR-122 Function in Hepatocellular Carcinoma Cells by using “Smart” Mesoporous Silica Nanoparticles. Angew. Chem. 2015, 127, 10720–10724. [Google Scholar] [CrossRef]
- Austin TM First microRNA mimic enters clinic. Nat Biotechnol 2013, 31, 577.
- Van Rooij, E.; Kauppinen, S. Development of micro RNA therapeutics is coming of age. EMBO Mol Med 2014, 6, 851–864. [Google Scholar] [CrossRef]
- Rupaimoole, R. ; Slack FJ MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Walayat, A.; Yang, M.; Xiao, D. Therapeutic Implication of miRNA in Human Disease. In Antisense Therapy; Sharad, S., Kapur, S., Eds.; IntechOpen: London, UK, 2019; ISBN 978-1-78984-532-7. [Google Scholar]
- Lennox, K.A. ; Behlke MA Chemical modification and design of anti-miRNA oligonucleotides. Gene Ther 2011, 18, 1111–1120. [Google Scholar] [CrossRef] [PubMed]
- Li, Z. ; Rana TM Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov 2014, 13, 622–638. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z. The principles of MiRNA-masking antisense oligonucleotides technology. MicroRNA Cancer Methods Protoc 2011, 43–49. [Google Scholar]
- Ebert, M.S. ; Sharp PA MicroRNA sponges: progress and possibilities. Rna 2010, 16, 2043–2050. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhao, H.; Tan, Z.; et al. Bottleneck limitations for microRNA-based therapeutics from bench to the bedside. Die Pharm Int J Pharm Sci 2015, 70, 147–154. [Google Scholar]
- Gavrilov, K. ; Saltzman WM Therapeutic siRNA: principles, challenges, and strategies. Yale J Biol Med 2012, 85, 187. [Google Scholar]
- Yang, N. An overview of viral and nonviral delivery systems for microRNA. Int. J. Pharm. Investig. 2015, 5, 179–81. [Google Scholar] [CrossRef]

| Type of Disease | Type of miRNA | Expression level | indication | reference |
| MS | Mir-193a | decreased | Prognostic, diagnostic marker, and act as therapeutic | [38] |
| MS | Let7b-5p, mir-143-3p | Decreased in CSF | Promising miRNA candidate to discriminate PPMS | [39] |
| MS | Mir-155a, mir-146a | Increased in level (serum) | Act as diagnostic, prognostic and therapeutic | [40] |
| Mir-34a, mir-143a,mir-373a | Decreased in level (serum) | |||
| MS | Mir-10,mir-21,mir-124 | Decreased (blood) | in the pathogenesis of MS | [41] |
| MS | MIR-146a | Decreased in whole blood and feces of RRMS in compression with CIS, decreased in female. | In fecal as Diagnostic, prognostic biomarker | [42] |
| PD | Mir -7-1-5p, mir-223-3p | Increased in serum and serum isolated exosome | play a role in inflammation in PD, a Potential biomarker to discriminate PD from HC | [43] |
| PD | MIR-24, MIR-195 | Increased serum-EVs |
Act as an active biomarker in the diagnosis of PD |
[44] |
| MIR-19b | Decreased Serum -EVs | |||
| PD | MIR-22-3P | Decreased |
As a diagnostic parameter in the early stage of PD |
[45] |
| 22-3P, MIR-10b-5p,mir-151a-3p | increased(CSF) | |||
| PD | Has-mir-144-3p | Decreased inserum | Play a role in the progression of the disease and as an Early marker of PD. | [46] |
| PD | Mir-27-a, | Increased plasma | Can act in early diagnosis of PD, Use of mir-27-a, mir 27-b as a potential therapeutic target |
[47,48] |
| Mir-142-3p, mir-222,let-7a,let-7f | Decreased (plasma) | |||
| PD | Mir-132 | Increased( in males than in females) in prephrial blood | Biomarker for PD as diagnostic and disease progression | [49] |
| ALS | miR-16-5p | increased in plasma, CSF | Neuroprotective role in ALS after administration of intrathecal linage negative cell, | [50] |
| mir-206 | Increased plasma |
|||
| ALS | miR-23a | Increase in skeletal muscle in ALS patients and mouse model | Therapeutic inhibition of mir-23a may be a strategy to rescue peroxisome proliferator-activated receptor-γ coactivator (PGC-1α )activity and ameliorate skeletal muscle mitochondrial function in ALS, and down-regulation of miR-23a-3p has been proven to alleviate neuronal cell death and ROS | [51,52] |
| ALS | Hsa-mir-4649-5p |
Increased (plasma) |
Acts as diagnostic markers | [53] |
| Has-mir-4299 | Decreased (plasma) | |||
| AD | MIR-155,mir-146a, mir-125b ,mir-9,mir-34a | Upregulation Brain tissue of AD, ECF and CSF |
Biomarker for AD | [54,55] |
| AD | Mir-455-3p | Increased levels in blood serum, CSF post-mortem brain tissues, AD fibroblasts, AD β-lymphocytes, AD cell lines, transgenic AD (TgAD) mouse models and AD CSF | Biomarker and therapeutic | [56,57] |
| AD | Mir146a-5p | Upregulated in AD brain, neocortex and hippocampus |
Act as Pathogenesis and therapeutic biomarker | [58,59] |
| AD | Mir-125b | Upregulated in AD brain tissue | Pathological role can be used as treating therapy by using anti-mir 125b, anti-NF-kB | [60] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





