Submitted:
16 February 2024
Posted:
22 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Methodology
3. Results
4. Discussion
5. Conclusion
Author Contributions
Funding
Competing Interests
Ethics approval
References
- Cazzola M: Myelodysplastic Syndromes. N Engl J Med 2020, 383, 1358–1374. [CrossRef]
- Patel SS: Pediatric Myelodysplastic Syndromes. Clin Lab Med 2021, 41, 517–528. [CrossRef] [PubMed]
- Balaian E, Wobus M, Bornhäuser M, Chavakis T, Sockel K: Myelodysplastic Syndromes and Metabolism. Int J Mol Sci 2021, 22, 11250. [CrossRef]
- Lauritsen TB, Norgaard JM, Dalton SO, Gronbaek K, El-Galaly TC, Ostgard LSG: 10-year nationwide trends in incidence, treatment patterns, and mortality of patients with myelodysplastic syndromes in Denmark. Leuk Res 2023, 128, 107056. [CrossRef]
- Hosono N: Genetic abnormalities and pathophysiology of MDS. Int J Clin Oncol 2019, 24, 885–892. [CrossRef]
- Ogawa S: Genetics of MDS. Blood 2019, 133, 1049–1059. [CrossRef] [PubMed]
- Cantor JR, Sabatini DM: Cancer cell metabolism: one hallmark, many faces. Cancer discovery 2012, 2, 881–898. [CrossRef]
- Folmes CD, Dzeja PP, Nelson TJ, Terzic A: Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell 2012, 11, 596–606. [CrossRef] [PubMed]
- Warburg O: On respiratory impairment in cancer cells. Science 1956, 124, 269–270. [CrossRef]
- Hoff FW, Madanat YF: Molecular Drivers of Myelodysplastic Neoplasms (MDS)-Classification and Prognostic Relevance. Cells 2023, 12, 627. [CrossRef]
- Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, Sultan C: Proposals for the classification of the myelodysplastic syndromes. Br J Haematol 1982, 51, 189–199. [CrossRef]
- Harris NL, Jaffe ES, Diebold J, Flandrin G, Muller-Hermelink HK, Vardiman J, Lister TA, Bloomfield CD: The World Health Organization classification of neoplastic diseases of the hematopoietic and lymphoid tissues. Report of the Clinical Advisory Committee meeting, Airlie House, Virginia, November, 1997. Ann Oncol 1999, 10, 1419–1432. [CrossRef]
- Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, Bloomfield CD, Cazzola M, Vardiman JW: The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [CrossRef] [PubMed]
- Pellagatti A, Armstrong RN, Steeples V, Sharma E, Repapi E, Singh S, Sanchi A, Radujkovic A, Horn P, Dolatshad H et al: Impact of spliceosome mutations on RNA splicing in myelodysplasia: dysregulated genes/pathways and clinical associations. Blood 2018, 132, 1225–1240. [CrossRef]
- Choudhary GS, Pellagatti A, Agianian B, Smith MA, Bhagat TD, Gordon-Mitchell S, Sahu S, Pandey S, Shah N, Aluri S et al: Activation of targetable inflammatory immune signaling is seen in myelodysplastic syndromes with SF3B1 mutations. Elife 2022, 11, e78136. [CrossRef]
- Dolatshad H, Pellagatti A, Fernandez-Mercado M, Yip BH, Malcovati L, Attwood M, Przychodzen B, Sahgal N, Kanapin AA, Lockstone H et al: Disruption of SF3B1 results in deregulated expression and splicing of key genes and pathways in myelodysplastic syndrome hematopoietic stem and progenitor cells. Leukemia 2015, 29, 1092–1103. [CrossRef]
- Berastegui N, Ainciburu M, Romero JP, Garcia-Olloqui P, Alfonso-Pierola A, Philippe C, Vilas-Zornoza A, San Martin-Uriz P, Ruiz-Hernández R, Abarrategi A et al: The transcription factor DDIT3 is a potential driver of dyserythropoiesis in myelodysplastic syndromes. Nat Commun 2022, 13, 7619. [CrossRef]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E et al: The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery 2012, 2, 401–404. [CrossRef]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E et al: Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling 2013, 6, l1. [CrossRef]
- Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, Potter NE, Heuser M, Thol F, Bolli N et al: Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med 2016, 374, 2209–2221. [CrossRef]
- Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL, Long N, Schultz AR, Traer E, Abel M et al: Functional genomic landscape of acute myeloid leukaemia. Nature 2018, 562, 526–531. [CrossRef]
- Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, Yoon CJ, Ellis P, Wedge DC, Pellagatti A et al: Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 2013, 122, 3616–3627. [CrossRef]
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M et al: Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011, 478, 64–69. [CrossRef]
- IDH1 isocitrate dehydrogenase (NADP(+)) 1 [ Homo sapiens (human) ]. In.; 27-Aug-2023.
- IDH2 isocitrate dehydrogenase (NADP(+)) 2 [ Homo sapiens (human) ]. In.; 18-Aug-2023.
- Welch JS, Petti AA, Miller CA, Fronick CC, O’Laughlin M, Fulton RS, Wilson RK, Baty JD, Duncavage EJ, Tandon B et al: TP53 and Decitabine in Acute Myeloid Leukemia and Myelodysplastic Syndromes. N Engl J Med 2016, 375, 2023–2036. [CrossRef]
- TET2 tet methylcytosine dioxygenase 2 [ Homo sapiens (human) ]. In.; 18-Aug-2023.
- DNMT3A DNA methyltransferase 3 alpha [ Homo sapiens (human) ]. In.; 18-Aug-2023.
- ASXL1 ASXL transcriptional regulator 1 [ Homo sapiens (human) ]. In.; 18-Aug-2023.
- SF3B1 splicing factor 3b subunit 1 [ Homo sapiens (human) ]. In.; 18-Aug-2023.
- SRSF2 serine and arginine rich splicing factor 2 [ Homo sapiens (human) ]. In.; 18-Aug-2023.
- RUNX1 RUNX family transcription factor 1 [ Homo sapiens (human) ]. In.; 18-Aug-2023.
- NPM1 nucleophosmin 1 [ Homo sapiens (human) ]. In.; 18-Aug-2023.
- IL1A interleukin 1 alpha [ Homo sapiens (human) ]. In.; 10-Oct-2023.
- OLR1 oxidized low density lipoprotein receptor 1 [ Homo sapiens (human) ]. In.; 10-Oct-2023.
- ADRA2B adrenoceptor alpha 2B [ Homo sapiens (human) ]. In.; 10-Oct-2023.
- ERFE erythroferrone [ Homo sapiens (human) ]. In.; 10-Oct-2023.
- SULT4A1 sulfotransferase family 4A member 1 [ Homo sapiens (human) ]. In.; 10-Oct-2023.
- Gangat N, Patnaik MM, Tefferi A: Myelodysplastic syndromes: Contemporary review and how we treat. Am J Hematol 2016, 91, 76–89. [CrossRef]
- Lu SC: Glutathione synthesis. Biochim Biophys Acta 2013, 1830, 3143–3153. [CrossRef]
- Xu P, Hu G, Luo C, Liang Z: DNA methyltransferase inhibitors: an updated patent review (2012-2015). Expert Opin Ther Pat 2016, 26, 1017–1030. [CrossRef]
- Guan Y, Tiwari AD, Phillips JG, Hasipek M, Grabowski DR, Pagliuca S, Gopal P, Kerr CM, Adema V, Radivoyevitch T et al: A Therapeutic Strategy for Preferential Targeting of TET2 Mutant and TET-dioxygenase Deficient Cells in Myeloid Neoplasms. Blood Cancer Discov 2021, 2, 146–161. [CrossRef]
- Simonetti G, Mengucci C, Padella A, Fonzi E, Picone G, Delpino C, Nanni J, De Tommaso R, Franchini E, Papayannidis C et al: Integrated genomic-metabolic classification of acute myeloid leukemia defines a subgroup with NPM1 and cohesin/DNA damage mutations. Leukemia 2021, 35, 2813–2826. [CrossRef]
- Kinnaird A, Zhao S, Wellen KE, Michelakis ED: Metabolic control of epigenetics in cancer. Nature reviews Cancer 2016, 16, 694–707. [CrossRef]
- Thakur C, Chen F: Connections between metabolism and epigenetics in cancers. Semin Cancer Biol 2019, 57, 52–58. [CrossRef] [PubMed]
- Morrison AJ: Cancer cell metabolism connects epigenetic modifications to transcriptional regulation. Febs j 2022, 289, 1302–1314. [CrossRef] [PubMed]
- Lelièvre P, Sancey L, Coll JL, Deniaud A, Busser B: Iron Dysregulation in Human Cancer: Altered Metabolism, Biomarkers for Diagnosis, Prognosis, Monitoring and Rationale for Therapy. Cancers (Basel) 2020, 12, 3524. [CrossRef]
- Garcia PL, Hossain MI, Andrabi SA, Falany CN: Generation and Characterization of SULT4A1 Mutant Mouse Models. Drug Metab Dispos 2018, 46, 41–45. [CrossRef] [PubMed]
- Hossain MI, Marcus JM, Lee JH, Garcia PL, Gagné JP, Poirier GG, Falany CN, Andrabi SA: SULT4A1 Protects Against Oxidative-Stress Induced Mitochondrial Dysfunction in Neuronal Cells. Drug Metab Dispos 2019, 47, 949–953. [CrossRef]
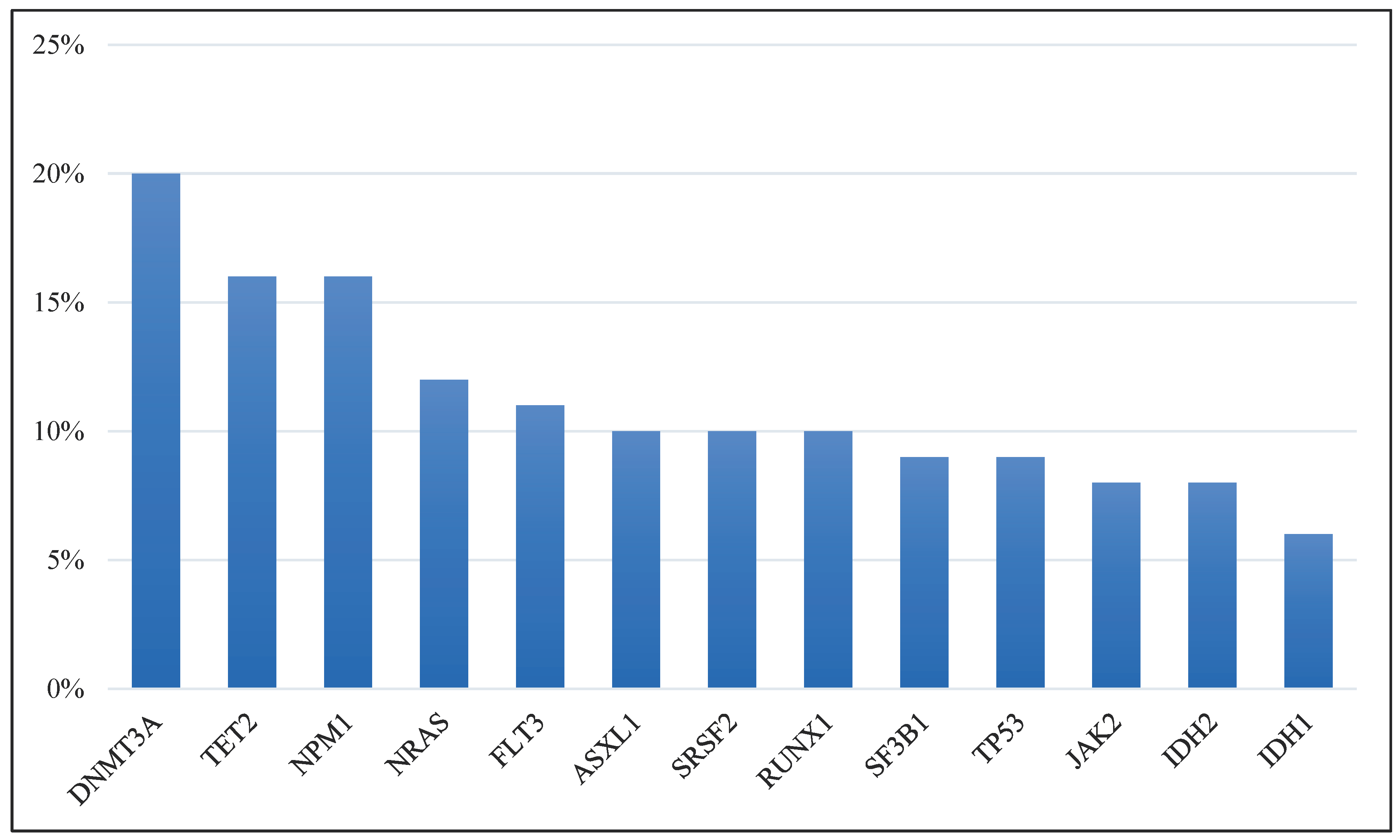
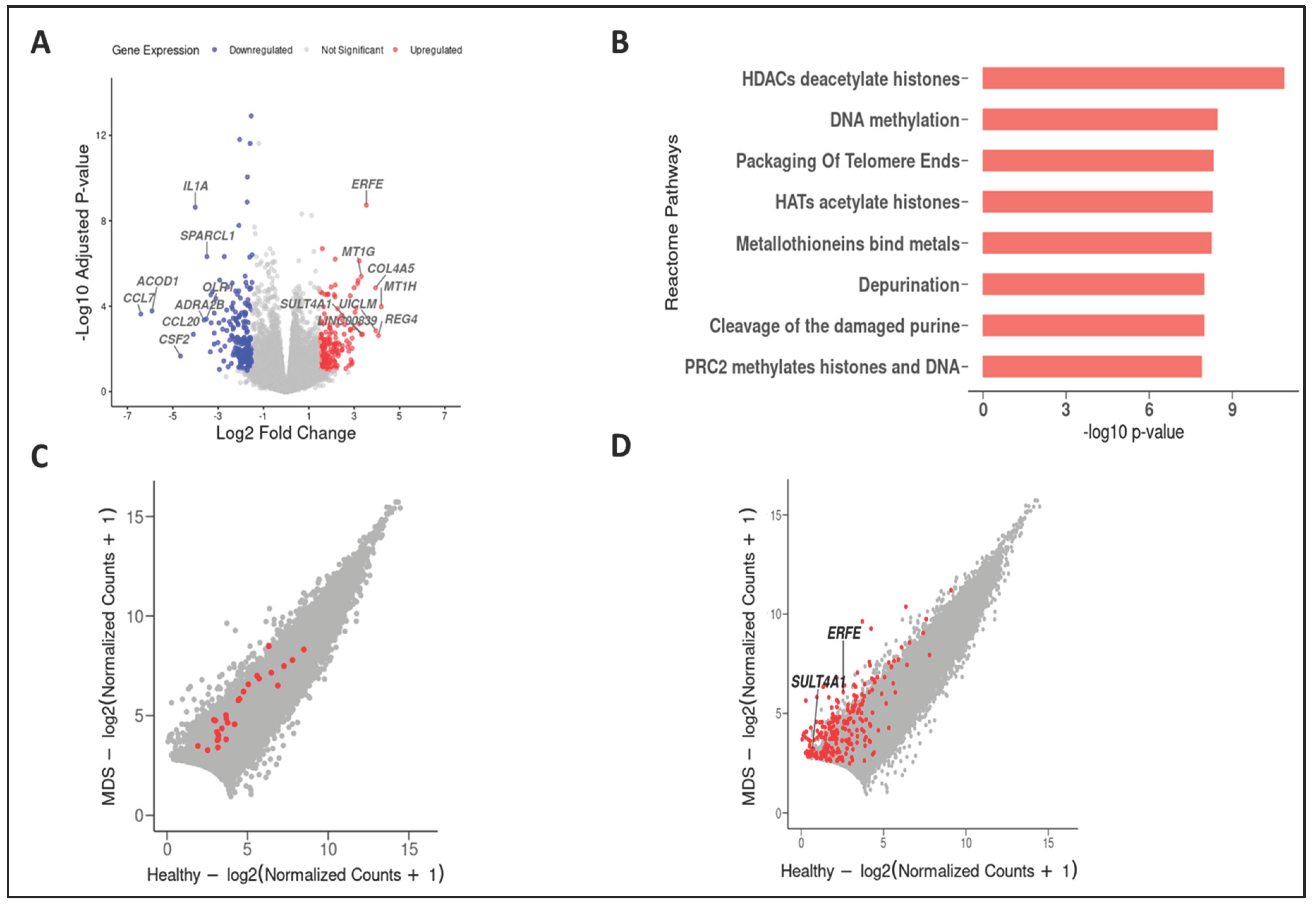
| A | B | Neither | A Not B | B Not A | Both | p-Value | Tendency |
|---|---|---|---|---|---|---|---|
| SRSF2 | DNMT3A | 3026 | 379 | 816 | 39 | <0.001 | Mutual exclusivity |
| ASXL1 | DNMT3A | 3014 | 391 | 810 | 45 | <0.001 | Mutual exclusivity |
| JAK2 | DNMT3A | 3110 | 295 | 814 | 41 | <0.001 | Mutual exclusivity |
| TP53 | DNMT3A | 3063 | 342 | 796 | 59 | 0.002 | Mutual exclusivity |
| NCT Number | Study Status | Conditions | Targeted Biological Process | Interventions/Drugs/Procedure |
|---|---|---|---|---|
| NCT04493164 | Recruiting | MDS/AML | Mutant IDH1 Inhibitor | Ivosidenib/Liposome-encapsulated Daunorubicin-Cytarabine |
| NCT03503409 | Recruiting | MDS/AML | Mutant IDH1 Inhibitor | AG-120 |
| NCT03744390 | Recruiting | MDS/AML | Mutant IDH2 Inhibitor | AG-221 |
| NCT04827719 | Recruiting | MDS/AML | High-dose cytarabine delivery | BST-236 |
| NCT04279847 | Recruiting | MDS | BET inhibitor and JAK inhibitor | INCB057643/Ruxolitinib |
| NCT04140487 | Recruiting | MDS/AML | FLT3 tyrosine kinase inhibitors and DNA Methylation and synthesis Inhibitor | Azacitidine/Gilteritinib/Venetoclax |
| NCT05010122 | Recruiting | MDS/AML | FLT3 tyrosine kinase inhibitor and DNMT1 Inhibitor | Decitabine and Cedazuridine/Gilteritinib/Venetoclax |
| NCT03661307 | Recruiting | MDS/AML | DNMT1 Inhibitor and FLT3 tyrosine kinase inhibitor | Decitabine/Quizartinib/Venetoclax |
| NCT03683433 | Recruiting | MDS/AML/CML | DNA Methylation and synthesis Inhibitor and Mutant IDH Inhibitor | Azacitidine/Enasidenib Mesylate |
| NCT05636514 | Recruiting | MDS/AML/CML | FLT3 tyrosine kinase inhibitor and FAK inhibitor | Decitabine-Cedazuridine 35 Mg-100 Mg ORAL TABLET/Defactinib |
| NCT04493138 | Recruiting | MDS/CML | DNMT1 Inhibitor and FLT3 tyrosine kinase inhibitor | Azacitidine/Quizartinib |
| NCT05817955 | Recruiting | MDS | DNA Methylation and synthesis Inhibitor and JAK Inhibitor | Azacitidine (AZA) with Ruxolitinib |
| NCT04803721 | Recruiting | MDS | ||
| NCT04250051 | Recruiting | MDS/AML | Mutant IDH1 Inhibitor | Cytarabine/Filgrastim/Fludarabine/Fludarabine Phosphate/Ivosidenib |
| NCT04167917 | Recruiting | MDS/AML/CML | DNA methyltransferase 1 (DNMT1) inhibition | NTX-301 |
| NCT04187703 | Recruiting | MDS | DNA methyltransferase (DNMT) inhibition | 5-azacytidine/Decitabine |
| NCT05282459 | Recruiting | MDS | Mutant IDH2 Inhibitor | Enasidenib mesylat dose escalation |
| NCT04741945 | Recruiting | MDS | Antihyperglycemic | Metformin |
| NCT04477291 | Recruiting | MDS/AML | FLT3 Inhibitor | CG-806 |
| NCT03839771 | Recruiting | MDS/AML | Mutant IDH1 Inhibitor and Mutant IDH2 Inhibitor | AG-120/Placebo for AG-120/AG-221/Placebo for AG-221 |
| NCT05030675 | Recruiting | MDS/CML | Tyrosine kinase inhibitor | Fostamatinib |
| NCT03953898 | Recruiting | MDS/AML | poly (ADP-ribose) polymerase (PARP) inhibitor | Biospecimen Collection/Bone Marrow Aspiration/Olaparib |
| NCT03999723 | Recruiting | MDS/AML/CML | Vitamin C/Placebo | |
| NCT04764474 | Recruiting | Hematological Malignancies With IDH mutations | Mutant IDH1/2 Inhibitor | HMPL-306 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





