Submitted:
02 April 2024
Posted:
04 April 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Immunohistochemistry Staining
2.3. Cell Culture
2.4. Transfection
2.5. Cell Viability
2.6. Western Blotting
2.7. Invasion Assay In Vitro
2.8. Migration Assay In Vitro
2.9. Animal Model
2.10. Data Analysis
3. Results
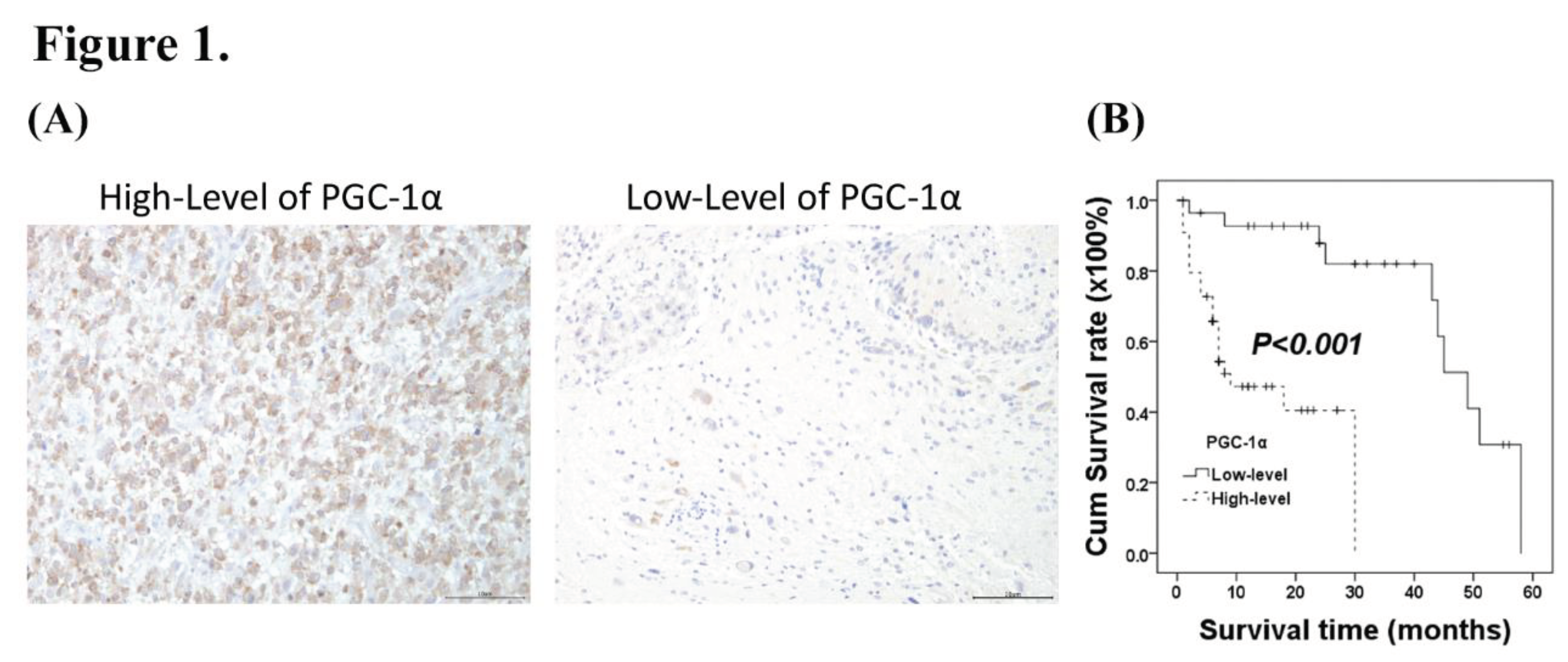
3.1. Associations of PGC-α Expression with Clinicopathological Characteristics in Patients with Intracranial Glioma
3.2. Prognostic Significance of PGC-1α Expression in Glioma
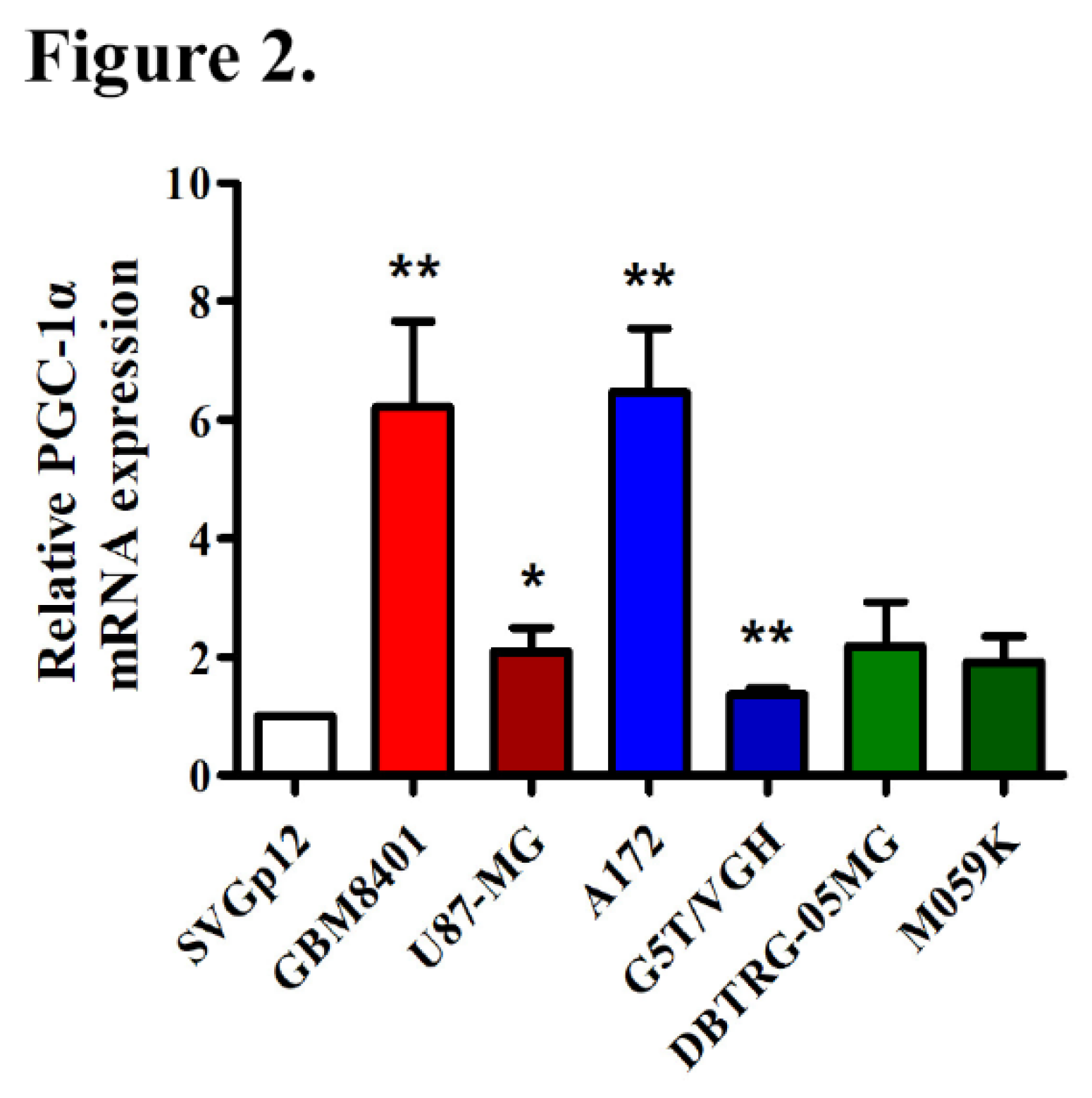
3.3. Elevated PGC-1α Protein Expression in GBM Cells Relative to Normal Cells
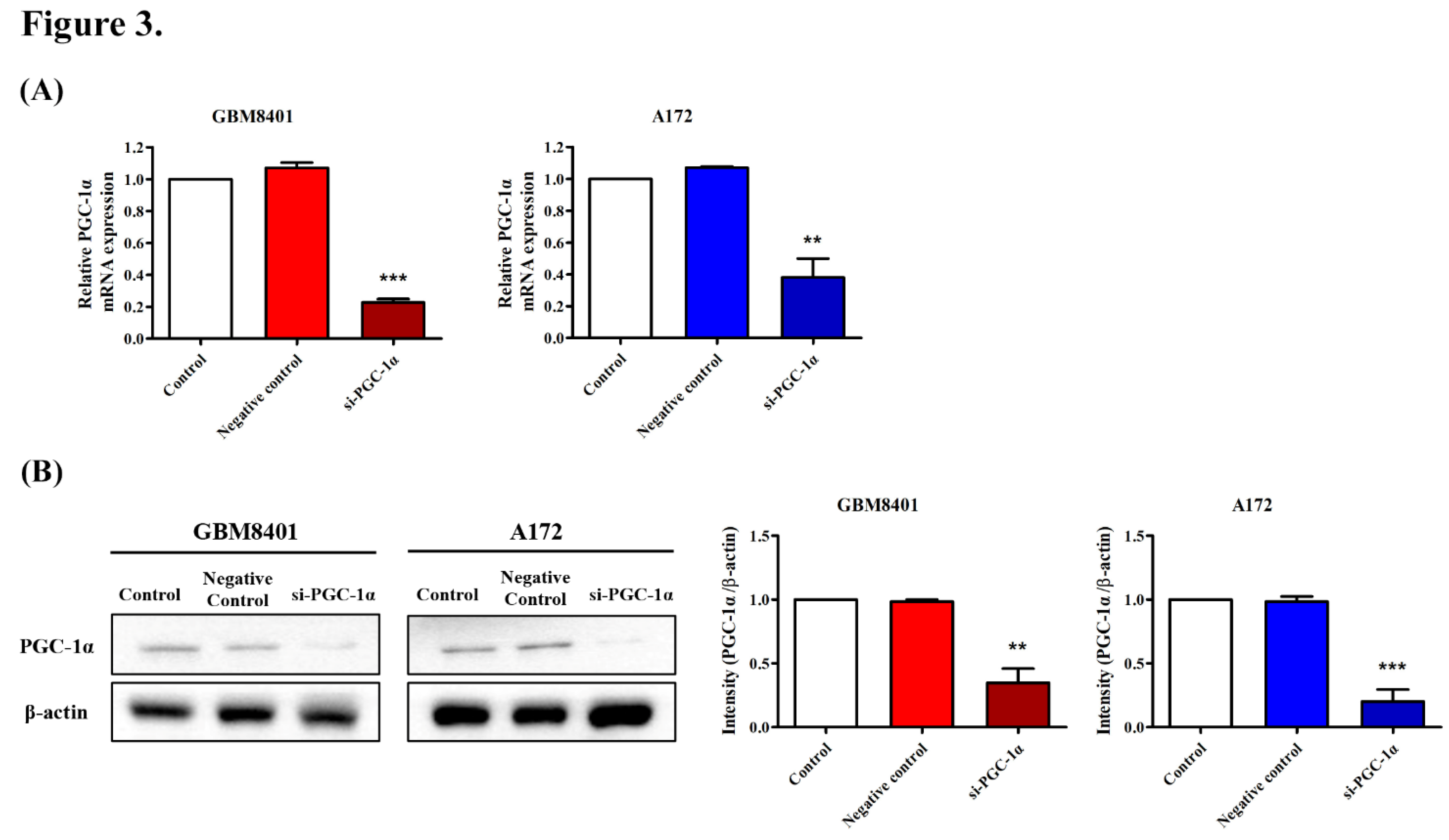
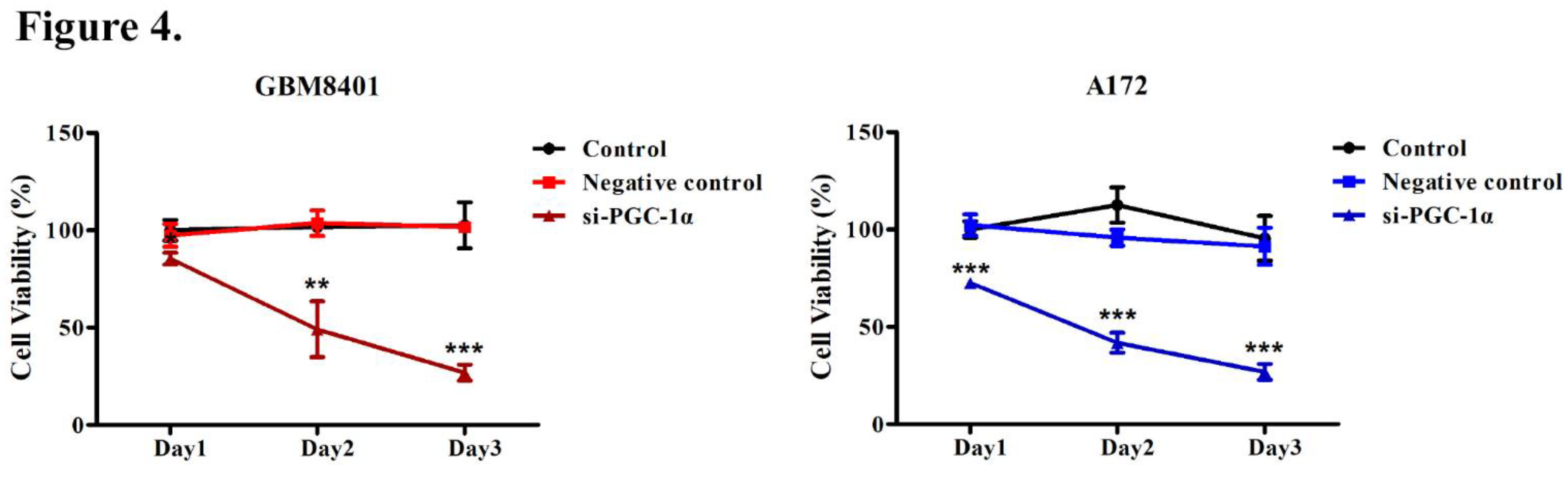
3.4. PGC-1α Knockdown Inhibited GBM Cell Proliferation
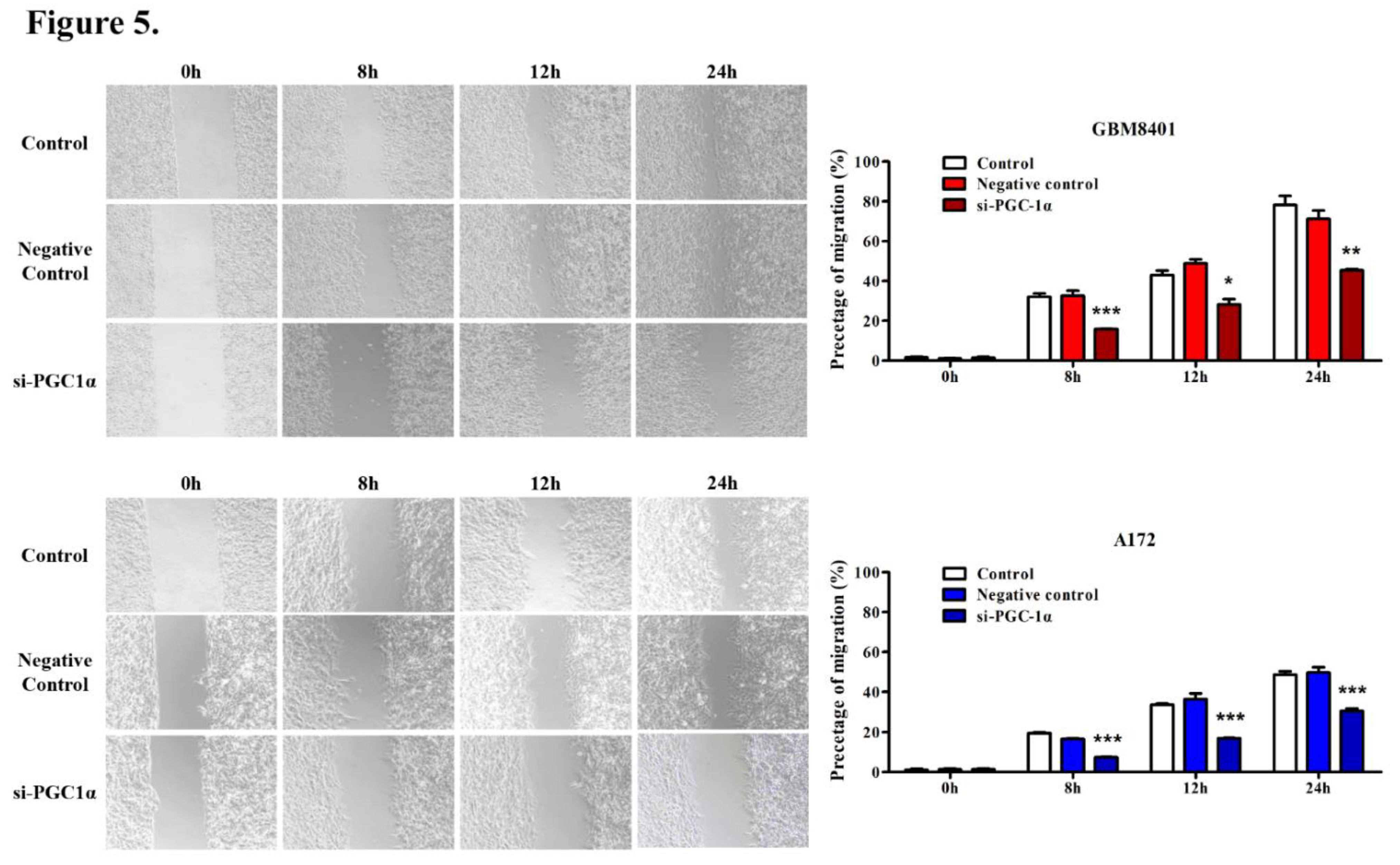
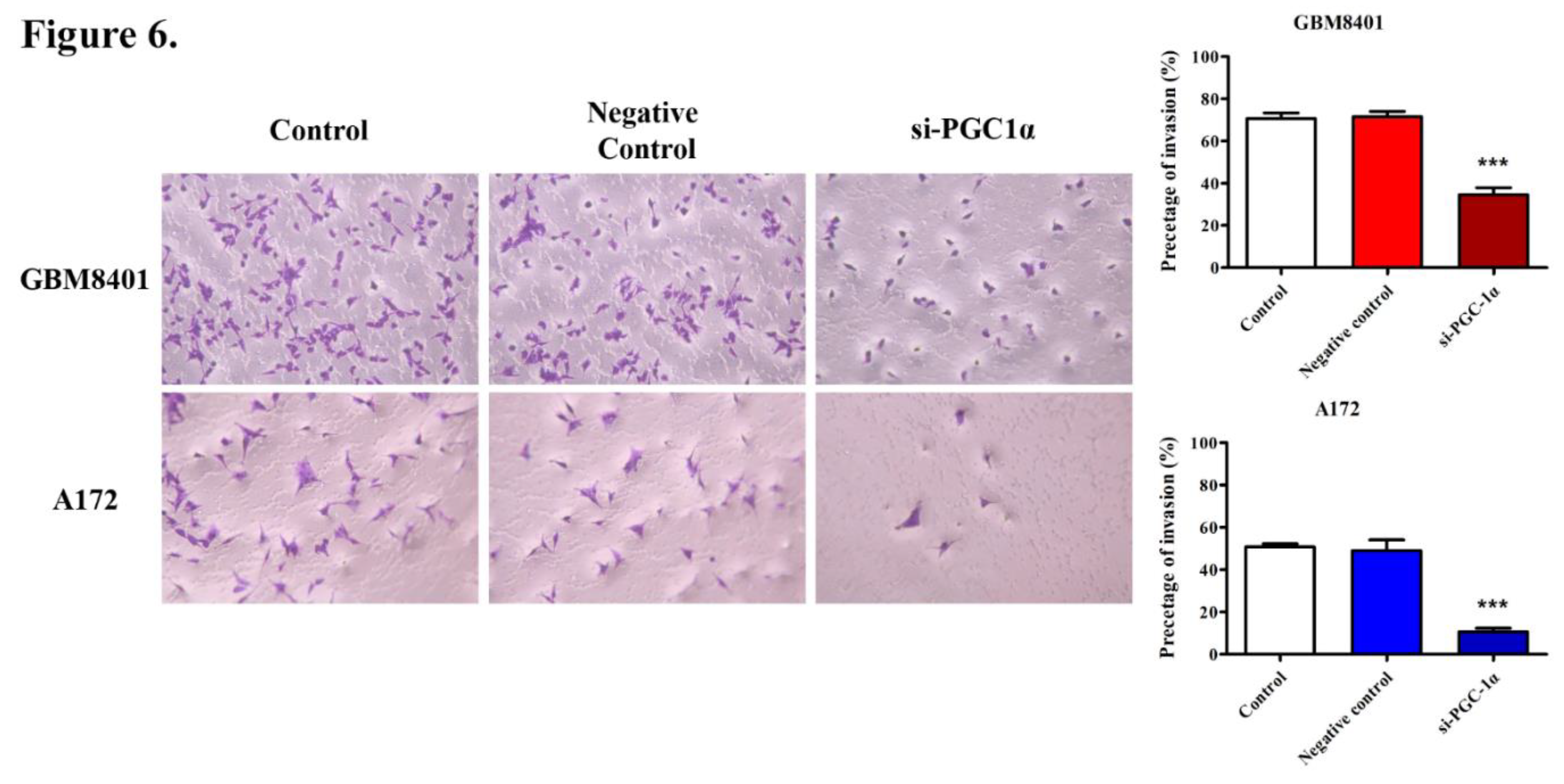
3.5. PGC-1α Knockdown Inhibited GBM Cell Invasion and Migration
3.6. Modulation of Carcinogenic Marker Expression by PGC-1α Knockdown in Glioma Cells
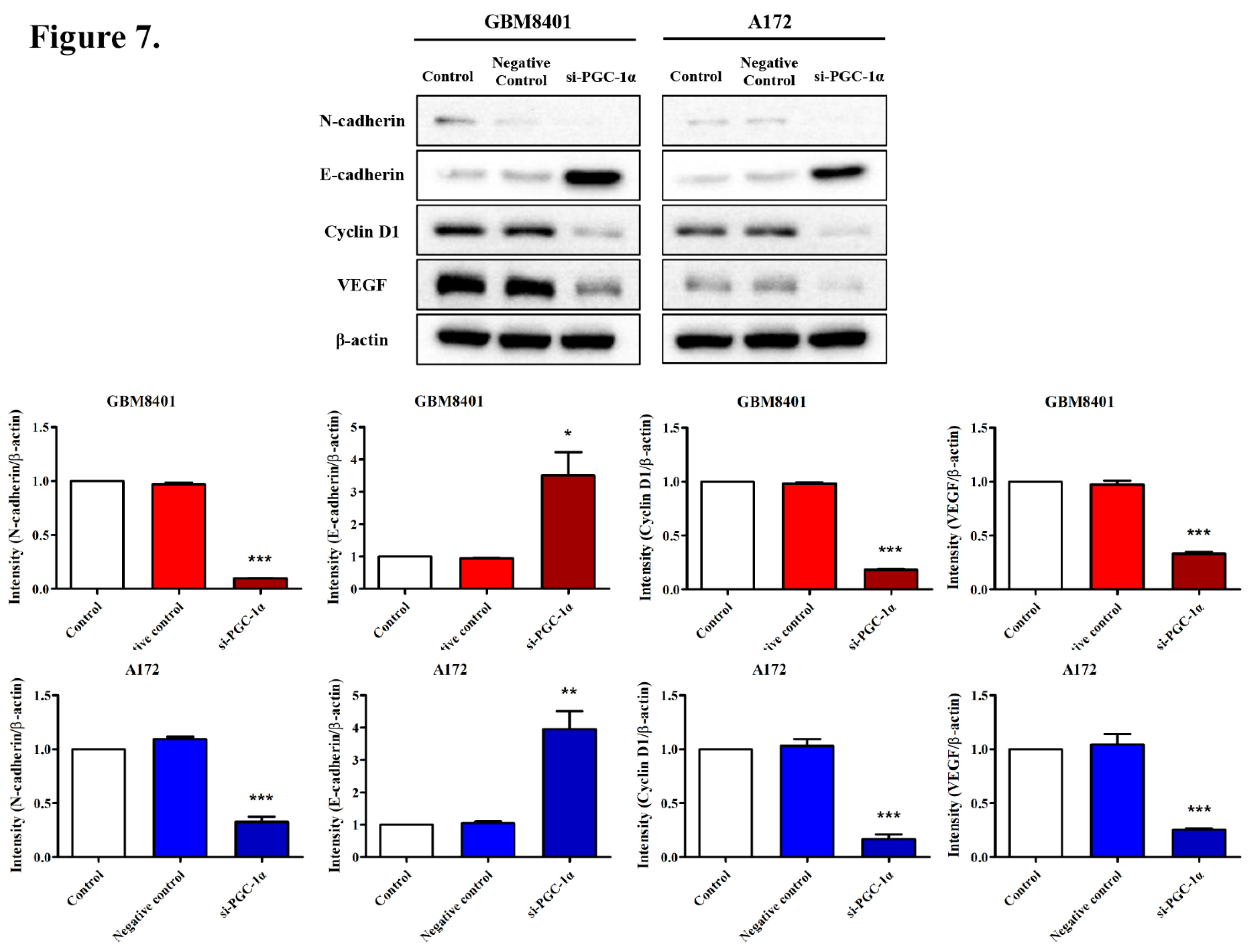
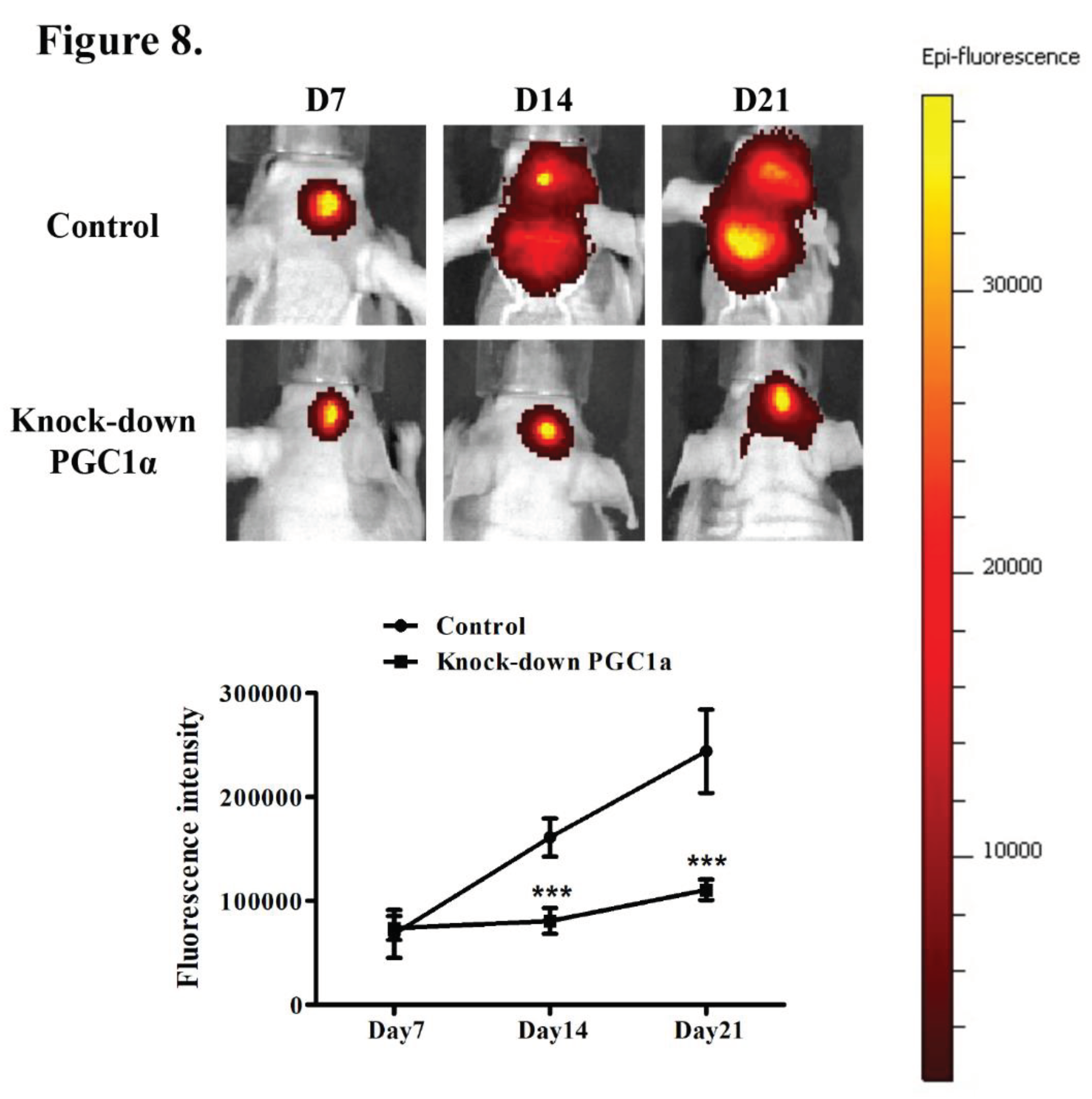
3.7. Effects of PGC-1α Knockdown on Tumor Growth and Survival in a Murine Glioblastoma Model
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Davis, F.G.; McCarthy, B.J. Current epidemiological trends and surveillance issues in brain tumors. Expert Rev. Anticancer Ther. 2001, 1, 395–401. [Google Scholar] [CrossRef]
- Ohgaki, H.; Kleihues, P. Epidemiology and etiology of gliomas. Acta Neuropathol. 2005, 109, 93–108. [Google Scholar] [CrossRef]
- Fuller GN, Scheithauer BW. The 2007 Revised World Health Organization (WHO) Classification of Tumours of the Central Nervous System: newly codified entities. Brain Pathol. 2007, 17, 304–307. [Google Scholar] [CrossRef] [PubMed]
- Wen, P.Y.; Kesari, S. Malignant gliomas in adults. N. Engl. J. Med. 2008, 359, 492–507. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Ducray, F.; Idbaih, A.; Wang, X.W.; Cheneau, C.; Labussiere, M.; Sanson, M. Predictive and prognostic factors for gliomas. Expert Rev. Anticancer Ther. 2011, 11, 781–789. [Google Scholar] [CrossRef]
- Tsai, H.P.; Tsai, T.H.; Hsieh, Y.J.; Chen, Y.T.; Lee, C.L.; Tsai, Y.C.; et al. Overexpression of Fli-1 in astrocytoma is associated with poor prognosis. Oncotarget. 2017, 8, 29174–29186. [Google Scholar] [CrossRef]
- Boroughs, L.K.; DeBerardinis, R.J. Metabolic pathways promoting cancer cell survival and growth. Nat. Cell Biol. 2015, 17, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Ward, P.S.; Thompson, C.B. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012, 21, 297–308. [Google Scholar] [CrossRef]
- Currie, E.; Schulze, A.; Zechner, R.; Walther, T.C.; Farese, R.V., Jr. Cellular fatty acid metabolism and cancer. Cell Metab. 2013, 18, 153–161. [Google Scholar] [CrossRef]
- Hensley, C.T.; Wasti, A.T.; DeBerardinis, R.J. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J. Clin. Invest. 2013, 123, 3678–3684. [Google Scholar] [CrossRef] [PubMed]
- Locasale, J.W. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat. Rev. Cancer. 2013, 13, 572–583. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, S.E.; Chandel, N.S. Targeting mitochondria metabolism for cancer therapy. Nat. Chem. Biol. 2015, 11, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Bobrovnikova-Marjon, E.; Hurov, J.B. Targeting metabolic changes in cancer: novel therapeutic approaches. Annu. Rev. Med. 2014, 65, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Seyfried, T.N.; Flores, R.E.; Poff, A.M.; D'Agostino, D.P. Cancer as a metabolic disease: implications for novel therapeutics. Carcinogenesis. 2014, 35, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Staubert, C.; Bhuiyan, H.; Lindahl, A.; Broom, O.J.; Zhu, Y.; Islam, S.; et al. Rewired metabolism in drug-resistant leukemia cells: a metabolic switch hallmarked by reduced dependence on exogenous glutamine. J. Biol. Chem. 2015, 290, 8348–8359. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Widlund, H.R.; Puigserver, P. PGC-1 coactivators: Shepherding the mitochondrial biogenesis of tumors. Trends Cancer. 2016, 2, 619–631. [Google Scholar] [CrossRef]
- Girnun, G.D. The diverse role of the PPARgamma coactivator 1 family of transcriptional coactivators in cancer. Semin. Cell Dev. Biol. 2012, 23, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.W.; Yao, Z.; Vicencio, J.M.; Karkucinska-Wieckowska, A.; Szabadkai, G. PGC-1 family coactivators and cell fate: roles in cancer, neurodegeneration, cardiovascular disease and retrograde mitochondria-nucleus signalling. Mitochondrion. 2012, 12, 86–99. [Google Scholar] [CrossRef]
- Austin, S.; St-Pierre, J. PGC1alpha and mitochondrial metabolism--emerging concepts and relevance in ageing and neurodegenerative disorders. J. Cell. Sci. 2012, 125, 4963–4971. [Google Scholar] [CrossRef]
- Wolf, D.A. Is reliance on mitochondrial respiration a "chink in the armor" of therapy-resistant cancer? Cancer Cell. 2014, 26, 788–795. [Google Scholar] [CrossRef] [PubMed]
- Bhalla, K.; Hwang, B.J.; Dewi, R.E.; Ou, L.; Twaddel, W.; Fang, H.B; et al. PGC1alpha promotes tumor growth by inducing gene expression programs supporting lipogenesis. Cancer Res. 2011, 71, 6888–6898. [Google Scholar] [CrossRef] [PubMed]
- Shiota, M.; Yokomizo, A.; Tada, Y.; Inokuchi, J.; Tatsugami, K.; Kuroiwa, K.; et al. Peroxisome proliferator-activated receptor gamma coactivator-1alpha interacts with the androgen receptor (AR) and promotes prostate cancer cell growth by activating the AR. Mol Endocrinol. 2010, 24, 114–127. [Google Scholar] [CrossRef]
- McGuirk, S.; Gravel, S.P.; Deblois, G.; Papadopoli, D.J.; Faubert, B.; Wegner, A.; et al. PGC-1alpha supports glutamine metabolism in breast cancer. Cancer Metab. 2013, 1, 22. [Google Scholar] [CrossRef]
- Vazquez, F.; Lim, J.H.; Chim, H.; Bhalla, K.; Girnun, G.; Pierce, K.; et al. PGC1alpha expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell. 2013, 23, 287–301. [Google Scholar] [CrossRef]
- Taguchi, A.; Delgado, O.; Celiktas, M.; Katayama, H.; Wang, H.; Gazdar, A.F; et al. Proteomic signatures associated with p53 mutational status in lung adenocarcinoma. Proteomics. 2014, 14, 2750–2759. [Google Scholar] [CrossRef]
- Haq, R.; Shoag, J.; Andreu-Perez, P.; Yokoyama, S.; Edelman, H.; Rowe, G.C.; et al. Oncogenic BRAF regulates oxidative metabolism via PGC1alpha and MITF. Cancer Cell. 2013, 23, 302–315. [Google Scholar] [CrossRef]
- LaGory, E.L.; Wu, C.; Taniguchi, C.M.; Ding, C.C.; Chi, J.T.; von Eyben, R.; et al. Suppression of PGC-1alpha is critical for reprogramming oxidative metabolism in renal cell carcinoma. Cell Rep. 2015, 12, 116–127. [Google Scholar] [CrossRef]
- Sancho, P.; Burgos-Ramos, E.; Tavera, A.; Bou Kheir, T.; Jagust, P.; Schoenhals, M.; et al. MYC/PGC-1alpha balance determines the metabolic phenotype and plasticity of pancreatic cancer stem cells. Cell Metab. 2015, 22, 590–605. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Luo, X.; Xiao, L.; Tang, M.; Bode, A.M.; Dong, Z.; et al. The role of PGC1alpha in cancer metabolism and its therapeutic implications. Mol. Cancer Ther. 2016, 15, 774–782. [Google Scholar] [CrossRef] [PubMed]
- Han, C.Y.; Patten, DA.; Richardson, R.B.; Harper, M.E.; Tsang, B.K. Tumor metabolism regulating chemosensitivity in ovarian cancer. Genes Cancer. 2018, 9, 155–175. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, N.; Das, G.M. Metabolic reprogramming in breast cancer and its therapeutic implications. Cells. 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Cassim, S.; Vucetic, M.; Zdralevic, M.; Pouyssegur, J. Warburg and beyond: the power of mitochondrial metabolism to collaborate or replace fermentative glycolysis in cancer. Cancers. 2020, 12, 1119. [Google Scholar] [CrossRef]
- Villena, J.A. New insights into PGC-1 coactivators: redefining their role in the regulation of mitochondrial function and beyond. Febs J. 2015, 282, 647–672. [Google Scholar] [CrossRef] [PubMed]
- LeBleu, V.S.; O'Connell, J.T.; Herrera, K.N.G.; Wikman, H.; Pantel, K.; Haigis, M.C.; et al. PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat. Cell. Biol. 2014, 16, 1125–1125. [Google Scholar] [CrossRef]
- Mastropasqua, F.; Girolimetti, G.; Shoshan, M. PGC1α: friend or foe in cancer? Genes-Basel, 2018; 9. [Google Scholar]
- Hipolito, A.; Martins, F.; Mendes, C.; Lopes-Coelho, F.; Serpa, J. Molecular and metabolic reprogramming: pulling the strings toward tumor metastasis. Front. Oncol. 2021, 11, 656851. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.; Chen, S.; Hu, X. Peroxisome proliferator-activated receptor gamma coactivator-1 alpha: a double-edged sword in prostate cancer. Curr. Cancer Drug Targets. 2022, 22, 541–559. [Google Scholar] [CrossRef]
- LeBleu, V.S.; O'Connell, J.T.; Gonzalez Herrera, K.N.; Wikman, H.; Pantel, K.; Haigis, M.C.; et al. PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat. Cell Biol. 2014, 16, 992–1003. [Google Scholar] [CrossRef]
- Rius-Perez, S.; Torres-Cuevas, I.; Millan, I.; Ortega, A.L.; Perez, S. PGC-1alpha, inflammation, and oxidative stress: an integrative view in metabolism. Oxid. Med. Cell Longev. 2020, 2020, 1452696. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, L.; Supuran, C.T.; Alfarouk, K.O. The Warburg effect and the hallmarks of cancer. Anticancer Agents Med. Chem. 2017, 17, 164–170. [Google Scholar] [CrossRef]
- Liang, H.; Ward, W.F. PGC-1alpha: a key regulator of energy metabolism. Adv. Physiol. Educ. 2006, 30, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.N.; Zhang, M.Q.; Yu, F.L.; Han, B.; Bao, M.Y.; Li, X.; et al. Peroxisom proliferator-activated receptor-γ coactivator-1α in neurodegenerative disorders: A promising therapeutic target. Biochem. Pharmacol. 2023, 215. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lou, L.; Li, D.; Wang, Y.; Jia, X.; Hao, X.; et al. Expression, clinicopathological significance, and prognostic potential of AMPK, p-AMPK, PGC-1α, and TFAM in astrocytomas. J. Neuropath. Exp. Neur. 2023, 83, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Bruns, I.; Sauer, B.; Burger, M.C.; Eriksson, J.; Hofmann, U.; Braun, Y.; et al. Disruption of peroxisome proliferator-activated receptor coactivator (PGC)-1 reverts key features of the neoplastic phenotype of glioma cells. J. Biol. Chem. 2019, 294, 3037–3050. [Google Scholar] [CrossRef] [PubMed]
| No. of patients | PGC-1α expression (n, %) | P-value | ||
|---|---|---|---|---|
| Low | High | |||
| Age | 0.342 | |||
| <60 | 71 | 29 (30.5%) | 42 (44.2%) | |
| >60 | 24 | 7 (7.4%) | 17 (17.9%) | |
| Gender | 1.000 | |||
| Male | 52 | 20 (21.1%) | 32 (33.7%) | |
| Female | 43 | 16 (16.8%) | 27 (28.4%) | |
| WHO Grade | 0.003** | |||
| II | 23 | 15 (15.8%) | 8 (8.4%) | |
| III/IV | 72 | 21 (22.1%) | 51 (53.7%) | |
| Tumor size | 1 | |||
| ≤3cm | 59 | 22 (23.2%) | 37 (38.9%) | |
| >3cm | 36 | 14 (14.7%) | 22 (23.2%) | |
| Radiotherapy | 1.000 | |||
| No | 54 | 20 (21.1%) | 34 (38.9%) | |
| Yes | 41 | 16 (16.8%) | 25 (26.3%) | |
| Chemotherapy | 0.052 | |||
| No | 59 | 27 (28.4%) | 32 (33.7%) | |
| Yes | 36 | 9 (9.5%) | 27 (28.4%) | |
| KPS | 0.643 | |||
| ≤70 | 67 | 24 (25.3%) | 43 (45.3%) | |
| >70 | 28 | 12 (12.6%) | 16 (16.8%) | |
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Relative risk | 95% CI | P | Relative risk | 95% CI | P | |
| Age | 0.648 | 0.311-1.351 | 0.247 | |||
| Gender | 0.892 | 0.473-1.682 | 0.724 | |||
| WHO grade | 0.382 | 0.171-0.851 | 0.019 | 0.546 | 0.223-1.335 | 0.185 |
| Tumor size | 1.114 | 0.572-2.171 | 0.751 | |||
| Radiotherapy | 1.845 | 0.965-3.526 | 0.064 | |||
| Chemotherapy | 1.542 | 0.776-3.065 | 0.217 | |||
| KPS | 1.167 | 0.586-2.324 | 0.660 | |||
| PGC-1α expression | 0.177 | 0.075-0.417 | <0.001 | 0.197 | 0.080-0.482 | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





