Submitted:
16 May 2024
Posted:
17 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
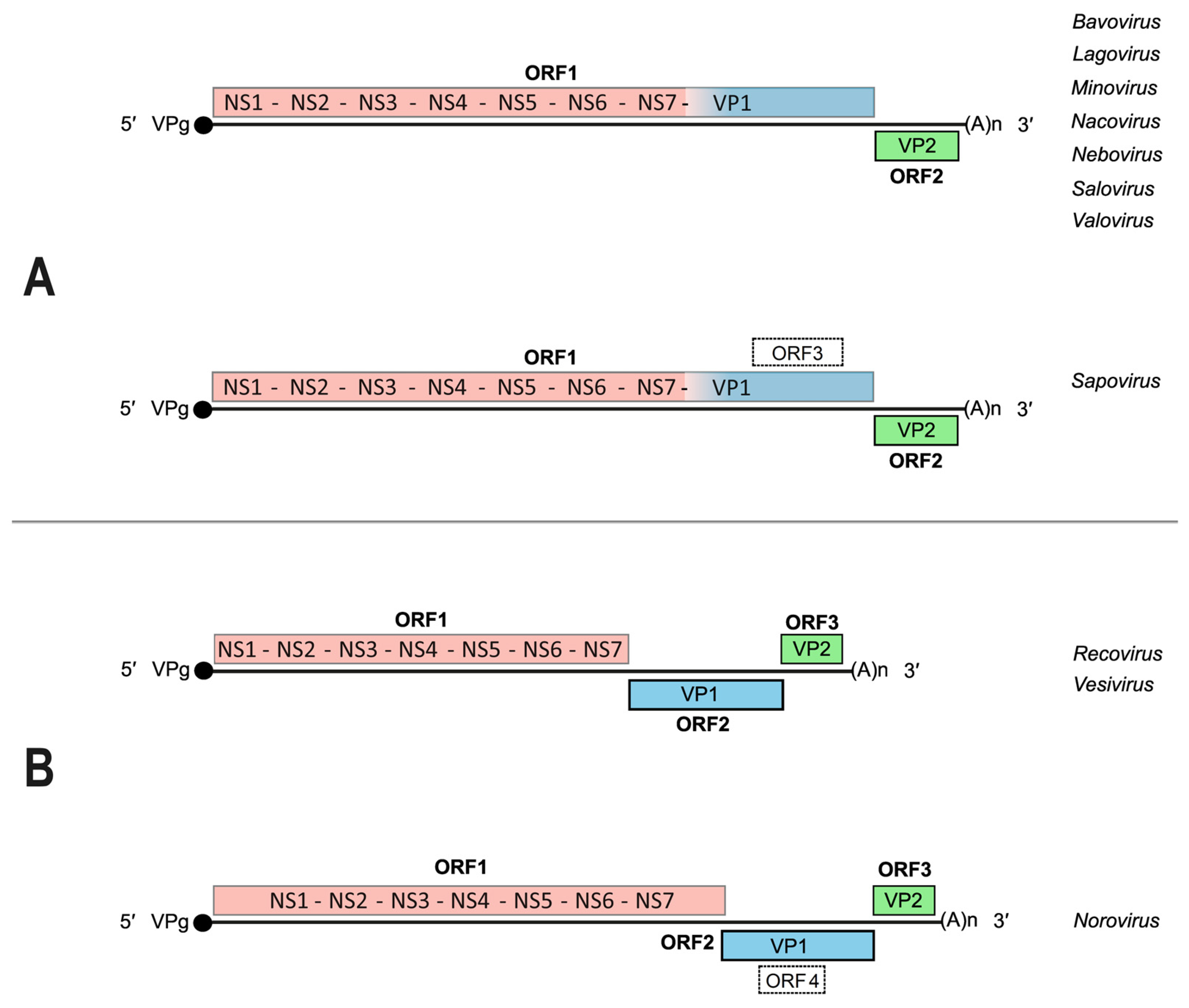
2. The Caliciviridae: Genome Organization, Gene Expression and Replication Strategies
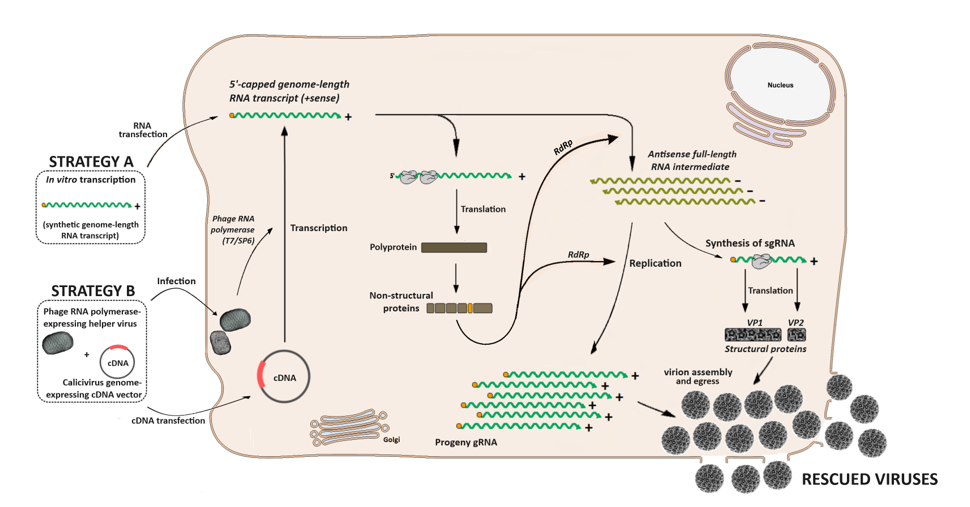
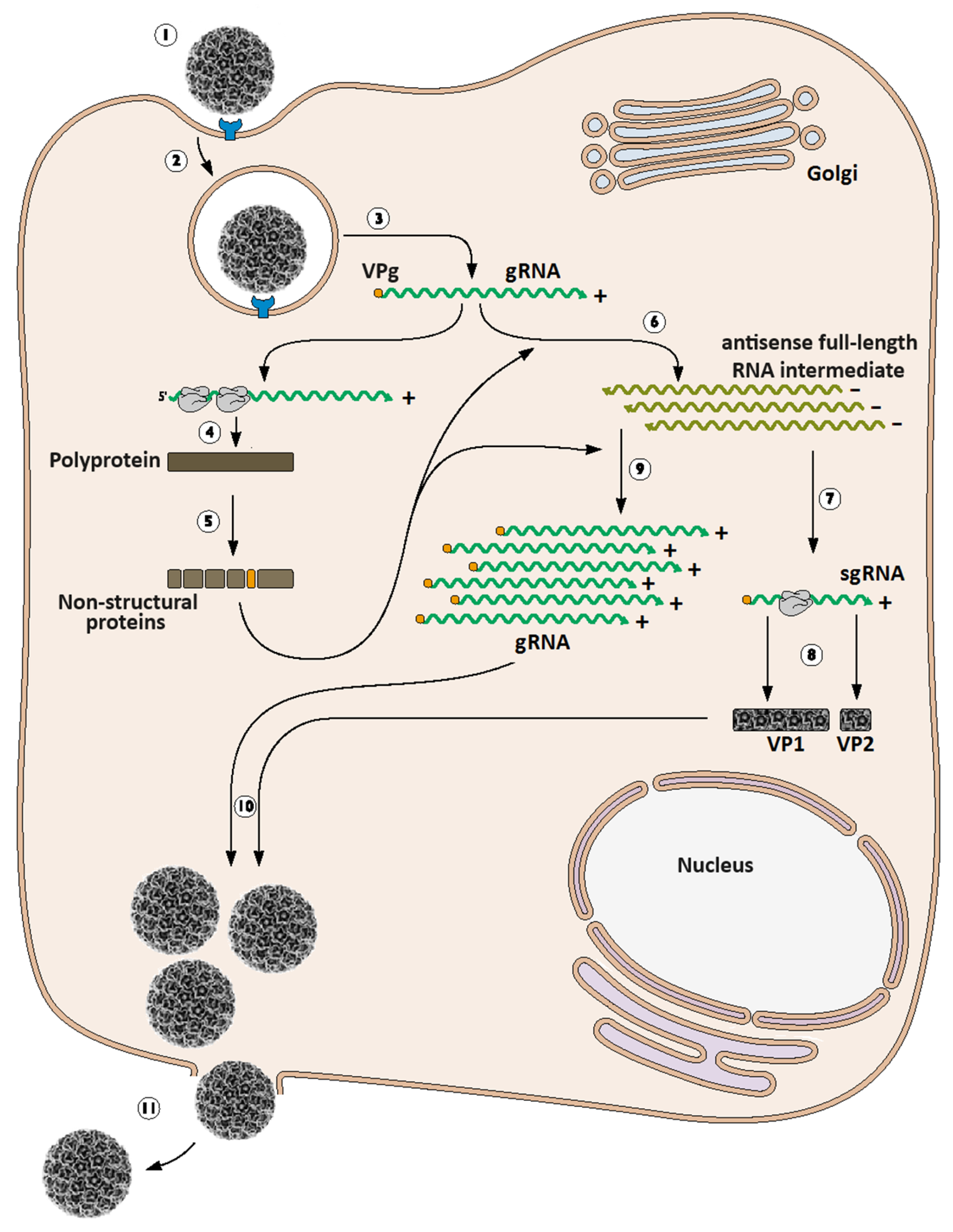
3. Calicivirus Replication Cycle
4. In Vitro Study of Caliciviruses
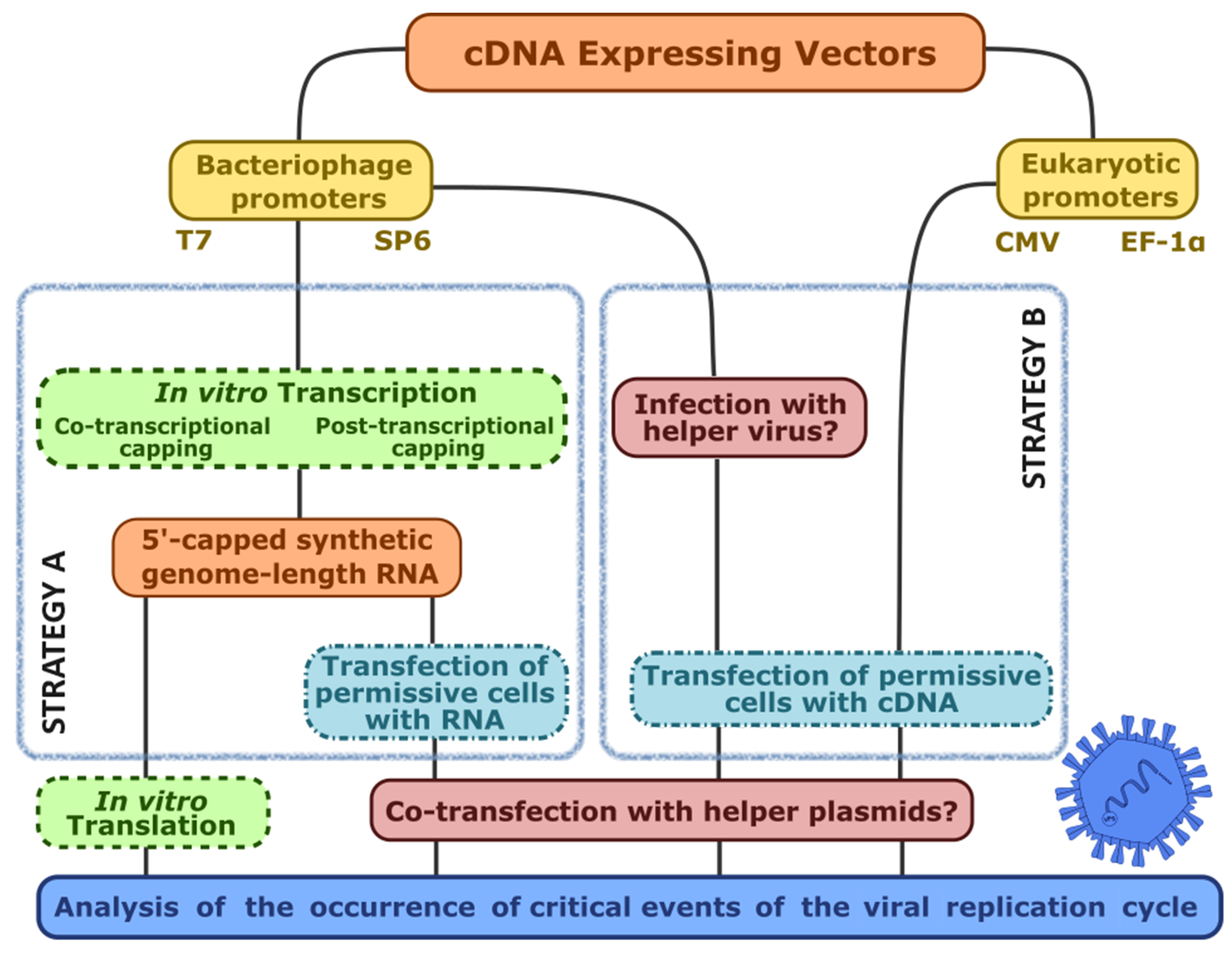
4.1. The Challenges of Reverse Genetics
4.2. The Hallmarks of a Promising Virus-Expressing cDNA
4.3. When Things Go Wrong: Interrogating the Viral Genome for the Occurrence of Replication-Critical Events
5. Chronology of the Calicivirus Reverse Genetics
5.1. The Rabbit Vesivirus Reverse Genetics Journey
| Virus | Recovery strategy and infectious clone features | Design | Year of publication [Reference] |
|---|---|---|---|
| Feline calicivirus | T7 RNA polymerase-driven IVT with co-transcriptional capping, followed by RNA transfection of CRFK cells | I |
1995 [2] |
| Feline calicivirus | T7 RNA polymerase-driven cDNA expression; Poly-(A)32. Two delivery methods:
|
I |
2002 [92] |
| Porcine enteric calicivirus | T7 RNA polymerase-driven cDNA expression; Poly-(A)35. Two delivery methods:
|
I |
2005 [76] |
| Human norovirus | T7 RNA polymerase promoter: transfection of rMVA-T7-infected 293T cells; Poly-(A)26 | III |
2005 [78] |
| Human norovirus | T7 RNA polymerase promoter: transfection of rVV-T7-infected 293T cells; Poly-(A)30 | III |
2006 [77] |
| Human norovirus | No virus rescue: neomycin-resistance gene replacing part of ORF2. Transfection of BHK21 and Huh7 cells with IVT-generated RNA led to the establishment of a VP1-defective replicon that persisted beyond cell passages. Apparently, the replicon further extracted from cells had covalently acquired the 5’–linked VPg. G418 was used for colony selection | IV |
2006 [106] |
| Murine norovirus–1 | Pol-II-driven: viral cDNA controlled by minCMV promoter; Poly-(A)31. Two delivery methods:
|
II |
2007 [95] |
| Murine norovirus–1 | T7 RNA polymerase-driven cDNA expression; Poly-(A)26. Two helper viruses tested for providing T7 pol:
|
II |
2007 [73] |
| Tulane virus | T7 RNA polymerase-driven IVT with co-transcriptional capping, followed by RNA transfection of LLC-MK2 cells; Poly-(A)17 | I |
2008 [93] |
| Murine norovirus–1 |
|
II |
2010, 2012 [32,118] |
| Human norovirus | Pol-II-driven cDNA expression: EF-1α promoter. cDNA plasmid was transfected into COS7 cells in the absence of helper virus; Poly-(A)26 | II |
2014 [105] |
| Feline calicivirus | Pol-II-driven cDNA expression: EF-1α promoter. cDNA plasmid was transfected into CRFK cells in the absence of helper virus; Poly-(A)30 | II |
2014 [96] |
| Human norovirus | Pol-II-driven cDNA expression: CMV promoter. cDNA plasmid constructed through Gibson assembly and transfected into Caco-2 cells; bovine growth hormone; poly-A signal | I |
2018 [97] |
| Rabbit vesivirus | T7 RNA polymerase-driven cDNA expression; Poly-(A)30. Two delivery methods:
|
III |
2020 [43] |
| Human norovirus Murine norovirus–1 |
Full-length cDNA with a linker fragment containing CMV promoter synthesized by CPER; transfected in NIH3T3 cells; Poly-(A)30 | II |
2021 [22] |
| Human sapovirus | T7 RNA polymerase-driven IVT with co-transcriptional capping, followed by RNA transfection of HuTu80 cells; Poly-(A)25 | I |
2022 [113] |
Concluding Remarks and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Acknowledgements
Conflicts of Interest
References
- [1] J.C. Boyer, and A.L. Haenni, Infectious transcripts and cDNA clones of RNA viruses. Virology 1994, 198, 415–426. [CrossRef] [PubMed]
- [2] S. Sosnovtsev, and K.Y. Green, RNA transcripts derived from a cloned full-length copy of the feline calicivirus genome do not require VpG for infectivity. Virology 1995, 210, 383–390. [CrossRef]
- 3. [3] I. Goodfellow, and S. Taube, Chapter 3.2 - Calicivirus Replication and Reverse Genetics. in: L. Svensson, U. Desselberger, H.B. Greenberg, and M.K. Estes, (Eds.), Viral Gastroenteritis, Academic Press, Boston, 2016, pp. 355-378.
- [4] U. Desselberger, Caliciviridae Other Than Noroviruses. Viruses 2019, 11.
- [5] H. Ebihara, A. Groseth, G. Neumann, Y. Kawaoka, and H. Feldmann, The role of reverse genetics systems in studying viral hemorrhagic fevers. Thromb Haemost 94 (2005) 240-53.
- [6] H.L. Cai, and Y.W. Huang, Reverse genetics systems for SARS-CoV-2: Development and applications. Virol Sin 38 (2023) 837-850.
- [7] T. Hoenen, J. Brandt, Y. Caì, J.H. Kuhn, and C. Finch, Reverse Genetics of Filoviruses. Curr Top Microbiol Immunol 411 (2017) 421-445.
- [8] D. Jackson, A. Cadman, T. Zurcher, and W.S. Barclay, A reverse genetics approach for recovery of recombinant influenza B viruses entirely from cDNA. J. Virol. 2002, 76, 11744–11747. [CrossRef] [PubMed]
- [9] G. Neumann, Influenza Reverse Genetics-Historical Perspective. Cold Spring Harb. Perspect. Cold Spring Harb. Perspect. Med. 2021, 11.
- [10] T. Kobayashi, A.A.R. Antar, K.W. Boehme, P. Danthi, E.A. Eby, K.M. Guglielmi, G.H. Holm, E.M. Johnson, M.S. Maginnis, S. Naik, W.B. Skelton, J.D. Wetzel, G.J. Wilson, J.D. Chappell, and T.S. Dermody, A Plasmid-Based Reverse Genetics System for Animal Double-Stranded RNA Viruses. Cell Host Microbe 2007, 2, 139.
- [11] T. Kobayashi, L.S. Ooms, M. Ikizler, J.D. Chappell, and T.S. Dermody, An improved reverse genetics system for mammalian orthoreoviruses. Virology 2010, 398, 194–200. [CrossRef] [PubMed]
- [12] A.M. Conradie, L. Stassen, H. Huismans, C.A. Potgieter, and J. Theron, Establishment of different plasmid only-based reverse genetics systems for the recovery of African horse sickness virus. Virology 2016, 499, 144–155. [CrossRef] [PubMed]
- [13] S. Komoto, J. Sasaki, and K. Taniguchi, Reverse genetics system for introduction of site-specific mutations into the double-stranded RNA genome of infectious rotavirus. Proc. Natl. Acad. Sci. USA 2006, 103, 4646–4651. [CrossRef]
- [14] Y. Kanai, and T. Kobayashi, Rotavirus reverse genetics systems: Development and application. Virus Res. 2021, 295, 198296. [CrossRef]
- [15] S. Komoto, S. Fukuda, T. Murata, and K. Taniguchi, Reverse genetics system for human rotaviruses. Microbiol. Immunol. 2020, 64, 401–406. [CrossRef] [PubMed]
- [16] T. Kanda, and K. Tomonaga, Reverse Genetics and Artificial Replication Systems of Borna Disease Virus 1. Viruses 2022, 14.
- [17] B. Tercero, and S. Makino, Reverse genetics approaches for the development of bunyavirus vaccines. Curr. Opin. Virol. 2020, 44, 16–25. [CrossRef] [PubMed]
- [18] R. Andino, D. Silvera, S.D. Suggett, P.L. Achacoso, C.J. Miller, D. Baltimore, and M.B. Feinberg, Engineering poliovirus as a vaccine vector for the expression of diverse antigens. Science 1994, 265, 1448–1451. [CrossRef] [PubMed]
- [19] A.A. Khromykh, and E.G. Westaway, Subgenomic replicons of the flavivirus Kunjin: construction and applications. J. Virol. 1997, 71, 1497–1505. [CrossRef] [PubMed]
- [20] P. Liljeström, Alphavirus expression systems. Current Opinion in Biotechnology 5 (1994) 495-500.
- [21] L. Enjuanes, I. Sola, F. Almazan, A. Izeta, J.M. Gonzalez, and S. Alonso, Coronavirus Derived Expression Systems. in: E. Lavi, S.R. Weiss, and S.T. Hingley, (Eds.), The Nidoviruses: Coronaviruses and Arteriviruses, Springer US, Boston, MA, 2001, pp. 309-321.
- [22] A.A. Amarilla, J.D.J. Sng, R. Parry, J.M. Deerain, J.R. Potter, Y.X. Setoh, D.J. Rawle, T.T. Le, N. Modhiran, X. Wang, N.Y.G. Peng, F.J. Torres, A. Pyke, J.J. Harrison, M.E. Freney, B. Liang, C.L.D. McMillan, S.T.M. Cheung, D. Guevara, J.M. Hardy, M. Bettington, D.A. Muller, F. Coulibaly, F. Moore, R.A. Hall, P.R. Young, J.M. Mackenzie, J. Hobson-Peters, A. Suhrbier, D. Watterson, and A.A. Khromykh, A versatile reverse genetics platform for SARS-CoV-2 and other positive-strand RNA viruses. Nat Commun 12 (2021) 3431.
- [23] X. Xie, K.G. Lokugamage, X. Zhang, M.N. Vu, A.E. Muruato, V.D. Menachery, and P.-Y. Shi, Engineering SARS-CoV-2 using a reverse genetic system. Nature Protocols 16 (2021) 1761-1784.
- [24] H. Jiang, T. Wang, L. Kong, B. Li, and Q. Peng, Reverse Genetics Systems for Emerging and Re-Emerging Swine Coronaviruses and Applications. Viruses 2023, 15.
- [25] K.Y. Green, Chapter 20. Caliciviridae: The Noroviruses. in: D.M. Knipe, Howley, P.M., (Ed.), Fields Virology, Philadelphia, PA, 2013, pp. 582-608.
- [26] J. Vinjé, M.K. Estes, P. Esteves, K.Y. Green, K. Katayama, N.J. Knowles, Y. L’Homme, V. Martella, H. Vennema, P.A. White, and I.R. Consortium, ICTV Virus Taxonomy Profile: Caliciviridae. Journal of General Virology 100 (2019) 1469-1470.
- I.N. Clarke, and P.R. Lambden, Organization and expression of calicivirus genes. J. Infect. Dis. 2000, 181 (Suppl. 2), S309–S316.
- [28] B. Alhatlani, S. Vashist, and I. Goodfellow, Functions of the 5’ and 3’ ends of calicivirus genomes. Virus Res. (2015).
- [29] T.P. Herbert, I. Brierley, and T.D. Brown, Identification of a protein linked to the genomic and subgenomic mRNAs of feline calicivirus and its role in translation. J. Gen. Virol. 1997, 78 Pt 5, 1033–1040. [CrossRef] [PubMed]
- [30] G. Meyers, C. Wirblich, and H.J. Thiel, Genomic and subgenomic RNAs of rabbit hemorrhagic disease virus are both protein-linked and packaged into particles. Virology 1991, 184, 677–686. [CrossRef] [PubMed]
- [31] S.V. Sosnovtsev, S.A. Sosnovtseva, and K.Y. Green, Cleavage of the feline calicivirus capsid precursor is mediated by a virus-encoded proteinase. J. Virol. 1998, 72, 3051–3059. [CrossRef]
- [32] M.A. Yunus, L.M. Chung, Y. Chaudhry, D. Bailey, and I. Goodfellow, Development of an optimized RNA-based murine norovirus reverse genetics system. J. Virol. Methods 2010, 169, 112–118. [CrossRef]
- [33] B. Boniotti, C. Wirblich, M. Sibilia, G. Meyers, H.J. Thiel, and C. Rossi, Identification and characterization of a 3C-like protease from rabbit hemorrhagic disease virus, a calicivirus. Journal of Virology 68 (1994) 6487-6495.
- [34] F. Parra, J.A. Boga, M.S. Marin, and R. Casais, The amino terminal sequence of VP60 from rabbit hemorrhagic disease virus supports its putative subgenomic origin. Virus Res. 1993, 27, 219–228. [CrossRef] [PubMed]
- [35] G. Meyers, Translation of the minor capsid protein of a calicivirus is initiated by a novel termination-dependent reinitiation mechanism. J.Biol.Chem. 278 (2003) 34051-34060.
- [36] T.A. Poyry, A. Kaminski, E.J. Connell, C.S. Fraser, and R.J. Jackson, The mechanism of an exceptional case of reinitiation after translation of a long ORF reveals why such events do not generally occur in mammalian mRNA translation. Genes Dev. 2007, 21, 3149–3162. [CrossRef] [PubMed]
- [37] I.N. Clarke, M.K. Estes, K.Y. Green, G.S. Hansman, N.J. Knowles, M.K. Koopmans, D.O. Matson, G. Meyers, J.D. Neill, A. Radford, A.W. Smith, M.J. Studdert, H.J. Thiel, and J. Vinje, Family Caliciviridae. in: A.M.Q. King, M.J. Adams, E.B. Carstens, and E.J. Lefkowitz, (Eds.), Virus Taxonomy. Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses, Elsevier, San Diego, 2011, pp. 977-986.
- [38] A. Machin, J.M. Martin Alonso, and F. Parra, Identification of the amino acid residue involved in rabbit hemorrhagic disease virus VPg uridylylation. J.Biol.Chem. 276 (2001) 27787-27792.
- [39] I. Goodfellow, Y. Chaudhry, I. Gioldasi, A. Gerondopoulos, A. Natoni, L. Labrie, J.F. Laliberte, and L. Roberts, Calicivirus translation initiation requires an interaction between VPg and eIF 4 E. Embo Rep. 2005, 6, 968–972.
- [40] Y. Chaudhry, A. Nayak, M.E. Bordeleau, J. Tanaka, J. Pelletier, G.J. Belsham, L.O. Roberts, and I.G. Goodfellow, Caliciviruses differ in their functional requirements for eIF4F components. J.Biol.Chem. 281 (2006) 25315-25325.
- [41] I. Goodfellow, The genome-linked protein VPg of vertebrate viruses - a multifaceted protein. Curr. Opin. Virol. 2011, 1, 355–362. [CrossRef] [PubMed]
- [42] K.F. Daughenbaugh, C.E. Wobus, and M.E. Hardy, VPg of murine norovirus binds translation initiation factors in infected cells. Virol. J. 2006, 3, 33. [CrossRef] [PubMed]
- [43] Á.L. Álvarez, A. García-Manso, K.P. Dalton, J.M. Martín-Alonso, I. Nicieza, A. Podadera, M. Acosta-Zaldívar, D. De Llano, and F. Parra, Reverse Genetics System for Rabbit vesivirus. Front. Microbiol. 2020, 11.
- [44] R. Casais, L.G. Molleda, A. Machin, B.G. del, A.G. Manso, K.P. Dalton, A. Coto, J.M. Alonso, M. Prieto, and F. Parra, Structural and functional analysis of virus factories purified from Rabbit vesivirus-infected Vero cells. Virus Res. 2008, 137, 112–121. [CrossRef] [PubMed]
- [45] E. Smertina, R.N. Hall, N. Urakova, T. Strive, and M. Frese, Calicivirus Non-structural Proteins: Potential Functions in Replication and Host Cell Manipulation. Front. Microbiol. 2021, 12, 712710.
- [46] Y. Matsuura, Y. Tohya, M. Onuma, F. Roerink, M. Mochizuki, and T. Sugimura, Expression and processing of the canine calicivirus capsid precursor. Microbiology 2000, 81, 195–199. [CrossRef]
- [47] K.O. Chang, D.W. George, J.B. Patton, K.Y. Green, and S.V. Sosnovtsev, Leader of the capsid protein in feline calicivirus promotes replication of Norwalk virus in cell culture. J. Virol. 2008, 82, 9306–9317. [CrossRef]
- [48] E.J. Abente, S.V. Sosnovtsev, C. Sandoval-Jaime, G.I. Parra, K. Bok, and K.Y. Green, The feline calicivirus leader of the capsid protein is associated with cytopathic effect. J. Virol. 2013, 87, 3003–3017. [CrossRef] [PubMed]
- [49] G.S. Hansman, T. Oka, K. Katayama, and N. Takeda, Human sapoviruses: genetic diversity, recombination, and classification. Rev. Med Virol. 2007, 17, 133–141. [CrossRef] [PubMed]
- [50] N. McFadden, D. Bailey, G. Carrara, A. Benson, Y. Chaudhry, A. Shortland, J. Heeney, F. Yarovinsky, P. Simmonds, A. Macdonald, and I. Goodfellow, Norovirus Regulation of the Innate Immune Response and Apoptosis Occurs via the Product of the Alternative Open Reading Frame 4. PLoS Pathog. 2011, 7, e1002413.
- [51] C.V. Subba-Reddy, M.A. Yunus, I.G. Goodfellow, and C.C. Kao, Norovirus RNA synthesis is modulated by an interaction between the viral RNA-dependent RNA polymerase and the major capsid protein, VP1. J. Virol. 2012, 86, 10138–10149. [CrossRef] [PubMed]
- [52] C.J. McCormick, O. Salim, P.R. Lambden, and I.N. Clarke, Translation termination reinitiation between open reading frame 1 (ORF1) and ORF2 enables capsid expression in a bovine norovirus without the need for production of viral subgenomic RNA. J. Virol. 2008, 82, 8917–8921. [CrossRef] [PubMed]
- [53] R. Wennesz, C. Luttermann, F. Kreher, and G. Meyers, Structure–function relationship in the ‘termination upstream ribosomal binding site’ of the calicivirus rabbit hemorrhagic disease virus. Nucleic Acids Research 47 (2019) 1920-1934.
- [54] K. Geissler, K. Schneider, A. Fleuchaus, C.R. Parrish, G. Sutter, and U. Truyen, Feline calicivirus capsid protein expression and capsid assembly in cultured feline cells. J. Virol. 1999, 73, 834–838. [CrossRef] [PubMed]
- [55] B. Di Martino, and F. Marsilio, Feline calicivirus VP2 is involved in the self-assembly of the capsid protein into virus-like particles. Res. Veter- Sci. 2010, 89, 279–281. [CrossRef] [PubMed]
- [56] L.G. Thorne, and I.G. Goodfellow, Norovirus gene expression and replication. The Journal of general virology 95 Pt 2 (2014) 278-91.
- [57] C.E. Wobus, S.M. Karst, L.B. Thackray, K.-O. Chang, S.V. Sosnovtsev, G. Belliot, A. Krug, J.M. Mackenzie, K.Y. Green, and H.W.I.V. Virgin, Replication of Norovirus in Cell Culture Reveals a Tropism for Dendritic Cells and Macrophages. PLOS Biology 2 (2004) e432.
- [58] C. Cox, S. Cao, and Y. Lu, Enhanced detection and study of murine norovirus-1 using a more efficient microglial cell line. Virol. J. 2009, 6, 196–196. [CrossRef]
- [59] L.C. Kreutz, B.S. Seal, and W.L. Mengeling, Early interaction of feline calicivirus with cells in culture. Arch. Virol. 1994, 136, 19–34. [CrossRef]
- [60] T. Farkas, K. Sestak, C. Wei, and X. Jiang, Characterization of a rhesus monkey calicivirus representing a new genus of Caliciviridae. J. Virol. 2008, 82, 5408–5416. [CrossRef]
- [61] J.M. Martín-Alonso, D.E. Skilling, L. González-Molleda, G. del Barrio, A. Machín, N.K. Keefer, D.O. Matson, P.L. Iversen, A.W. Smith, and F. Parra, Isolation and characterization of a new Vesivirus from rabbits. Virology 2005, 337, 373–383.
- [62] M.K. Jones, K.R. Grau, V. Costantini, A.O. Kolawole, M. de Graaf, P. Freiden, C.L. Graves, M. Koopmans, S.M. Wallet, S.A. Tibbetts, S. Schultz-Cherry, C.E. Wobus, J. Vinjé, and S.M. Karst, Human norovirus culture in B cells. Nat. Protoc. 2015, 10, 1939–1947.
- [63] S.J. Flint, L.W. Enquist, R.M. Krug, V.R. Racaniello, and A.M. Skalka, Picornaviruses, Principles of Virology. Molecular Biology, Pathogenesis and Control, ASM, Washington, 2000, pp. 752.
- 64] L.C. van Dinten, J.A. den Boon, A.L. Wassenaar, W.J. Spaan, and E.J. Snijder, An infectious arterivirus cDNA clone: identification of a replicase point mutation that abolishes discontinuous mRNA transcription. Proc Natl Acad Sci U S A 94 (1997) 991-6.
- [65] G. van Marle, L.C. van Dinten, W.J. Spaan, W. Luytjes, and E.J. Snijder, Characterization of an equine arteritis virus replicase mutant defective in subgenomic mRNA synthesis. J. Virol. 1999, 73, 5274–5281. [CrossRef] [PubMed]
- [66] F. Almazan, I. Sola, S. Zuniga, S. Marquez-Jurado, L. Morales, M. Becares, and L. Enjuanes, Coronavirus reverse genetic systems: infectious clones and replicons. Virus Res. 2014, 189, 262–270.
- [67] R. Casais, V. Thiel, S.G. Siddell, D. Cavanagh, and P. Britton, Reverse genetics system for the avian coronavirus infectious bronchitis virus. J Virol 75 (2001) 12359-69.
- [68] C.J. Lai, B.T. Zhao, H. Hori, and M. Bray, Infectious RNA transcribed from stably cloned full-length cDNA of dengue type 4 virus. Proc.Natl.Acad.Sci.U.S.A 88 (1991) 5139-5143. 5139–5143.
- [69] N. Ruggli, J.D. Tratschin, C. Mittelholzer, and M.A. Hofmann, Nucleotide sequence of classical swine fever virus strain Alfort/187 and transcription of infectious RNA from stably cloned full-length cDNA. J. Virol. 1996, 70, 3478–3487. [CrossRef] [PubMed]
- [70] M. Yanagi, R.H. Purcell, S.U. Emerson, and J. Bukh, Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Natl. Acad. Sci. USA 1997, 94, 8738–8743. [CrossRef]
- [71] D.G. Gibson, Enzymatic assembly of overlapping DNA fragments. Methods Enzymol 498 (2011) 349-61.
- [72] D.G. Gibson, L. Young, R.-Y. Chuang, J.C. Venter, C.A. Hutchison, and H.O. Smith, Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods 6 (2009) 343-345.
- [73] Y. Chaudhry, M.A. Skinner, and I.G. Goodfellow, Recovery of genetically defined murine norovirus in tissue culture by using a fowlpox virus expressing T7 RNA polymerase. J. Gen. Virol. 2007, 88, 2091–2100. [CrossRef]
- [74] C. Sandoval-Jaime, K.Y. Green, and S.V. Sosnovtsev, Recovery of murine norovirus and feline calicivirus from plasmids encoding EMCV IRES in stable cell lines expressing T7 polymerase. J. Virol. Methods 2015, 217, 1–7. [CrossRef]
- [75] A. Bridgen, and R.M. Elliot, Chapter 9. Reverse genetics of RNA viruses. in: A.J. Cann, (Ed.), RNA viruses: a practical approach, Oxford University Press, New York, 2000, pp. 201-227.
- [76] K.O. Chang, S.V. Sosnovtsev, G. Belliot, Q. Wang, L.J. Saif, and K.Y. Green, Reverse genetics system for porcine enteric calicivirus, a prototype sapovirus in the Caliciviridae. J. Virol. 2005, 79, 1409–1416. [CrossRef]
- [77] K. Katayama, G.S. Hansman, T. Oka, S. Ogawa, and N. Takeda, Investigation of norovirus replication in a human cell line. Arch. Virol. 2006, 151, 1291–1308. [CrossRef]
- [78] M. Asanaka, R.L. Atmar, V. Ruvolo, S.E. Crawford, F.H. Neill, and M.K. Estes, Replication and packaging of Norwalk virus RNA in cultured mammalian cells. Proc. Natl. Acad. Sci. USA 2005, 102, 10327–10332. [CrossRef]
- [79] S.V. Sosnovtsev, S.A. Sosnovtseva, and K.Y. Green, Recovery of feline calicivirus from plasmid DNA containing a full-length copy of the genome. in: D. Chasey, R.M. Gaskell, and I.N. Clarke, (Eds.), The first international symposium on calicivirus, European Society for Veterinary Virology and Central Veterinary Laboratory, Reading, United Kingdom, 1996, pp. 125-130.
- [80] T. Mitra, S.V. Sosnovtsev, and K.Y. Green, Mutagenesis of tyrosine 24 in the VPg protein is lethal for feline calicivirus. J. Virol. 2004, 78, 4931–4935. [CrossRef] [PubMed]
- [81] C. Morgan, S.A. Ellison, H.M. Rose, and D.H. Moore, Structure and development of viruses observed in the electron microscope: II. vaccinia and fowl pox viruses. The Journal of Experimental Medicine 100 (1954) 301-310.
- [82] P. Somogyi, J. Frazier, and M.A. Skinner, Fowlpox virus host range restriction: gene expression, DNA replication, and morphogenesis in nonpermissive mammalian cells. Virology 1993, 197, 439–444. [CrossRef] [PubMed]
- [83] E. Scotto-Lavino, G. Du, and M.A. Frohman, 5’ end cDNA amplification using classic RACE. Nat Protoc 1 (2006) 2555-62.
- [84] C.T. Ranjith-Kumar, Y. Wen, N. Baxter, K. Bhardwaj, and C.C. Kao, A cell-based assay for RNA synthesis by the HCV polymerase reveals new insights on mechanism of polymerase inhibitors and modulation by NS5A. PLoS ONE 2011, 6, e22575.
- [85] C.V. Subba-Reddy, I. Goodfellow, and C.C. Kao, VPg-primed RNA synthesis of norovirus RNA-dependent RNA polymerases by using a novel cell-based assay. J. Virol. 2011, 85, 13027–13037. [CrossRef]
- [86] R.B. Seth, L. Sun, and Z.J. Chen, Antiviral innate immunity pathways. Cell Res. 2006, 16, 141–147. [CrossRef]
- [87] S.V. Sosnovtsev, M. Garfield, and K.Y. Green, Processing map and essential cleavage sites of the nonstructural polyprotein encoded by ORF1 of the feline calicivirus genome. J. Virol. 2002, 76, 7060–7072. [CrossRef] [PubMed]
- [88] J.D. Neill, S.V. Sosnovtsev, and K.Y. Green, Recovery and altered neutralization specificities of chimeric viruses containing capsid protein domain exchanges from antigenically distinct strains of feline calicivirus. J. Virol. 2000, 74, 1079–1084. [CrossRef]
- [89] S.V. Sosnovtsev, G. Belliot, K.O. Chang, O. Onwudiwe, and K.Y. Green, Feline calicivirus VP2 is essential for the production of infectious virions. J. Virol. 2005, 79, 4012–4024. [CrossRef]
- [90] E.J. Abente, S.V. Sosnovtsev, K. Bok, and K.Y. Green, Visualization of feline calicivirus replication in real-time with recombinant viruses engineered to express fluorescent reporter proteins. Virology 2010, 400, 18–31. [CrossRef]
- [91] I. Karakasiliotis, S. Vashist, D. Bailey, E.J. Abente, K.Y. Green, L.O. Roberts, S.V. Sosnovtsev, and I.G. Goodfellow, Polypyrimidine tract binding protein functions as a negative regulator of feline calicivirus translation. PLoS ONE 2010, 5, e9562.
- [92] J.R.O. Thumfart, and G. Meyers, Feline Calicivirus: Recovery of Wild-Type and Recombinant Viruses after Transfection of cRNA or cDNA Constructs. J. Virol. 2002, 76, 6398–6407. [CrossRef] [PubMed]
- [93] C. Wei, T. Farkas, K. Sestak, and X. Jiang, Recovery of infectious virus by transfection of in vitro-generated RNA from tulane calicivirus cDNA. J. Virol. 2008, 82, 11429–11436. [CrossRef]
- [94] S. Guix, M. Asanaka, K. Katayama, S.E. Crawford, F.H. Neill, R.L. Atmar, and M.K. Estes, Norwalk Virus RNA Is Infectious in Mammalian Cells. J. Virol. 2007, 81, 12238–12248.
- [95] V.K. Ward, C.J. McCormick, I.N. Clarke, O. Salim, C.E. Wobus, L.B. Thackray, H.W. Virgin, and P.R. Lambden, Recovery of infectious murine norovirus using pol II-driven expression of full-length cDNA. Proc. Natl. Acad. Sci. USA 2007, 104, 11050–11055. [CrossRef] [PubMed]
- [96] T. Oka, H. Takagi, and Y. Tohya, Development of a novel single step reverse genetics system for feline calicivirus. J. Virol. Methods 2014, 207, 178–181. [CrossRef]
- [97] L.M. Oliveira, R. Blawid, A.F. Orílio, B.Y.G. Andrade, A.C.A. Souza, and T. Nagata, Development of an infectious clone and replicon system of norovirus GII.4. J. Virol. Methods 2018, 258, 49–53. [CrossRef]
- [98] P. Simmonds, I. Karakasiliotis, D. Bailey, Y. Chaudhry, D.J. Evans, and I.G. Goodfellow, Bioinformatic and functional analysis of RNA secondary structure elements among different genera of human and animal caliciviruses. Nucleic Acids Res. 2008, 36, 2530–2546. [CrossRef]
- [99] D. Bailey, I. Karakasiliotis, S. Vashist, L.M. Chung, J. Rees, N. McFadden, A. Benson, F. Yarovinsky, P. Simmonds, and I. Goodfellow, Functional analysis of RNA structures present at the 3’ extremity of the murine norovirus genome: the variable polypyrimidine tract plays a role in viral virulence. J. Virol. 2010, 84, 2859–2870.
- [100] D. Bailey, L.B. Thackray, and I.G. Goodfellow, A single amino acid substitution in the murine norovirus capsid protein is sufficient for attenuation in vivo. J. Virol. 2008, 82, 7725–7728. [CrossRef]
- [101] N. McFadden, A. Arias, I. Dry, D. Bailey, J. Witteveldt, D.J. Evans, I. Goodfellow, and P. Simmonds, Influence of genome-scale RNA structure disruption on the replication of murine norovirus—similar replication kinetics in cell culture but attenuation of viral fitness in vivo. Nucleic Acids Res. 2013, 41, 6316–6331.
- [102] L. Thorne, D. Bailey, and I. Goodfellow, High-resolution functional profiling of the norovirus genome. J. Virol. 2012, 86, 11441–11456. [CrossRef] [PubMed]
- [103] E. Lopez-Manriquez, S. Vashist, L. Urena, I. Goodfellow, P. Chavez, J.E. Mora-Heredia, C. Cancio-Lonches, E. Garrido, and A.L. Gutierrez-Escolano, Norovirus Genome Circularization and Efficient Replication Are Facilitated by Binding of PCBP2 and hnRNP A1. J. Virol. 2013, 87, 11371–11387.
- [104] J. Cheng, A. Tang, J. Chen, D. Zhang, C. Meng, C. Li, H. Wei, and G. Liu, A cDNA-based reverse genetics system for feline calicivirus identifies the 3’ untranslated region as an essential element for viral replication. Arch Virol 168 (2023) 33.
- [105] K. Katayama, K. Murakami, T.M. Sharp, S. Guix, T. Oka, R. Takai-Todaka, A. Nakanishi, S.E. Crawford, R.L. Atmar, and M.K. Estes, Plasmid-based human norovirus reverse genetics system produces reporter-tagged progeny virus containing infectious genomic RNA. Proc. Natl. Acad. Sci. USA 2014, 111, E4043–E4052.
- [106] K.O. Chang, S.V. Sosnovtsev, G. Belliot, A.D. King, and K.Y. Green, Stable expression of a Norwalk virus RNA replicon in a human hepatoma cell line. Virology 2006, 353, 463–473. [CrossRef]
- [107] K.-O. Chang, and D.W. George, Interferons and Ribavirin Effectively Inhibit Norwalk Virus Replication in Replicon-Bearing Cells. J. Virol. 2007, 81, 12111–12118. [CrossRef] [PubMed]
- [108] K.-O. Chang, Role of Cholesterol Pathways in Norovirus Replication. J. Virol. 2009, 83, 8587–8595. [CrossRef] [PubMed]
- [109] K. Bok, V.G. Prikhodko, K.Y. Green, and S.V. Sosnovtsev, Apoptosis in murine norovirus-infected RAW264.7 cells is associated with downregulation of survivin. J. Virol. 2009, 83, 3647–3656. [CrossRef]
- [110] K.C. Tiew, G. He, S. Aravapalli, S.R. Mandadapu, M.R. Gunnam, K.R. Alliston, G.H. Lushington, Y. Kim, K.O. Chang, and W.C. Groutas, Design, synthesis, and evaluation of inhibitors of Norwalk virus 3C protease. Bioorganic Med. Chem. Lett. 2011, 21, 5315–5319.
- [111] S. Torii, C. Ono, R. Suzuki, Y. Morioka, I. Anzai, Y. Fauzyah, Y. Maeda, W. Kamitani, T. Fukuhara, and Y. Matsuura, Establishment of a reverse genetics system for SARS-CoV-2 using circular polymerase extension reaction. Cell Rep 35 (2021) 109014.
- [112] W. Wang, X. Peng, Y. Jin, J.A. Pan, and D. Guo, Reverse genetics systems for SARS-CoV-2. J Med Virol 94 (2022) 3017-3031.
- [113] T.C. Li, M. Kataoka, Y.H. Doan, H. Saito, H. Takagi, M. Muramatsu, and T. Oka, Characterization of a Human Sapovirus Genotype GII.3 Strain Generated by a Reverse Genetics System: VP2 Is a Minor Structural Protein of the Virion. Viruses 2022, 14.
- [114] G. Liu, Y. Zhang, Z. Ni, T. Yun, Z. Sheng, H. Liang, J. Hua, S. Li, Q. Du, and J. Chen, Recovery of infectious rabbit hemorrhagic disease virus from rabbits after direct inoculation with in vitro-transcribed RNA. J. Virol. 2006, 80, 6597–6602. [CrossRef] [PubMed]
- [115] G. Liu, Z. Ni, T. Yun, B. Yu, L. Chen, W. Zhao, J. Hua, and J. Chen, A DNA-launched reverse genetics system for rabbit hemorrhagic disease virus reveals that the VP2 protein is not essential for virus infectivity. J. Gen. Virol. 2008, 89, 3080–3085. [CrossRef]
- [116] G. Liu, Z. Ni, T. Yun, B. Yu, J.M. Zhu, J.G. Hua, and J.P. Chen, Rabbit hemorrhagic disease virus poly(A) tail is not essential for the infectivity of the virus and can be restored in vivo. Arch. Virol. 2008, 153, 939–944. [CrossRef]
- [117] S. González-Reyes, A. García-Manso, G. del Barrio, K.P. Dalton, L. Gonzalez-Molleda, J. Arrojo-Fernández, I. Nicieza, and F. Parra, Role of annexin A2 in cellular entry of rabbit vesivirus. J. Gen. Virol. 2009, 90, 2724–2730.
- [118] A. Arias, L. Urena, L. Thorne, M.A. Yunus, and I. Goodfellow, Reverse genetics mediated recovery of infectious murine norovirus. J.Vis.Exp. (2012).
- [119] G. Euller-Nicolas, C.L. Mennec, J. Schaeffer, X.-L. Zeng, K. Ettayebi, R.L. Atmar, F.S.L. Guyader, M.K. Estes, and M. Desdouits, Human Sapovirus Replication in Human Intestinal Enteroids. J. Virol. 2023, 97, e0038323.
- [120] K. Ettayebi, V.R. Tenge, N.W. Cortes-Penfield, S.E. Crawford, F.H. Neill, X.L. Zeng, X. Yu, B.V. Ayyar, D. Burrin, S. Ramani, R.L. Atmar, and M.K. Estes, New Insights and Enhanced Human Norovirus Cultivation in Human Intestinal Enteroids. mSphere 2021, 6.
- [121] T. Hayashi, K. Murakami, J. Hirano, Y. Fujii, Y. Yamaoka, H. Ohashi, K. Watashi, M.K. Estes, and M. Muramatsu, Dasabuvir Inhibits Human Norovirus Infection in Human Intestinal Enteroids. mSphere 2021, 6, e0062321.
- [122] E. Kardia, O. Fakhri, M. Pavy, H. Mason, N. Huang, E. Smertina, M. Jenckel, N.Y.G. Peng, M.K. Estes, T. Strive, M. Frese, I. Smith, and R.N. Hall, Hepatobiliary organoids derived from leporids support the replication of hepatotropic lagoviruses. J. Gen. Virol. 2023, 104.

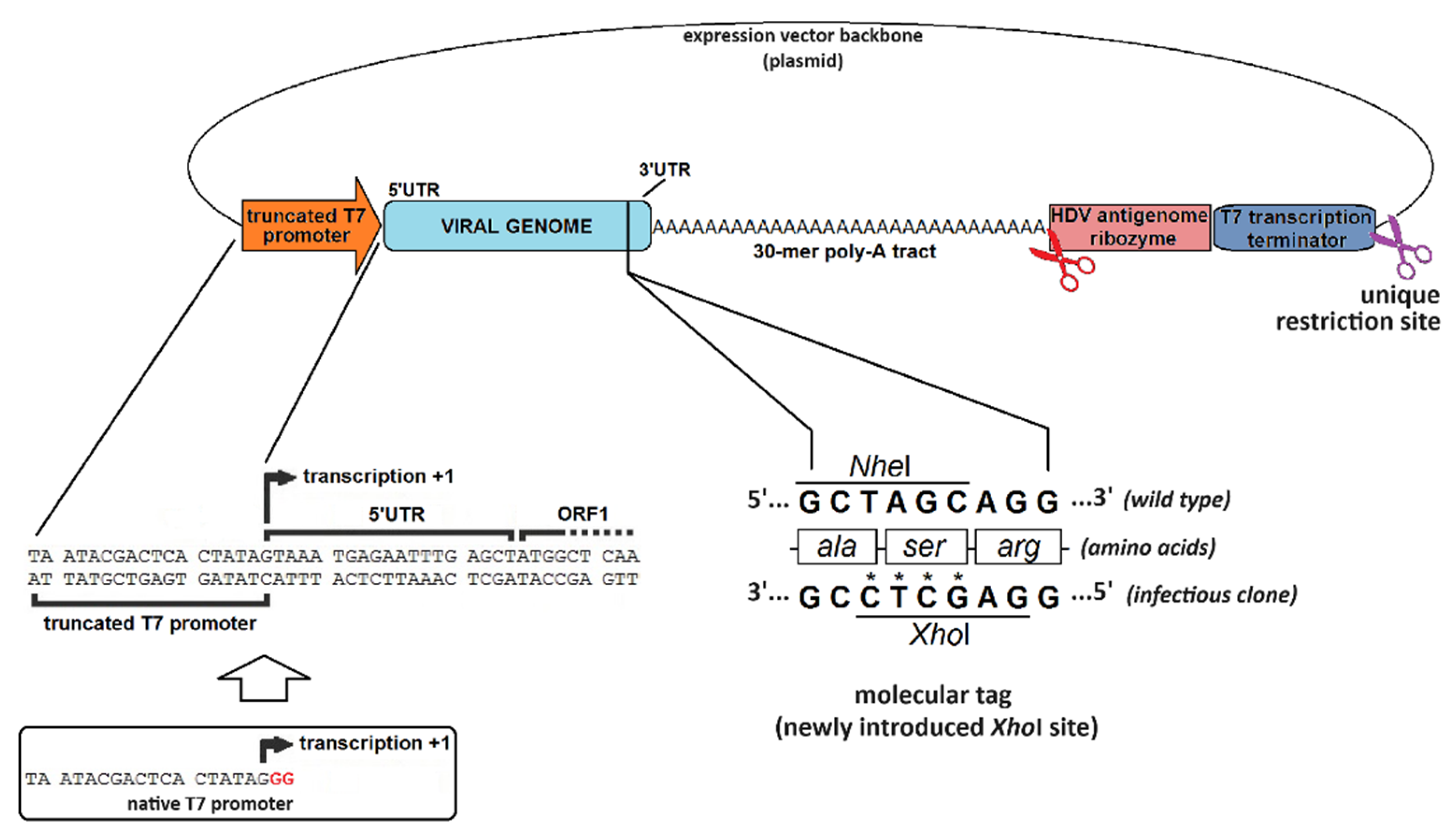
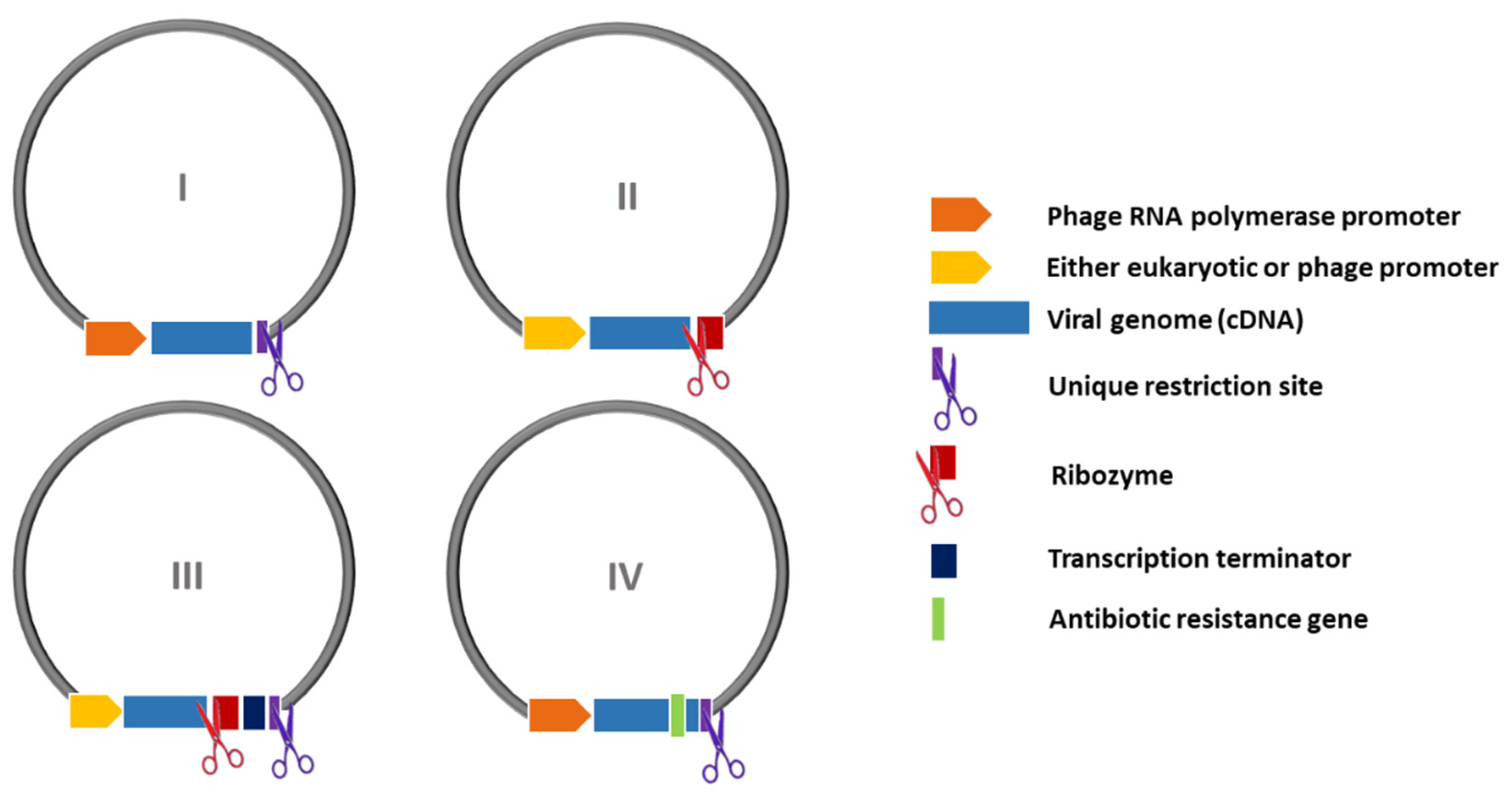
| Promoter | Bacteriophage (T3, T7, SP6, etc.) | Eukaryotic (CMV, EF-1α, SV40, etc.) |
||
|---|---|---|---|---|
| Reverse genetics strategy | Transfection of in vitro-transcribed RNA | cDNA transfection of helper virus-infected cells | cDNA transfection of phage RNA pol-expressing cell line (no helper virus) |
RNA pol II-driven nuclear transcription of cDNA |
| Is there virus-induced CPE in transfected cell monolayers (passage 0)? | • Microscopic examination of transfected cells | |||
| Does the supernatant from transfected cells (passage 0) contain infectious virions? | • Blind passage of the supernatant from transfected cells and microscopic examination of inoculated (passage 1) monolayers | |||
| Is the genome-emulating RNA transcript present in the cytosol? | • Extraction of total cell RNA followed by detection of viral RNA through RT-PCR; emphasis should be given to RNA integrity (intactness and overall quality; lack of RNA degradation) | |||
| Is there any non-viral sequence inadvertently added to the 5’ or 3’–end? | • 5’–RACE / 3’–RACE assays | |||
| Is ORF1 being expressed? | • Western blot (WB) or Immunofluorescence (IF) using antibodies specific to viral proteins | |||
| Is the viral protease functional? | • WB, focusing on the expected sizes of ORF1 derived mature peptides • Radioactive labeling and autoradiography |
|||
| Is the viral RdRp functional? | • 5BR assay | |||
| Is the negative strand being synthesized? | • Northern blot • RT-PCR with primers specific for negative strand |
|||
| Are VP1 or VP2 being synthesized? | • WB with specific antibodies for the detection of VP1 or VP2 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





