1. Introduction
Rhizobia are nitrogen fixing bacteria which are known to be free living in the soil and are an integral part of the soil micro-flora ( Fox
et al., 1990; Thomas, 1993; Franco and Faria, 1997). They are able to make inert nitrogen available for leguminous plants by forming symbiotic relationships with them and in turn are able to utilize certain nutrients produced by the plants (Franco and Dorbereiner, 1994; Woomer
et al., 2011, Ajayi
et al., 2019) and this is known as biological nitrogen fixation. Antibiotics are substances which are able to inhibit the growth of bacteria cells, they are usually produced by other bacteria cells in a bid to out-compete or ensure that nutrient that are available in limited quantities is sufficient enough for their own use (Reller
et al., 2009; Lee
et al., 2013; Veins and Liittmann, 2015; Ayukekbong
et al., 2017) (
Table 1). These antibiotics are therefore important for treatment of infections caused by various micro-organisms and thus are produced in large quantities as drugs by pharmaceutical companies (Tacconelli
et al., 2018). These antibiotics are sometimes released indiscriminately into the environment as industrial waste and also in hospital environment thereby affecting the natural soil micro floral which enhances plant growth and also partially expose the soil micro-organisms to the antibiotics thereby allowing them to build/form resistance to these antibiotics (Veins and Liittmann, 2015; Ventola, 2015). The soil is therefore rendered useless for farming activities and later becomes abandoned as the antibiotics ravage and destroys or incapacitates soil micro-flora that are not resistant to them.
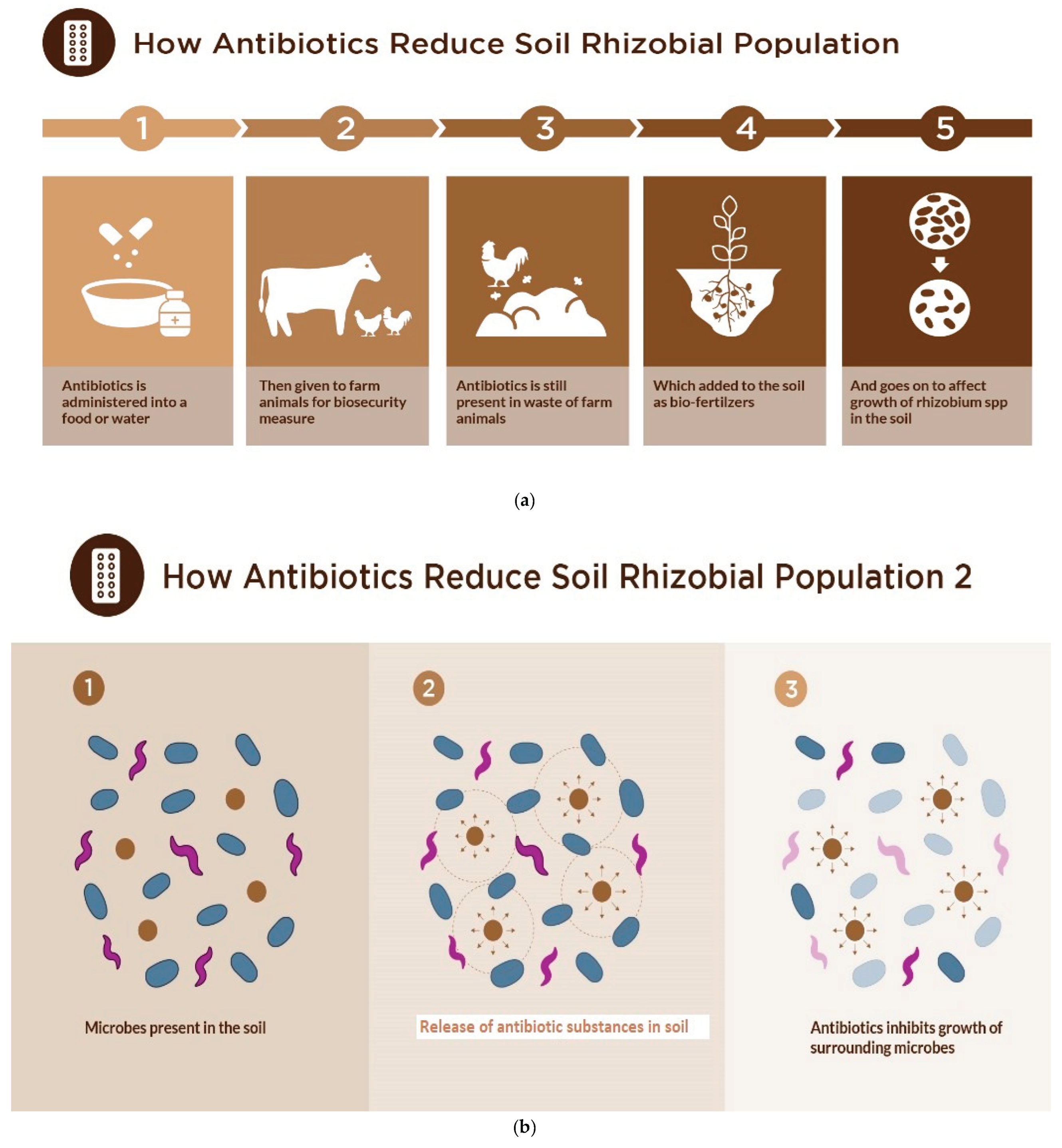
In addition, antibiotics are usually abused and misused in overdosed quantities in the treatment of animals, with half of the antibiotics being released back into the environment (Martinez, 2008). Animal waste are also commonly collected and used for fertilizers for plants or released into the environment and water bodies and they usually contain high levels of antibiotics which have previously been used to treat the animals to keep them healthy (Van
et al., 2014; Hong
et al., 2022). They are either applied in their water, food, or injected in the body of the animals (and the excess is released along with the animal faeces) (Emilia
et al., 2023) (
Figure 1a).
Antibiotics when released into the environment indiscriminately (Martinez, 2008; Van
et al., 2014) affects the soil micro flora making it impossible for them to carry out their various activities which help plants and thus rendering the soil useless for farming activities (Dommerguet 1997; Dong
et al., 2022) (
Figure 1b). Also, rhizobia are found in association with several micro-organisms in the rhizosphere which naturally produce antibiotics which can be lethal or harmful to the soil rhizobia population rendering them useless, thus there is a need to study the intrinsic resistance of commonly used rhizobial strains as this is a desirable trait required for them to increase their chances of growth, multiplication, persistence and survival in the soil (Naamala
et al., 2016). Antibiotic resistance describes the genetic abilities of bacteria to encode resistance genes that assists with helping to counteract the inhibition effects of potential antibiotics substances to ensure their survival when subjected to these substances (Blair
et al., 2015; Zeeshan
et al., 2019). Bacteria mostly develop these resistance either intrinsically by using natural integrated insertions or recombinations into the bacteria genetic make-up, or acquired by horizontal genetic mutations processes including transformations, conjugations and transductions (Martinez
et al., 2008). Antibiotic susceptibility testing (AST) is commonly used for specifying what dosages of antibiotic are most effective and help in the formulation of profiles for standard therapy to ensure proper management of deadly infections occurring as a result of particular bacteria types and can be used as a rapid diagnostic method where it plays an important roles for treatment of infection caused by bacteria.
In the current emergence of multi drug resistance, and micro-organisms increasing occurrence of resistance of micro-organisms to antibiotics (Ventola, 2015; Meng et al., 2023; Song et al., 2023) there is a need for proffering preventive precaution by bio-remediation of these soils an example of which is growing legumes which are inoculated with rhizobia spp which can grow in such abandoned or waste land there by revitalizing the soils and rebuilding the soil micro-flora making it once again useful for normal agricultural activities.
2. Materials and Methods
2.1. Antibiotics susceptibility Test
Viable Rhiziobia strains were plated on yeast mannitol agar using spread plate method. The antibiotics (Oxoid Aura (Oxoid Ltd., Basingstoke, UK) were purchased and stored until use. The discs were placed on the plate aseptically using sterile forcep (Poupard
et al., 1994; Kassim
et al., 2016; Zeeshan
et al., 2019). The antibiotics used include Ceftazamine (CAZ), Florfenicol (FFC), Cefpodoxime (CPD), Sulphamethoxazole (SXT), Ciprofloxacin (CIP), Gentamycin (CN), Kanamycin (K), Ertapenem (ETP), Tetracycline (TE), Meropenem (MEM) and Carbapenem (
Table 1.). The plates were sealed with laboratory parafilm and placed in an incubator at 28°C for 3-7 days to allow for the slow growing Bradyrhizobia
spp. This was done using modified methods (Woomer
et al., 2011; Reller
et al., 2018; Zeeshan
et al., 2019).
2.2. Preparation of test isolates
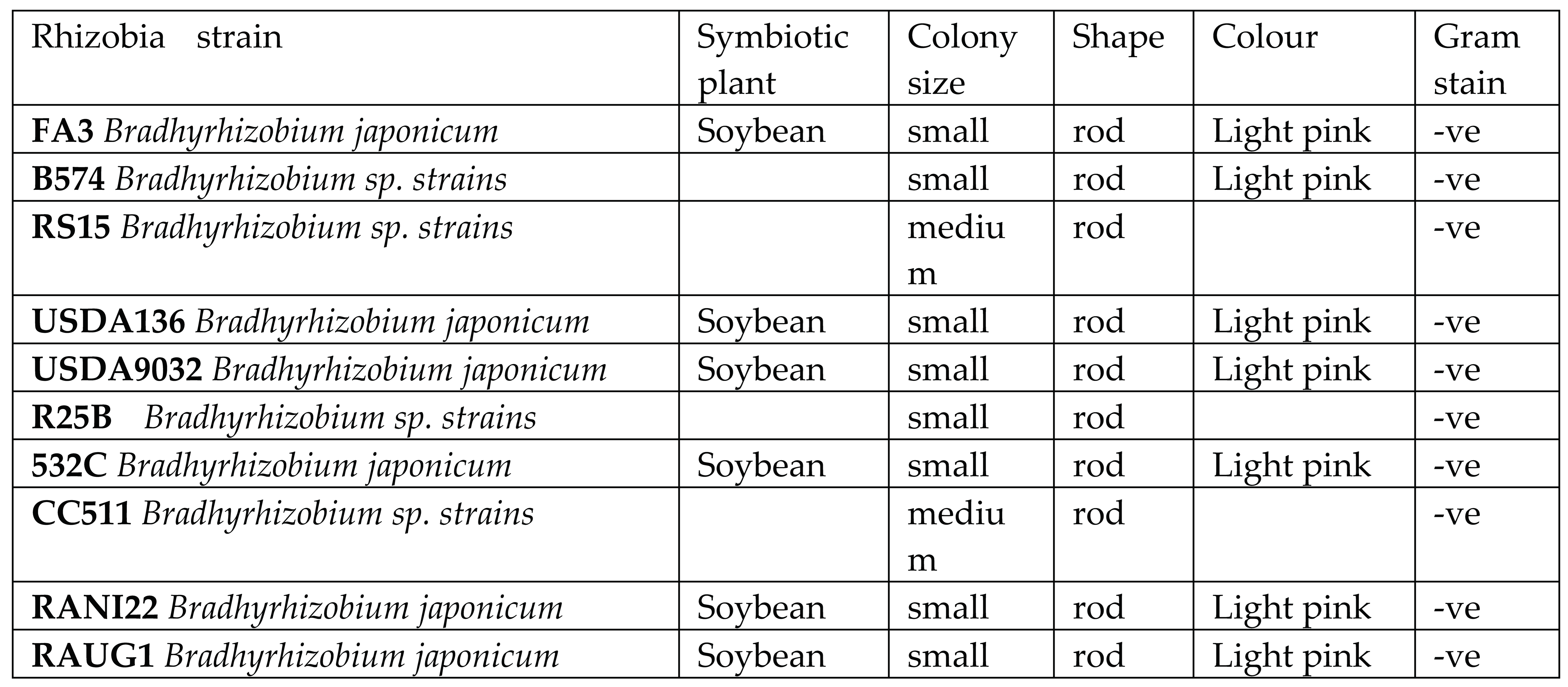
The strains tested include FA3, B574, RS15, USDA136, USDA9032, R25B, 532C, CC511, RANI22, RAUG1 (
Table 2). These typed strains were obtained from the collection of rhizobia isolated at the soil microbiology laboratory of the International Institute for Tropical Agriculture. rhizobia strains collected were plated on yeast mannitol agar (YMA was prepared by adding 15 g of agar, 10 g of mannitol, 0.5 g of K
2HPO
4, 0.2 g of MgSO
4.7H
2O
, 0.1 g of NaCl, 3 g of CaCO
3 and 10 g of yeast extract to 1 litre of water in a conical flask, pH 6.8 sterilised at 121° C for 15 mins) using streaking method to obtain viable cells. These were incubated at 28°C for 3 to 7 days depending on their growth rate (Woomer
et al., 2011).
2.3. McFarland Standard
McFarland’s standard suspensions are prepared using particles of latex or barium sulfate, (the Kirby-Bauer procedure was used (Bauer et al., 1959; 1966)) using a vortex machine this allows for comparison visually to determine bacterial density. A given aliquot (0.5-ml) of BaCl2 (1.175% wt/vol BaCl2 . 2H2O) added to 99.5 ml of 0.18 mol/liter H2SO4 (1% vol/vol) and was stirred constantly to obtain distribution for adequate comparison. The given densities were verified using a measurement of its absorbance using a spectrophotometer at an absorbency of 625 nm which was between 0.10 for 0.5 McFarland standard, this were kept in sealed tubes (stored at room temperature) and used for standardizing prepared bacterial inoculums.
Sterile diluent solution (K2HPO4 and MgSO4) was prepared to which bacterial inoculum was added under sterile conditions. Prepared bacterial suspension were then compared with the standard and more inoculum added when it appeared to be lighter than the 0.5 McFarland standard (Bauer et al., 1959; 1966, Zeeshan et al., 2019).
2.4. Disc diffusion test
The standardized disk diffusion susceptibility method which is a highly practical and simple method was used (Jorgensen and Turnidge, 2007; EUCAST, 2011; Hombach
et al., 2013). The test was carried out using a bacterial inoculum of about 1–2 × 10
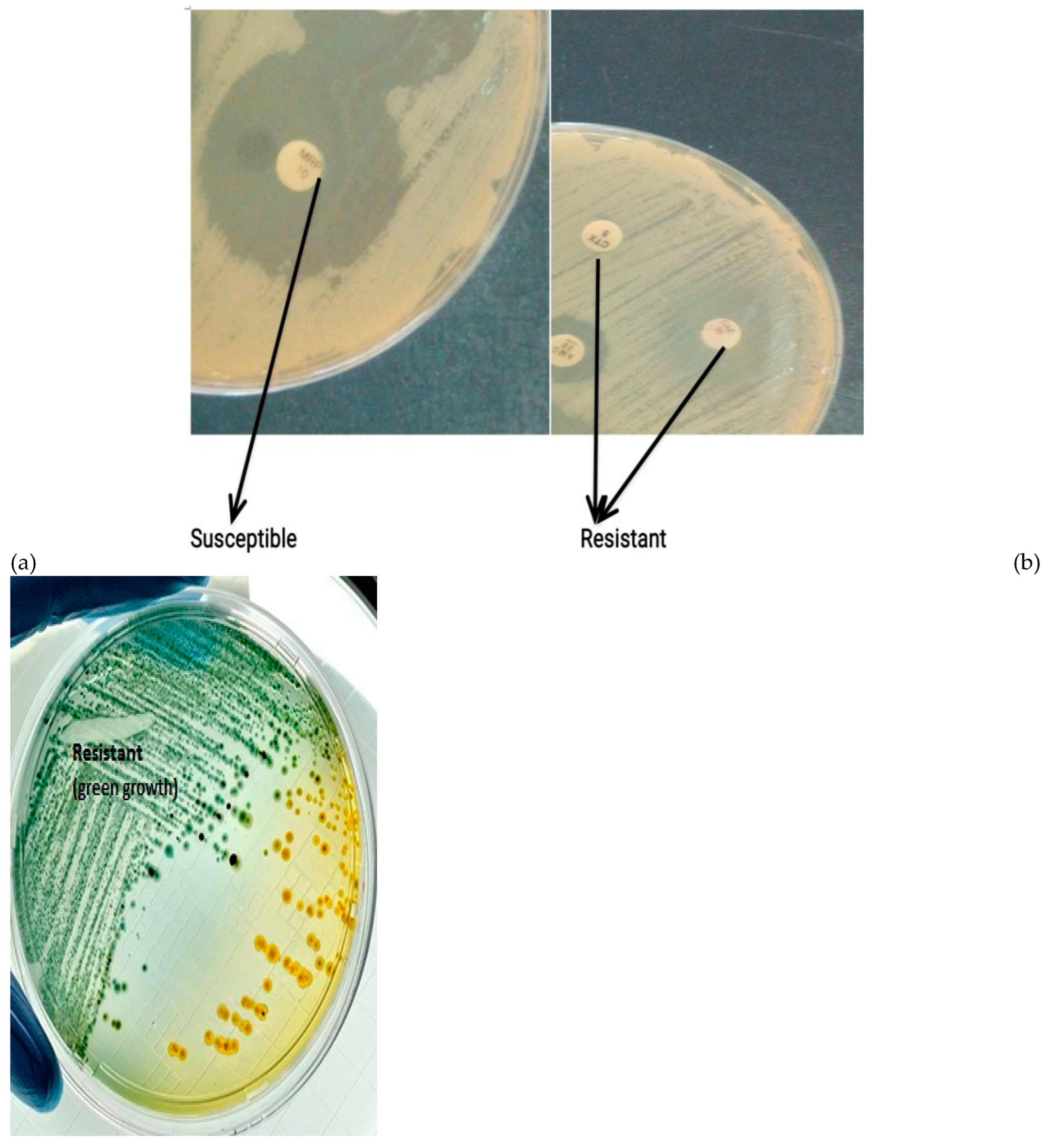
8CFU/mL which was added to the surface of a Yeast Mannitol agar plate (9cm diameter) and spread evenly. Five commercially-prepared, paper antibiotic disks (with known concentrations) were placed on the inoculated agar surface (
Figure 2 a,b). The Petri dishes were then are incubated for 3-7 days at 28°C prior to determination of results, because rhizobia are slow growers. The zones of inhibition of growth around the antibiotic disks were measured in millimeter using a ruler. The width of the diameter tells about the level of susceptibility of the isolate and also about the rate at which the drug diffusions through the agar medium.
The zone diameters of each antibiotics were interpreted using the size of diameter as a criteria, the results of the disk diffusion test were “qualitative,” and the categories of susceptibility used were susceptible, intermediate, or resistant (Kirby et al., 1957; Difco 1984; Winn et al., 2006).
2.5. Disk Placement
Disks was placed on plate so that it was not closer than 24 mm (center to center) on the Yeast mannitol agar plate. Five (5) disks were used in each plate as the size of the plate was 90mm. The disks were picked and placed on the agar using sterile forceps a separate forceps was used for each antibiotics (Jorgensen et al., 2007; Mayrhofer et al., 2008; Patel et al., 2011). The edge of the plate was avoided to ensure zones were fully round and could be measured. Each disk was carefully pressed down with sterile forceps to ensure complete contact with the agar surface. It was ensured that the surface of the agar was smooth and well dried before use to ensure that the shape of the zone was not affected (Mayrhofer et al., 2008; Hombach et al., 2013).
2.5. Determination of resistance property
Plates were observed for inhibition/ susceptibility or growth which was measured using a meter rule while resistance was recorded as R. Inhibition was shown as restricted growth around the antibiotic disc while resistance was shown as undisturbed bacteria grow on the antibiotic disc (
Figure 2a). For Cabapenem, resistance was shown by the production of a green coloration on the plate while susceptibility showed no colour change (
Figure 2b) (Lalitha, 2004; CLSI, 2009; 2021). After 7 days, plates were observed for growth or inhibition. Susceptibility was measured in Millimetres (mm) while resistance were recorded as
S- Suseptible (>20mm),
I- intermediate (<20mm),
R- resistant while for Cabapenem, susceptibility showed no colour change while resistance showed green colouration. Zones of inhibition were then classified as
S- Suseptible where zone of inhibition was >20mm,
I- intermediate where zone of inhibition was <20mm) and
R- resistant where no zone of inhibition was formed.
3. Results
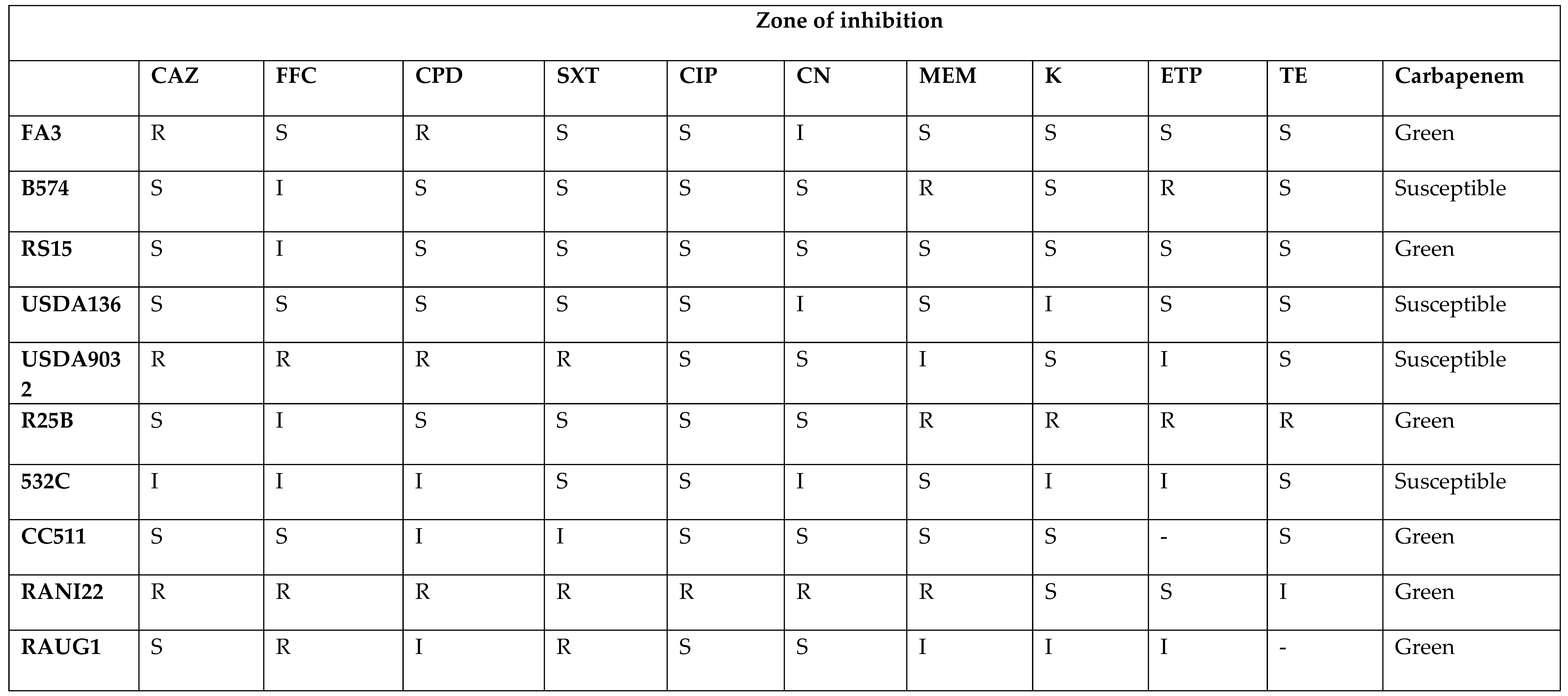
3.1. Ceftazamine (CAZ)
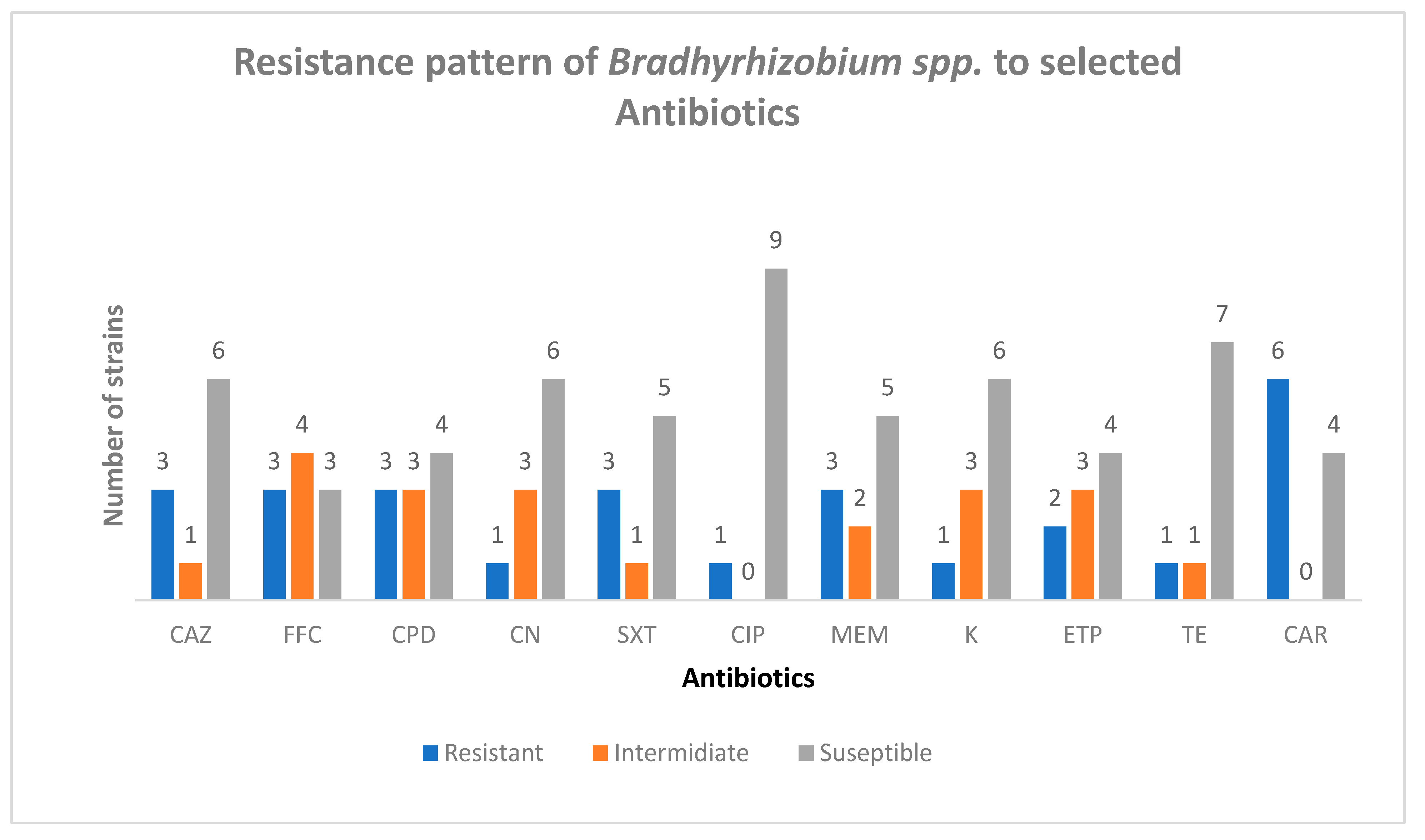
Three strains were resistant to Ceftazamine (CAZ), FA3, USDA9032, RANI 22 and the highest susceptibility was observed in B574 and RS15 with zones of inhibition of 40mm while the least zone of inhibition was 19 mm which was seen in 532C (see
Table 3,
Figure 4). 30% of the strains were resistant to
Ceftazamine (CAZ)
3.2. Florfenicol (FFC)
Three strains were resistant to Florfenicol (FFC), USDA 9032, RANI 22 and RAUG 1. Highest susceptibility was in CC511 with a zone of inhibition of 40mm, while the least zone of inhibition was obtained in RS15 of about 9mm (see
Table 3,
Figure 4), this was also the smallest zone of inhibition amongst all the antibiotics tested. 70% of the strains were susceptible to
Florfenicol (FFC).
3.3. Cefpodoxime (CPD)
FA3, USDA9032 and RANI 22 were resistant to Cefpodoxime (CPD) while all others were susceptible to Cefpodoxime (CPD). The Zone of inhibition was between 19 mm obtained in CC511 and RAUG 1 and 34 mm observed in RS15 (see
Table 3,
Figure 4). 30% of the strains were resistant to Cefpodoxime (CPD)
3.4. Sulphamethoxazole (SXT)
USDA9032, RANI22 and RAUG 1 were resistant to Sulphamethoxazole (SXT) while all others were suspetible to Sulphamethoxazole (SXT). The zone of inhibition obtained was between 18 mm in CC511 and 37 mm seen in FA3 (see
Table 3,
Figure 4). 70 % of the strains were susceptible to Sulphamethoxazole (SXT).
3.5. Ciprofloxacin (CIP)
All strains were susceptible to Ciprofloxacin (CIP) except RANI22 and the largest zone of inhibition was 45 mm in CC 511 while the least of 22 mm was observed in USDA 136 (see
Table 3,
Figure 4). 90% of the strains were susceptible to Ciprofloxacin (CIP).
3.6. Gentamycin (CN)
All strains were susceptible to Gentamycin (CN) except RANI 22 and the highest Zone of inhibition was 52mm which was obtained in RS15 while the least was 19 mm as seen in FA3 (see
Table 3,
Figure 4). Gentamycin was highly effective in inhibiting the growth of
Bradhyrhizobium strains tested. 90% of the strains were susceptible to Gentamycin (CN).
3.7. Meropenem (MEM)
B574, R25B and Rani 2 were resistant to Meropenem (MEM) while other tested strains were suscepyible to Meropenem (MEM). The zone of inhibition was between 16mm which was obtained in USDA136 and 55mm obtained in RS15 (see
Table 3). The largest zone of inhibition which was 55mm was observed in Meropenem (MEM) against CC551. It was the most effective against the
Bradhyrhizobium strains tested. 30% of the strains were resistant to Meropenem (MEM).
3.8. Kanamycin (K)
All strains were susceptible to Kanamycin (K) except R25B the zones of inhibition observed were between 17mm and 45 mm (see
Table 3,
Figure 4) and showed high activity against the test
Bradyrhizobium strains used. 90% of the strains were susceptible to Kanamycin (K).
3.9. Ertapenem (ETP)
B574 and R25B were resistant to Ertapenem (ETP) but all others were susceptible Ertapenem (ETP) showed high activity against the test strains with zones of inhibition between 12mm and 45mm (see
Table 3,
Figure 4). 80% of the strains were susceptible to Ertapenem (ETP)
3.10. Tetracycline (TE)
only R25 B was resistant to tetracycline while others were found to be susceptible to Tetracycline (TE) which showed a average activity against the bacteria having zones of inhibition between 15mm and 33mm (see
Table 3,
Figure 4). 90% of the strains were susceptible to Tetracycline (TE)
.
3.11. Carbapenem
The least susceptibility was obtained to Carbapenem (see
Table 3,
Figure 4), with six of the strains showing resistance to Carbapenem and producing green colouration on plate. Carbapenem had the most minimal activity against the test strains.
3.12. Percentage resistance
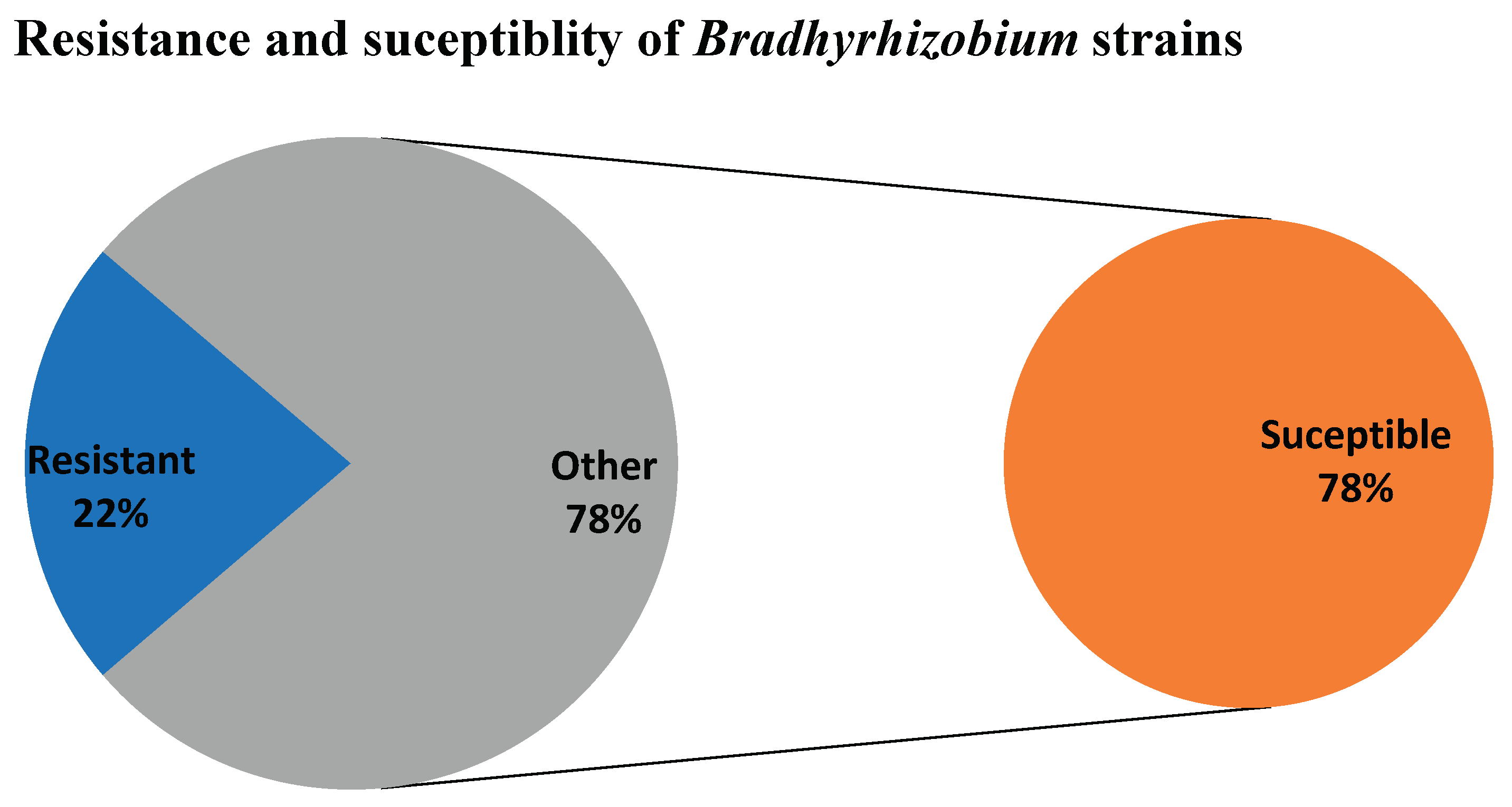
The higher percentage of the strains were susceptible to the antibiotics used sowing about 78% susceptibility to the antibiotics, while 22% of the strains showed resistance to the different antibiotics used in this study. This implies that less resistance to antibiotics was observed in the rhizobia strains used and that although they are not pathogenic in nature, their growth can easily be controlled using antibiotics (
Figure 3).
Figure 3.
Percentage resistance and susceptibility in the Bradhyrhizobium sp.
Figure 3.
Percentage resistance and susceptibility in the Bradhyrhizobium sp.
Figure 4.
Susceptibility and resistance responses of some Bradhyrhizobium spp. to selected antibiotics. Ceftazamine (CAZ), Florfenicol (FFC), Cefpodoxime (CPD), Sulphamethoxazole (SXT), Ciprofloxacin (CIP), Gentamycin (CN), Kanamycin (K), Ertapenem (ETP), Tetracycline (TE), Meropenem (MEM) and Carbapenem.
Figure 4.
Susceptibility and resistance responses of some Bradhyrhizobium spp. to selected antibiotics. Ceftazamine (CAZ), Florfenicol (FFC), Cefpodoxime (CPD), Sulphamethoxazole (SXT), Ciprofloxacin (CIP), Gentamycin (CN), Kanamycin (K), Ertapenem (ETP), Tetracycline (TE), Meropenem (MEM) and Carbapenem.
4. Discussion
The least resistance were observed in Ciprofloxacin, Tetracycline, kanamycin and Gentamycin which had the largest zones of inhibition between 45 mm – 55mm while the least zone of inhibition obtained in them were between 16mm- 23mm, a similar result was obtained in the findings of Rahimi et al. 2013 where their study showed that the pattern of resistance in methicillin resistant Staphylococcus aureus, the resistance to gentamicin, SXT were less than 30% a similar observation was made for SXT where 30% of the gram-negative Bradhyrhizobium spp. were resistant, but only 10% of the gram-negative Bradhyrhizobium spp. was resistant to Gentamycin, with Gentamycin being one of the most effective drugs showing strong activity against S. aureus. In the work of Tooke et al. 2019, gram negative bacteria including E. coli, K. pneumoniae, P. aeruginosa, A. baumannii were found to be resistant to Carbapenem and gram-negative bacteria according to the work of Bush et al. 2018 were resistant to Carbapenem on the other hand our findings show that 60% of the gram-negative Bradhyrhizobium spp. were resistant and 40% of the gram-negative Bradhyrhizobium spp. were Susceptible to Carbapenem.
The findings of Tamma et al. 2012 and Sharma et al. 2017 showed that gram-negative bacteria were resistant to Ciprofloxacin and this was attributed to their ability to produce β-lactamase enzyme, this differed from our findings where 90% of the gram-negative Bradhyrhizobium spp. were susceptible to Ciprofloxacin showing that Bradhyrhizobium spp. may not be capable of producing β-lactamase enzyme (John, 2008; Sageerabanoo et al., 2015), this may be as a result of their non-pathogenic nature. 60% of the gram-negative Bradhyrhizobium spp. were susceptible to kanamycin, showing similar responses to Psuedomonas, Ancinebacter, K. pneumoniae, A. bauannii, Proteus and Bacillus spp. were found to be susceptible to Kanamycin in the report of Palmer et al. 2020, Simpson et al. 2021; Christiaan et al. 2022. In the findings of Carbello et al. 2013; it was observed that 66.71% of the gram-negative bacteria were resistant to tetracycline while our report show that 70% of the gram-negative Bradhyrhizobium spp. were susceptible to tetracycline. Also, in the work of Doubra et al. 2022, Ecoli. isolated from soil and stool 80.7% of the isolates showed resistance to tetracyline also differing from our findings. Ertapenem showed minimal activity against Burkholderia cepacia having an MIC of 8mg/L, differing from our findings where 70% of the gram-negative Bradhyrhizobium spp. were suceptible to Ertapenem as seen in the study carried out by David et al. 2003. The suceptibility of gram- negative strains E.coli, K. pneumoniae and K. oxytoca stains were 99.3%, 96.4% and 90.9% respectively when subjected to Ertapenem as observed in the work of Kuzucu et al. 2011 which was highly similar to our report where only 20% were resistant to Ertapenem.
RANI22 was the most resistant to the antibiotics used and was only susceptible to Kanamycin, Ertapenem, Tetracycline and Carbapenem. This implies that it is a most likely strain to be recommended for use to plant soybean in soils or farming land contaminated with antibiotics or for their recovery. After RANI 22, R25B and USDA 9032 showed most resistance to the selected antibiotics. This was similar to the findings of Bamidele et al. 2022 where S. epidermis (associated with neonatal hospital diseases) had highest resistance to Cefotaxime, Imipenem, Penicillin G, Meropenem and Gentamycin but differed from the work of Tessema et al. 2021 where gram- negative E.coli (associated with neonatal disease) showed highest resistance to Ampicilin, Ampicillin-Sulbactam and Piperacillin. 532C was the most susceptible strain (being susceptible to all the antibiotics used) and was followed by CC511 and RS15 which only showed resistance to Carbapenem. A similar report was obtained in the work of Tessema et al. 2021 where E.coli showed no resistance and was susceptible to Gentamicin, Imipenem, Amikacin and Colistin.
The 78% susceptibility pattern (
Figure 3) showed in these test strains shows or suggests that rhizobia strains do not readily pick or transfer resistance genes and those in which resistance is observed may be as a result of having genes for other characteristics that are co-expressed with the resistance they showed to these antibiotics or also their non-pathogenic nature. It also suggests that resistance are not intrinsic or conserved in their genetic conformation (Humphries
et al., 2021). This implies that these antibiotics are the most effective against the
Brahyrhizobium strains tested and may hamper or deter the growth and activity of these strains in soils where they are present or have been indiscriminately released particularly around hospitals or industries where they are being manufactured or dump-sites close to these places where they are regularly in use. This may make recovering such land areas for farming activities very difficult.
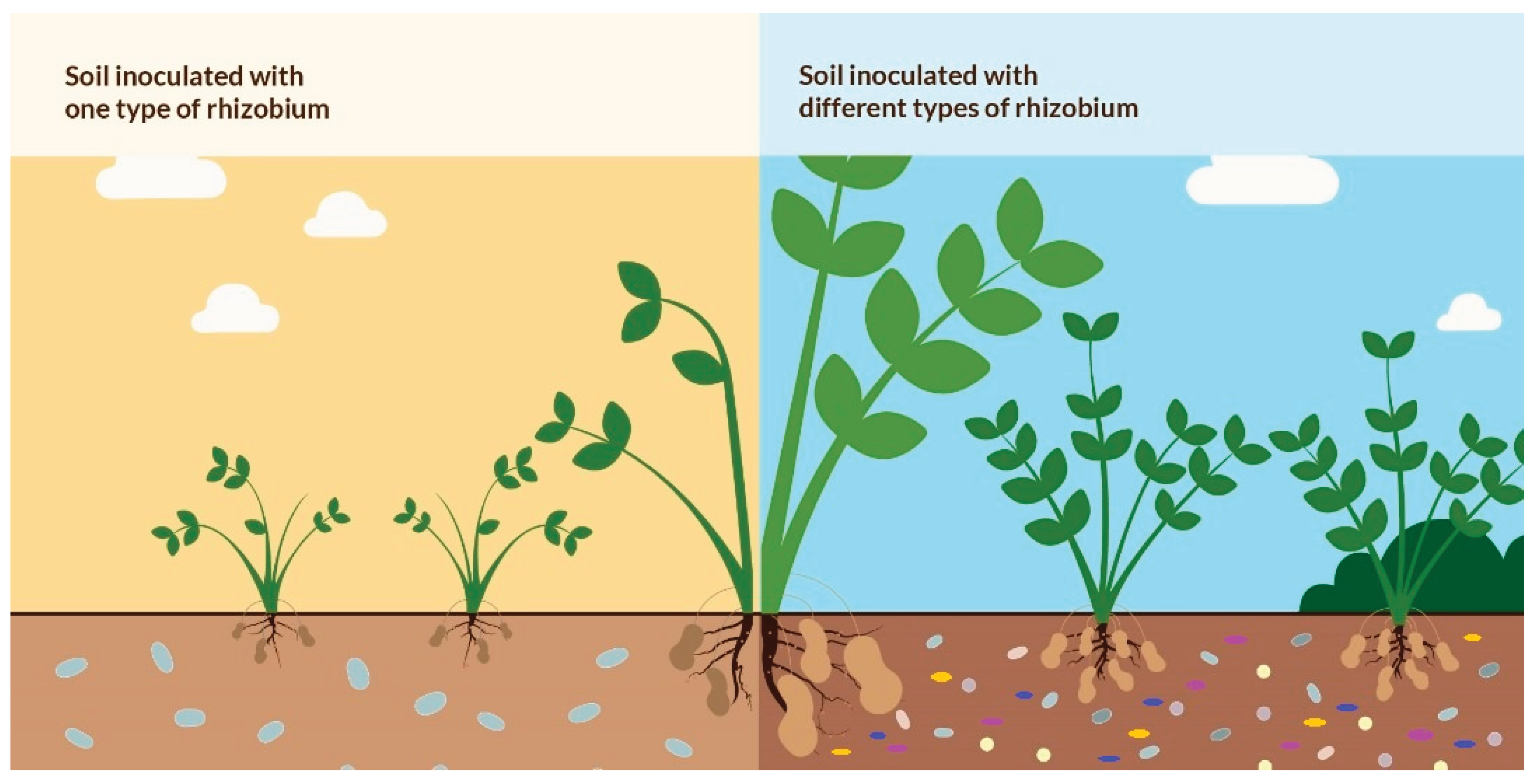
This is particularly important for farmers that collect animal waste for use as plant boosters or practise both animal husbandry and crop farming together, and also release waste from animals indiscriminately to the environs particularly farming areas, lands close by or into water bodies or streams that are common sources of providing water for irrigation for farmers around. Our findings suggests for the recovery of such antibiotic polluted sites or soils, it may be advisable to use a consortium of different symbiotic rhizobia strains as the strains showed differences in their susceptibility pattern (
Figure 5) to achive adequate yields and crop production when planting legumes in such areas.
This may suggest that rhizobia strains do not readily pick or transfer resistance genes and those in which resistance is observed may be conferred on them or connected with other genes which they carry and are co-expressed with the resistance they possess against these antibiotics (Humphries et al., 2021).
5. Conclusion
This may also suggest that in recovery of such sites or soils, it may be advisable to use a consortium of different symbiotic rhizobia strains as the strains showed differences in their susceptibility pattern. Also, the antibiotics produce or contain active compounds and substances that are detrimental for the growth of rhizobia as plant growth promoting bacteria and are may therefore be a hindrance to agricultural practises using these plant growth promoting bacteria during agricultural practises. Most importantly the presence of antibiotics in the soil through indiscriminate release and unregulated use pose a major problem for the use of rhizobia in promoting the growth of legumes especially soybean, particularly in areas where poultry farming and livestock production is being practised. There is therefore a need to ensure proper regulation of the use of antibiotics by poultry farmers and livestock farmers to avoid the indiscriminate release of such antibiotics into the soil particularly and also the need to provide proper education for farmers about the misuse of antibiotics.
References
- Ajayi O. O., Dianda M., Fagade O. E., Nwadike B. (2019). Effect of co-inoculation of Non- Rhizobial Microorganisms (NRMs) with Rhizobia on Nodulation and Leaf chlorophyll content on Cowpea Vigna Ungigulata [(L.) Walp.] (Tvx3236). International Journal of Research and Innovation in Applied Science (IJRIAS) Volume IV, Issue XII, December 2019ISSN 2454-6194. 20 December.
- Ayukekbong J.A., Ntemgwa M., Atabe A.N. (2017). The threat of antimicrobial resistance in developing countries: Causes and control strategies. Antimicrob. Resist. Infect. Control. 6:47. [CrossRef]
- Bamidele O.; Yakubu A.;Joseph, E.B.; Amole, T.A. (2022). Antibiotic Resistance of Bacterial Isolates from Smallholder Poultry Droppings in the Guinea Savanna Zone of Nigeria. Antibiotics. 11: 973. 11, 973. [CrossRef]
- Bauer A.W., Kirby W. M., Sherris J. C., Turck M. (1966). Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 36: 493-496.
- Bauer, A. W., D. M. Perry, and W. M. M. Kirby. (1959). Single disc antibiotic sensitivity testing of Staphylococci. A.M.A. Arch. Intern. Med. 104: 208–216. [CrossRef]
- Blair J.M., Webber M.A., Baylay A.J., Ogbolu D.O., Piddock L.J. (2015). Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 13:42–51. [CrossRef]
- Bush K. (2018). Past and Present Perspectives on β-Lactamases. Antimicrob Agents. Chemother 62:10.1128/aac.01076-18. [CrossRef]
- Carballo, M., Esperón F., Sacristán C., González M., Vázquez B., Aguayo S. and Torre A. (2013). Occurrence of tetracycline residues and antimicrobial resistance in gram negative bacteria isolates from cattle farms in Spain. Advances in Bioscience and Biotechnology. 4: 295-303. [CrossRef]
- Card R. M., Mafura M., Hunt T., Kirchner M., Weile J., Rashid M., Weintraub A., Nord C. E., Anjum MF.2015. Impact of Ciprofloxacin and Clindamycin Administration on Gram-Negative Bacteria Isolated from Healthy Volunteers and Characterization of the Resistance Genes They Harbor. Antimicrob Agents Chemother59. [CrossRef]
- Christiaan D. M. W., Ly P., Martin V. D., Eric P S., Lauren D P., Michael J N. (2022). Gram-negative bacteria act as a reservoir for aminoglycoside antibiotics that interact with host factors to enhance bacterial killing in a mouse model of pneumonia, FEMS Microbes, xtac016. [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). (2009)Methods for dilution antimicrobial susceptibility testing for bacteria that grew aerobically. Approved Standard M7-A10, 2009 Wayne, PAClinical and Laboratory Standards Institute.
- CLSI. (2021). Performance standards for antimicrobial susceptibility testing, M100, 31st ed. Clinical and Laboratory Standards Institute, Wayne, PA.
- David M. Livermore, Armine M. Sefton, Geoffrey M. Scott, Properties and potential of ertapenem. (2003). Journal of Antimicrobial Chemotherapy. 52: 331–344. [CrossRef]
- Difco. (1984). Difco manual, 10th ed. Difco Laboratories, Detroit, MI.
- DOMMERGUES, Y. R. (1997). Contribution of actinorrhizal plant to tropical soil productivity and rehabilitation. Soil Biology Biochemistry. 29 : 931-941. [CrossRef]
- Dong Liu, Yao-Yang X., Muhammad J. Yong-Guan Zhu, Jun W. (2022). Distribution, transfer, ecological and human health risks of antibiotics in bay ecosystems Environment International 158 106949 . [CrossRef]
- Doubra O. P., Kome O., Obakpororo E. A. (2022) “Tetracycline-Resistant Genes in Escherichia coli from Clinical and Nonclinical Sources in Rivers State, Nigeria”, International Journal of Microbiology. [CrossRef]
- Emilia T., Constantin C., Diana O., Sergiu D. Z., Marina S., Adrian V. P., Emoke P., and Florinel B. (2023). Old Antibiotics Can Learn New Ways: A Systematic Review of Florfenicol Use in Veterinary Medicine and Future Perspectives Using Nanotechnology. Animals (Basel). 13: 1695. 13, 1695. [CrossRef]
- Fox, R.H.; Myers, R.J.K. e Vallis, I. (1990). The nitrogen mineralization rate of legumes residues in soil as influenced by polyphenol, lignin and nitrogen contents. Plant and soil. 129: 251-259. [CrossRef]
- Franco, A. A. and Dobereiner, J. A. (1994). biologia do solo e a sustentabilidade dos solos tropicais. Summa Phitopatologica. 20: 68-74.
- Franco, A. A. and Faria, S. M. (1997). The contribution of N2-fixing tree legumes to land reclamation and sustainability in the tropics. Soil Biol. Biochem. 29 : 897-903. [CrossRef]
- George M. H., Pentii H. (2001). Resistance to Trimethprim-Sulfamethoxazole. Clinical infectious Dieseases. 32: 1608-1614. [CrossRef]
- Hombach, M., Zbinden, R. and Böttger, E.C. (2013). Standardisation of disk diffusion results for antibiotic susceptibility testing using the sirscan automated zone reader. BMC Microbiol. 13: 225. [CrossRef]
- Hong B., Liang-Ying H., Dai-Ling W., Fang-Zhou G., Min Z., Hai-Yan Z., Mao-Sheng Y., Guang-Guo Y. (2022). Spread of airborne antibiotic resistance from animal farms to the environment: Dispersal pattern and exposure risk. Environment International 158: 106927. 158, 106927. [CrossRef]
- Humphries R., Bobenchik A. M., Hindler J. A., Schuetz A. N. (2021). Overview of Changes to the Clinical and Laboratory Standards Institute Performance Standards for Antimicrobial Susceptibility Testing, M100, 31st Edition. J Clin Microbiol. 18;59(12):e0021321. https://doi.org/10.1128/JCM.00213-21. Epub 2021 Sep 22. PMID: 34550809; PMCID: PMC8601225. [CrossRef]
- John F. M., Update on the Efficacy and Tolerability of Meropenem in the Treatment of Serious Bacterial Infections, Clinical Infectious Diseases, Volume 47, Issue Supplement_1, September 2008, Pages S41–S51. 20 September. [CrossRef]
- Jorgensen J. H., Turnidge J. D. Murray P. R., Baron E. J., Jorgensen J. H., Landry M. L., Pfaller M. A. (2007). Antibacterial susceptibility tests: dilution and disk diffusion methods, Manual of clinical microbiolog, 9th ed.Washington, DCAmerican Society for Microbiology (pg. 1152-72).
- Jorgensen, J. H. Jorgensen, J. H., and Turnidge J. D. (2007). Susceptibility test methods: dilution and disk diffusion methods, p. 1152–1172. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. L. Landry, and M. A. Pfaller (ed.), Manual of clinical microbiology, 9th ed. ASM Press, Washington, D.C.
- Kassim A., Omuse G., Premji Z., Revathi G. (2016). Comparison of Clinical Laboratory Standards Institute and European Committee on Antimicrobial Susceptibility Testing guidelines for the interpretation of antibiotic susceptibility at a University teaching hospital in Nairobi, Kenya: A cross-sectional study. Ann. Clin. Microbiol. Antimicrob. 15:21. [CrossRef]
- Kirby, W. M. M., Yoshihara G. M., Sundsted K. S., and Warren J. H. (1957). Clinical usefulness of a single disc method for antibiotic sensitivity testing. Antibiotics Annu. 1956-1957:892.
- Knothe, H., Shah, P. M. & Eckardt, O. Cefpodoxime: Comparative antibacterial activity, influence of growth conditions, and bactericidal activity. Infection 19, 370–376 (1991). [CrossRef]
- Kuzucu C., Yetkin F., Görgeç S., Ersoy Y. (2011). Genişlemiş Spektrumlu Beta-Laktamaz Üreten Escherichia coli ve Klebsiella spp. Suşlarının Ertapenem ve Diğer Karbapenemlere Karşı Duyarlılıklarının Araştırılması [Investigation of the susceptibilities of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella spp. strains to ertapenem and other carbapenems]. Mikrobiyol Bul. 45: 28-35. Turkish. PMID: 21341156.
- Lalitha M. (2004). Performance Standards for Antimicrobial Testing: Twelfth Informational Supplement. Volume 56238. CLSI; Wayne, PA, USA. Manual on antimicrobial susceptibility testing; pp: 454–456.
- Lee C.-R., Cho I. H., Jeong B. C., Lee S. H. Strategies to Minimize Antibiotic Resistance. Int. J. Environ. Res. Public Health. 2013;10:4274–4305. [CrossRef]
- Martínez J. L. (2008) Antibiotics and Antibiotic Resistance Genes in Natural Environments. Science. 321: 365–367. [CrossRef]
- Mayrhofer S., Domig K. J., Mair C., Zitz U., Huys G., Kneifel W. (2008). Comparison of broth microdilution, Etest, and agar disk diffusion methods for antimicrobial susceptibility testing of Lactobacillus acidophilus group members. Appl. Environ. Microbiol. 74: 3745–3748. [CrossRef]
- Meng H., Zhao Y, An Q., Zhu B., Cao Z., Lu J. (2023). Use of Ceftazidime-Avibactam for Suspected or Confirmed Carbapenem-Resistant Organisms in Children: A Retrospective Study. Infect Drug Resist. 16: 5815-5824. PMID: 37692471; PMCID: PMC10492574. [CrossRef]
- Mylotte JM. (1987). Gentamicin Resistance Among Gram-negative Bacillary Blood Isolates in a Hospital With Long-term Use of Gentamicin. Arch Intern Med. 147:1642–1644. [CrossRef]
- Naamala J., Jaiswal S.K., and Dakora F.D. (2016). ANTIBIOTIC RESISTANCE IN RHIZOBIUM: type, process, MECHANISM AND Benefits for agriculture. Current Microbiology. 72: 804-816. [CrossRef]
- Palmer LD, Minor KE, Mettlach JA et al. (2020). Modulating isoprenoid biosynthesis increases lipooligosaccharides and restores acinetobacterbaumannii resistance to host and antibiotic stress. Cell Rep. 32 :108129. [CrossRef]
- Patel J.B., Tenover F.C., Turnidge J.D., Jorgensen J.H. (2011). Manual of Clinical Microbiology. 10th ed. American Society of Microbiology; Sterling, VA, USA. Susceptibility test methods: Dilution and disk diffusion methods; pp. 1122–1143.
- Poupard J.A., Rittenhouse S.F. (1994). Walsh L.R. Antimicrobial Susceptibility Testing. Springer, Plenum Publishing; New York, NY, USA:. The evolution of antimicrobial susceptibility testing methods; pp. 3–14.
- Rahimi F., Bouzari M., Katouli M., Pourshafie M. R. (2013). Antibiotic Resistance Pattern of Methicillin Resistant and Methicillin Sensitive Staphylococcus aureus Isolate in Tehran, Iran. Jundishapur J Microbiol.; 6: 144-149. [CrossRef]
- Reller L. B., Weinstein M., Jorgensen J. H., Ferraro M. J. (2009). Antimicrobial Susceptibility Testing: A Review of General Principles and Contemporary Practices, Clinical Infectious Diseases. 49: 1749–1755. [CrossRef]
- Sageerabanoo S., Malini A., Mangaiyarkarasi T., Hemalatha G. (2015). Phenotypic detection of extended spectrum β-lactamase and Amp-C β-lactamase producing clinical isolates in a Tertiary Care Hospital: A preliminary study. J. Nat. Sci. Biol. Med. 6: 383–387. [CrossRef]
- Sharma D., Patel R. P., Zaidi S. T. R., Sarker M. M. R., Lean Q. Y., Ming L. C. (2017). Interplay of the Quality of Ciprofloxacin and Antibiotic Resistance in Developing Countries. Frontiers in pharmacology. 8: 546-546. [CrossRef]
- Simpson BW, Nieckarz M, Pinedo V et al. Acinetobacter baumannii can survive with an outer membrane lacking lipooligosaccharide due to structural support from elongasome peptidoglycan synthesis. Mbio. 2021;12 :e0309921. [CrossRef]
- Song S, Zhao S, Wang W, Jiang F, Sun J, Ma P, Kang H. (2023). Characterization of ST11 and ST15 Carbapenem-Resistant Hypervirulent Klebsiella pneumoniae from Patients with Ventilator-Associated Pneumonia. Infect Drug Resist. 8;16:6017-6028. PMID: 37705511; PMCID: PMC10496924. [CrossRef]
- Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D.L., Pulcini C., Kahlmeter G., Kluytmans J., Carmeli Y., et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018;18:318–327. [CrossRef]
- Tamma P. D., Cosgrove S. E., Maragakis L. L. (2012). Combination therapy for treatment of infections with gram-negative bacteria. Clin. Microbiol. Rev. 25: 450–470. [CrossRef]
- Tang Y.-W., Stratton C.W. Advanced Techniques in Diagnostic Microbiology. Springer; Nashville, TN, USA: 2012.
- Tessema, B.; Lippmann, N.; Knüpfer, M.; Sack, U.; König, B. (2021). Antibiotic Resistance Patterns of Bacterial Isolates from Neonatal Sepsis Patients at University Hospital of Leipzig, Germany. Antibiotics, 10; 323. [CrossRef]
- THOMAS R. J., ASAKAWA N. M. (1993). Decomposition of leaf litter from tropical forage grasses and legumes. Soil Biology and Biochemistry. 25: 1351-1361. [CrossRef]
- Tooke C. L., Hinchliffe P.,† Bragginton E. C., Colenso C. K., Hirvonen V. H. A., Takebayashi Y, and Spencer J. (2019). β-Lactamases and β-Lactamase Inhibitors in the 21st Century. J Mol Biol. 431: 3472–3500. PMCID: PMC6723624PMID: 30959050. [CrossRef]
- Van Boeckel T.P., Gandra S., Ashok A., Caudron Q., Grenfell B.T., Levin S.A., Laxminarayan R. (2014)Global antibiotic consumption 2000 to 2010: An analysis of national pharmaceutical sales data. Lancet Infect. Dis. 14:742–750. [CrossRef]
- Ventola C.L. (2015).The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 40:277–283.
- Viens A.M., Littmann J. (2015). Is Antimicrobial Resistance a Slowly Emerging Disaster? Public Health Ethics. 8:255–265. [CrossRef]
- Winn, Jr., W., et al. (2006). Konemann’s color atlas and diagnostic text of microbiology, 6th ed., p. 945–1021. Lippencott Williams & Wilkins Publishers, Philadelphia, PA.
- Woomer, P. L., Karanja. N., Stanley M. (2011). A revised manual for rhizobium methods and standard protocol. Available online: www.n2africa.org.
- Zeeshan A. K., Mohd F. S., and Seungkyung P. (2019). Current and Emerging Methods of Antibiotic Susceptibility Testing. Diagnostics (Basel) 9: 49. PMCID: PMC6627445PMID: 31058811. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).