1. Introduction
Fishmeal (FM) is undoubtedly an excellent source of protein in aquafeeds due to its essential amino acid (EAA) profile, palatability, and other remarkable properties [
1]. However, in recent years, we have observed a steady decline in the inclusion levels of FM in aquaculture feeds due to limited supply, increasing price, and ethical concerns [
1,
2]. Currently, FM is widely regarded as no longer a sustainable and essential component of fish feed.
Therefore, alternative, and more sustainable protein sources are needed for the development and promotion of sustainable aquaculture. Since the beginning of this century, considerable research efforts have been made to find alternative proteins, focusing mainly on plant-based ingredients such as soybean meal, corn gluten meal and rapeseed meal. Plant proteins are used extensively in feed formulations and will continue to be important raw materials in aquafeeds, although they contain several antinutritional factors (ANFs), complex indigestible carbohydrates, and low EAAs, which can have negative side effects on feed intake, digestion and absorption of nutrients, and fish health [
3].
A real alternative to vegetable proteins as FM substitutes are animal by-products, which have similar properties to FM in terms of AA content, digestibility, palatability, and lack of ANFs. Since June 1, 2013, the European Commission has approved processed animal proteins from non-ruminants in aquafeed [
4]. Before this date, their use was prohibited due to restrictive regulations to prevent the spread of bovine spongiform encephalopathy [
5]. Among the by-products of land animals, poultry by-product meal (PBM), which consists of ground, rendered, and cleaned parts of the carcasses of slaughtered poultry including legs, necks, intestines, and undeveloped eggs, is the most economical and widely used component of aquafeed [
6]. PBM is a high quality palatable and digestible protein source due to its content of EAA (except lysine and methionine), fatty acids, vitamins, and minerals [
7].
To date, the suitability of PBM inclusion has been reported for several marine species [
8]. PBM has successfully partially or completely replaced FM in the diet of a number of marine fish species, including gilthead seabream (
Sparus aurata) [
9,
10,
11,
12], black seabass (Centropristis striata) [
13], and red seabream (
Pagrus major) [
14]. In particular, in gilthead seabream, up to 100% of FM replacement (corresponding to 38% of inclusion) was achieved with lysine and methionine supplementation [
12,
15].
Optimal growth performance has even been obtained in salmonids, including rainbow trout (Oncorhynchus mykiss) and Atlantic salmon (Salmo salar), fed feeds with high PBM content [
16,
17,
18,
19,
20]. However, it should be noted that differences in maximum dietary PBM inclusion rate are highly dependent on fish species, PBM quality, and overall feed formulation. Although some studies suggest that PBM can completely replace conventional protein sources, most studies recommend partial replacement of FM to maintain the nutritional balance, palatability, and digestibility of the feed [
8,
9,
10,
11,
17,
21,
22].
Moreover, mixtures of alternative protein sources are usually preferred over a single protein source to replace FM in fish feed, especially in FM-free formulations. For example, mixtures of poultry and insect meal are being investigated as alternatives to FM in aquafeeds. Like PBM, insect meal is a promising alternative to FM in aquafeed as it has several advantages. Insects have a high protein (34-74% DM) and lipid (10-30% DM) content, balanced EAA, and a good content of vitamins (B12) and minerals (iron and zinc); they also contain several bioactive compounds such as chitin, fatty acids, and antimicrobial peptides [
23].
The use of insects in animal feed is strictly regulated by the European Union (EU). Regulation no 2001/999 (Annex IV), amended by Regulation 2017/893 (Annex X), authorises the use of seven insect species in feed for aquaculture, namely the common housefly (Musca domestica), the black soldier fly (Hermetia illucens), lesser mealworm (Alphitobius diaperinus), mealworm (Tenebrio molitor), house cricket (Acheta domesticus), field cricket (Gryllus assimilis), and banded cricket (Gryllodes sigillatus). As of November 2021 (Regulation 2021/1925), the list was extended to eight authorized species with the inclusion of the silkworm (Bombyx mori). Among them, the black soldier fly and the mealworm are the most commonly used insect species in aquafeed [
24,
25].
Recent studies have shown that an effective replacement of FM in aquafeed can be achieved by combining black soldier fly (BSF) larval meal with PBM. In rainbow trout fed a plant-based diet without FM, a high proportion of plant protein-rich components in the diet with both BSF and PBM resulted in better growth and gut/liver health outcomes than BSF and PBM alone [
20]. Similarly, BSF and PBM successfully replaced plant proteins in a diet without FM in gilthead seabream, improving nutrient uptake and promoting gut health [
26].
In addition, the combination of BSF and PBM in particular restored the gut microbiota of fish negatively affected by a plant-based diet by improving the richness of bacterial species and the abundance of beneficial bacteria [
27,
28].
The prebiotic effect of chitin, the indigestible polysaccharide that makes up the exoskeleton of insects, and insect meal containing chitin has been widely demonstrated in fish [
24,
29]. The addition of insect meal usually increases the amount of lactic acid bacteria (LAB) and the genus Bacillus, which are commonly used as probiotics in aquaculture. Finally, the gut microbiota of fish is known to be very sensitive to dietary manipulations [
30], and since proteins are the most important nutrient, they have a major impact on the composition of the gut microbiota.
Accordingly, in the present study we tested the effect of partial replacement of FM by PBM alone or in combination with insect meal or exuviae meal, a chitin source from insect exoskeletons, in European seabass. A multidisciplinary approach was used in which growth performance, gut and liver morphology, gut microbiota and gut volatile short chain fatty acid production, were evaluated.
4. Discussion
The drive to find alternative and more sustainable protein sources to replace FM stems from the need to alleviate the pressure on global marine resource depletion and also to contain the cost of aquaculture production, as protein is the most expensive nutrient in fish diets, accounting for 30-50% of the total cost.
Various plant protein sources have traditionally been used to partially or completely replace FM in aquafeeds. However, nowadays animal protein sources are considered the best alternative to FM due to their higher protein and lipid content, better amino acid profile, and palatability [
47,
48].
The results of this study seem to go in this direction, confirming that PBM together with insect or exuviae meal successfully meet the requirements for adequate growth in seabass. This result confirms what has already been reported in the literature, according to which the optimal substitution rate of FM by PBM is between 25% and 50% for most marine carnivorous animals [
8]. Indeed, both formulations containing PBM were well accepted and at the end of the feeding trial, the fish doubled their weight, regardless of the diet.
However, only minor differences were found between the control group and the experimental feeding groups PM20 and PM25. The growth performance determined from the final weight showed that the fish group fed with the 20% PBM diet performed worse than the group fed with the control diet, resulting in a lower final weight compared to the control, but did not differ from the PM25 group. In fact, in marine fish, such as seabream (
Sparus aurata) and red seabream (
Pagrus major), there is evidence that even the complete replacement of FM with PBM had no negative effect on the growth parameters and productivity of the fish [
10,
14,
15].
The inclusion of PBM in the diet of freshwater fish has been shown to be successful as well. In particular, the present study is consistent with our previous results in rainbow trout. The growth data showed that trout fed a diet rich in PBM (55-70%) grew as well as fish fed a control diet rich in FM (37.3%) and free of PBM [
18].
The formulation of optimal diets for farmed fish species requires the application of different types of analyzes to verify their effects on the health status of the specimens, which cannot be limited to growth performance and feed efficiency alone. Histomorphological examination is a good biomarker for the assessment of fish welfare, especially in gut and liver histomorphology [
49]. No potential changes related to intestinal inflammation or hepatic lipid accumulation in response to FM replacement were detected in the histological examinations, suggesting that PBM in combination with insect and exuviae meal was well tolerated by seabass. In agreement with our results, Pleić and colleagues [
28] showed that partial replacement of plant protein diet with insect and PBM could even mitigate the negative effects of plant proteins on the proximal and distal gut of seabass by significantly improving all gut morphometric parameters, the degree of vacuolization and cellular infiltration. The health and integrity of the intestinal epithelium are critical to nutrient absorption and fish health, as damage to the gut can lead to immune dysfunction, reduced disease resistance, loss of appetite and slow growth.
In addition to the gut, the liver is also considered a valuable marker for nutritional pathologies, as it plays a key role in energy metabolism and storage, as well as in immune defense and detoxification [
50,
51]. As for the present study, the histological results indicate a favorable liver health status, with a moderate accumulation of lipids in hepatocytes in all fish, regardless of diet. In contrast, Donadelli et al. [
52] recently observed a significant accumulation of hepatocyte lipids associated with marked histological changes indicative of an incipient steatotic state in seabass fed a FM-free diet in which 40% of the plant protein was replaced with insect or poultry by-product. These changes appeared to be slightly attenuated when insect meal and PBM were combined to partially replace plant proteins in the diet. The data available in the literature indicate species-specific differences in the responses to alternative protein-rich feed components. For example, the inclusion of
Hermetia illucens and PBM in diets containing no FM resulted in improved gut and liver health in gilthead seabream and rainbow trout [
20,
26,
52].
Interestingly, a higher amount of SCFAs was found in the fecal samples of fish fed experimental diets containing insect-derived raw material compared to controls. Insect meal and especially exuviae are considered valuable sources of chitin. Chitin is an insoluble dietary fiber consisting of β-1,4-poly-N-acetyl-D-glucosamine and can be used as a substrate for bacterial fermentations leading to the production of acetate, propionate, and butyrate as the main end products with positive effects on gut health [
24,
53]. Indeed, chitin and its deacetylated derivative chitosan are bioactive compounds which can positively influence the health and performance of farmed fish. In the present study, the highest amount of propionate and butyrate was found in the gut of seabass fed with exuviae meal. Accordingly, trout fed with pupal exuviae meal had the highest content of SCFAs, especially butyrate, in their feces [
29]. Butyrate is well known to have anti-inflammatory properties and to promote fish intestinal health, barrier function, and mucosal immunity in fish. Therefore, its increased production in the fish gut should be considered a desirable effect [
54,
55,
56].
As a prebiotic, chitin may also increase the biodiversity of the gut microbiota by promoting the proliferation of beneficial chitin-degrading bacteria, such as
Bacillus and
Paenibacillus, which have recently been isolated from the gastrointestinal tract of European seabass fed chitin-enriched diets [
57]. It is known that replacing FM with insect meal affects the biodiversity and number of gut bacteria. The partial replacement of FM with at least 10% insect meal had an important effect in modulating the transient gut microbial communities by increasing both butyrate-producing bacteria and beneficial lactic acid bacteria [
37,
58,
59,
60,
61]. Similarly, the ingestion of
H. illucens exuviae meal led to an enrichment of the gut microbiota with the families Bacillaceae, Staphylococcaceae, Paenibacillaceae, and Brevibacteriaceae in seabass and rainbow trout [
29,
58]. Unlike previous studies, the proportion of insect or exuviae meal utilized in the present study was not sufficient to promote the proliferation of beneficial bacteria. In fact, the gut microbiota of PM20 and PM25 seabass was characterized by an increase in the Proteobacteria/Fimicutes ratio compared to the FM diet-fed controls. A similar increase in the Proteobacteria fraction, mainly represented by Gammaproteobacteria, was previously reported in trout fed diets containing a high proportion of alternative terrestrial animal proteins (>50%), mainly represented by PBM [
18]. Accordingly, the trout gut microbiota was characterized by a high abundance of bacterial genera belonging to the class of Gammaproteobacteria, such as
Vibrio,
Pasteurella, and
Proteus. Also, at the genus level, PM20 and PM25 fish showed an enrichment of
Vibrio and
Photobacterium genera in their gut, which are normally considered potential pathogens for fish. Similarly, we found an increase of the genus
Photobacterium in the gut microbiota of trout fed a diet containing 20% of head shrimp meal, another chitin-rich ingredient [
29]. However, it is also true that this genus includes several chitinase-producing bacterial species [
62,
63]. In contrast, it was recently reported that seabass fed a plant-based diet supplemented with a combination of 10% H.
illucens meal and 30% PBM showed increased abundance of the phylum Firmicutes, particularly the beneficial genera
Lactobacillus and
Bacillus in their gut [
28].
In relation to the present study, the genus
Lactobacillus decreased significantly in the gut of PM20 seabass, but not in PM25 compared to controls. In contrast to the exuviae meal, the amount of insect meal was not sufficient to attenuate the negative effects of PBM on the abundance of lactic acid bacteria. Similarly, in gilthead seabream, a 10% content of
H. illucens meal was not sufficient to increase the abundance of lactic acid bacteria in the gut [
37,
59].
The comparison of microbial profiles in the gut of PM20 and PM25 fish showed a positive association of the genus
Paenibacillus with insect meal. This result confirmed what was previously reported in rainbow trout and gilthead seabream in response to dietary
H. illucens meal ingestion [
37,
39,
59]. In contrast, Rangel et al. [
58] found that consumption of diets enriched with
H. illucens exuviae, but not insect meal, led to an increase in the genus
Paenibacillus in the gut of European seabass. This genus is of interest because it is considered a good probiotic candidate. In fact,
Paenibacillus shares several characteristics with members of the genus
Bacillus. It can produce antimicrobial and volatile organic compounds and degrade non-starch polysaccharides, and last, but not least, the bacteria of this genus have chitinolytic activity [
57,
58,
64,
65,
66]. The presence of chitinolytic bacteria is particularly important when feeding insect-derived ingredients to fish, as bacterial chitinases help to improve the digestibility of the feed [
53]. In rainbow trout, on the other hand, the addition of 1.6% pupal exuviae meal of
H. illucens to a diet containing 20% FM resulted in an enrichment of various bacteria genera, such as
Corynebacterium,
Bacillus,
Facklamia, and
Brevibacterium. These divergent results suggest that the relationship between dietary components and the gut microbiota are complex and species-specific.
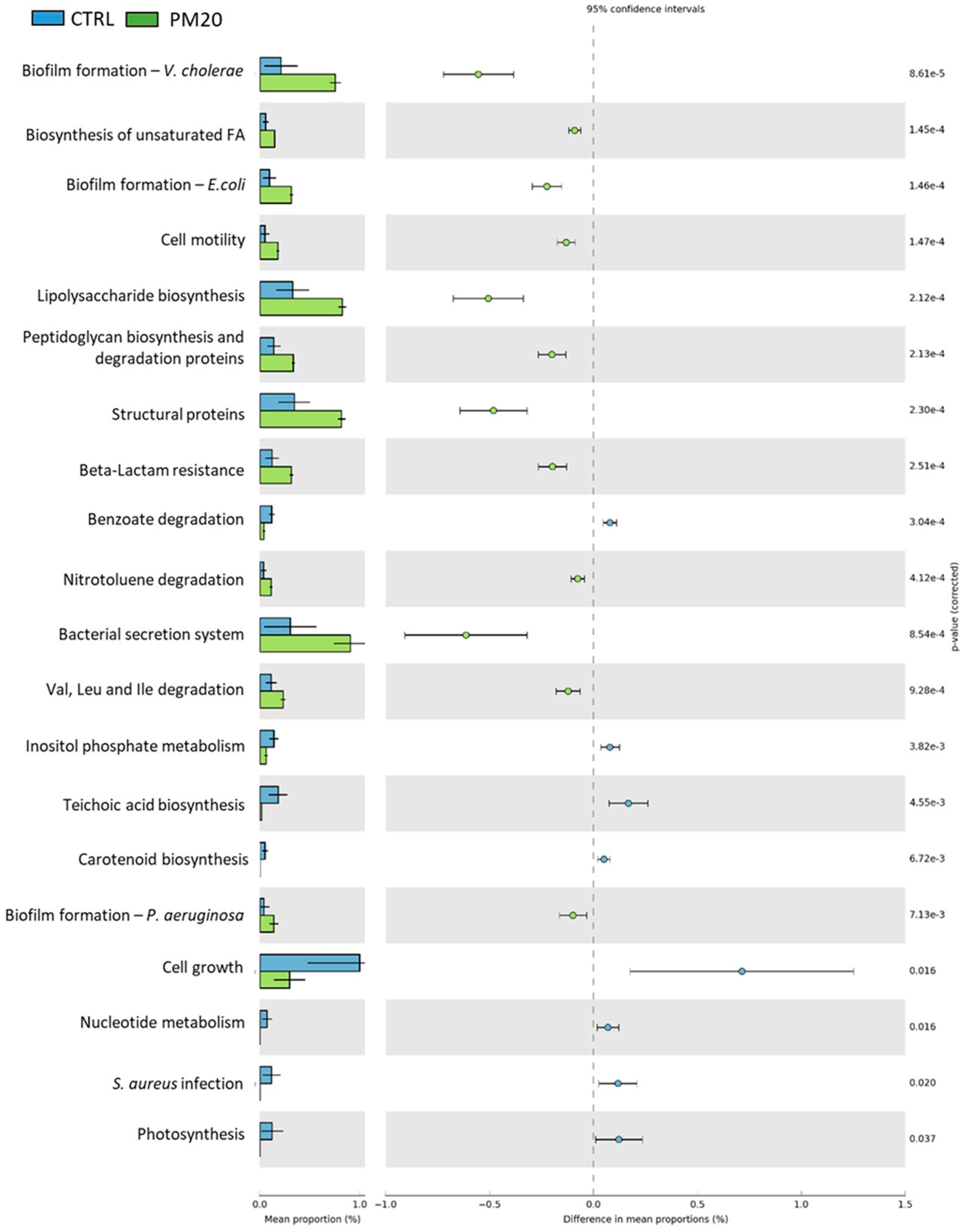
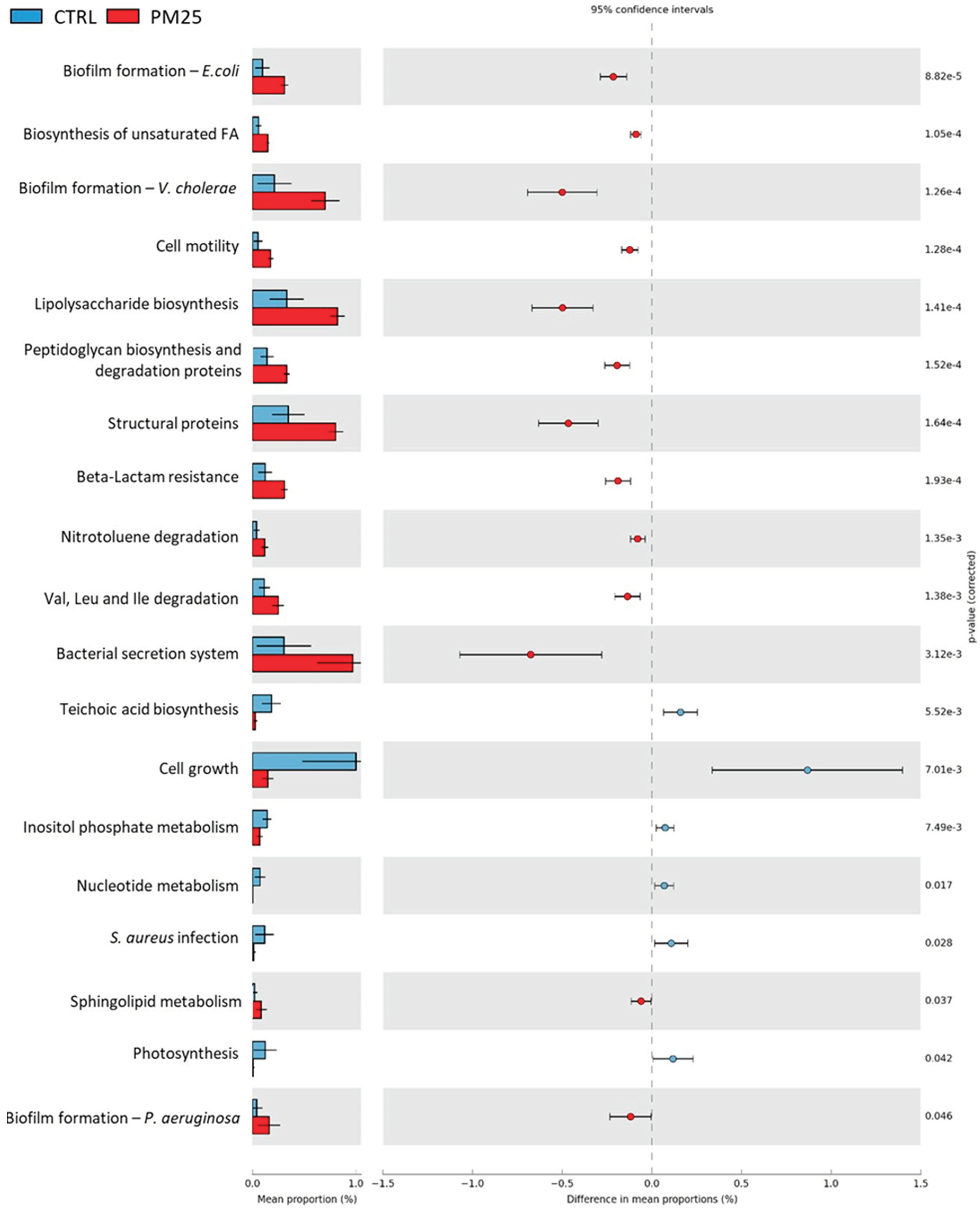
The predictive functional analysis PICRUSt showed large differences between the controls and the PBM-fed experimental groups. Compared to controls, fish fed PBM diets were associated with biofilm formation, peptidoglycan and lipopolysaccharide biosynthesis, structural proteins, bacterial secretion system, and unsaturated fatty acid biosynthesis. Much more interesting was the reduction of signaling pathways related to Staphylococcus aureus infection in PM20 and PM25 fish. This result is consistent with the lower amount of the genus Staphyloccoccus observed in these samples.
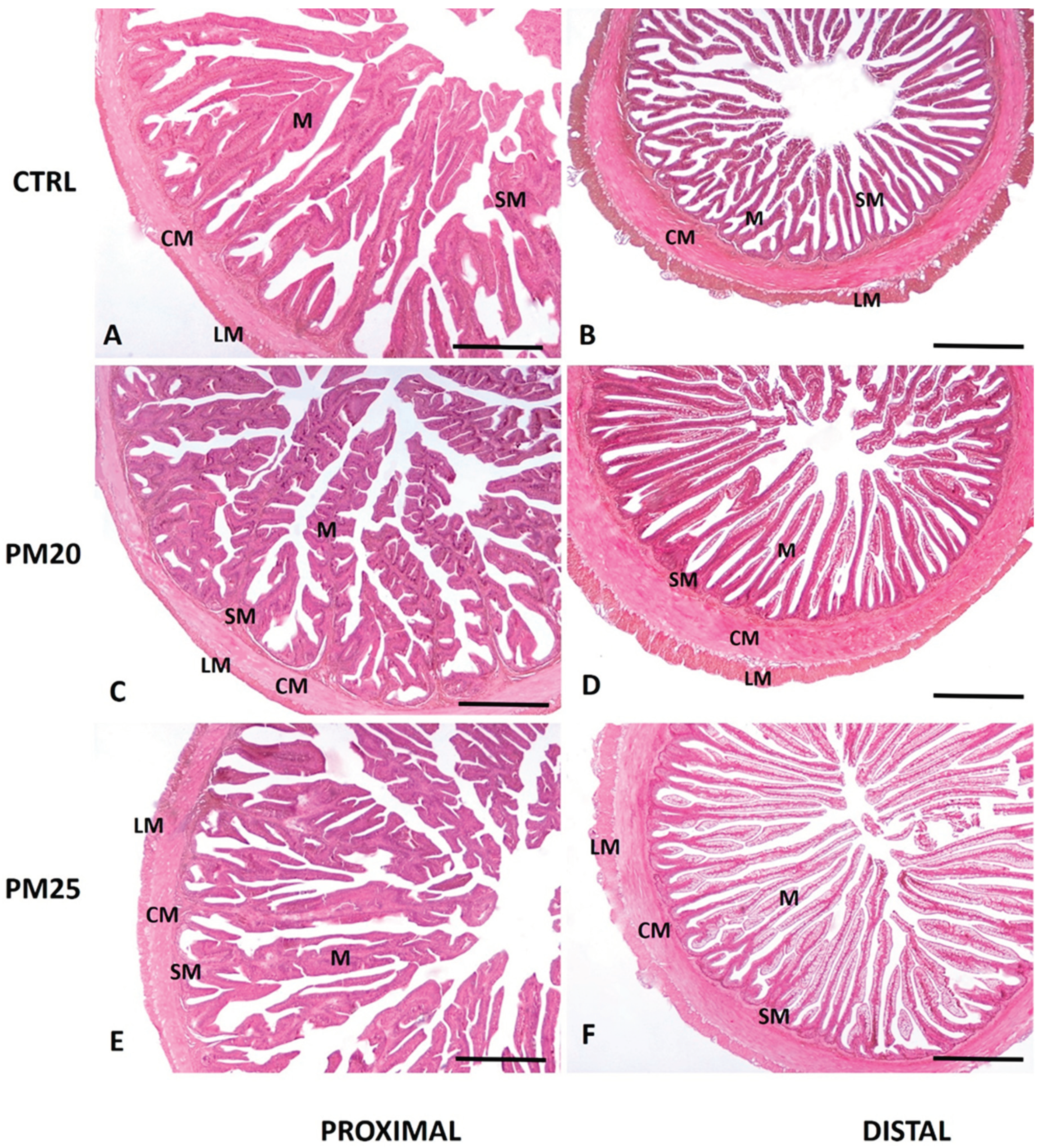
Figure 1.
Standard hematoxylin-eosin (H&E) histochemical analysis of proximal (panels A, C, E), and distal (panels B, D, F) seabass intestine. 5 µm cross sections were obtained from the proximal and distal paraffin-embedded intestine of CTRL (panels A and B), PM20 (panels C and D), and PM25 (panels E and F) fish. M, mucosa; SM, submucosa; CM, circular muscle layer; LM, longitudinal muscle layer. Scale bar = 1000 µm.
Figure 1.
Standard hematoxylin-eosin (H&E) histochemical analysis of proximal (panels A, C, E), and distal (panels B, D, F) seabass intestine. 5 µm cross sections were obtained from the proximal and distal paraffin-embedded intestine of CTRL (panels A and B), PM20 (panels C and D), and PM25 (panels E and F) fish. M, mucosa; SM, submucosa; CM, circular muscle layer; LM, longitudinal muscle layer. Scale bar = 1000 µm.
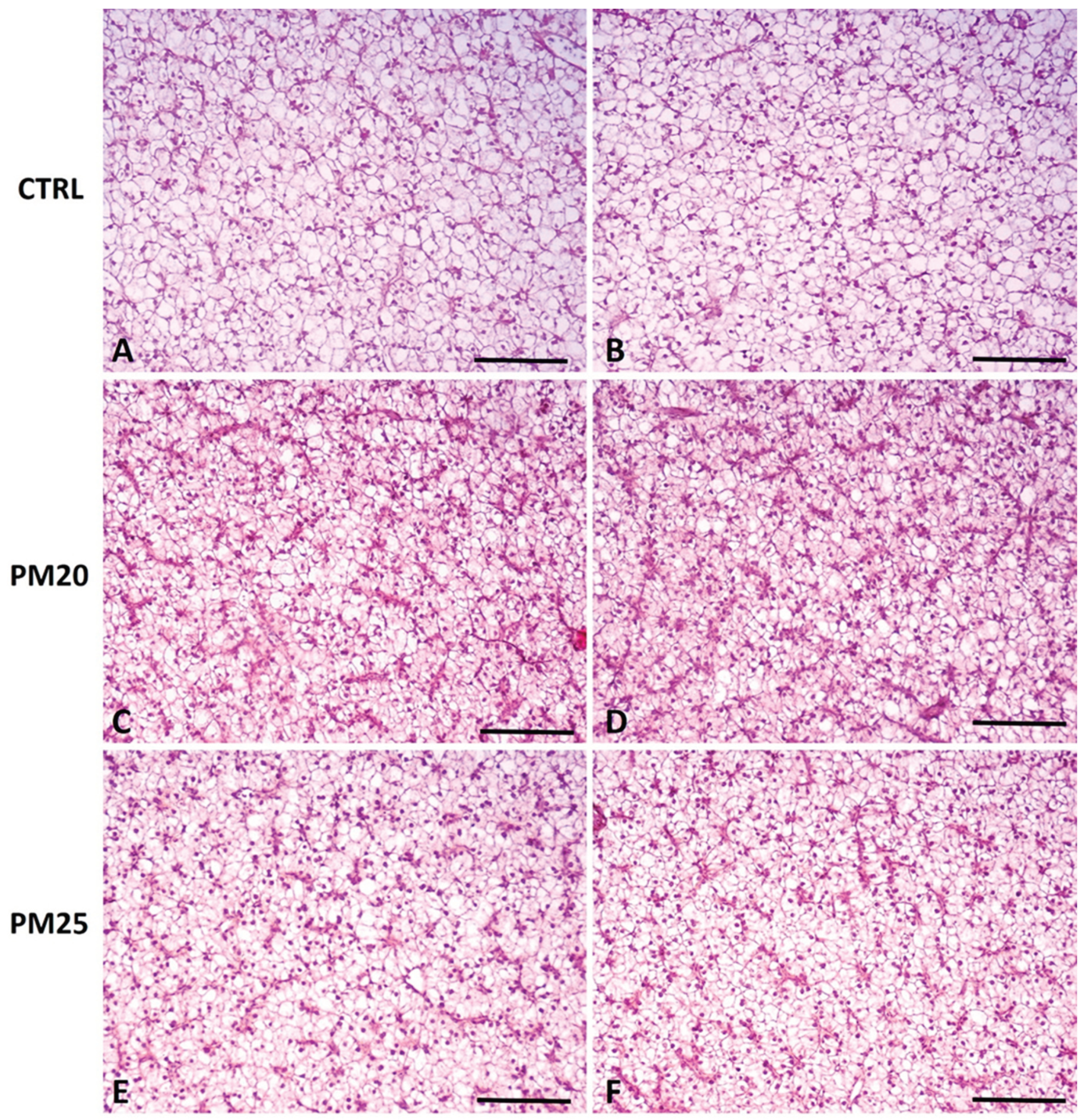
Figure 2.
Standard hematoxylin-eosin (H&E) histochemical analysis of liver of representative images of fish fed Control, PM20, and PM25 diets. 5 µm cross sections were obtained from the liver paraffin-embedded intestine of CTRL (panels A and B), PM20 (panels C and D), and PM25 (panels E and F) fish. Scale bar = 100 µm.
Figure 2.
Standard hematoxylin-eosin (H&E) histochemical analysis of liver of representative images of fish fed Control, PM20, and PM25 diets. 5 µm cross sections were obtained from the liver paraffin-embedded intestine of CTRL (panels A and B), PM20 (panels C and D), and PM25 (panels E and F) fish. Scale bar = 100 µm.
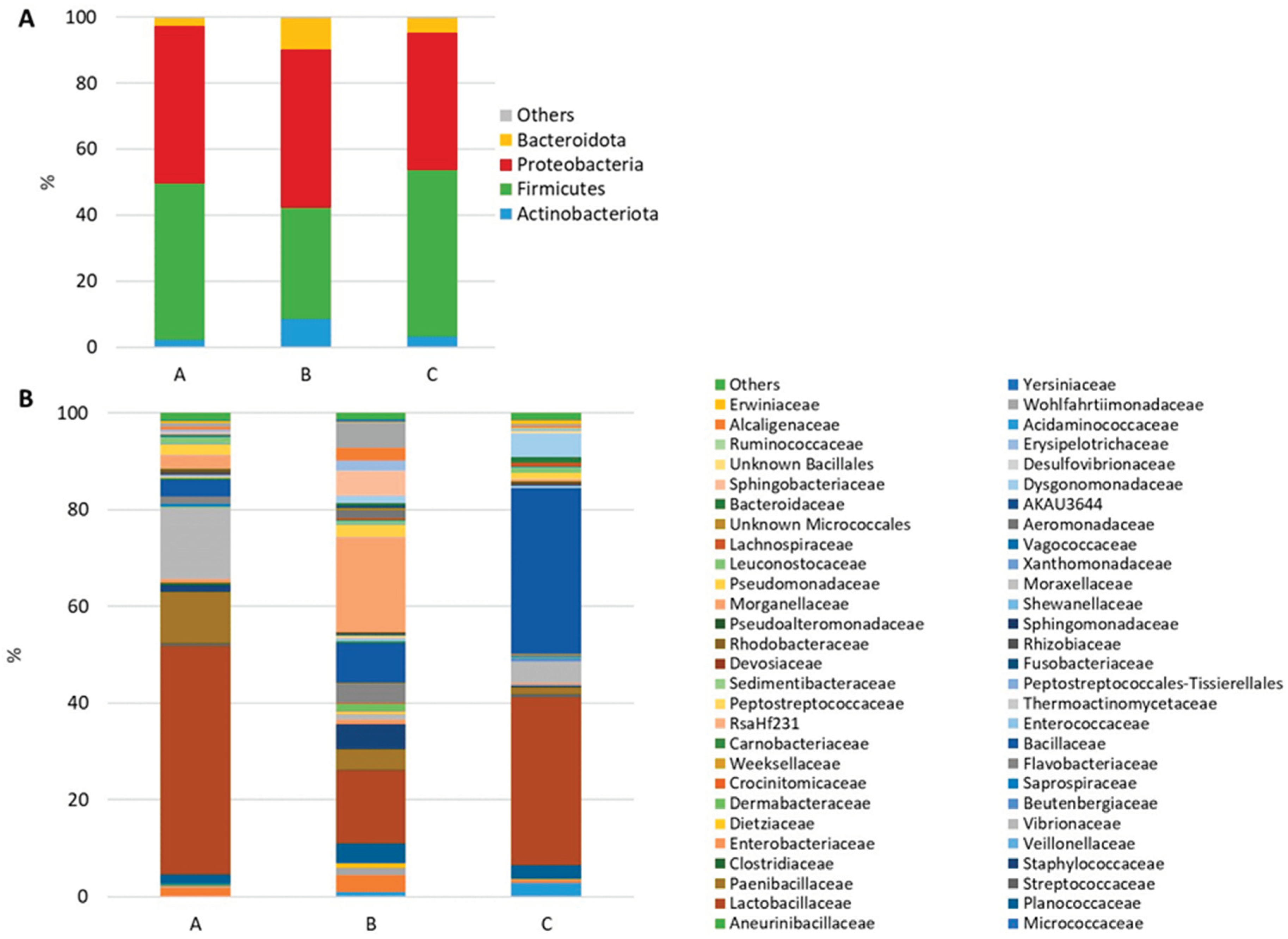
Figure 3.
Stacked bar chart of the mean relative abundances (%) of the most abundant classified bacterial phyla (A) and families (B) most frequently found in feed samples.
Figure 3.
Stacked bar chart of the mean relative abundances (%) of the most abundant classified bacterial phyla (A) and families (B) most frequently found in feed samples.
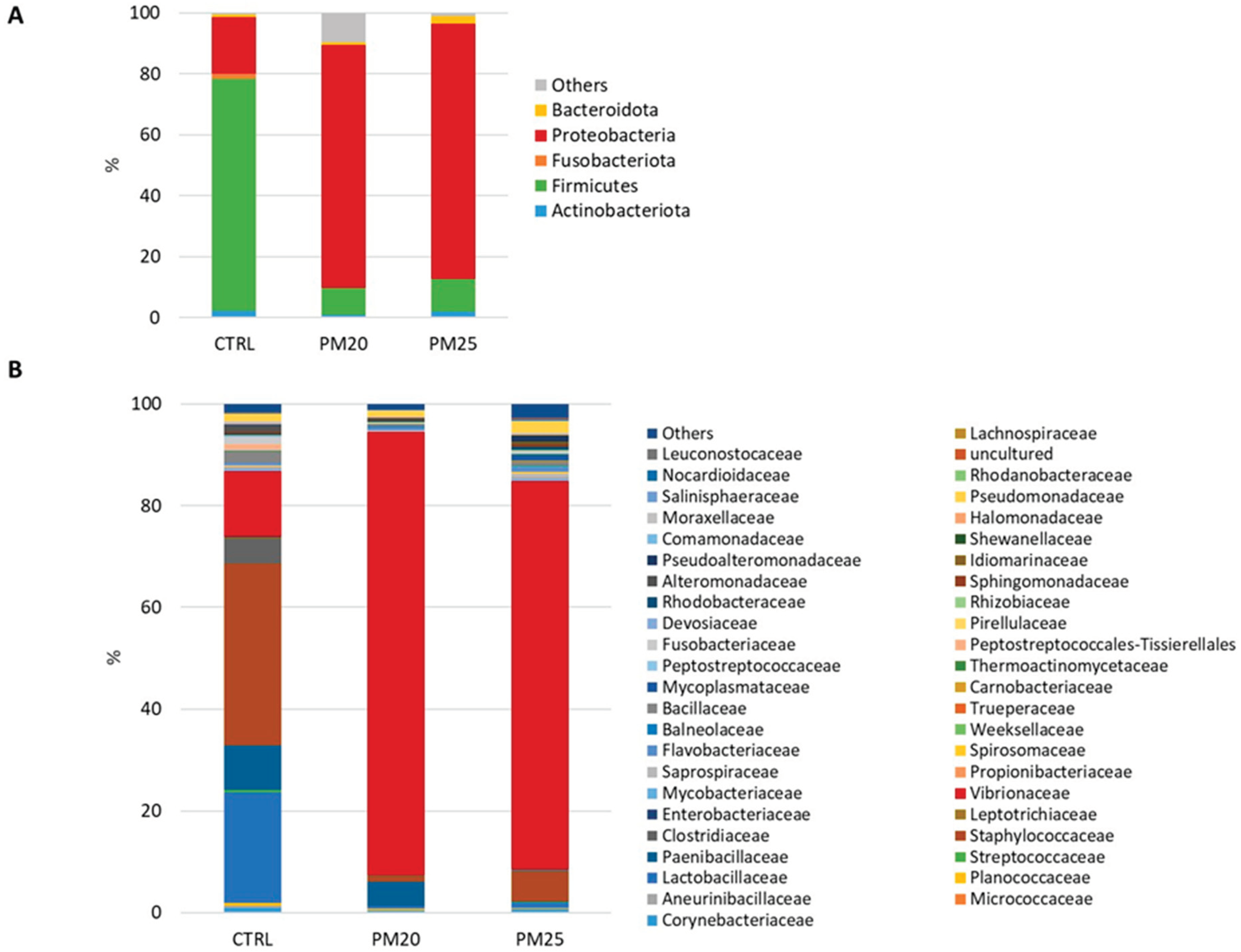
Figure 4.
Stacked bar chart of the mean relative abundances (%) of the most abundant classified bacterial phyla (A) and families (B) found in gut samples.
Figure 4.
Stacked bar chart of the mean relative abundances (%) of the most abundant classified bacterial phyla (A) and families (B) found in gut samples.
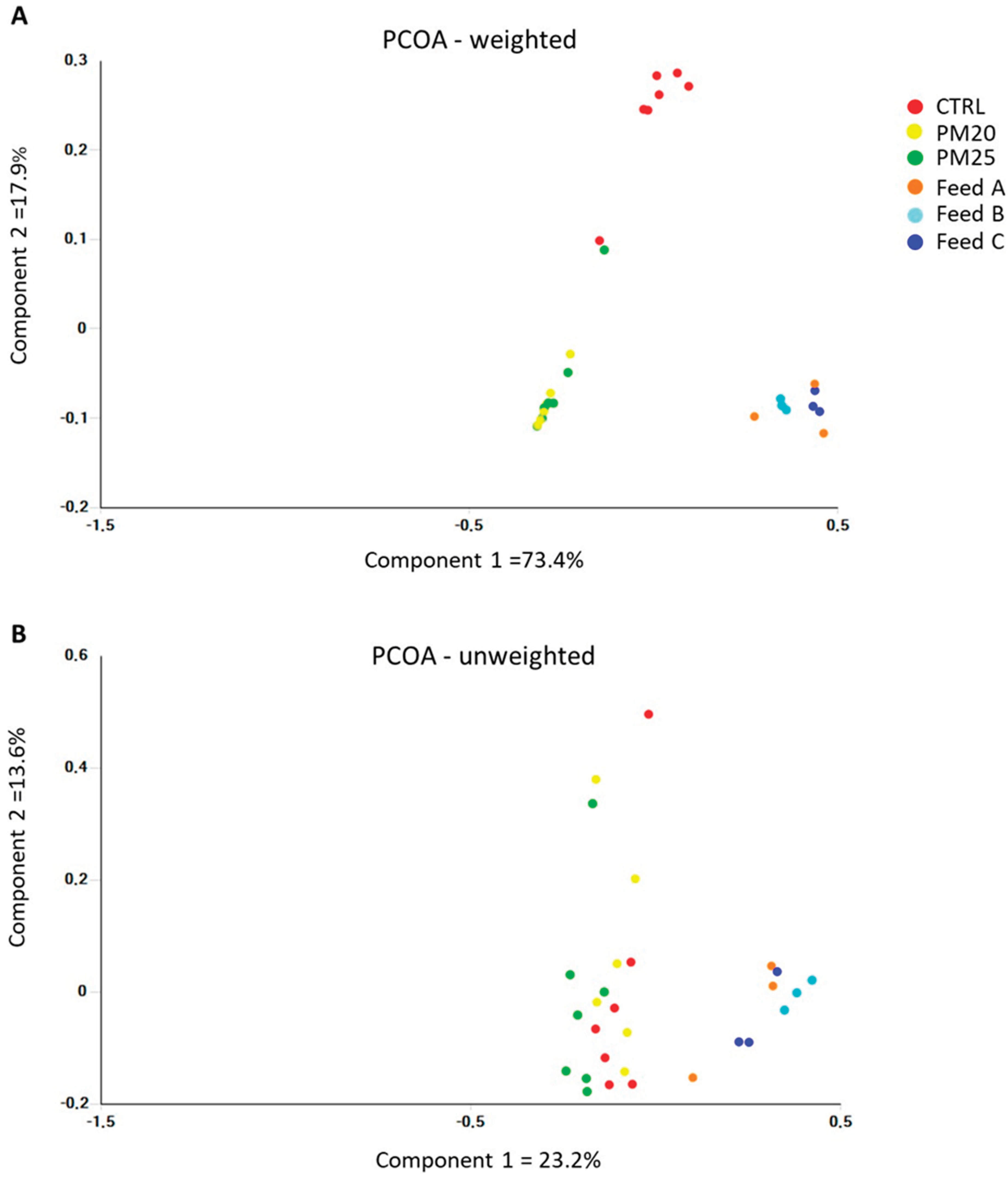
Figure 5.
Plot of principal coordinate analysis (PCoA) using weighted (A) and unweigheted (B) UniFrac distance matrices of gut microbial communities at the genus level.
Figure 5.
Plot of principal coordinate analysis (PCoA) using weighted (A) and unweigheted (B) UniFrac distance matrices of gut microbial communities at the genus level.
Figure 6.
Comparison of the relative abundance of the PICRUSt functional profile of the gut microbiota between the control and PM20 experimental groups. Only the predicted functional pathways that differ significantly (p < 0.05) are shown.
Figure 6.
Comparison of the relative abundance of the PICRUSt functional profile of the gut microbiota between the control and PM20 experimental groups. Only the predicted functional pathways that differ significantly (p < 0.05) are shown.
Figure 7.
Comparison of the relative abundance of PICRUSt functional profile of the gut microbiota between the control and PM25 experimental groups. Only the predicted functional pathways that differ significantly (p < 0.05) are shown.
Figure 7.
Comparison of the relative abundance of PICRUSt functional profile of the gut microbiota between the control and PM25 experimental groups. Only the predicted functional pathways that differ significantly (p < 0.05) are shown.
Table 1.
Diet formulations.
Table 1.
Diet formulations.
| INGREDIENTS (%) |
A |
B |
C |
| Fishmeal |
20 |
10 |
10 |
| Poultry by-product |
0 |
20 |
25 |
| Fish oil |
3 |
3 |
3 |
| Soybean meal |
11 |
11 |
11 |
| Exuviae meal |
0 |
0 |
0.5 |
| Insect meal |
0 |
5.0 |
0 |
| Corn gluten meal |
21.9 |
7.9 |
7.9 |
| Vital wheat gluten |
6 |
6 |
6 |
| Soy Protein Concentrate |
6 |
4 |
2 |
| Rapeseed meal |
11 |
11 |
12.5 |
| Soybean oil |
2 |
2 |
2 |
| Minerals and Vitamins |
2 |
2 |
2 |
| Premix antioxidants |
0.1 |
0.1 |
0.1 |
Table 2.
Proximate composition of diets.
Table 2.
Proximate composition of diets.
| |
A |
B |
C |
| Gross Energy (MJ/kg) |
17.34 |
17.55 |
17.58 |
| DE (MJ/kg) |
15.60 |
15.66 |
15.66 |
| DE (%) |
89.98 |
89.23 |
89.09 |
| Crude Fat (g/100 g) |
10.63 |
10.77 |
10.82 |
| Crude Protein (g/100 g) |
45.08 |
45.00 |
45.04 |
| DP (%) |
40.17 |
39.35 |
39.28 |
| Digestible Protein (%) |
89.12 |
87.44 |
87.22 |
| Fish Protein (%) |
13.20 |
6.60 |
6.60 |
| Animal Protein (%) |
13.20 |
22.98 |
23.85 |
| FP/TP (%) |
29.28 |
14.67 |
14.65 |
| DP/DE (mg/Kj or g/MJ) |
25.75 |
25.14 |
25.09 |
| AP/TP (%) |
29.28 |
51.06 |
52.96 |
| Fiber (g/100 g) |
3.03 |
3.41 |
3.04 |
| EI (g/100 g) |
35.48 |
32.29 |
33.01 |
| Amido (g/100 g) |
15.95 |
15.55 |
15.55 |
| NSP (g/100 g) |
22.57 |
21.15 |
20.51 |
| Protein-to-lipid ratio |
4.50 |
4.18 |
4.16 |
| Ash (g/100 g) |
6.38 |
7.53 |
8.09 |
| DE (kcal/kg) |
3729.34 |
3741.83 |
3742.61 |
| Crude En (kcal/kg) |
4304.66 |
4243.40 |
4233.13 |
Table 3.
Growth performance of seabass fed with three experimental diets. Different letters in the same row indicate a significant difference between the mean values (p<0.05; N(A) = 70; N(B) = 67; N(C) = 69).
Table 3.
Growth performance of seabass fed with three experimental diets. Different letters in the same row indicate a significant difference between the mean values (p<0.05; N(A) = 70; N(B) = 67; N(C) = 69).
| |
CTRL |
PM20 |
PM25 |
| IBW (g) |
52.88 ± 7.24 |
52.16 ± 6.14 |
52.04 ± 5.13 |
| FBW (g) |
135.88 ± 20.59a
|
120.26 ± 19.76b
|
128.37 ± 23.01ab
|
| SGR (% day-1) |
1.1 |
0.96 |
1.03 |
| FCR |
1.76 |
2.17 |
1.94 |
Table 4.
Intestinal morphometric parameters of seabass fed experimental diets.
Table 4.
Intestinal morphometric parameters of seabass fed experimental diets.
| |
CTRL |
PM20 |
PM25 |
p-value |
| Proximal intestine |
| ViH (μm) |
1008.78 ± 21.50 |
1064.90 ± 37.14 |
1035.87 ± 31.11 |
0.435 |
| ViW (μm) |
78.35 ± 2.57 |
83.18 ± 3.05 |
84.79 ± 3.42 |
0.300 |
| LPW (μm) |
28.44 ± 1.00 |
29.64 ± 1.42 |
27.3 ± 1.78 |
0.585 |
| SMT (μm) |
352.65 ± 14.93 |
365.94 ± 18.56 |
332.02 ± 9.22 |
0.266 |
| Distal intestine |
| ViH (μm) |
1008.78 ± 21.50 |
1064.90 ± 3 7.14 |
1035.87 ± 31.11 |
0.435 |
| ViW (μm) |
78.35 ± 2.57 |
83.18 ± 3.05 |
84.79 ± 3.42 |
0.300 |
| LPW (μm) |
28.44 ± 1.00 |
29.64 ± 1.42 |
27.3 ± 1.78 |
0.585 |
| SMT (μm) |
352.65 ± 14.93 |
365.94 ± 18.56 |
332.02 ± 9.22 |
0.266 |
Table 5.
Results of histological scoring of the liver.
Table 5.
Results of histological scoring of the liver.
| |
CTRL |
PM 20% |
PM 25% |
| HV |
2.00 |
1.67 |
2.00 |
| ND |
2.00 |
1.33 |
1.67 |
| NS |
1.00 |
1.00 |
1.00 |
| CH |
1.00 |
1.00 |
1.00 |
| Total |
6.00 |
5.00 |
5.67 |
Table 6.
Volatile SCFA content in fecal samples from three test groups. The results are given in mmol/L, mean ± SD (N = 6).
Table 6.
Volatile SCFA content in fecal samples from three test groups. The results are given in mmol/L, mean ± SD (N = 6).
| |
Acetate (C 2:0) |
Propionate (C 3:0) |
Butyrate (C 4:0) |
| CTRL |
14.67 ± 0.66b
|
5.93 ± 0.62c
|
2.13 ± 0.27b
|
| PM20 |
17.05 ± 0.94a
|
6.43 ± 0.46b
|
2.00 ± 0.13b
|
| PM25 |
15.19 ± 0.42b
|
7.41 ± 0.18a
|
3.03 ± 0.13a
|
Table 7.
Alpha diversity analysis of bacterial communities associated with feeds.
Table 7.
Alpha diversity analysis of bacterial communities associated with feeds.
| |
A |
B |
C |
p-value |
| Observed OTUs |
1325 ± 216b
|
1721 ± 33a
|
1224 ± 168b
|
0.019 |
| Chao1 |
1541 ± 250ab
|
1893 ± 47a
|
1382 ± 226b
|
0.047 |
| Faith-PD |
21.7 ± 4.9 |
23.9 ± 1.3 |
21.8 ± 3.3 |
> 0.05 |
| Shannon |
5.8 ± 0.4b
|
7.1 ± 0.1a
|
5.9 ± 0.2b
|
0.004 |
| Simpson |
0.92 ± 0.02 |
0.95 ± 0.00 |
0.92 ± 0.00 |
> 0.05 |
Table 8.
The list of bacterial genera that differ between experimental feeds A (control) and B.
Table 8.
The list of bacterial genera that differ between experimental feeds A (control) and B.
| PHYLUM |
CLASS |
ORDER |
FAMILY |
GENUS |
A (%) |
SD (%) |
B (%) |
SD (%) |
p-value |
| Actinobacteriota |
Actinobacteria |
Micrococcales |
AKAU3644 |
AKAU3644 |
0.01 |
0.01 |
0.64 |
0.02 |
0.0000 |
| Actinobacteriota |
Actinobacteria |
Micrococcales |
Dermabacteraceae |
Brachybacterium |
0.16 |
0.06 |
1.44 |
0.07 |
0.0001 |
| Actinobacteriota |
Actinobacteria |
Actinomycetales |
Actinomycetaceae |
Actinomyces |
0.19 |
0.11 |
1.20 |
0.10 |
0.0006 |
| Actinobacteriota |
Actinobacteria |
Micrococcales |
Brevibacteriaceae |
Brevibacterium |
0.28 |
0.13 |
1.70 |
0.06 |
0.0012 |
| Actinobacteriota |
Actinobacteria |
Micrococcales |
Microbacteriaceae |
Leucobacter |
0.06 |
0.02 |
0.43 |
0.04 |
0.0019 |
| Actinobacteriota |
Actinobacteria |
Micrococcales |
Micrococcaceae |
Enteractinococcus |
0.05 |
0.02 |
0.19 |
0.01 |
0.0023 |
| Actinobacteriota |
Actinobacteria |
Corynebacteriales |
Dietziaceae |
Dietzia |
0.06 |
0.01 |
0.66 |
0.07 |
0.0070 |
| Actinobacteriota |
Actinobacteria |
Corynebacteriales |
Corynebacteriaceae |
Corynebacterium |
1.62 |
0.74 |
3.72 |
0.19 |
0.0497 |
| Bacteroidota |
Bacteroidia |
Bacteroidales |
Dysgonomonadaceae |
Dysgonomonadaceae |
0.03 |
0.01 |
0.36 |
0.02 |
0.0000 |
| Bacteroidota |
Bacteroidia |
Sphingobacteriales |
Sphingobacteriaceae |
Sphingobacterium |
0.30 |
0.13 |
2.62 |
0.10 |
0.0001 |
| Bacteroidota |
Bacteroidia |
Flavobacteriales |
Flavobacteriaceae |
Myroides |
1.06 |
0.23 |
3.89 |
0.13 |
0.0004 |
| Bacteroidota |
Bacteroidia |
Bacteroidales |
Dysgonomonadaceae |
Dysgonomonas |
0.17 |
0.12 |
1.33 |
0.07 |
0.0009 |
| Bacteroidota |
Bacteroidia |
Sphingobacteriales |
Sphingobacteriaceae |
Pedobacter |
0.02 |
0.02 |
0.13 |
0.02 |
0.0034 |
| Bacteroidota |
Bacteroidia |
Flavobacteriales |
Flavobacteriaceae |
Ulvibacter |
0.02 |
0.01 |
0.38 |
0.05 |
0.0059 |
| Bacteroidota |
Bacteroidia |
Bacteroidales |
Bacteroidaceae |
Bacteroides |
0.04 |
0.02 |
0.51 |
0.07 |
0.0066 |
| Desulfobacterota |
Desulfovibrionia |
Desulfovibrionales |
Desulfovibrionaceae |
Desulfovibrio |
0.00 |
0.01 |
0.14 |
0.01 |
0.0002 |
| Firmicutes |
Clostridia |
Peptostreptococcales-Tissierellales |
Sedimentibacteraceae |
Sedimentibacter |
0.01 |
0.01 |
0.11 |
0.00 |
0.0007 |
| Firmicutes |
Bacilli |
Bacillales |
Bacillaceae |
Pseudogracilibacillus |
0.70 |
0.23 |
2.72 |
0.18 |
0.0008 |
| Firmicutes |
Bacilli |
Lactobacillales |
Enterococcaceae |
Enterococcus |
0.10 |
0.05 |
0.63 |
0.06 |
0.0009 |
| Firmicutes |
Bacilli |
Staphylococcales |
Staphylococcaceae |
Staphylococcus |
1.43 |
0.44 |
5.38 |
0.23 |
0.0014 |
| Firmicutes |
Bacilli |
RsaHf231 |
RsaHf231 |
RsaHf231 |
0.01 |
0.01 |
0.16 |
0.02 |
0.0020 |
| Firmicutes |
Bacilli |
Bacillales |
Bacillaceae |
Bacillus |
1.15 |
0.12 |
2.09 |
0.16 |
0.0032 |
| Firmicutes |
Bacilli |
Erysipelotrichales |
Erysipelotrichaceae |
Erysipelothrix |
0.41 |
0.32 |
2.20 |
0.21 |
0.0043 |
| Firmicutes |
Bacilli |
Bacillales |
Bacillaceae |
Amphibacillus |
0.08 |
0.04 |
0.31 |
0.02 |
0.0062 |
| Firmicutes |
Clostridia |
Peptostreptococcales-Tissierellales |
Peptostreptococcaceae |
Clostridioides |
0.07 |
0.04 |
0.25 |
0.01 |
0.0153 |
| Firmicutes |
Clostridia |
Lachnospirales |
Lachnospiraceae |
Lachnoclostridium |
0.02 |
0.02 |
0.11 |
0.03 |
0.0273 |
| Firmicutes |
Clostridia |
Oscillospirales |
Ruminococcaceae |
Incertae_Sedis |
0.01 |
0.01 |
0.08 |
0.02 |
0.0300 |
| Proteobacteria |
Gammaproteobacteria |
Enterobacterales |
Morganellaceae |
Providencia |
1.14 |
0.62 |
12.29 |
0.56 |
0.0001 |
| Proteobacteria |
Gammaproteobacteria |
Pseudomonadales |
Pseudomonadaceae |
Oblitimonas |
0.19 |
0.12 |
1.32 |
0.14 |
0.0010 |
| Proteobacteria |
Gammaproteobacteria |
Cardiobacteriales |
Wohlfahrtiimonadaceae |
Ignatzschineria |
0.83 |
0.38 |
5.12 |
0.54 |
0.0013 |
| Proteobacteria |
Gammaproteobacteria |
Enterobacterales |
Yersiniaceae |
Serratia |
0.09 |
0.06 |
0.64 |
0.03 |
0.0017 |
| Proteobacteria |
Gammaproteobacteria |
Burkholderiales |
Alcaligenaceae |
Paenalcaligenes |
0.24 |
0.11 |
1.28 |
0.15 |
0.0019 |
| Proteobacteria |
Gammaproteobacteria |
Aeromonadales |
Aeromonadaceae |
Aeromonas |
0.15 |
0.09 |
1.97 |
0.20 |
0.0022 |
| Proteobacteria |
Gammaproteobacteria |
Burkholderiales |
Alcaligenaceae |
Alcaligenes |
0.26 |
0.02 |
1.47 |
0.12 |
0.0035 |
| Proteobacteria |
Gammaproteobacteria |
Enterobacterales |
Morganellaceae |
Proteus |
1.49 |
0.83 |
7.20 |
0.24 |
0.0068 |
| Proteobacteria |
Gammaproteobacteria |
Enterobacterales |
Morganellaceae |
Morganella |
0.12 |
0.07 |
1.99 |
0.33 |
0.0122 |
| Proteobacteria |
Alphaproteobacteria |
Rhizobiales |
Rhizobiaceae |
Paenochrobactrum |
0.08 |
0.05 |
0.41 |
0.06 |
0.0044 |
Table 9.
The list of bacterial genera that differ between experimental feeds A (control) and C.
Table 9.
The list of bacterial genera that differ between experimental feeds A (control) and C.
| PHYLUM |
CLASS |
ORDER |
FAMILY |
GENUS |
A (%) |
SD (%) |
C (%) |
SD (%) |
p-value |
| Actinobacteriota |
Actinobacteria |
Actinomycetales |
Actinomycetaceae |
Actinomyces |
0.19 |
0.11 |
3.01 |
0.22 |
0.0006 |
| Bacteroidota |
Bacteroidia |
Bacteroidales |
Bacteroidaceae |
Bacteroides |
0.04 |
0.02 |
1.27 |
0.05 |
0.0001 |
| Bacteroidota |
Bacteroidia |
Bacteroidales |
Dysgonomonadaceae |
Dysgonomonadaceae |
0.03 |
0.01 |
1.93 |
0.06 |
0.0002 |
| Bacteroidota |
Bacteroidia |
Bacteroidales |
Dysgonomonadaceae |
Dysgonomonas |
0.17 |
0.12 |
2.90 |
0.44 |
0.0090 |
| Bacteroidota |
Bacteroidia |
Flavobacteriales |
Flavobacteriaceae |
Myroides |
1.06 |
0.23 |
0.10 |
0.04 |
0.0244 |
| Firmicutes |
Clostridia |
Oscillospirales |
Ruminococcaceae |
Incertae_Sedis |
0.01 |
0.01 |
0.23 |
0.01 |
0.0000 |
| Firmicutes |
Bacilli |
Bacillales |
Planococcaceae |
Sporosarcina |
0.41 |
0.17 |
2.54 |
0.21 |
0.0004 |
| Firmicutes |
Clostridia |
Lachnospirales |
Lachnospiraceae |
Lachnoclostridium |
0.02 |
0.02 |
0.17 |
0.02 |
0.0019 |
| Firmicutes |
Clostridia |
Peptostreptococcales-Tissierellales |
Sedimentibacteraceae |
Sedimentibacter |
0.01 |
0.01 |
0.11 |
0.01 |
0.0040 |
| Firmicutes |
Bacilli |
Bacillales |
Bacillaceae |
Bacillus |
1.15 |
0.12 |
21.97 |
1.98 |
0.0044 |
| Firmicutes |
Bacilli |
Bacillales |
Bacillaceae |
Ureibacillus |
0.23 |
0.10 |
12.00 |
1.37 |
0.0065 |
| Firmicutes |
Negativicutes |
Acidaminococcales |
Acidaminococcaceae |
Phascolarctobacterium |
0.01 |
0.01 |
0.14 |
0.04 |
0.0303 |
| Firmicutes |
Clostridia |
Clostridiales |
Clostridiaceae |
Clostridium_sensu_stricto_18 |
0.10 |
0.03 |
0.02 |
0.02 |
0.0416 |
| Firmicutes |
Bacilli |
RsaHf231 |
RsaHf231 |
RsaHf231 |
0.01 |
0.01 |
0.06 |
0.02 |
0.0443 |
| Firmicutes |
Bacilli |
Staphylococcales |
Staphylococcaceae |
Staphylococcus |
1.43 |
0.44 |
0.16 |
0.12 |
0.0460 |
| Proteobacteria |
Gammaproteobacteria |
Burkholderiales |
Alcaligenaceae |
Alcaligenes |
0.26 |
0.02 |
0.05 |
0.01 |
0.0027 |
Table 10.
The list of bacterial genera that differ between experimental feeds B and C.
Table 10.
The list of bacterial genera that differ between experimental feeds B and C.
| PHYLUM |
CLASS |
ORDER |
FAMILY |
GENUS |
B (%) |
SD (%) |
C (%) |
SD (%) |
p-value |
| Actinobacteriota |
Actinobacteria |
Micrococcales |
Brevibacteriaceae |
Brevibacterium |
1.70 |
0.06 |
0.07 |
0.03 |
4.41E-05 |
| Actinobacteriota |
Actinobacteria |
Corynebacteriales |
Corynebacteriaceae |
Corynebacterium |
3.72 |
0.19 |
0.46 |
0.17 |
5.54E-05 |
| Actinobacteriota |
Actinobacteria |
Micrococcales |
Dermabacteraceae |
Brachybacterium |
1.44 |
0.07 |
0.08 |
0.03 |
0.0003 |
| Actinobacteriota |
Actinobacteria |
Micrococcales |
Micrococcaceae |
Enteractinococcus |
0.19 |
0.01 |
0.01 |
0.01 |
0.0004 |
| Actinobacteriota |
Actinobacteria |
Micrococcales |
AKAU3644 |
AKAU3644 |
0.64 |
0.02 |
0.00 |
0.00 |
0.0004 |
| Actinobacteriota |
Actinobacteria |
Micrococcales |
Microbacteriaceae |
Leucobacter |
0.43 |
0.04 |
0.10 |
0.03 |
0.0015 |
| Actinobacteriota |
Actinobacteria |
Actinomycetales |
Actinomycetaceae |
Actinomyces |
1.20 |
0.10 |
3.01 |
0.22 |
0.0025 |
| Actinobacteriota |
Actinobacteria |
Corynebacteriales |
Dietziaceae |
Dietzia |
0.66 |
0.07 |
0.02 |
0.03 |
0.0032 |
| Bacteroidota |
Bacteroidia |
Flavobacteriales |
Crocinitomicaceae |
Fluviicola |
0.16 |
0.01 |
0.00 |
0.00 |
2.09E-05 |
| Bacteroidota |
Bacteroidia |
Sphingobacteriales |
Sphingobacteriaceae |
Sphingobacterium |
2.62 |
0.10 |
0.09 |
0.04 |
0.0001 |
| Bacteroidota |
Bacteroidia |
Bacteroidales |
Dysgonomonadaceae |
Dysgonomonadaceae |
0.36 |
0.02 |
1.93 |
0.06 |
0.0002 |
| Bacteroidota |
Bacteroidia |
Flavobacteriales |
Flavobacteriaceae |
Myroides |
3.89 |
0.13 |
0.10 |
0.04 |
0.0003 |
| Bacteroidota |
Bacteroidia |
Bacteroidales |
Bacteroidaceae |
Bacteroides |
0.51 |
0.07 |
1.27 |
0.05 |
0.0005 |
| Bacteroidota |
Bacteroidia |
Flavobacteriales |
Flavobacteriaceae |
Flavobacterium |
0.17 |
0.02 |
0.03 |
0.02 |
0.0021 |
| Bacteroidota |
Bacteroidia |
Sphingobacteriales |
Sphingobacteriaceae |
Pedobacter |
0.13 |
0.02 |
0.01 |
0.01 |
0.0041 |
| Bacteroidota |
Bacteroidia |
Flavobacteriales |
Flavobacteriaceae |
Ulvibacter |
0.38 |
0.05 |
0.09 |
0.06 |
0.0055 |
| Bacteroidota |
Bacteroidia |
Bacteroidales |
Dysgonomonadaceae |
Dysgonomonas |
1.33 |
0.07 |
2.90 |
0.44 |
0.0343 |
| Firmicutes |
Clostridia |
Peptostreptococcales-Tissierellales |
Peptostreptococcaceae |
Clostridioides |
0.25 |
0.01 |
0.01 |
0.01 |
0.0001 |
| Firmicutes |
Bacilli |
Staphylococcales |
Staphylococcaceae |
Staphylococcus |
5.38 |
0.23 |
0.16 |
0.12 |
0.0001 |
| Firmicutes |
Bacilli |
Bacillales |
Bacillaceae |
Pseudogracilibacillus |
2.72 |
0.18 |
0.85 |
0.12 |
0.0005 |
| Firmicutes |
Bacilli |
Bacillales |
Bacillaceae |
Amphibacillus |
0.31 |
0.02 |
0.05 |
0.01 |
0.0005 |
| Firmicutes |
Bacilli |
Bacillales |
Bacillaceae |
Virgibacillus |
0.23 |
0.01 |
0.14 |
0.01 |
0.0008 |
| Firmicutes |
Bacilli |
Bacillales |
Planococcaceae |
Sporosarcina |
0.74 |
0.11 |
2.54 |
0.21 |
0.0015 |
| Firmicutes |
Clostridia |
Oscillospirales |
Ruminococcaceae |
Incertae_Sedis |
0.08 |
0.02 |
0.23 |
0.01 |
0.0031 |
| Firmicutes |
Clostridia |
Clostridiales |
Clostridiaceae |
Clostridium_sensu_stricto_7 |
0.00 |
0.00 |
0.04 |
0.00 |
0.0033 |
| Firmicutes |
Bacilli |
Bacillales |
Bacillaceae |
Geobacillus |
0.04 |
0.01 |
0.10 |
0.01 |
0.0038 |
| Firmicutes |
Bacilli |
Lactobacillales |
Enterococcaceae |
Enterococcus |
0.63 |
0.06 |
0.14 |
0.02 |
0.0038 |
| Firmicutes |
Bacilli |
Bacillales |
Bacillaceae |
Bacillus |
2.09 |
0.16 |
21.97 |
1.98 |
0.0047 |
| Firmicutes |
Bacilli |
Erysipelotrichales |
Erysipelotrichaceae |
Erysipelothrix |
2.20 |
0.21 |
0.40 |
0.05 |
0.0047 |
| Firmicutes |
Bacilli |
Bacillales |
Planococcaceae |
Savagea |
2.62 |
0.27 |
0.16 |
0.04 |
0.0052 |
| Firmicutes |
Bacilli |
Lactobacillales |
Streptococcaceae |
Streptococcus |
0.30 |
0.03 |
0.72 |
0.07 |
0.0057 |
| Firmicutes |
Bacilli |
RsaHf231 |
RsaHf231 |
RsaHf231 |
0.16 |
0.02 |
0.06 |
0.02 |
0.0060 |
| Firmicutes |
Bacilli |
Bacillales |
Bacillaceae |
Ureibacillus |
0.19 |
0.05 |
12.00 |
1.37 |
0.0066 |
| Firmicutes |
Bacilli |
Lactobacillales |
Leuconostocaceae |
Weissella |
0.69 |
0.02 |
1.17 |
0.07 |
0.0070 |
| Firmicutes |
Bacilli |
Lactobacillales |
Carnobacteriaceae |
Atopostipes |
0.36 |
0.05 |
0.07 |
0.02 |
0.0077 |
| Firmicutes |
Bacilli |
Bacillales |
Bacillaceae |
Cerasibacillus |
0.14 |
0.01 |
0.07 |
0.01 |
0.0094 |
| Firmicutes |
Bacilli |
Lactobacillales |
Lactobacillaceae |
Lactobacillus |
16.56 |
0.76 |
35.81 |
3.29 |
0.0111 |
| Firmicutes |
Bacilli |
Staphylococcales |
Staphylococcaceae |
Nosocomiicoccus |
0.13 |
0.02 |
0.00 |
0.00 |
0.0159 |
| Firmicutes |
Bacilli |
Bacillales |
Bacillaceae |
Oceanobacillus |
0.24 |
0.04 |
0.09 |
0.01 |
0.0251 |
| Fusobacteriota |
Fusobacteriia |
Fusobacteriales |
Fusobacteriaceae |
Cetobacterium |
0.06 |
0.01 |
0.13 |
0.01 |
0.0042 |
| Proteobacteria |
Gammaproteobacteria |
Enterobacterales |
Yersiniaceae |
Serratia |
0.64 |
0.03 |
0.03 |
0.02 |
6.01E-05 |
| Proteobacteria |
Gammaproteobacteria |
Enterobacterales |
Morganellaceae |
Proteus |
7.20 |
0.24 |
0.07 |
0.02 |
0.0005 |
| Proteobacteria |
Gammaproteobacteria |
Enterobacterales |
Morganellaceae |
Providencia |
12.29 |
0.56 |
0.17 |
0.06 |
0.0009 |
| Proteobacteria |
Gammaproteobacteria |
Pseudomonadales |
Moraxellaceae |
Acinetobacter |
0.23 |
0.02 |
0.05 |
0.02 |
0.0010 |
| Proteobacteria |
Gammaproteobacteria |
Burkholderiales |
Alcaligenaceae |
Alcaligenes |
1.47 |
0.12 |
0.05 |
0.01 |
0.0032 |
| Proteobacteria |
Gammaproteobacteria |
Burkholderiales |
Alcaligenaceae |
Paenalcaligenes |
1.28 |
0.15 |
0.31 |
0.08 |
0.0038 |
| Proteobacteria |
Gammaproteobacteria |
Aeromonadales |
Aeromonadaceae |
Aeromonas |
1.97 |
0.20 |
0.06 |
0.02 |
0.0050 |
| Proteobacteria |
Gammaproteobacteria |
Pseudomonadales |
Pseudomonadaceae |
Oblitimonas |
1.32 |
0.14 |
0.04 |
0.01 |
0.0052 |
| Proteobacteria |
Gammaproteobacteria |
Cardiobacteriales |
Wohlfahrtiimonadaceae |
Ignatzschineria |
5.12 |
0.54 |
0.46 |
0.02 |
0.0066 |
| Proteobacteria |
Gammaproteobacteria |
Enterobacterales |
Morganellaceae |
Morganella |
1.99 |
0.33 |
0.08 |
0.04 |
0.0134 |
| Proteobacteria |
Alphaproteobacteria |
Rhizobiales |
Rhizobiaceae |
Paenochrobactrum |
0.41 |
0.06 |
0.07 |
0.01 |
0.0139 |
| Proteobacteria |
Gammaproteobacteria |
Xanthomonadales |
Xanthomonadaceae |
Stenotrophomonas |
0.18 |
0.04 |
0.05 |
0.02 |
0.0169 |
Table 11.
Result of the alpha diversity analysis of the microbial communities in the gut.
Table 11.
Result of the alpha diversity analysis of the microbial communities in the gut.
| |
CTRL |
PM20 |
PM25 |
p-value |
| Observed OTUs |
1144 ± 380 |
756 ± 329 |
945 ± 445 |
> 0.05 |
| Chao1 |
1276 ± 440 |
914 ± 386 |
1086 ± 471 |
> 0.05 |
| Faith-PD |
18.5 ± 6.7 |
15.8 ± 5.6 |
18.5 ± 5.9 |
> 0.05 |
| Shannon |
6.20 ± 1.06 |
4.69 ± 0.65 |
5.18 ± 1.23 |
0.048 |
| Simpson |
0.93 ± 0.04 |
0.89 ± 0.02 |
0.89 ± 0.05 |
> 0.05 |
Table 12.
Validation of the PCoA analysis.
Table 12.
Validation of the PCoA analysis.
| Unweighted UniFrac PCoA |
|---|
| Anosim |
|
|
|
|
|
| |
|
Sample size |
Permutations |
R |
p-value |
| CTRL |
PM20 |
13 |
999 |
0.316 |
0.005 |
| CTRL |
PM25 |
14 |
999 |
0.312 |
0.005 |
| PM20 |
PM25 |
13 |
999 |
0.184 |
0.037 |
| CTRL |
A |
10 |
999 |
0.496 |
0.030 |
| PM20 |
B |
9 |
999 |
0.728 |
0.014 |
| PM25 |
C |
10 |
999 |
0.639 |
0.007 |
| Permanova |
|
|
|
|
| |
|
Sample size |
Permutations |
pseudo-F |
p-value |
| CTRL |
PM20 |
13 |
999 |
1.922 |
0.010 |
| CTRL |
PM25 |
14 |
999 |
1.979 |
0.003 |
| PM20 |
PM25 |
13 |
999 |
1.567 |
0.024 |
| CTRL |
A |
10 |
999 |
2.486 |
0.013 |
| PM20 |
B |
9 |
999 |
4.473 |
0.012 |
| PM25 |
C |
10 |
999 |
3.749 |
0.007 |
| Weighted UniFrac PCoA |
| Anosim |
|
|
|
|
|
| |
|
Sample size |
Permutations |
R |
p-value |
| CTRL |
PM20 |
13 |
999 |
0.947 |
0.003 |
| CTRL |
PM25 |
14 |
999 |
0.892 |
0.002 |
| PM20 |
PM25 |
13 |
999 |
-0.009 |
0.437 |
| CTRL |
A |
10 |
999 |
1 |
0.009 |
| PM20 |
B |
9 |
999 |
1 |
0.013 |
| PM25 |
C |
10 |
999 |
1 |
0.009 |
| Permanova |
|
|
|
|
| |
|
Sample size |
Permutations |
pseudo-F |
p-value |
| CTRL |
PM20 |
13 |
999 |
38.427 |
0.002 |
| CTRL |
PM25 |
14 |
999 |
26.332 |
0.001 |
| PM20 |
PM25 |
13 |
999 |
1.441 |
0.233 |
| CTRL |
A |
10 |
999 |
23.772 |
0.012 |
| PM20 |
B |
9 |
999 |
268.127 |
0.012 |
| PM25 |
C |
10 |
999 |
75.875 |
0.011 |
Table 13.
The list of genera of intestinal bacteria that differ between CTRL and PM20 fish.
Table 13.
The list of genera of intestinal bacteria that differ between CTRL and PM20 fish.
| PHYLUM |
CLASS |
ORDER |
FAMILY |
GENUS |
CTRL (%) |
SD (%) |
PM20 (%) |
SD (%) |
p-values |
| Actinobacteriota |
Actinobacteria |
Corynebacteriales |
Mycobacteriaceae |
Mycobacterium |
0.69 |
0.51 |
0.04 |
0.05 |
0.0210 |
| Bacteroidota |
Bacteroidia |
Flavobacteriales |
Flavobacteriaceae |
Ulvibacter |
0.11 |
0.08 |
0.01 |
0.02 |
0.0196 |
| Firmicutes |
Bacilli |
Staphylococcales |
Staphylococcaceae |
Staphylococcus |
36.87 |
24.78 |
1.14 |
1.04 |
0.0123 |
| Firmicutes |
Bacilli |
Bacillales |
Bacillaceae |
Geobacillus |
0.84 |
0.62 |
0.00 |
0.00 |
0.0168 |
| Firmicutes |
Clostridia |
Clostridiales |
Clostridiaceae |
Clostridium_sensu_stricto_7 |
1.23 |
0.96 |
0.01 |
0.01 |
0.0208 |
| Firmicutes |
Clostridia |
Clostridiales |
Clostridiaceae |
Clostridium_sensu_stricto_1 |
0.53 |
0.36 |
0.07 |
0.14 |
0.0228 |
| Firmicutes |
Clostridia |
Peptostreptococcales-Tissierellales |
Peptostreptococcales-Tissierellales |
Anaerosalibacter |
1.08 |
0.89 |
0.02 |
0.03 |
0.0259 |
| Firmicutes |
Clostridia |
Clostridiales |
Clostridiaceae |
Clostridium_sensu_stricto_18 |
0.28 |
0.24 |
0.01 |
0.01 |
0.0364 |
| Firmicutes |
Bacilli |
Bacillales |
Bacillaceae |
Oceanobacillus |
0.25 |
0.21 |
0.04 |
0.04 |
0.0494 |
| Firmicutes |
Bacilli |
Lactobacillales |
Lactobacillaceae |
Lactobacillus |
22.47 |
22.09 |
0.35 |
0.36 |
0.0496 |
| Fusobacteriota |
Fusobacteriia |
Fusobacteriales |
Fusobacteriaceae |
Cetobacterium |
1.47 |
0.81 |
0.00 |
0.01 |
0.0044 |
| Proteobacteria |
Gammaproteobacteria |
Vibrionales |
Vibrionaceae |
Photobacterium |
6.66 |
5.30 |
43.79 |
10.80 |
0.0002 |
| Proteobacteria |
Gammaproteobacteria |
Vibrionales |
Vibrionaceae |
Vibrio |
6.70 |
11.52 |
44.16 |
13.29 |
0.0006 |
Table 14.
The list of genera of intestinal bacteria that differ between CTRL and PM25 fish.
Table 14.
The list of genera of intestinal bacteria that differ between CTRL and PM25 fish.
| PHYLUM |
CLASS |
ORDER |
FAMILY |
GENUS |
CTRL (%) |
SD (%) |
PM25 (%) |
SD (%) |
p-value |
| Firmicutes |
Clostridia |
Clostridiales |
Clostridiaceae |
Clostridium sensu stricto 1 |
0.53 |
0.36 |
0.06 |
0.08 |
0.0188 |
| Firmicutes |
Bacilli |
Paenibacillales |
Paenibacillaceae |
Paenibacillus |
9.33 |
7.21 |
0.06 |
0.07 |
0.0197 |
| Firmicutes |
Bacilli |
Bacillales |
Bacillaceae |
Geobacillus |
0.84 |
0.62 |
0.04 |
0.05 |
0.0209 |
| Firmicutes |
Bacilli |
Staphylococcales |
Staphylococcaceae |
Staphylococcus |
36.87 |
24.78 |
6.14 |
8.80 |
0.0226 |
| Firmicutes |
Bacilli |
Bacillales |
Bacillaceae |
Oceanobacillus |
0.25 |
0.21 |
0.00 |
0.01 |
0.0288 |
| Firmicutes |
Clostridia |
Peptostreptococcales-Tissierellales |
Peptostreptococcales-Tissierellales |
Anaerosalibacter |
1.08 |
0.89 |
0.06 |
0.09 |
0.0300 |
| Firmicutes |
Bacilli |
Mycoplasmatales |
Mycoplasmataceae |
Mycoplasma |
0.02 |
0.05 |
1.23 |
1.14 |
0.0413 |
| Firmicutes |
Clostridia |
Clostridiales |
Clostridiaceae |
Clostridium sensu stricto 18 |
0.28 |
0.24 |
0.03 |
0.05 |
0.0452 |
| Fusobacteriota |
Fusobacteriia |
Fusobacteriales |
Fusobacteriaceae |
Cetobacterium |
1.47 |
0.81 |
0.08 |
0.16 |
0.0052 |
| Proteobacteria |
Gammaproteobacteria |
Vibrionales |
Vibrionaceae |
Vibrio |
6.70 |
11.52 |
43.21 |
30.77 |
0.0273 |
| Proteobacteria |
Gammaproteobacteria |
Vibrionales |
Vibrionaceae |
Photobacterium |
6.66 |
5.30 |
34.77 |
26.22 |
0.0393 |