1. Introduction
Nucleoside compounds [
1] include natural nucleosides as well as nucleoside analogs modified at multiple positions on the sugar or base. The structure of nucleoside compounds is highly variable [
2], and most nucleoside drugs currently are chemically modified derivatives of natural nucleosides [
3]. These modified nucleosides enter the human body and interact with viral polymerases or tumor cells’ nucleic acids, leading to mismatch formation and competitive inhibition, effectively inhibiting viral replication and interrupting tumor cell division and growth [
4]. Particularly, following the global COVID-19 pandemic, the development and application of nucleoside compounds have become a significant focus of attention [
5].Non-natural nucleoside compounds are primarily synthesized through chemical methods involving base modification and protection, which can be intricate and lead to environmental pollution, and the synthesis efficiency is not ideal. Compared to organic synthesis, using enzyme-catalyzed reactions in nucleoside compound synthesis is not only environmentally friendly but also significantly enhances the reaction’s stereo-selectivity and regio-selectivity, providing multiple possibilities for efficient production [
6,
7,
8].
Nucleoside Deoxyribosyltransferase (NDT), also known as deoxyribosyltransferase, is a polymerase that mediates ribose nucleobase transfers. It primarily exhibits two types of activities. Type I NDT specifically catalyzes the conversion between different purine bases on deoxyribose (Pur→Pur). Type II NDT has a broader catalytic range, as it can catalyze conversions between different purine bases, as well as between purine and pyrimidine bases (Pur→Pur, Pur→Pyr, Pyr→Pyr) [
9,
10]. These enzymes were first discovered in Lactobacillus strains. Subsequent research has shown that other types of Lactobacillus bacteria can also synthesize NDT, such as
L. helveticus,
L. leichmannii,
L. fermentum, and
L. reuteri, among others [
11].
In biocatalytic processes, it is common to use immobilization methods to fix the target enzyme on a carrier, creating a bioreactor that minimizes enzyme damage and is easy to operate [
12,
13,
14]. This method offers several advantages over batch-type enzymatic reactions. Immobilization allows for enzyme recovery and reuse, significantly improving enzyme stability in industrial production processes. It also meets the economic practicality required for industrial production, thereby reducing production costs. In recent years, immobilized metal affinity chromatography (IMAC) has gained widespread application as one of the directional immobilization methods. This technology is based on the highly specific interaction between tagged proteins and metals. The most common strategy involves immobilizing fusion proteins with a His tag onto a Ni
2+ chelating scaffold [
15]. This method preserves higher enzyme activity and can achieve one-step purification effects in industrial production.
This study utilizes the principle of specific binding between nickel chelating agarose affinity chromatography medium and Nucleoside deoxyribosyltransferase (referred to as
LrNDT) with histidine tags. This forms an immobilized enzyme reactor (IMER) and conducts a comparative study of the immobilized enzyme’s properties. The N-Deoxyribose Transferase derived from
Lactobacillus reuteri (
LrNDT) demonstrates high catalytic activity towards deoxyribose compounds. To firmly attach the target protein to the immobilized carrier, the N-Deoxyribose Transferase gene from the
Lactobacillus reuteri strain was recombinantly modified with a 6×His tag. Subsequently, it was covalently linked to Ni2+ chelating agarose affinity chromatography medium (Ni2+-NTA) with immobilized imidazole groups on the His tag, resulting in a stable covalent bond. This process is used to prepare an efficient, simple, and recyclable immobilized enzyme reactor (IMER) for the enzymatic synthesis of nucleoside precursor compounds with therapeutic value [
16]. The study then explores the impact of temperature, pH, and reusability on the relative activity of immobilized
LrNDT in catalyzing the production of 2′-deoxyribonucleoside. This research provides a preliminary method for preparing highly active immobilized
LrNDT and lays the groundwork for practical industrial applications of Nucleoside deoxyribosyltransferase.
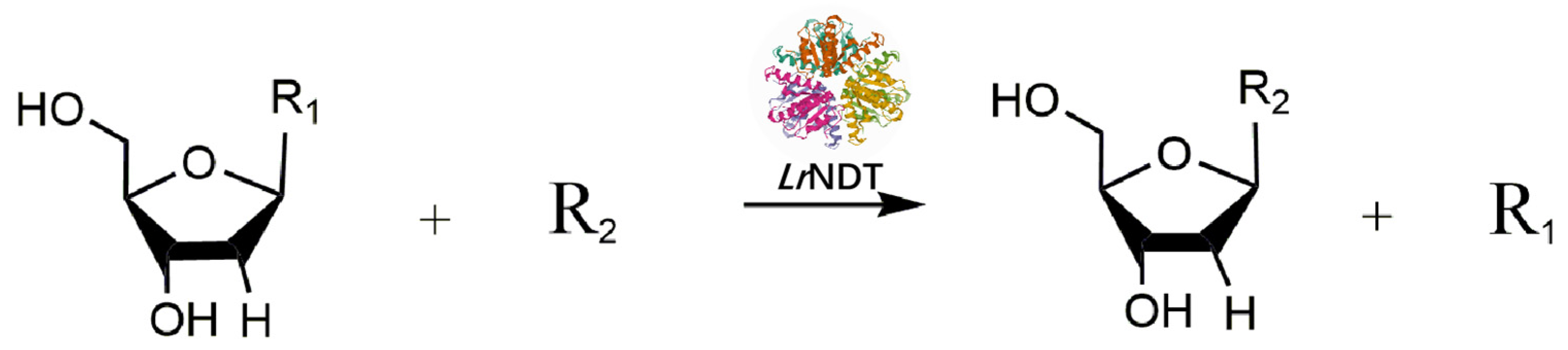
Scheme 1.
The reaction scheme of 2′-Deoxytheymidine catalyzed by LrNDT (R1=Thd, R2=Ado/Ura/Cyt).
Scheme 1.
The reaction scheme of 2′-Deoxytheymidine catalyzed by LrNDT (R1=Thd, R2=Ado/Ura/Cyt).
2. Results
2.1. Characterization and Identification of LrNDT
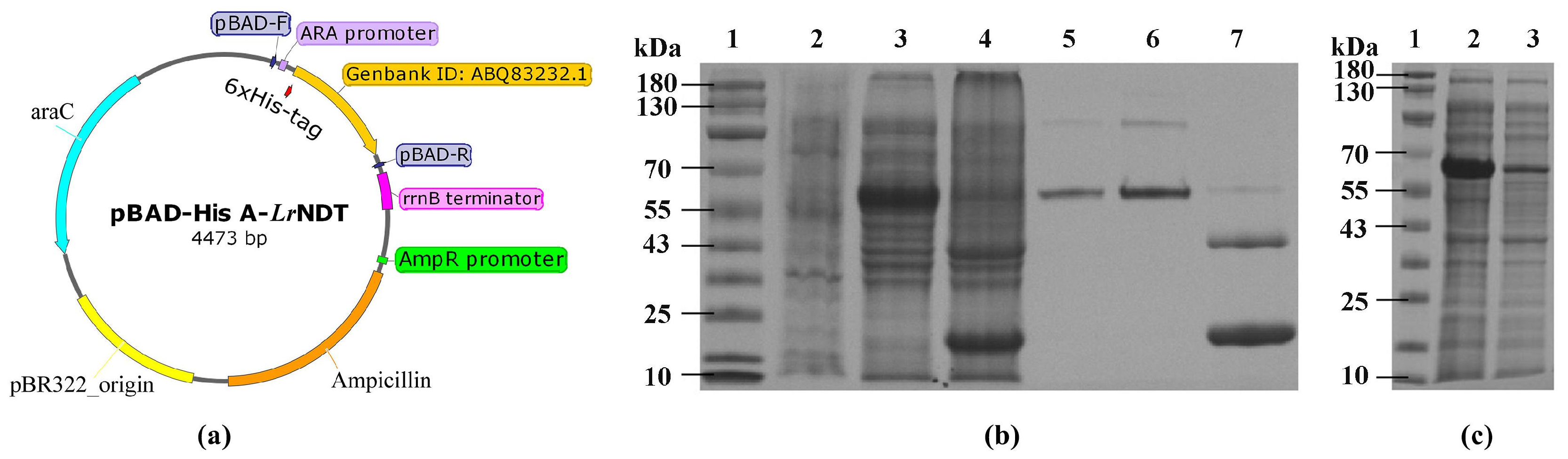
The
E. coli pBAD (TOP10) strain containing the recombinant His-tagged
LrNDT was activated and expanded at 37°C with shaking at 220 rpm (
Figure 1a). When the OD600 of the bacterial culture was between 0.6-0.8, induction was carried out using 20% L-arabinose as an inducer at 28°C with shaking at 220 rpm overnight. This resulted in the successful expression of the target gene (
Figure 1b). After an initial centrifugation at 8000 rpm for 5 minutes, the cells were resuspended in 50 mmol/L pH 7.0 phosphate buffer. The resuspended cells were then disrupted under high pressure (1000 bar), followed by a second centrifugation at 20,000 rpm for 30 minutes, and the supernatant was retained. The Ni-NTA agarose medium in the protein purification Ni column chelated with Ni
2+ ions. These chelated Ni
2+ ions could bind to the imidazole ring on the histidine residues in the supernatant. A gradient elution was performed using imidazole solutions ranging from 0.02 to 1 mol/L. Lower concentration imidazole 0.02 to 0.05 mol/L was used for washing to remove weakly bound target proteins and impurities from the supernatant, while the target proteins were subsequently eluted with 500 mmol/L imidazole. The order of elution was related to the protein binding capacity. Samples were collected from induced pre-culture, initial centrifugation supernatant, and post-centrifugation precipitate. Samples were also taken from the flow-through during the purification process after binding to the Ni resin and the elution with 500 mmol/L imidazole. These samples were processed and subjected to analysis via 15% SDS-PAGE. As shown in
Figure 1b, there was a noticeable difference in band patterns around 55 kDa, indicating significant expression of
LrNDT after induction in the E. coli TOP10 host strain (
Figure 1b). According to the literature, the monomeric size of N-Deoxyribosyltransferase II is approximately 20 kDa. The sample after heated at 85°C (
Figure 1b) showed similar sizes to the expected
LrNDT. The intensity of the target band in Line 5 significantly decreased compared to Line 3, demonstrating that the His-tagged
LrNDT was bound to the Ni resin. Line 6 indicates that the elution with 500 mmol/L imidazole showed a prominent band at 55 kDa, similar to the heated sample in Line 4, confirming the presence of a soluble expression of
LrNDT in E. coli in this concentration of imidazole elution.
2.2. Evaluation of Immobilization Effect
Free
LrNDT enzyme activity can be determined by a reaction involving the exchange of bases between 2′-deoxyribosylthymidine and adenine [
17]. In this study, we first tested the immobilization capacity of Ni
2+-NTA resin for free
LrNDT. A 1 mL Ni
2+-NTA carrier was placed in a gravity flow column for protein purification. Then, 3 mL of crude enzyme was slowly shaken with the carrier at 4°C for 1 hour to facilitate better binding of the carrier to the enzyme. The solution obtained after binding was collected and analyzed by SDS-PAGE. The results showed a clear target protein band in the crude enzyme solution at 55 kDa. Contrarily, the
LrNDT protein band of the flow-through after immobilization compared to the non-immobilized sample indicated a significant binding capacity of free
LrNDT with the immobilized carrier of Ni
2+-NTA resin (
Figure 1c). The immobilized
LrNDT using Ni
2+-NTA reached a concentration of 2 mg/mL and had an enzyme activity of approximately 235 U/mL, also retained about of a 97% of initial activity with 99% yield (
Table 1). These results proved that free nickel chelating agarose affinity chromatography medium for
LrNDT achieved one-step purification and immobilization process.
Table 1.
The effect of immobilization NDT with different carriers.
Table 1.
The effect of immobilization NDT with different carriers.
| Enzymes |
Carrier |
Immobilization (%) |
Recovery (%) |
Reference |
|
BpNDT |
PEI-agarose |
100 |
100 |
[18] |
|
TbPDT |
Ni2+-Fe2O3 |
99±1 |
56 |
[19] |
|
LdNDT |
SiGPEI1200-1300 |
100 |
97 |
[20] |
|
LrNDT |
Ni2+-NTA |
99 |
97 |
this study |
The effect of temperature on enzyme activity is relatively straightforward. Higher temperatures can increase the efficiency of enzyme-catalyzed reactions but may also alter the spatial conformation of the enzyme, potentially leading to reduced enzyme activity. Immobilized enzyme on a carrier can also affect its temperature tolerance to some extent [
21,
22,
23]. To estimate the thermostability, the immobilized
LrNDT were incubated at various temperatures for 10 minutes, 20 minutes, 40 minutes, and 60 minutes, and then, the residual enzyme activity was determined. As shown in
Figure 2, immobilized
LrNDT had good temperature tolerance of in the temperature range of 20°C-45°C with above 95% activity remained. The best reaction temperature of immobilized
LrNDT was at 40°C, remaining 97% of its relative activity.
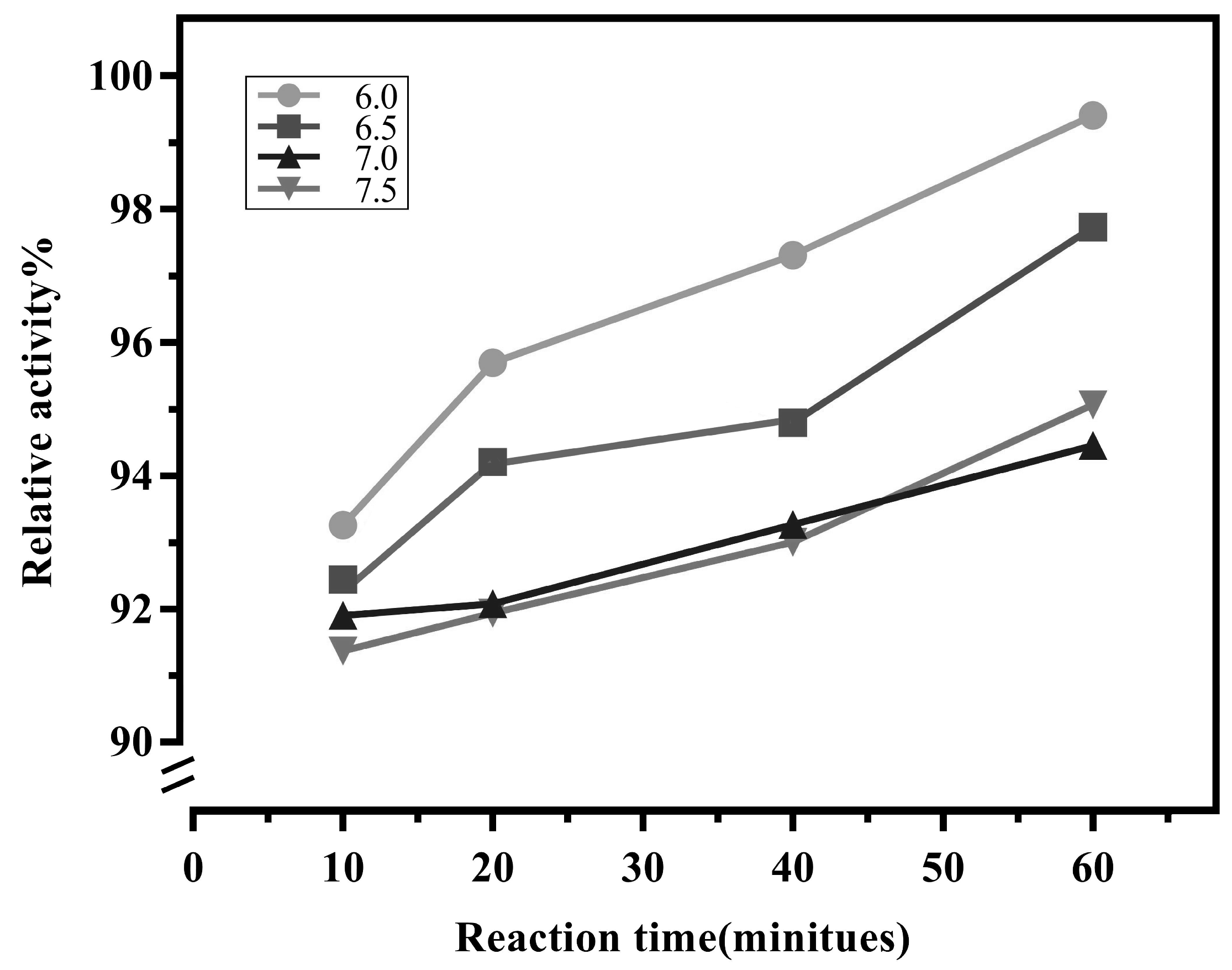
Previous research has shown that
LrNDT is sensitive to the pH of the reaction environment, with the highest activity observed at pH 7.1 [
24,
25]. The optimum reaction pH value and pH stability of free and immobilized
LrNDT were determined. The pH of buffer too high or too low may lead to changes in spatial structure of the enzyme, thus reducing its catalytic activity. we conducted experiments in four different pH buffer environments ranging from pH 6.0 to pH 7.5. As shown in
Figure 3, the immobilized
LrNDT remained above 90% activity from pH 6.0 to pH 7.5. The pH stability of immobilized
LrNDT was better than that of the free enzyme as reported. It also showed that immobilized
LrNDT retained 97% of its enzyme activity at pH 6.0 with a reaction time of 20 minutes.
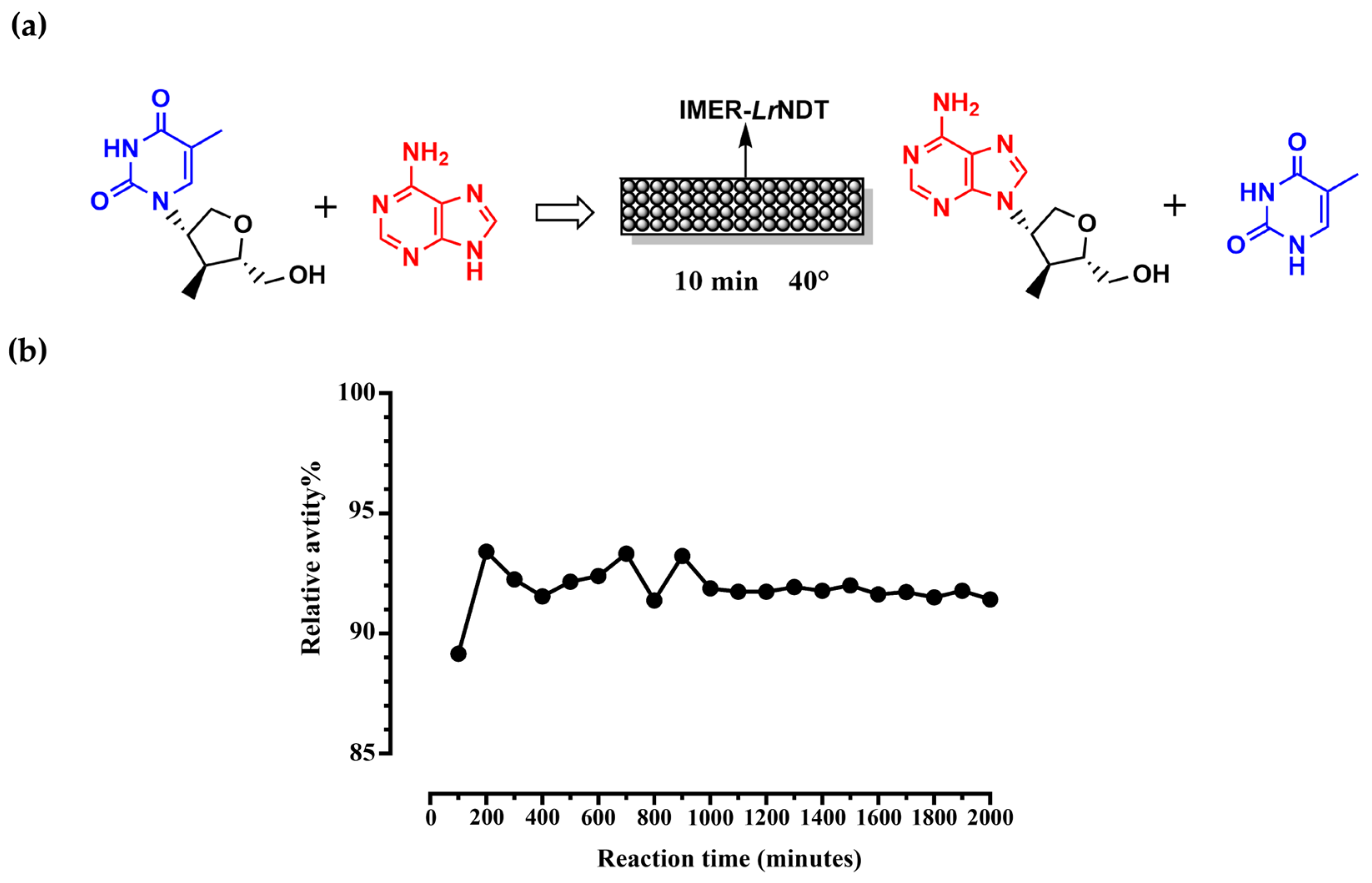
2.3. Reusability of Immobilized LrNDT
Although the immobilized
LrNDT enzyme has improved temperature tolerance and pH stability compared with the free enzyme, the current immobilization mode uses the
LrNDT enzyme for batch production, and the immobilized enzyme still encounters problems such as inefficient catalysis and intermediate degradation in batch production [
26]. Here, we made a packed-bed bioreactor (PBR), which composed by the immobilized
LrNDT as IMER-
LrNDT (
Figure 4a). The biocatalysis reaction was performed at 40°C, and the enzyme activity was measured for 24 h with a continuous flow rate of 1 mL/min and a column reactor volume of 20 mL and a column residence time of 20 min. As shown in
Figure 4, the PBR reactor remain above 90% activity after 24h (
Figure 4b). The IMER-
LrNDT enzyme also has good operational stability and reusability, and can still maintain a high activity level after long time, which has good potential for industrial application.
3. Discussion
The research on nucleoside drugs is gaining widespread attention as enzymatic synthesis of these drugs is not only more cost-effective but also environmentally friendly [
27,
28,
29]. Nucleoside diphosphate transferase II (NDT) has drawn interest due to its broad base recognition range and high catalytic efficiency, making it suitable for catalyzing various nucleoside substrates in a one-step reaction to produce target compounds.
Although NDT has tremendous potential for producing nucleoside analogs, most current enzyme application methods are limited to batch production, increasing the costs associated with industrial-scale production and limiting enzyme utility. Employing immobilization in enzyme production not only simplifies the purification process but also allows for enzyme reuse, greatly enhancing the efficiency of enzyme utilization [
30]. In this study, we expressed NDT from
Lactobacillus reuteri and utilized nickel-chelating agarose gel as the immobilization resin. The specific binding between NDT and amino acids carrying imidazole rings on the matrix served the dual purpose of immobilization and purification, which achieved one-step purification and immobilization of free
LrNDT.
We investigated the enzymatic properties of the immobilized LrNDT at different temperatures and pH levels. Using 2′-deoxythymidine and adenine as substrates, the immobilized LrNDT exhibited a relative activity of over 90% under the conditions of 40°C and pH 6.0, retaining over 97% of its original activity. This significant improvement in the reusability of the packed-bed reactor with immobilized LrNDT reduces the cost associated with using the free enzyme and enables more environmentally friendly and energy-efficient biocatalysis.
In previous papers, different types of carriers for NDT immobilization were compared. The use of Ni-NTA as a metal-chelating matrix preparation framework enriched the immobilization sites, ensuring a strong bond with the enzyme fusion tag. The immobilized enzyme exhibited high stability, requiring the use of high-concentration imidazole solutions for successful elution. Furthermore, the nickel in the Ni2+-NTA medium, after elution, could be removed by ethylenediaminetetraacetic acid (EDTA) and then regenerated using nickel sulfate, serving the purpose of immobilized matrix recycling and reducing the enzyme production costs.
4. Materials and Methods
4.1. Strain and Culture Media
Recombinant Escherichia coli TOP10/pBAD/His-A, carrying a recombinant plasmid pBAD/His-A. This plasmid contains the coding gene for N-Deoxyribose Transferase (
LrNDT) derived from
Lactobacillus reuteri strain (GenBank accession number: ABQ83232.1), which was constructed in our laboratory as shown in
Figure 1a.
Seed liquid culture medium and liquid fermentation medium: LB medium (tryptone 10 g/L, yeast extract 5 g/L, NaCl 10 g/L, pH 7.0), sterilized, and then supplemented with ampicillin to a final concentration of 50 µg/ml.
4.2. Reagents and Equipment
Nickel chelating agarose chromatography medium (Ni2+-NTA) was purchased from Jiangsu Qian Chun Biotech Co., Ltd. Nucleoside compounds were obtained from Wuhu Huaren Technology Co., Ltd. Other common reagents were of analytical grade and purchased from Shanghai. The high-performance liquid chromatography detector used was from Shimadzu (model DGU-20A3R).
4.3. Cultivation Conditions
Seed Cultivation: Take 10 µl of the stored culture at 4°C and inoculate it into 10 mL of seed liquid culture medium. Incubate at 37°C with agitation at 200 rpm for 14 hours. Fermentation Cultivation: Transfer the seed culture into a 1 L Erlenmeyer flask containing 500 mL of fermentation medium. Incubate at 37°C with agitation at 200 rpm.
4.4. Enzyme Expression and Purification
When the fermentation culture reaches an OD600 of 0.6-0.8, induce with a final concentration of 20% L-arabinose. Continue cultivation at 28°C for an additional 12 hours. After induction, collect the cells by centrifugation at 8000 rpm for 5 minutes. Resuspend the cell pellet in phosphate buffer (50 mmol/L, pH 6.5). Disrupt the resuspended cells using a high-pressure cell disruptor. Centrifuge at 20,000 rpm for 30 minutes and retain the supernatant. Purify the enzyme using a His-Trap HP column, followed by overnight dialysis in phosphate buffer containing 1 mol/L NaCl. Measure protein concentration using a microspectrophotometer. Samples are collected before and after induction and analyzed using SDS-PAGE to confirm the successful expression of the target protein. A 15% SDS-PAGE gel is used for this purpose.
4.5. LrNDT Enzyme Activity Assay
Dissolve 2′-deoxyribonucleoside at a concentration of 6 mmol/L and adenine at a concentration of 9 mmol/L in phosphate buffer (50 mmol/L, pH 6.5). Add 500 µl of purified enzyme and prepare a 3 mL reaction system. Place it in a 40°C shaking incubator for 20 minutes. After the reaction, heat the reaction mixture in an 85°C metal bath for 20 minutes to inactivate the enzyme. Cool to room temperature, filter the reaction mixture through a 0.22µm microfilter membrane, and dilute the supernatant for HPLC analysis. HPLC conditions: Ultimate XB-C18 (4.6×250 mm, 5 µm) column, with acetonitrile/TEAA (5/95, V/V) as the mobile phase, a flow rate of 1 mL/min, column temperature at 40°C, and detection at 254 nm. Enzyme activity is defined as the amount of enzyme required to convert 1 mmol of substrate into product in one unit of time (1 U). The formula for calculating enzyme activity (U) is as follows:
In the formula:
- -
C represents the concentration of the substrate 2′-deoxyribonucleoside (mmol/L).
- -
V1 is the volume of the reaction system (mL).
- -
V2 is the volume of enzyme added (mL).
- -
T is the reaction time (min).
The calculation method for relative activity (Relative activity%) is as follows:
In the formula:
- -
P1 represents the peak area of the substrate 2′-deoxyribonucleoside after the reaction.
- -
C represents the peak area of the substrate 2′-deoxyribonucleoside in the standard.
- -
M1 and M2 are the dilution factors for the substrate (dNTP) standard during sample preparation.
- -
V3 and V4 are the respective volumes for sample injection (mL).
The reaction is as depicted in
Figure 4.
4.6. Immobilization Condition
1 mL of activated Ni2+-NTA affinity chromatography medium is packed into a 10 mL gravity column. Mix it with 3 mL of crude LrNDT enzyme solution in immobilization buffer (50 mmol/L PBS buffer). Incubate gently on a shaker at 4°C for 1 hour to allow better binding of the enzyme to the carrier. After incubation, collect the flow-through liquid. Check if the carrier is saturated with the enzyme using SDS-PAGE. Wash off other non-specific proteins on the carrier with a 1 mol/L NaCl solution, and then wash the immobilized enzyme with immobilization buffer three times to remove loosely bound target proteins. Store the immobilized enzyme at 4°C.
4.7. Measurement of Immobilized LrNDT Activity
Using the detection conditions mentioned above, add 3 mL of reaction solution to the immobilized carrier with LrNDT. Place it on a shaker at 40°C with gentle shaking for 10 minutes to allow the reaction to proceed fully. The activity of immobilized enzyme units (U) is defined as the amount of enzyme required to produce 1 mmol of 2′-deoxyadenosine from 1 mmol of 2′-deoxyribonucleoside under reaction conditions in 1 minute (mL). Use the formula from section 3 to calculate immobilized enzyme activity and further determine the comparison of enzyme activity before and after immobilization.
4.8. Effect of Temperature on Immobilized LrNDT Activity
In 2 mL of immobilized enzyme, add 6 mmol 2′-deoxyribonucleoside and 9 mmol adenine as substrate. Add 50 mmol/L PBS buffer to the reaction system. Place the reaction systems in parallel on a shaker at 20°C, 25°C, 30°C, 35°C, 40°C, and 45°C, and shake gently for 20 minutes. Then, take the supernatant for HPLC analysis to check the synthesis of the product, 2′-deoxyadenosine.
4.9. Effect of pH on Immobilized LrNDT Activity
With the same reaction concentrations as mentioned above, after adding 50 mmol/L PBS buffer, adjust the pH of the reaction systems to 6.0, 6.5, 7.0, and 7.5. Place them on a shaker at 40°C and shake gently for 10 minutes. Analyze the results using HPLC.
4.10. Reusability of Immobilized LrNDT
Prepare a reaction solution containing 6 mmol 2′-deoxyribonucleoside and 9 mmol adenine in pH 6.0 PBS buffer. The biocatalysis reaction was performed at 40°C, and the enzyme activity was measured for 24 h with a continuous flow rate of 1 mL/min and a column reactor volume of 20 mL and a column residence time of 20 minutes. Collect the reaction buffer each 100 minutes, dilute, and monitor the results using HPLC.
5. Conclusions
In summary, this article successfully achieved the immobilization of LrDNT enzyme, which not only achieved purification and efficient reuse of the enzyme, but also significantly improved its stability, enabling it to maintain excellent performance under various reaction conditions. By introducing the histidine tag, we successfully combined the LrDNT enzyme with the nickel chelated agarose affinity gel medium. Through the one-step purification and immobilization process, we achieved an enzyme recovery rate of up to 99%, while retaining about 97% of the initial enzyme activity, opening up a new way for the synthesis of nucleoside analogs.
Further studies showed that the immobilized LrDNT enzyme maintained high enzyme activity at temperatures ranging from 20-45°C and pH values ranging from 6.0-7.5, providing broad space for its application in industrial production. In addition, we also applied the immobilized enzyme reactor IMER to the continuous flow of 2′-deoxyadenosine synthesis reaction. The results showed that even after 24 hours of continuous operation, the enzyme activity remained above 90%, fully demonstrating its excellent stability and durability.
This research not only provides a practical method for the industrial production of nucleoside analogues, but also provides valuable experience and beneficial references for the application of other enzyme immobilization methods. Through immobilization technology, we can achieve efficient utilization and improved stability of enzymes, thus promoting the industrialization of biocatalytic reactions. Looking forward to the future, we have reason to believe that with the continuous development and improvement of immobilization technology, more enzymes will be successfully immobilized and applied in industrial production, making greater contributions to the sustainable development of human society.
Author Contributions
Conceptualization, M.L. and T.F.; methodology, C.D.; formal analysis, M.Q.; investigation, X.H.,Y.L., R.L. and Y.X.; writing—original draft preparation, M.L.; writing—review and editing, K.Z.; All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Natural Science Foundation of China (31601191).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- N. Li, T. J. Smith, M. H. Zong. Biocatalytic transformation of nucleoside derivatives. Biotechnol Adv 2010, 28, 348–366. [Google Scholar] [CrossRef] [PubMed]
- J. Müller, F. A. Polonius, E. Freisinger, E. Gil Bardají. X-ray crystallographic study of several 2′-deoxy-beta-D-ribonucleosides with 1-deazapurine-derived aglycones. Carbohydrate research 2008, 343, 397–403. [Google Scholar] [CrossRef] [PubMed]
- X. Jia, D. Schols, C. Meier. Antiviral Activity of Lipophilic Nucleoside Tetraphosphate Compounds. J Med Chem 2024, 67, 2864–2883. [Google Scholar] [CrossRef] [PubMed]
- N. Higashi-Kuwata et al. Identification of a novel long-acting 4’-modified nucleoside reverse transcriptase inhibitor against HBV. J Hepatol 2021, 74, 1075–1086. [Google Scholar] [CrossRef] [PubMed]
- D. Das et al. Can plant-derived anti-HIV compounds be used in COVID-19 cases? Med Hypotheses 2022, 110926, 166. [Google Scholar]
- J. R. Hanrahan, D. W. Hutchinson. The enzymatic synthesis of antiviral agents. J Biotechnol 1992, 23, 193–210. [Google Scholar] [CrossRef] [PubMed]
- W. Ye et al. Ethenoguanines undergo glycosylation by nucleoside 2′-deoxyribosyltransferases at non-natural sites. PLoS ONE 2014, 9, e115082. [Google Scholar]
- K. F. Hellendahl et al. Optimized Biocatalytic Synthesis of 2-Selenopyrimidine Nucleosides by Transglycosylation*. Chembiochem: A European journal of chemical biology 2021, 22, 2002–2009. [Google Scholar] [CrossRef] [PubMed]
- Y. J. Yoo, K. H. Choi, B. K. Kim, S. S. Choi, E. S. Kim. Isolation and Characterization of Engineered Nucleoside Deoxyribosyltransferase with Enhanced Activity Toward 2′-Fluoro-2′-Deoxynucleoside. Journal of microbiology and biotechnology 2022, 32, 1041–1046. [Google Scholar] [CrossRef] [PubMed]
- R. Cardinaud, J. Holguin. Nucleoside deoxyribosyltransferase-II from Lactobacillus helveticus Substrate specificity studied. Pyrimidine bases as acceptors. Biochimica et biophysica acta 1979, 568, 339–347. [Google Scholar] [CrossRef] [PubMed]
- W. S. Macnutt. The enzymically catalysed transfer of the deoxyribosyl group from one purine or pyrimidine to another. Biochem J 1952, 50, 384–397. [Google Scholar] [CrossRef] [PubMed]
- R. A. Sheldon, S. van Pelt. Enzyme immobilisation in biocatalysis: Why, what and how. Chemical Society reviews 2013, 42, 6223–6235. [Google Scholar] [CrossRef] [PubMed]
- R. A. Sheldon, A. Basso, D. Brady. New frontiers in enzyme immobilisation: Robust biocatalysts for a circular bio-based economy. Chemical Society reviews 2021, 50, 5850–5862. [Google Scholar] [CrossRef] [PubMed]
- N. Laurent, R. Haddoub, S. L. Flitsch. Enzyme catalysis on solid surfaces. Trends in biotechnology 2008, 26, 328–337. [Google Scholar] [CrossRef] [PubMed]
- S.D. Glover, C. Tommos, A. Quick and Colorful Method to Measure Low-Level Contaminations of Paramagnetic Ni(2+) in Protein Samples Purified by Immobilized Metal Ion Affinity Chromatography. Methods Enzymol 2019, 614, 87–106. [Google Scholar]
- F. Rinaldi et al. Immobilized enzyme reactors based on nucleoside phosphorylases and 2′-deoxyribosyltransferase for the in-flow synthesis of pharmaceutically relevant nucleoside analogues. Bioresour Technol 2020, 123258, 307. [Google Scholar]
- J. Fernandez-Lucas et al. Magnetic chitosan beads for covalent immobilization of nucleoside 2′-deoxyribosyltransferase: Application in nucleoside analogues synthesis. J Ind Microbiol Biotechnol 2013, 40, 955–966. [Google Scholar] [CrossRef] [PubMed]
- Fresco-Taboada et al. Nucleoside 2′-deoxyribosyltransferase from psychrophilic bacterium Bacillus psychrosaccharolyticus--preparation of an immobilized biocatalyst for the enzymatic synthesis of therapeutic nucleosides. Molecules (Basel, Switzerland) 2014, 19, 11231–11249. [Google Scholar] [CrossRef]
- J. Del Arco, J. Jordaan, V. Moral-Dardé, J. Fernández-Lucas. Sustainable production of nucleoside analogues by a high-efficient purine 2′-deoxyribosyltransferase immobilized onto Ni(2+) chelate magnetic microparticles. Bioresource technology 2019, 121772, 289. [Google Scholar]
- C. W. Rivero et al. Green Production of Cladribine by Using Immobilized 2′-Deoxyribosyltransferase from Lactobacillus delbrueckii Stabilized through a Double Covalent/Entrapment Technology. Biomolecules 2021, 11. [Google Scholar]
- R. M. Daniel, M. J. Danson, R. Eisenthal, C. K. Lee, M. E. Peterson. The effect of temperature on enzyme activity: New insights and their implications. Extremophiles 2008, 12, 51–59. [Google Scholar] [CrossRef] [PubMed]
- B. L. Vallee, J. F. Riordan, Dynamics of local conformation and enzyme function. Ciba Found Symp 1977, 197-223.
- Q. Xiao et al. Characterization and immobilization of arylsulfatase on modified magnetic nanoparticles for desulfation of agar. International journal of biological macromolecules 2017, 94, 576–584. [Google Scholar] [CrossRef] [PubMed]
- J. Fernandez-Lucas, A. Fresco-Taboada, C. Acebal, I. de la Mata, M. Arroyo. Enzymatic synthesis of nucleoside analogues using immobilized 2′-deoxyribosyltransferase from Lactobacillus reuteri. Appl Microbiol Biotechnol 2011, 91, 317–327. [Google Scholar] [CrossRef]
- J. Fernandez-Lucas, C. Acebal, J. V. Sinisterra, M. Arroyo, I. de la Mata. Lactobacillus reuteri 2′-deoxyribosyltransferase, a novel biocatalyst for tailoring of nucleosides. Appl Environ Microbiol 2010, 76, 1462–1470. [Google Scholar] [CrossRef]
- Y. Asanomi, H. Yamaguchi, M. Miyazaki, H. Maeda. Enzyme-immobilized microfluidic process reactors. Molecules (Basel, Switzerland) 2011, 16, 6041–6059. [Google Scholar] [CrossRef] [PubMed]
- M. M. F. Ismail, M. S. Ayoup. Review on fluorinated nucleoside/non-nucleoside FDA-approved antiviral drugs. RSC Adv 2022, 12, 31032–31045. [Google Scholar] [CrossRef]
- X. Lin et al. Advance of structural modification of nucleosides scaffold. Eur J Med Chem 2021, 113233, 214. [Google Scholar]
- S. Nascimento et al. Developments in the chemistry and biology of 1,2,3-triazolyl-C-nucleosides. Arch Pharm (Weinheim) 2024, 357, e2300580. [Google Scholar] [CrossRef]
- Z. Ashkan et al. Immobilization of enzymes on nanoinorganic support materials: An update. Int J Biol Macromol 2021, 168, 708–721. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).