Submitted:
21 February 2024
Posted:
22 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
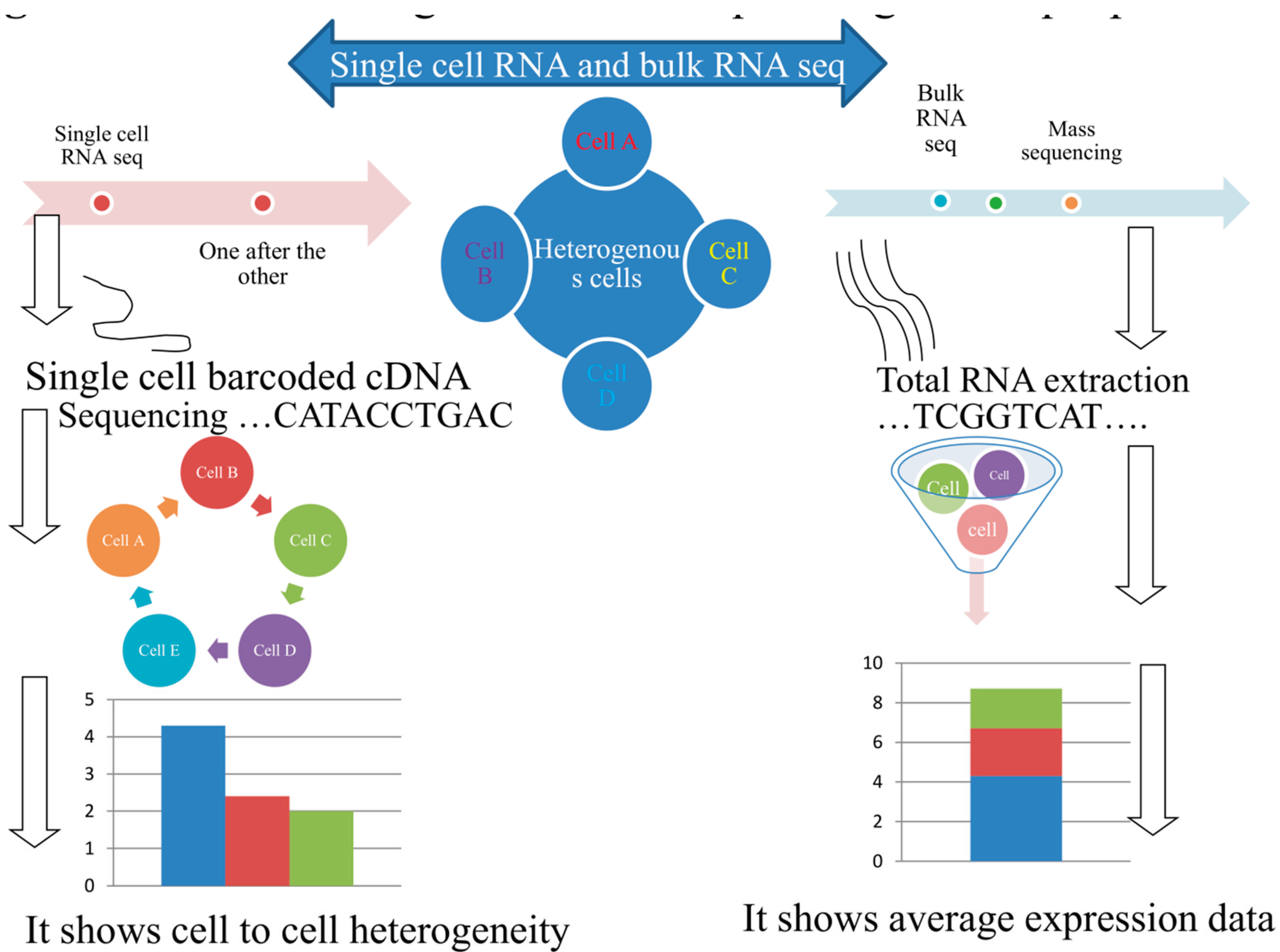
2. From bulk to single-cell transcriptomic dissection
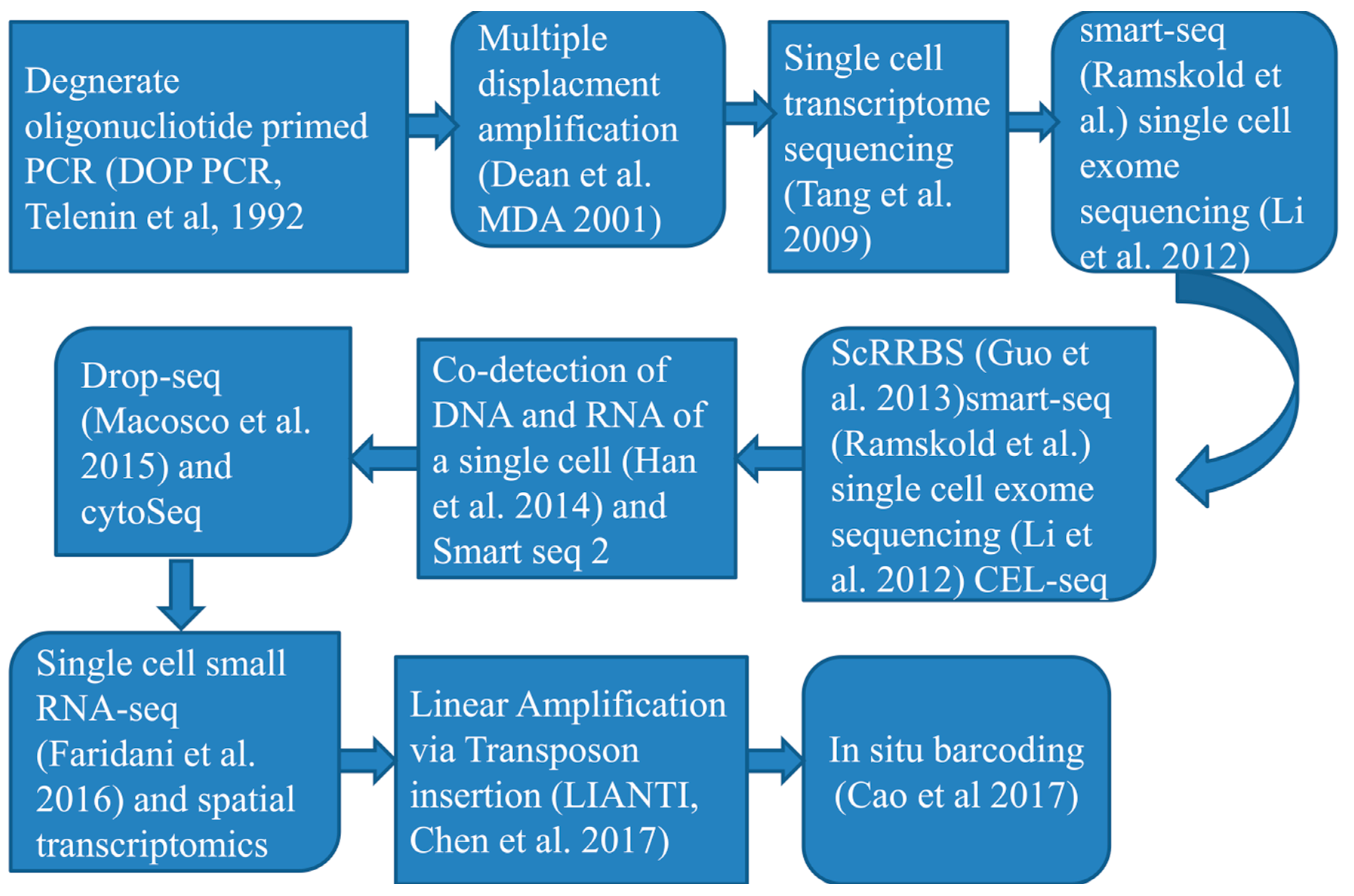
3. Progresses in single-cell RNA profiling techniques and innovation
3.1. Current progresses in single-cell RNA sequencing tools
3.2. Single- cell isolation process and library preparation
3.3. Single-cell transcriptomic sequence data analysis
3.3.1. Data preprocessing
3.3.2. Exploratory analysis
4. Spatial Single-Cell RNA sequencing
5. Single-cell sequencing Applications in the field of biomedical research
5.1. Applications in cancer research
5.1. Implications in the area of immunology
5.2. Implications in the gastro-intestinal system and urinary tract system
5.3. Implications in the neurology
5.4. Implications in the area of reproductive and embryonic medicine
6. Challenges in single-cell RNA sequencing technologies
7. Future perspectives and Concluding Remarks
Funding
Authors’ Contributions
Availability of Data and Materials
Acknowledgements
Competing Interests
Ethics Approval and Consent to Participate
Consent for Publication
Authors’ Information
Notations
| cDNA | Complementary Deoxyribonucleic Acid |
| CNV | Copy number variation |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| DNA | Deoxyribonucleic Acid |
| GSVA | Gene set variation analysis |
| NK | Natural killer cell |
| NOA | Non-obstructive azoospermia |
| PCR | Polymerase chain reaction |
| RNA | Ribonucleic acid |
| scRNA-seq | Single-cell RNA sequencing |
| UMIs | Unique molecular identifiers |
References
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Soon, W.W.; Hariharan, M.; Snyder, M.P. High-throughput sequencing for biology and medicine. Molecular systems biology 2013, 9, 640. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: a revolutionary tool for transcriptomics. Nature reviews. Genetics 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Ozsolak, F.; Milos, P.M. RNA sequencing: advances, challenges and opportunities. Nature reviews. Genetics 2011, 12, 87–98. [Google Scholar] [CrossRef]
- Tang, F.; Barbacioru, C.; Wang, Y.; Nordman, E.; Lee, C.; Xu, N.; Wang, X.; Bodeau, J.; Tuch, B.B.; Siddiqui, A.; et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nature methods 2009, 6, 377–382. [Google Scholar] [CrossRef]
- Wills, Q.F.; Livak, K.J.; Tipping, A.J.; Enver, T.; Goldson, A.J.; Sexton, D.W.; Holmes, C. Single-cell gene expression analysis reveals genetic associations masked in whole-tissue experiments. Nature biotechnology 2013, 31, 748–752. [Google Scholar] [CrossRef]
- Trapnell, C.; Cacchiarelli, D.; Grimsby, J.; Pokharel, P.; Li, S.; Morse, M.; Lennon, N.J.; Livak, K.J.; Mikkelsen, T.S.; Rinn, J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nature biotechnology 2014, 32, 381–386. [Google Scholar] [CrossRef]
- Shapiro, E.; Biezuner, T.; Linnarsson, S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nature reviews. Genetics 2013, 14, 618–630. [Google Scholar] [CrossRef]
- Snijder, B.; Sacher, R.; Rämö, P.; Damm, E.M.; Liberali, P.; Pelkmans, L. Population context determines cell-to-cell variability in endocytosis and virus infection. Nature 2009, 461, 520–523. [Google Scholar] [CrossRef]
- Sharma, S.V.; Lee, D.Y.; Li, B.; Quinlan, M.P.; Takahashi, F.; Maheswaran, S.; McDermott, U.; Azizian, N.; Zou, L.; Fischbach, M.A.; et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 2010, 141, 69–80. [Google Scholar] [CrossRef]
- Helaine, S.; Cheverton, A.M.; Watson, K.G.; Faure, L.M.; Matthews, S.A.; Holden, D.W. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science (New York, N.Y.) 2014, 343, 204–208. [Google Scholar] [CrossRef]
- Baccelli, I.; Schneeweiss, A.; Riethdorf, S.; Stenzinger, A.; Schillert, A.; Vogel, V.; Klein, C.; Saini, M.; Bäuerle, T.; Wallwiener, M.; et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nature biotechnology 2013, 31, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Rinke, C.; Schwientek, P.; Sczyrba, A.; Ivanova, N.N.; Anderson, I.J.; Cheng, J.F.; Darling, A.; Malfatti, S.; Swan, B.K.; Gies, E.A.; et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature 2013, 499, 431–437. [Google Scholar] [CrossRef]
- Raj, A.; van Oudenaarden, A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 2008, 135, 216–226. [Google Scholar] [CrossRef] [PubMed]
- Munsky, B.; Neuert, G.; van Oudenaarden, A. Using gene expression noise to understand gene regulation. Science (New York, N.Y.) 2012, 336, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.; Tang, F. Boosting the power of single-cell analysis. 2018, 36, 408–409. [Google Scholar] [CrossRef]
- The biology of genomes. Single-cell sequencing tackles basic and biomedical questions. Science (New York, N.Y.) 2012, 336, 976–977. [CrossRef]
- Metzker, M.L. Sequencing technologies - the next generation. Nature reviews. Genetics 2010, 11, 31–46. [Google Scholar] [CrossRef]
- Bendall, S.C.; Nolan, G.P. From single cells to deep phenotypes in cancer. Nature biotechnology 2012, 30, 639–647. [Google Scholar] [CrossRef]
- Kalisky, T.; Blainey, P.; Quake, S.R. Genomic analysis at the single-cell level. Annual review of genetics 2011, 45, 431–445. [Google Scholar] [CrossRef]
- Brady, G.; Barbara, M.A.M.; Iscove, N.N. Representative in Vitro cDNA Amplification From Individual Hemopoietic Cells and Colonies. Corpus ID: 39940906.
- 1999 Sheet 2 of 3 5, 958, 688 xxx xx : rir rrrrr.
- Conlin, D.G.; Piffat, K.A. SYNTHESIS OF NUCLEC ACDS USINGA SOLID SUPPORT. 2017.
- Dulac, C.; Axel, R. A novel family of genes encoding putative pheromone receptors in mammals. Cell 1995, 83, 195–206. [Google Scholar] [CrossRef]
- Shumyatsky, G.P.; Tsvetkov, E.; Malleret, G.; Vronskaya, S.; Hatton, M.; Hampton, L.; Battey, J.F.; Dulac, C.; Kandel, E.R.; Bolshakov, V.Y. Identification of a signaling network in lateral nucleus of amygdala important for inhibiting memory specifically related to learned fear. Cell 2002, 111, 905–918. [Google Scholar] [CrossRef]
- Sul, J.Y.; Wu, C.W.; Zeng, F.; Jochems, J.; Lee, M.T.; Kim, T.K.; Peritz, T.; Buckley, P.; Cappelleri, D.J.; Maronski, M.; et al. Transcriptome transfer produces a predictable cellular phenotype. Proceedings of the National Academy of Sciences of the United States of America 2009, 106, 7624–7629. [Google Scholar] [CrossRef]
- Kumar, R.M.; Cahan, P.; Shalek, A.K.; Satija, R.; DaleyKeyser, A.; Li, H.; Zhang, J.; Pardee, K.; Gennert, D.; Trombetta, J.J.; et al. Deconstructing transcriptional heterogeneity in pluripotent stem cells. Nature 2014, 516, 56–61. [Google Scholar] [CrossRef] [PubMed]
- de Vargas Roditi, L.; Claassen, M. Computational and experimental single cell biology techniques for the definition of cell type heterogeneity, interplay and intracellular dynamics. Current opinion in biotechnology 2015, 34, 9–15. [Google Scholar] [CrossRef]
- Islam, S.; Zeisel, A.; Joost, S.; La Manno, G.; Zajac, P.; Kasper, M.; Lönnerberg, P.; Linnarsson, S. Quantitative single-cell RNA-seq with unique molecular identifiers. Nature methods 2014, 11, 163–166. [Google Scholar] [CrossRef]
- Sanchez, A.; Golding, I. Genetic determinants and cellular constraints in noisy gene expression. Science (New York, N.Y.) 2013, 342, 1188–1193. [Google Scholar] [CrossRef]
- Marguerat, S.; Schmidt, A.; Codlin, S.; Chen, W.; Aebersold, R.; Bähler, J. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell 2012, 151, 671–683. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, Y.; Choi, P.J.; Li, G.W.; Chen, H.; Babu, M.; Hearn, J.; Emili, A.; Xie, X.S. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science (New York, N.Y.) 2010, 329, 533–538. [Google Scholar] [CrossRef]
- Luo, Y.; Coskun, V.; Liang, A.; Yu, J.; Cheng, L.; Ge, W.; Shi, Z.; Zhang, K.; Li, C.; Cui, Y.; et al. Single-cell transcriptome analyses reveal signals to activate dormant neural stem cells. Cell 2015, 161, 1175–1186. [Google Scholar] [CrossRef]
- Mahata, B.; Zhang, X.; Kolodziejczyk, A.A.; Proserpio, V.; Haim-Vilmovsky, L.; Taylor, A.E.; Hebenstreit, D.; Dingler, F.A.; Moignard, V.; Göttgens, B.; et al. Single-cell RNA sequencing reveals T helper cells synthesizing steroids de novo to contribute to immune homeostasis. Cell reports 2014, 7, 1130–1142. [Google Scholar] [CrossRef]
- Bendall, S.C.; Davis, K.L.; Amir el, A.D.; Tadmor, M.D.; Simonds, E.F.; Chen, T.J.; Shenfeld, D.K.; Nolan, G.P.; Pe'er, D. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell 2014, 157, 714–725. [Google Scholar] [CrossRef]
- Chang, H.H.; Hemberg, M.; Barahona, M.; Ingber, D.E.; Huang, S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature 2008, 453, 544–547. [Google Scholar] [CrossRef] [PubMed]
- Shalek, A.K.; Satija, R.; Adiconis, X.; Gertner, R.S.; Gaublomme, J.T.; Raychowdhury, R.; Schwartz, S.; Yosef, N.; Malboeuf, C.; Lu, D.; et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 2013, 498, 236–240. [Google Scholar] [CrossRef]
- Deng, Q.; Ramsköld, D.; Reinius, B.; Sandberg, R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science (New York, N.Y.) 2014, 343, 193–196. [Google Scholar] [CrossRef]
- Vitak, S.A.; Torkenczy, K.A. Sequencing thousands of single-cell genomes with combinatorial indexing. 2017, 14, 302–308. [Google Scholar] [CrossRef]
- Chen, C.; Xing, D. Single-cell whole-genome analyses by Linear Amplification via Transposon Insertion (LIANTI). 2017, 356, 189–194. [Google Scholar] [CrossRef]
- Guo, F.; Li, L.; Li, J.; Wu, X.; Hu, B.; Zhu, P.; Wen, L.; Tang, F. Single-cell multi-omics sequencing of mouse early embryos and embryonic stem cells. Cell research 2017, 27, 967–988. [Google Scholar] [CrossRef] [PubMed]
- Casasent, A.K.; Schalck, A.; Gao, R.; Sei, E.; Long, A.; Pangburn, W.; Casasent, T.; Meric-Bernstam, F.; Edgerton, M.E.; Navin, N.E. Multiclonal Invasion in Breast Tumors Identified by Topographic Single Cell Sequencing. Cell 2018, 172, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Demaree, B.; Weisgerber, D.; Lan, F.; Abate, A.R. An Ultrahigh-throughput Microfluidic Platform for Single-cell Genome Sequencing. Journal of visualized experiments : JoVE 2018. [Google Scholar] [CrossRef]
- Han, X.; Wang, R.; Zhou, Y.; Fei, L.; Sun, H.; Lai, S.; Saadatpour, A.; Zhou, Z.; Chen, H.; Ye, F.; et al. Mapping the Mouse Cell Atlas by Microwell-Seq. Cell 2018, 172, 1091–1107. [Google Scholar] [CrossRef]
- Rosenberg, A.B.; Roco, C.M. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. 2018, 360, 176–182. [CrossRef]
- Chen, G.; Ning, B.; Shi, T. Single-Cell RNA-Seq Technologies and Related Computational Data Analysis. Frontiers in genetics 2019, 10, 317. [Google Scholar] [CrossRef]
- Sheng, K.; Cao, W.; Niu, Y.; Deng, Q.; Zong, C. Effective detection of variation in single-cell transcriptomes using MATQ-seq. Nature methods 2017, 14, 267–270. [Google Scholar] [CrossRef]
- Picelli, S.; Björklund, Å.K.; Faridani, O.R.; Sagasser, S.; Winberg, G.; Sandberg, R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nature methods 2013, 10, 1096–1098. [Google Scholar] [CrossRef]
- Goldstein, L.D.; Chen, Y.J.; Dunne, J.; Mir, A.; Hubschle, H.; Guillory, J.; Yuan, W.; Zhang, J.; Stinson, J.; Jaiswal, B.; et al. Massively parallel nanowell-based single-cell gene expression profiling. BMC genomics 2017, 18, 519. [Google Scholar] [CrossRef]
- Fan, X.; Zhang, X.; Wu, X.; Guo, H.; Hu, Y.; Tang, F.; Huang, Y. Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome biology 2015, 16, 148. [Google Scholar] [CrossRef]
- Islam, S.; Kjällquist, U.; Moliner, A.; Zajac, P.; Fan, J.B.; Lönnerberg, P.; Linnarsson, S. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome research 2011, 21, 1160–1167. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.X.; Terry, J.M.; Belgrader, P.; Ryvkin, P.; Bent, Z.W.; Wilson, R.; Ziraldo, S.B.; Wheeler, T.D.; McDermott, G.P.; Zhu, J.; et al. Massively parallel digital transcriptional profiling of single cells. 2017, 8, 14049. [CrossRef]
- DeLaughter, D.M. The Use of the Fluidigm C1 for RNA Expression Analyses of Single Cells. Current protocols in molecular biology 2018, 122, e55. [Google Scholar] [CrossRef]
- Macosko, E.Z.; Basu, A.; Satija, R.; Nemesh, J.; Shekhar, K.; Goldman, M.; Tirosh, I.; Bialas, A.R.; Kamitaki, N.; Martersteck, E.M.; et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 2015, 161, 1202–1214. [Google Scholar] [CrossRef]
- Klein, A.M.; Mazutis, L.; Akartuna, I.; Tallapragada, N.; Veres, A.; Li, V.; Peshkin, L.; Weitz, D.A.; Kirschner, M.W. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 2015, 161, 1187–1201. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Zhang, W.; Xin, H.; Deng, G. Single Cell Isolation and Analysis. Frontiers in cell and developmental biology 2016, 4, 116. [Google Scholar] [CrossRef] [PubMed]
- Saliba, A.E.; Westermann, A.J.; Gorski, S.A.; Vogel, J. Single-cell RNA-seq: advances and future challenges. Nucleic acids research 2014, 42, 8845–8860. [Google Scholar] [CrossRef]
- Menon, V. Clustering single cells: a review of approaches on high-and low-depth single-cell RNA-seq data. Briefings in functional genomics 2018, 17, 240–245. [Google Scholar] [CrossRef]
- Cembrowski, M.S. Single-cell transcriptomics as a framework and roadmap for understanding the brain. Journal of neuroscience methods 2019, 326, 108353. [Google Scholar] [CrossRef]
- Hwang, B.; Lee, J.H.; Bang, D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Experimental & molecular medicine 2018, 50, 1–14. [Google Scholar] [CrossRef]
- Kolodziejczyk, A.A.; Kim, J.K.; Svensson, V.; Marioni, J.C.; Teichmann, S.A. The technology and biology of single-cell RNA sequencing. Molecular cell 2015, 58, 610–620. [Google Scholar] [CrossRef]
- Gross, A.; Schoendube, J.; Zimmermann, S.; Steeb, M.; Zengerle, R.; Koltay, P. Technologies for Single-Cell Isolation. International journal of molecular sciences 2015, 16, 16897–16919. [Google Scholar] [CrossRef]
- Boeynaems, S.; Bogaert, E.; Kovacs, D.; Konijnenberg, A.; Timmerman, E.; Volkov, A.; Guharoy, M.; De Decker, M.; Jaspers, T.; Ryan, V.H. Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Molecular cell 2017, 65, 1044–1055. [Google Scholar] [CrossRef]
- Hashimshony, T.; Senderovich, N.; Avital, G.; Klochendler, A.; De Leeuw, Y.; Anavy, L.; Gennert, D.; Li, S.; Livak, K.J.; Rozenblatt-Rosen, O. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome biology 2016, 17, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Jaitin, D.A.; Kenigsberg, E.; Keren-Shaul, H.; Elefant, N.; Paul, F.; Zaretsky, I.; Mildner, A.; Cohen, N.; Jung, S.; Tanay, A.; et al. Massively Parallel Single-Cell RNA-Seq for Marker-Free Decomposition of Tissues into Cell Types. Science (New York, N.Y.) 2014, 343, 776–779. [Google Scholar] [CrossRef] [PubMed]
- Ramsköld, D.; Luo, S.; Wang, Y.C.; Li, R.; Deng, Q.; Faridani, O.R.; Daniels, G.A.; Khrebtukova, I.; Loring, J.F.; Laurent, L.C.; et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nature biotechnology 2012, 30, 777–782. [Google Scholar] [CrossRef]
- Ziegenhain, C.; Vieth, B.; Parekh, S.; Reinius, B.; Guillaumet-Adkins, A.; Smets, M.; Leonhardt, H.; Heyn, H.; Hellmann, I.; Enard, W. Comparative Analysis of Single-Cell RNA Sequencing Methods. Molecular cell 2017, 65, 631–643. [Google Scholar] [CrossRef]
- Proposed methods for testing and selecting the ERCC external RNA controls. BMC genomics 2005, 6, 150. [CrossRef]
- Jaitin, D.A.; Kenigsberg, E.; Keren-Shaul, H.; Elefant, N.; Paul, F.; Zaretsky, I.; Mildner, A.; Cohen, N.; Jung, S.; Tanay, A.; et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science (New York, N.Y.) 2014, 343, 776–779. [Google Scholar] [CrossRef] [PubMed]
- Arendt, D.; Musser, J.M.; Baker, C.V.H.; Bergman, A.; Cepko, C.; Erwin, D.H.; Pavlicev, M.; Schlosser, G.; Widder, S.; Laubichler, M.D.; et al. The origin and evolution of cell types. Nature reviews. Genetics 2016, 17, 744–757. [Google Scholar] [CrossRef]
- Murphy, D. Gene expression studies using microarrays: principles, problems, and prospects. Advances in physiology education 2002, 26, 256–270. [Google Scholar] [CrossRef]
- Jovic, D.; Liang, X.; Zeng, H.; Lin, L.; Xu, F.; Luo, Y. Single-cell RNA sequencing technologies and applications: A brief overview. 2022, 12, e694. [CrossRef]
- Rodriques, S.G.; Stickels, R.R. Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. 2019, 363, 1463–1467. [CrossRef]
- Shahbazi, M.N.; Scialdone, A.; Skorupska, N.; Weberling, A.; Recher, G.; Zhu, M.; Jedrusik, A.; Devito, L.G.; Noli, L.; Macaulay, I.C.; et al. Pluripotent state transitions coordinate morphogenesis in mouse and human embryos. Nature 2017, 552, 239–243. [Google Scholar] [CrossRef]
- Enge, M.; Arda, H.E.; Mignardi, M.; Beausang, J.; Bottino, R.; Kim, S.K.; Quake, S.R. Single-Cell Analysis of Human Pancreas Reveals Transcriptional Signatures of Aging and Somatic Mutation Patterns. Cell 2017, 171, 321–330. [Google Scholar] [CrossRef]
- Puram, S.V.; Tirosh, I.; Parikh, A.S.; Patel, A.P.; Yizhak, K.; Gillespie, S.; Rodman, C.; Luo, C.L.; Mroz, E.A.; Emerick, K.S.; et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 2017, 171, 1611–1624. [Google Scholar] [CrossRef]
- Macaulay, I.C.; Voet, T. Single cell genomics: advances and future perspectives. PLoS genetics 2014, 10, e1004126. [Google Scholar] [CrossRef]
- Sanz, E.; Yang, L.; Su, T.; Morris, D.R.; McKnight, G.S.; Amieux, P.S. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proceedings of the National Academy of Sciences of the United States of America 2009, 106, 13939–13944. [Google Scholar] [CrossRef]
- Zeb, Q.; Wang, C.; Shafiq, S.; Liu, L. An overview of single-cell isolation techniques. Single-cell omics 2019, 101-135.
- van den Brink, S.C.; Sage, F.; Vértesy, Á. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. 2017, 14, 935–936. [CrossRef]
- Zappia, L.; Phipson, B. Exploring the single-cell RNA-seq analysis landscape with the scRNA-tools database. 2018, 14, e1006245. [CrossRef]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M., 3rd; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902. [Google Scholar] [CrossRef]
- Lun, A.T.; McCarthy, D.J.; Marioni, J.C. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Research 2016, 5, 2122. [Google Scholar] [CrossRef]
- Wolf, F.A.; Angerer, P.; Theis, F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome biology 2018, 19, 15. [Google Scholar] [CrossRef] [PubMed]
- Lambrechts, D.; Wauters, E.; Boeckx, B.; Aibar, S. Phenotype molding of stromal cells in the lung tumor microenvironment. 2018, 24, 1277–1289. [CrossRef]
- Fan, X.; Bialecka, M.; Moustakas, I. Single-cell reconstruction of follicular remodeling in the human adult ovary. 2019, 10, 3164. [CrossRef]
- Lytal, N.; Ran, D.; An, L. Normalization Methods on Single-Cell RNA-seq Data: An Empirical Survey. Frontiers in genetics 2020, 11, 41. [Google Scholar] [CrossRef]
- Yip, S.H.; Sham, P.C.; Wang, J. Evaluation of tools for highly variable gene discovery from single-cell RNA-seq data. Briefings in bioinformatics 2019, 20, 1583–1589. [Google Scholar] [CrossRef] [PubMed]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC bioinformatics 2013, 14, 7. [Google Scholar] [CrossRef]
- Aibar, S.; González-Blas, C.B.; Moerman, T.; Huynh-Thu, V.A.; Imrichova, H.; Hulselmans, G.; Rambow, F.; Marine, J.-C.; Geurts, P.; Aerts, J.; et al. SCENIC: single-cell regulatory network inference and clustering. Nature methods 2017, 14, 1083–1086. [Google Scholar] [CrossRef]
- Aibar, S.; González-Blas, C.B. SCENIC: single-cell regulatory network inference and clustering. 2017, 14, 1083–1086. [CrossRef]
- Van de Sande, B.; Flerin, C. A scalable SCENIC workflow for single-cell gene regulatory network analysis. 2020, 15, 2247–2276. [CrossRef]
- Ma, A.; Wang, C.; Chang, Y.; Brennan, F.H.; McDermaid, A.; Liu, B.; Zhang, C.; Popovich, P.G.; Ma, Q. IRIS3: integrated cell-type-specific regulon inference server from single-cell RNA-Seq. Nucleic acids research 2020, 48, W275–w286. [Google Scholar] [CrossRef]
- Lewis, J. From signals to patterns: space, time, and mathematics in developmental biology. Science (New York, N.Y.) 2008, 322, 399–403. [Google Scholar] [CrossRef] [PubMed]
- Cremisi, F.; Philpott, A.; Ohnuma, S. Cell cycle and cell fate interactions in neural development. Current opinion in neurobiology 2003, 13, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.G.; Lee, H.J.; Asatsuma, T.; Vento-Tormo, R.; Haque, A. An introduction to spatial transcriptomics for biomedical research. Genome medicine 2022, 14, 68. [Google Scholar] [CrossRef] [PubMed]
- Haque, A.; Engel, J.; Teichmann, S.A.; Lönnberg, T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. 2017, 9, 75. [CrossRef]
- Terabayashi, T.; Germino, G.G.; Menezes, L.F. Pathway identification through transcriptome analysis. Cellular signalling 2020, 74, 109701. [Google Scholar] [CrossRef] [PubMed]
- Ståhl, P.L.; Salmén, F.; Vickovic, S.; Lundmark, A.; Navarro, J.F.; Magnusson, J.; Giacomello, S.; Asp, M.; Westholm, J.O.; Huss, M.; et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science (New York, N.Y.) 2016, 353, 78–82. [Google Scholar] [CrossRef]
- Asp, M.; Bergenstråhle, J.; Lundeberg, J. Spatially Resolved Transcriptomes-Next Generation Tools for Tissue Exploration. 2020, 42, e1900221. [CrossRef]
- Zhuang, X. Spatially resolved single-cell genomics and transcriptomics by imaging. Nature methods 2021, 18, 18–22. [Google Scholar] [CrossRef]
- Larsson, L.; Frisén, J.; Lundeberg, J. Spatially resolved transcriptomics adds a new dimension to genomics. 2021, 18, 15–18. [CrossRef]
- Crosetto, N.; Bienko, M.; van Oudenaarden, A. Spatially resolved transcriptomics and beyond. Nature reviews. Genetics 2015, 16, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Moor, A.E.; Itzkovitz, S. Spatial transcriptomics: paving the way for tissue-level systems biology. Current opinion in biotechnology 2017, 46, 126–133. [Google Scholar] [CrossRef]
- Armingol, E.; Officer, A. Deciphering cell-cell interactions and communication from gene expression. 2021, 22, 71–88. [CrossRef]
- Rompolas, P.; Mesa, K.R.; Greco, V. Spatial organization within a niche as a determinant of stem-cell fate. Nature 2013, 502, 513–518. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-E.; Jo, S.H.; Lee, R.H.; Macks, C.P.; Ku, T.; Park, J.; Lee, C.W.; Hur, J.K.; Sohn, C.H. Spatial Transcriptomics: Technical Aspects of Recent Developments and Their Applications in Neuroscience and Cancer Research. Advanced Science n/a, 2206939. [CrossRef]
- Shaw, R.; Tian, X.; Xu, J. Single-Cell Transcriptome Analysis in Plants: Advances and Challenges. Molecular plant 2021, 14, 115–126. [Google Scholar] [CrossRef]
- Denyer, T.; Ma, X.; Klesen, S.; Scacchi, E.; Nieselt, K.; Timmermans, M.C.P. Spatiotemporal Developmental Trajectories in the Arabidopsis Root Revealed Using High-Throughput Single-Cell RNA Sequencing. Developmental cell 2019, 48, 840–852. [Google Scholar] [CrossRef]
- Shulse, C.N.; Cole, B.J.; Ciobanu, D.; Lin, J.; Yoshinaga, Y.; Gouran, M.; Turco, G.M.; Zhu, Y.; O'Malley, R.C.; Brady, S.M.; et al. High-Throughput Single-Cell Transcriptome Profiling of Plant Cell Types. Cell reports 2019, 27, 2241–2247. [Google Scholar] [CrossRef]
- Zhang, T.Q.; Xu, Z.G.; Shang, G.D.; Wang, J.W. A Single-Cell RNA Sequencing Profiles the Developmental Landscape of Arabidopsis Root. Molecular plant 2019, 12, 648–660. [Google Scholar] [CrossRef]
- Levitin, H.M.; Yuan, J.; Sims, P.A. Single-Cell Transcriptomic Analysis of Tumor Heterogeneity. Trends in cancer 2018, 4, 264–268. [Google Scholar] [CrossRef]
- Cao, J.; Spielmann, M.; Qiu, X.; Huang, X.; Ibrahim, D.M.; Hill, A.J.; Zhang, F.; Mundlos, S.; Christiansen, L.; Steemers, F.J.; et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 2019, 566, 496–502. [Google Scholar] [CrossRef] [PubMed]
- Haghverdi, L.; Lun, A.T.L. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. 2018, 36, 421–427. [CrossRef]
- Lavin, Y.; Kobayashi, S.; Leader, A.; Amir, E.D.; Elefant, N.; Bigenwald, C.; Remark, R.; Sweeney, R.; Becker, C.D.; Levine, J.H.; et al. Innate Immune Landscape in Early Lung Adenocarcinoma by Paired Single-Cell Analyses. Cell 2017, 169, 750–765. [Google Scholar] [CrossRef]
- Ji, A.L.; Rubin, A.J.; Thrane, K.; Jiang, S.; Reynolds, D.L.; Meyers, R.M.; Guo, M.G.; George, B.M.; Mollbrink, A.; Bergenstråhle, J.; et al. Multimodal Analysis of Composition and Spatial Architecture in Human Squamous Cell Carcinoma. Cell 2020, 182, 1661–1662. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.T.; Lu, A.; Craessaerts, K.; Pavie, B.; Sala Frigerio, C.; Corthout, N.; Qian, X.; Laláková, J.; Kühnemund, M.; Voytyuk, I.; et al. Spatial Transcriptomics and In Situ Sequencing to Study Alzheimer's Disease. Cell 2020, 182, 976–991. [Google Scholar] [CrossRef] [PubMed]
- Asp, M.; Giacomello, S.; Larsson, L.; Wu, C.; Fürth, D.; Qian, X.; Wärdell, E.; Custodio, J.; Reimegård, J.; Salmén, F.; et al. A Spatiotemporal Organ-Wide Gene Expression and Cell Atlas of the Developing Human Heart. Cell 2019, 179, 1647–1660. [Google Scholar] [CrossRef] [PubMed]
- Bansal, M.; Belcastro, V.; Ambesi-Impiombato, A.; di Bernardo, D. How to infer gene networks from expression profiles. Molecular systems biology 2007, 3, 78. [Google Scholar] [CrossRef]
- Xue, Z.; Huang, K.; Cai, C.; Cai, L.; Jiang, C.Y.; Feng, Y.; Liu, Z.; Zeng, Q.; Cheng, L.; Sun, Y.E.; et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 2013, 500, 593–597. [Google Scholar] [CrossRef] [PubMed]
- Darmanis, S.; Sloan, S.A.; Croote, D.; Mignardi, M.; Chernikova, S.; Samghababi, P.; Zhang, Y.; Neff, N.; Kowarsky, M.; Caneda, C.; et al. Single-Cell RNA-Seq Analysis of Infiltrating Neoplastic Cells at the Migrating Front of Human Glioblastoma. Cell reports 2017, 21, 1399–1410. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhou, X. Applications of Single-Cell Sequencing for Multiomics. Methods in molecular biology (Clifton, N.J.) 2018, 1754, 327–374. [Google Scholar] [CrossRef]
- Ley, T.J.; Mardis, E.R.; Ding, L.; Fulton, B.; McLellan, M.D.; Chen, K.; Dooling, D.; Dunford-Shore, B.H.; McGrath, S.; Hickenbotham, M.; et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 2008, 456, 66–72. [Google Scholar] [CrossRef]
- Lawson, D.A.; Kessenbrock, K. Tumour heterogeneity and metastasis at single-cell resolution. 2018, 20, 1349–1360. [CrossRef]
- Caswell, D.R.; Swanton, C. The role of tumour heterogeneity and clonal cooperativity in metastasis, immune evasion and clinical outcome. BMC medicine 2017, 15, 133. [Google Scholar] [CrossRef]
- Alderton, G.K. Tumour evolution: Epigenetic and genetic heterogeneity in metastasis. Nature reviews. Cancer 2017, 17, 141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yu, X.; Zheng, L.; Zhang, Y.; Li, Y.; Fang, Q.; Gao, R.; Kang, B.; Zhang, Q.; Huang, J.Y.; et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 2018, 564, 268–272. [Google Scholar] [CrossRef]
- Bian, S.; Hou, Y. Single-cell multiomics sequencing and analyses of human colorectal cancer. 2018, 362, 1060–1063. [CrossRef]
- Li, J.; Wang, R.; Zhou, X.; Wang, W. Genomic and transcriptomic profiling of carcinogenesis in patients with familial adenomatous polyposis. 2020, 69, 1283–1293. [CrossRef]
- Chen, Y.C.; Sahoo, S.; Brien, R.; Jung, S.; Humphries, B.; Lee, W.; Cheng, Y.H.; Zhang, Z.; Luker, K.E.; Wicha, M.S.; et al. Single-cell RNA-sequencing of migratory breast cancer cells: discovering genes associated with cancer metastasis. The Analyst 2019, 144, 7296–7309. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.W.; Chung, W.; Lee, H.-O.; Jeong, D.E.; Jo, A.; Lim, J.E.; Hong, J.H.; Nam, D.-H.; Jeong, B.C.; Park, S.H. Single-cell RNA sequencing reveals the tumor microenvironment and facilitates strategic choices to circumvent treatment failure in a chemorefractory bladder cancer patient. Genome medicine 2020, 12, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Narayanan, S.P.; Mannan, R.; Raskind, G.; Wang, X.; Vats, P.; Su, F.; Hosseini, N.; Cao, X.; Kumar-Sinha, C. Single-cell analyses of renal cell cancers reveal insights into tumor microenvironment, cell of origin, and therapy response. Proceedings of the National Academy of Sciences 2021, 118, e2103240118. [Google Scholar] [CrossRef]
- Shalek, A.K.; Benson, M. Single-cell analyses to tailor treatments. 2017, 9. [CrossRef]
- Mustachio, L.M.; Roszik, J. Single-Cell Sequencing: Current Applications in Precision Onco-Genomics and Cancer Therapeutics. Cancers 2022, 14, 657. [Google Scholar] [CrossRef]
- Crinier, A.; Milpied, P.; Escalière, B.; Piperoglou, C.; Galluso, J.; Balsamo, A.; Spinelli, L.; Cervera-Marzal, I.; Ebbo, M.; Girard-Madoux, M.; et al. High-Dimensional Single-Cell Analysis Identifies Organ-Specific Signatures and Conserved NK Cell Subsets in Humans and Mice. Immunity 2018, 49, 971–986. [Google Scholar] [CrossRef]
- Villani, A.C.; Satija, R. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. 2017, 356. [CrossRef]
- Xin, G.; Zander, R.; Schauder, D.M.; Chen, Y.; Weinstein, J.S.; Drobyski, W.R.; Tarakanova, V.; Craft, J. Single-cell RNA sequencing unveils an IL-10-producing helper subset that sustains humoral immunity during persistent infection. 2018, 9, 5037. [CrossRef]
- Papalexi, E.; Satija, R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nature reviews. Immunology 2018, 18, 35–45. [Google Scholar] [CrossRef]
- Martinez-Jimenez, C.P.; Eling, N.; Chen, H.C.; Vallejos, C.A.; Kolodziejczyk, A.A.; Connor, F.; Stojic, L.; Rayner, T.F.; Stubbington, M.J.T.; Teichmann, S.A.; et al. Aging increases cell-to-cell transcriptional variability upon immune stimulation. Science (New York, N.Y.) 2017, 355, 1433–1436. [Google Scholar] [CrossRef]
- Haber, A.L.; Biton, M.; Rogel, N.; Herbst, R.H.; Shekhar, K.; Smillie, C.; Burgin, G.; Delorey, T.M.; Howitt, M.R.; Katz, Y.; et al. A single-cell survey of the small intestinal epithelium. Nature 2017, 551, 333–339. [Google Scholar] [CrossRef]
- Gao, S.; Yan, L. Tracing the temporal-spatial transcriptome landscapes of the human fetal digestive tract using single-cell RNA-sequencing. 2018, 20, 721–734. [CrossRef]
- McConnell, M.J.; Lindberg, M.R.; Brennand, K.J.; Piper, J.C.; Voet, T.; Cowing-Zitron, C.; Shumilina, S.; Lasken, R.S.; Vermeesch, J.R.; Hall, I.M.; et al. Mosaic copy number variation in human neurons. Science (New York, N.Y.) 2013, 342, 632–637. [Google Scholar] [CrossRef]
- Li, H.; Horns, F.; Wu, B.; Xie, Q.; Li, J.; Li, T.; Luginbuhl, D.J.; Quake, S.R.; Luo, L. Classifying Drosophila Olfactory Projection Neuron Subtypes by Single-Cell RNA Sequencing. Cell 2017, 171, 1206–1220. [Google Scholar] [CrossRef]
- Luo, C.; Keown, C.L. Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. 2017, 357, 600–604. [CrossRef]
- Lake, B.B.; Ai, R.; Kaeser, G.E.; Salathia, N.S.; Yung, Y.C.; Liu, R.; Wildberg, A.; Gao, D.; Fung, H.L.; Chen, S.; et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science (New York, N.Y.) 2016, 352, 1586–1590. [Google Scholar] [CrossRef] [PubMed]
- Carter, R.A.; Bihannic, L.; Rosencrance, C.; Hadley, J.L.; Tong, Y.; Phoenix, T.N.; Natarajan, S.; Easton, J.; Northcott, P.A.; Gawad, C. A Single-Cell Transcriptional Atlas of the Developing Murine Cerebellum. Current biology : CB 2018, 28, 2910–2920. [Google Scholar] [CrossRef]
- Hou, Y.; Fan, W.; Yan, L.; Li, R.; Lian, Y.; Huang, J.; Li, J.; Xu, L.; Tang, F.; Xie, X.S.; et al. Genome analyses of single human oocytes. Cell 2013, 155, 1492–1506. [Google Scholar] [CrossRef]
- Li, W.; Ma, Y.; Yu, S.; Sun, N.; Wang, L.; Chen, D.; Yang, G.; Lu, S.; Li, Y.; Yang, B.; et al. The mutation-free embryo for in vitro fertilization selected by MALBAC-PGD resulted in a healthy live birth from a family carrying PKD 1 mutation. Journal of assisted reproduction and genetics 2017, 34, 1653–1658. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Yang, M.; Guo, H.; Yang, L.; Wu, J.; Li, R.; Liu, P.; Lian, Y.; Zheng, X.; Yan, J.; et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nature structural & molecular biology 2013, 20, 1131–1139. [Google Scholar] [CrossRef]
- Briggs, J.A.; Weinreb, C.; Wagner, D.E. The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. 2018, 360. [CrossRef]
- Li, L.; Guo, F. Publisher Correction: Single-cell multi-omics sequencing of human early embryos. 2018, 20, 1227. [CrossRef]
- Vento-Tormo, R.; Efremova, M.; Botting, R.A.; Turco, M.Y.; Vento-Tormo, M.; Meyer, K.B.; Park, J.E.; Stephenson, E.; Polański, K.; Goncalves, A.; et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 2018, 563, 347–353. [Google Scholar] [CrossRef]
- Chen, Y.; Zheng, Y.; Gao, Y.; Lin, Z.; Yang, S.; Wang, T.; Wang, Q.; Xie, N.; Hua, R.; Liu, M.; et al. Single-cell RNA-seq uncovers dynamic processes and critical regulators in mouse spermatogenesis. 2018, 28, 879–896. [CrossRef]
- Wang, M.; Liu, X.; Chang, G.; Chen, Y.; An, G.; Yan, L.; Gao, S.; Xu, Y.; Cui, Y.; Dong, J.; et al. Single-Cell RNA Sequencing Analysis Reveals Sequential Cell Fate Transition during Human Spermatogenesis. Cell stem cell 2018, 23, 599–614. [Google Scholar] [CrossRef]
- Marinov, G.K.; Williams, B.A.; McCue, K.; Schroth, G.P.; Gertz, J.; Myers, R.M.; Wold, B.J. From single-cell to cell-pool transcriptomes: stochasticity in gene expression and RNA splicing. Genome research 2014, 24, 496–510. [Google Scholar] [CrossRef] [PubMed]
- Brennecke, P.; Anders, S.; Kim, J.K.; Kołodziejczyk, A.A.; Zhang, X.; Proserpio, V.; Baying, B.; Benes, V.; Teichmann, S.A.; Marioni, J.C.; et al. Accounting for technical noise in single-cell RNA-seq experiments. Nature methods 2013, 10, 1093–1095. [Google Scholar] [CrossRef]
- Ding, B.; Zheng, L.; Zhu, Y.; Li, N.; Jia, H.; Ai, R.; Wildberg, A.; Wang, W. Normalization and noise reduction for single cell RNA-seq experiments. Bioinformatics (Oxford, England) 2015, 31, 2225–2227. [Google Scholar] [CrossRef]
- Grün, D.; Kester, L.; van Oudenaarden, A. Validation of noise models for single-cell transcriptomics. Nature methods 2014, 11, 637–640. [Google Scholar] [CrossRef]
- Grindberg, R.V.; Yee-Greenbaum, J.L.; McConnell, M.J.; Novotny, M.; O'Shaughnessy, A.L.; Lambert, G.M.; Araúzo-Bravo, M.J.; Lee, J.; Fishman, M.; Robbins, G.E.; et al. RNA-sequencing from single nuclei. Proceedings of the National Academy of Sciences of the United States of America 2013, 110, 19802–19807. [Google Scholar] [CrossRef]
- Lee, J.H.; Daugharthy, E.R.; Scheiman, J.; Kalhor, R. Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. 2015, 10, 442–458. [CrossRef]
- Kang, Y.; Norris, M.H.; Zarzycki-Siek, J.; Nierman, W.C.; Donachie, S.P.; Hoang, T.T. Transcript amplification from single bacterium for transcriptome analysis. Genome research 2011, 21, 925–935. [Google Scholar] [CrossRef]
- Armour, C.D.; Castle, J.C.; Chen, R.; Babak, T.; Loerch, P.; Jackson, S.; Shah, J.K.; Dey, J.; Rohl, C.A.; Johnson, J.M.; et al. Digital transcriptome profiling using selective hexamer priming for cDNA synthesis. Nature methods 2009, 6, 647–649. [Google Scholar] [CrossRef] [PubMed]
- Fincher, C.T.; Wurtzel, O.; de Hoog, T.; Kravarik, K.M.; Reddien, P.W. Cell type transcriptome atlas for the planarian Schmidtea mediterranea. 2018, 360. 360. [CrossRef]
- Cao, J.; Packer, J.S. Comprehensive single-cell transcriptional profiling of a multicellular organism. 2017, 357, 661–667. [CrossRef]
- Replogle, J.M.; Norman, T.M.; Xu, A. Combinatorial single-cell CRISPR screens by direct guide RNA capture and targeted sequencing. 2020, 38, 954–961. [CrossRef]
- Zafar, H.; Lin, C.; Bar-Joseph, Z. Single-cell lineage tracing by integrating CRISPR-Cas9 mutations with transcriptomic data. 2020, 11, 3055. [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Perkel, J.M. Single-cell proteomics takes centre stage. Nature 2021, 597, 580–582. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



