2.2. Methods
Outline of ISM and comparison with other Latent Space approaches
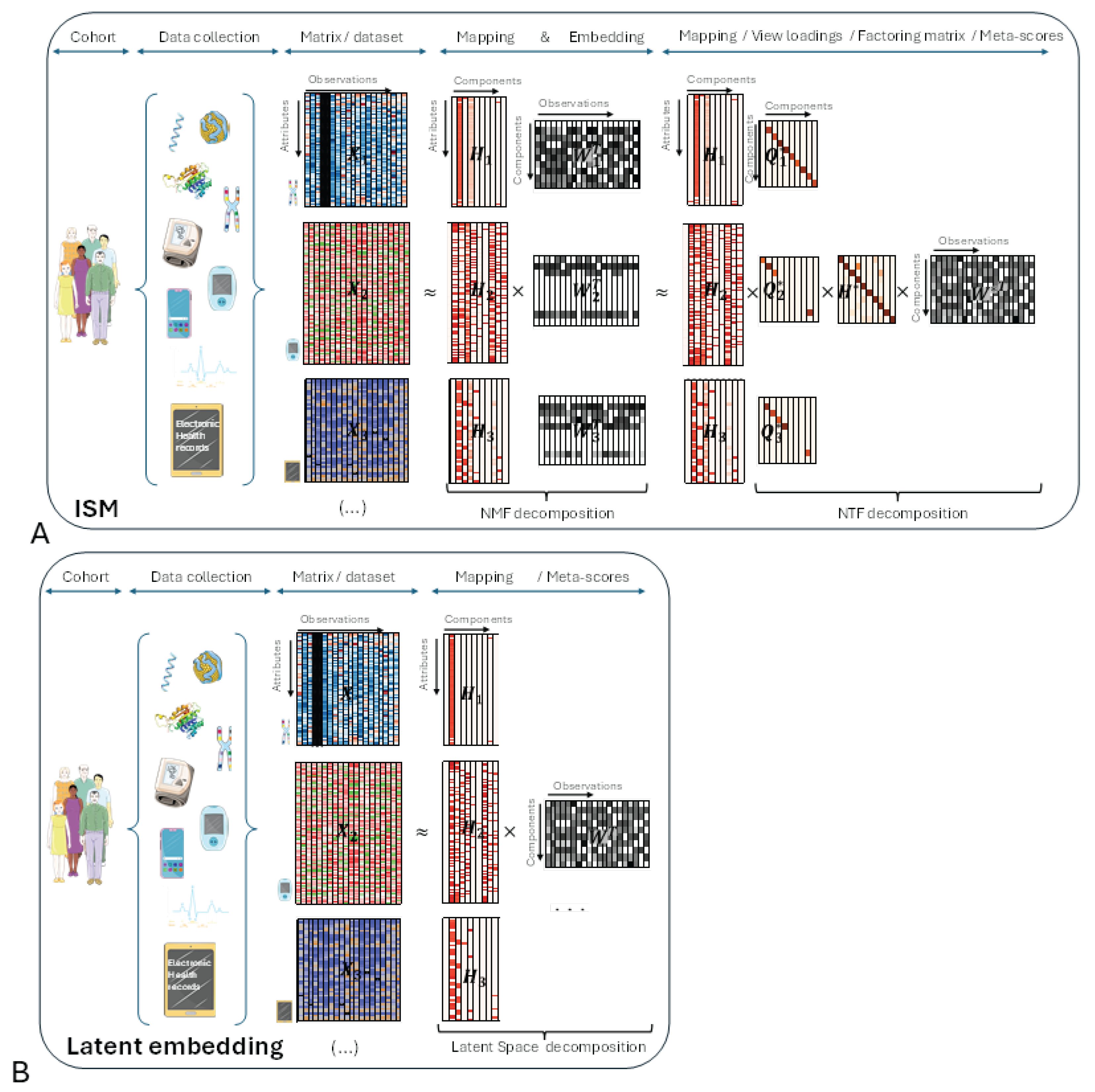
Before delving into the details of the ISM workflow, let’s introduce the main underlying ideas with an illustrative figure (
Figure 1, panel A) and try to compare with other Latent Space approaches (
Figure 2, panel B). The different views are represented by heatmaps on the left side of both panels, with attributes on the vertical axis and observations on the horizontal axis, respectively.
In the central part of the figure, each non-negative view has been decomposed into the product of two non-negative matrices and using NMF. Each matrix corresponds to the transformation of a particular view to a latent space that is common to all transformed views. ISM ensures that the transformed views share the same number and type of latent attributes, called components, as explained in the detailed description. This transforming process, which we call embedding, results in a three-dimensional array, or tensor. The corresponding matrices contain the loadings of the original attributes on each component. We call these matrices the mapping between the original and transformed views.
In the right part of the figure, the three-dimensional array is decomposed into the tensor product of three matrices: using NTF. contains the meta-scores – the single transformation to the latent space common to all views. and contain the loadings of NMF latent attributes and views, respectively, on each NTF component. Each row of is represented by a diagonal matrix, where the diagonal contains the loadings for a particular view. This allows, for each view of the tensor, to translate in the figure the tensor product into a simple matrix product
Other Latent Space approaches (
Figure 1B):
In the right part of the figure, each view has been decomposed into the product of two matrices and using the latent space method algorithm. As with ISM, contains the meta-scores – the single transformation in the latent space common to all views.
Figure 1.
Comparison between ISM, (panel A) and other Latent Space approaches (panel B).
Figure 1.
Comparison between ISM, (panel A) and other Latent Space approaches (panel B).
Comparison between ISM and other Latent Space approaches:
If we multiply in
Figure 1A each mapping matrix
by
, we obtain a representation that is similar to
Figure 1B. This shows that ISM belongs to the family of Latent Space decomposition methods. However, view loadings are a constitutive part of ISM, whereas they are derived in other models, e.g., using variance decomposition by factor as in the MOFA article [
3].
Important implications of ISM’s preliminary embedding:
As will be seen in the detailed description of the workflow, ISM begins by applying NMF to the concatenated views. Importantly, NMF can be applied to each view independently, leading to view-specific decompositions before ISM itself is applied to the NMF transformed views . In this case, the view mapping returned by ISM, , refers to the NMF components of each . However, by embedding the in a 3-dimensional array, ISM allows to be mapped back to the original views by simple chained matrix multiplication so that: with . This “meta-ISM” approach has important consequences in several respects, which will be presented in the Discussion.
Detailed workflows
In this section, we present three workflows. The first workflow consists of training the ISM model to generate a latent space representation and view-mapping. The second workflow enables the projection of new observations obtained in multiple views into the latent space. The third workflow contains the detailed analysis steps for each example.
Workflow 1: Latent space representation and view-mapping
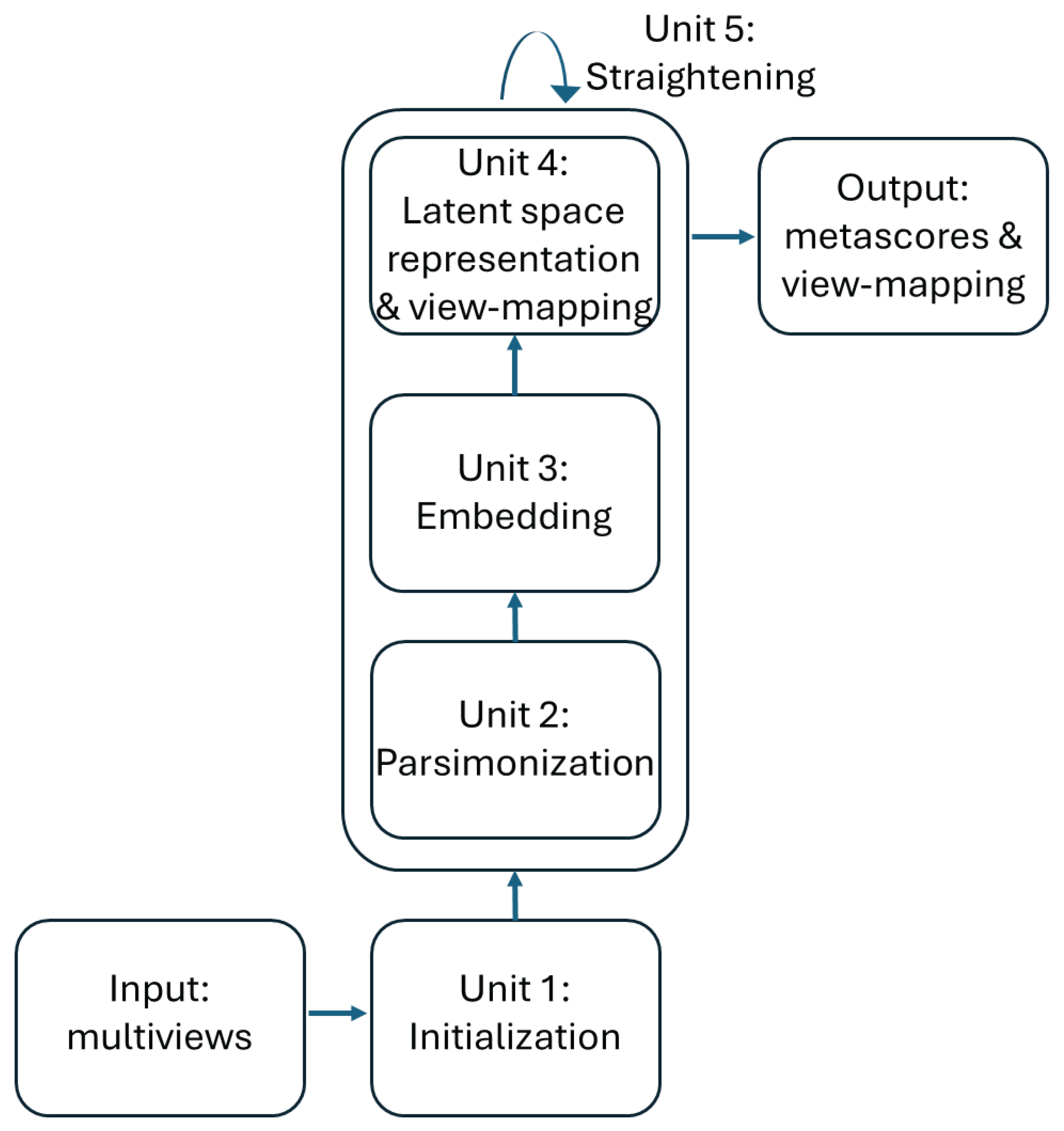
The training of the ISM model can be divided into 5 units as described in
Figure 2. The first 4 process units enable the discovery of the latent space in an “embedding” space. Once the latent space has been found, it is assimilated with the embedding space. During the fifth “straightening” unit, the latent space remains fixed, while the sequence of units 3, 4 and 2 is repeated to further parsimonize the view-mapping until the degree of sparsity remains unchanged. The sizes of the embedding space and the latent space are discussed in the section describing the third workflow.
Figure 2.
Training of the ISM model.
Figure 2.
Training of the ISM model.
Unit 1: Initialization
A non-negative matrix factorization is first performed on the matrix
of the
concatenated views
, resulting in the decomposition:
where
represents the transformed data, the columns of
contain the loadings of the
attributes across all views on each component,
is the embedding size and
is the total number of observations.
|
Unit 1 Initialization |
| |
Input: views where is the number of rows common to all views and is the number of columns in the view (it is assumed for each column that its values lie between 0 and 1 after normalization by the maximum row value). |
| |
Output: Factoring matrices where is the embedding dimension and is the sum of the number of columns in all views, and the matrix of concatenated views . |
| |
1: Concatenate the views: ; |
| |
2: Factorize using NMF with components: |
| |
|
|
Unit 2: Parsimonization
The initial degree of sparsity in
is crucial to the embedding dimensions from being overly distorted between the different views during the embedding process, as will be seen in the next section. This is achieved by applying a hard-threshold to each column of the
matrix. The threshold is based on the reciprocal of the Herfindhal-Hirschman index [
16], which provides an estimate of the number of non-negligible values in a non-negative vector. For columns with strongly positively skewed values, the use of the L2 norm for the estimate’s denominator can lead to excessively sparse factors, which in turn can lead to an overly large approximation error during embedding. Therefore, the estimate is multiplied by a coefficient whose default value was set at 0.8 after extensive testing with various data sets.
|
Unit 2 Parsimonization |
| |
Input: Factoring matrix
|
| |
Output: Parsimonized factoring matrix (since the initial is not used outside parsimonization, we use the same symbol for the sake of simplicity) |
| |
1: for each component of do
|
| |
2: |
Calculate Herfindahl-Hirschman Inverse Index to estimate the number of non-negligible entries in :
; |
| |
3: |
Enforce sparsity on using hard-thresholding:
0 where is a sparsity parameter (01, the default value 8 was chosen as it led in many trials to better results than the original index , which may be a too strict filter); |
| |
4: end for
|
Unit 3: Embedding
and are further updated along each view, yielding matrices of common shape (number of observations factorization rank ) corresponding to the transformed views.
NMF multiplicative updates are used during view matching to leave the zeros in the primary
matrix unchanged. Further optimizations of the simplicial cones
for each view
are therefore limited to the non-zero loadings so that they remain tightly connected. This ensures that the transformed views
form a tensor. Multiplicative updates usually start with a linear rate of convergence, which becomes sublinear after a few hundred iterations [
17]. By default, the number of iterations is set to 200 to ensure a reasonable approximation to each view, as required for the latent space representation described in the next section.
|
Unit 3 Embedding |
| |
Input: views and factoring matrices
|
| |
Output: view-specific factoring matrices and tensor . |
| |
1: for each view do
|
| |
2: |
Define as the part of corresponding to view ; |
| |
3: |
Factorize into view-specific using NMF multiplicative updating rules and initialization matrices :
|
| |
4: |
Normalize each component of by its maximum value and update accordingly; |
| |
5: |
Define tensor slice: |
| |
6: end for
|
Unit 4: Latent space representation and view-mapping
The resulting tensor
is analyzed using NTF, which leads to the decomposition:
where
and
is the dimension of the latent space. The components
,
and
enable the reconstruction of the horizontal, lateral and frontal slices of the embedding tensor: the loadings of the views on each component are contained in the matrix
; the integrated multiple views, or
meta-scores, are contained in the matrix
; and the matrix
represents the latent space in the form of a simplicial cone contained in the embedding space. Finally, the view-mapping matrix
is updated by applying steps 3-8 of unit 4. Its sparsity is ensured by further applying the parsimonization unit 2.
|
Unit 4 Latent space representation & View-mapping |
| |
Input: view-specific factoring matrices and tensor
|
| |
Output: NTF factors and view-mapping matrix . |
| |
1: Define view-mapping matrix as the concatenation of |
| |
2: Factorize using NTF with components:
where |
| |
3: Update view-mapping matrix : ; |
| |
4: for each view do
|
| |
5: |
Update : ; |
| |
6 end for
|
| |
7: Update view-mapping matrix as the concatenation of updated ; |
| |
8: Parsimonize view-mapping matrix by applying Unit 2; |
Unit 5: Straightening
The sparsity of the view-mapping matrix
can be further optimized together with the meta-scores
and the view-loadings
by repeating units 3, 4 and 2 until the number of 0-entries in
remains unchanged. To achieve this, the embedding is restricted to the latent space defined by the simplicial cone formed by
. In this simplified embedding space,
becomes the Identity matrix
when the updating process of
and
starts. In other words, embedding and latent spaces are being assimilated during the straightening process. Optionally, for faster convergence,
can be fixed to
, at the cost of a slightly higher approximation error, as observed in numerous experiments, due to only small deviations from
.
|
Unit 5 Straightening |
| |
Input: . |
| |
Output: NTF factors and updated view-mapping matrix . |
| |
1: Set where is the size of the latent space; |
| |
2: do until number of zero entries in remains unchanged |
| |
3: |
Apply Unit 3 to embed using embedding size , initialization matrices and view-mapping matrix found in previous iteration; |
| |
4: |
Apply Unit 4 to factorize and update view-mapping matrix , using embedding size initialization matrices obtained in previous iteration, and fixed ; |
| |
5: end for
|
Workflow 2: Projection of new observations
For new observations
comprising
views,
, ISM parameters
,
and view-mapping matrix
can be used to project
on the latent ISM components, as described in workflow 2.
|
Workflow 2 Projection of new observations |
| |
Input: New observations ( views, ), |
| |
|
NTF factors and mapping matrix . |
| |
Output: Estimation . |
| |
1: Disregard any views in that are absent in ; |
| |
2: Apply Unit 3 of workflow 1 to embed with initialized with ones and with fixed mapping matrix ; |
| |
3: Apply step 2 of Unit 4 of workflow 1 to calculate with fixed NTF factors and define the projection of on the latent space as ; |
Workflow 3: Data analysis
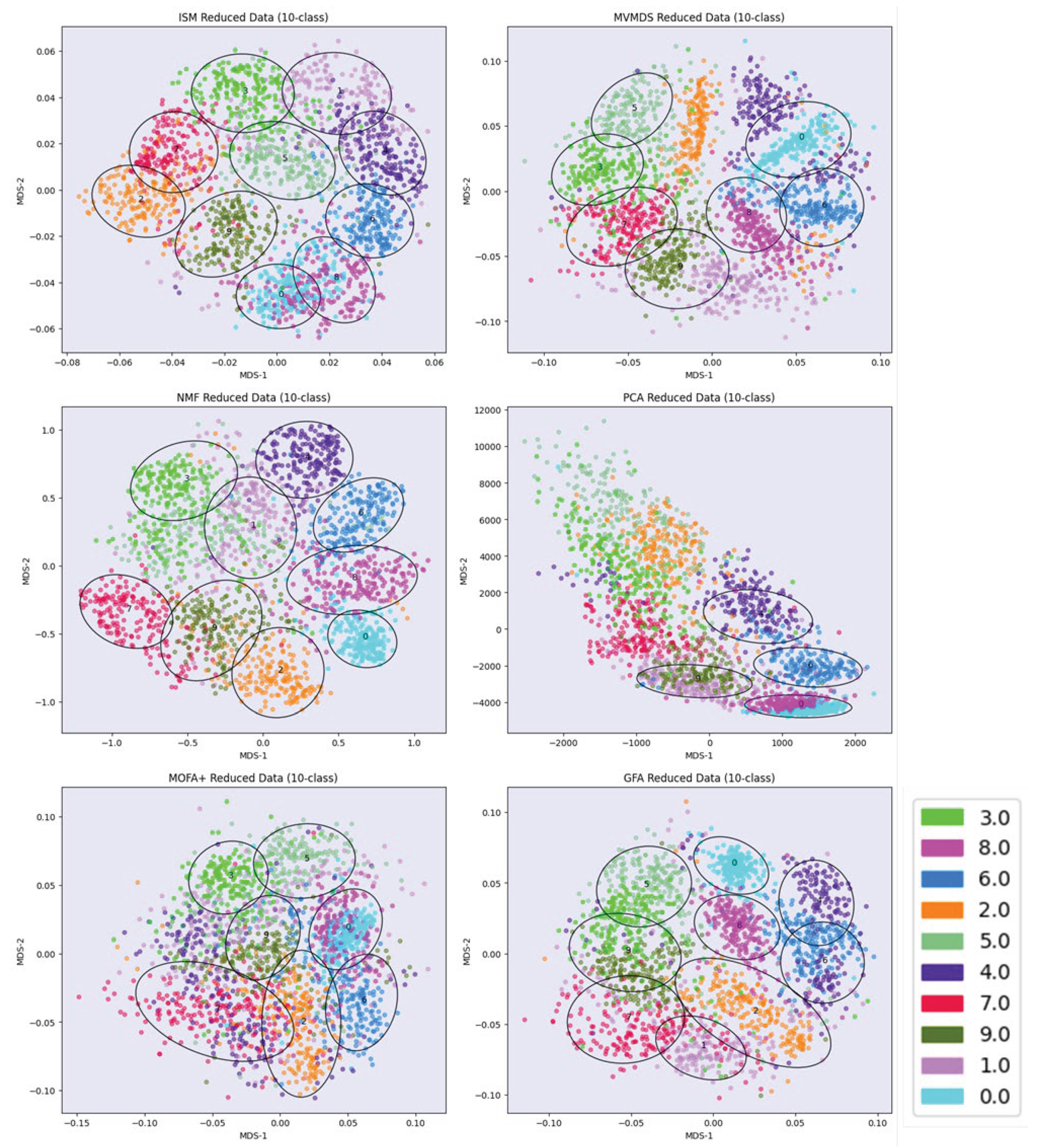
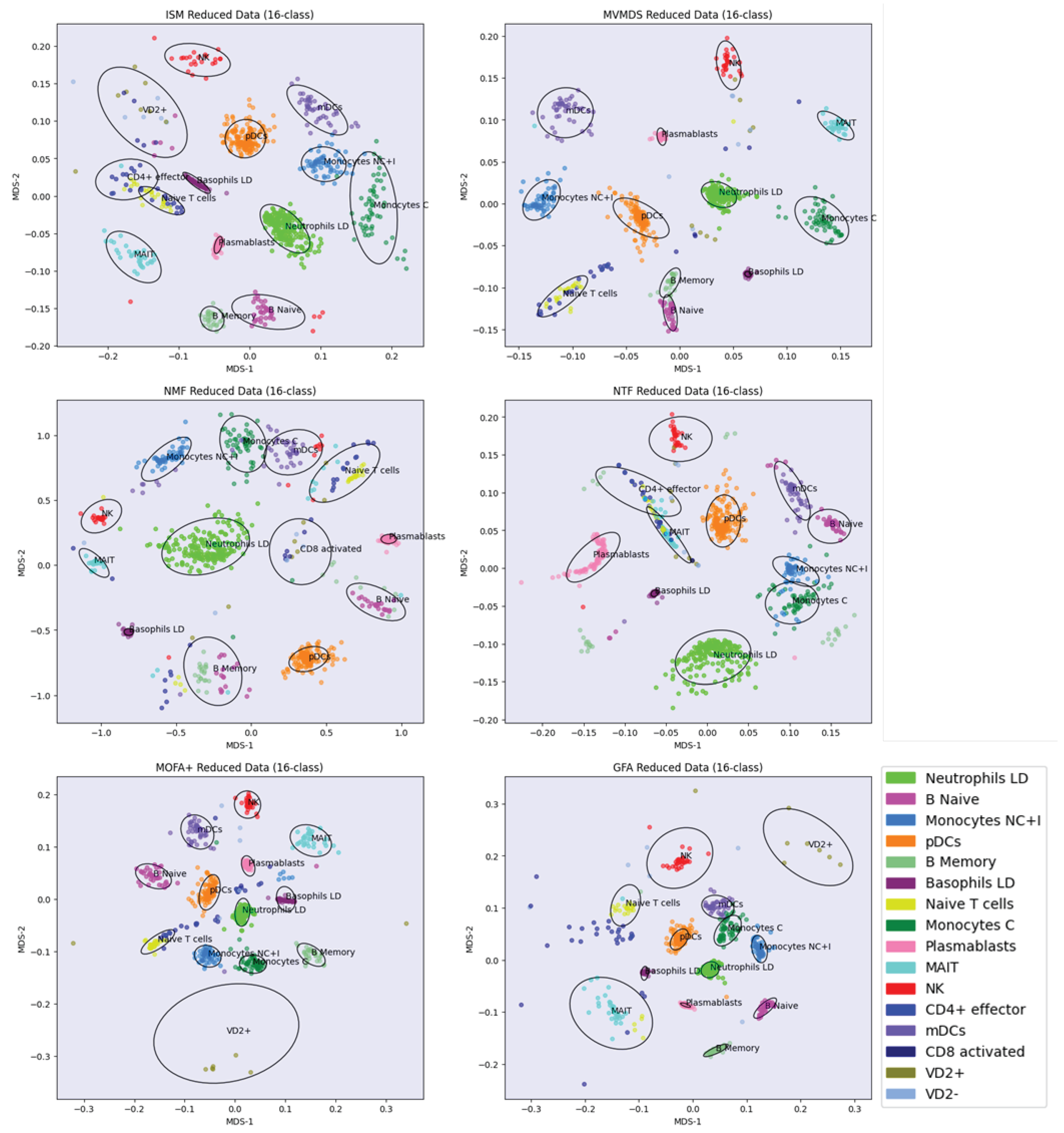
The data from the UCI Digits and Signature 915 datasets are analyzed using several alternative approaches: ISM, Multi-View Multi-Dimensional Scaling (MVMDS), NMF, NTF, Principal Components Analysis (PCA), Group Factor Analysis (GFA), and Multi-Omic Factor Analysis (MOFA+). Because PCA and NMF are not multi-view approaches, they are applied to the concatenated views. To facilitate interpretation, the transformed data is projected onto a 2D map before being subjected to k-means clustering, where k is the known number of classes (k-means clustering was chosen for its versatility and simplicity, as it only requires the number of clusters to be found, and this number is known in both our example datasets). Within each cluster, the class that contains the majority of the points, i.e., the main class, is identified. If two clusters share the same main class, they are merged, unless they are not contiguous (ratio of the distance between the centroids to the intra-cluster distance between points > 1). In this case, the non-contiguous clusters are excluded because they are assigned to the same class, which should appear homogeneous in the representation. Similarly, any cluster that does not contain an absolute majority is not considered clearly representative of the class to which it is assigned and is excluded. A global purity index is then calculated for the remaining clusters. To enhance clarity, the clusters are visualized using 95% confidence ellipses, while the classes are represented using as distinct colors as possible. In addition to the global purity index defined above, the adjusted Rand index [
18] is also included.
Multidimensional scaling (MDS) is applied to the 2D map projection. MDS uses a simple metric objective to find a low-dimensional embedding that accurately represents the distances between points in the latent space [
19]. MDS is therefore agnostic to the intrinsic clustering performances of the methods that we want to evaluate. Effective embedding methods, e.g., UMAP or t-SNE, are not as optimal for preserving the global geometric structure in the latent space [
20]. For example, a resolution parameter needs to be defined for the UMAP embedding of single-cell data, whereby a higher resolution leads to a higher number of clusters. In addition, the subtle differences between some cell types from one family can be smoothed out if the dataset contains transcriptionally distinct cell types from multiple families, as is the case with immune cells (second dataset in the article).
Latent Space methods require that the rank of the factorization is determined in advance. ISM benefits from the advantages of the NMF and NTF workflow components, i.e., the choice of the correct rank is less critical than with other methods (we will come back to this point in the Results and Discussion). This allows, even if we expect some redundancy in the latent factors due to the proximity of certain digits or cell types, to set the rank to the number of known classes. However, the dimension of the ISM embedding space must also be determined during the discovery step. The choice of close dimensions for the embedding and latent spaces is consistent with the fact that both spaces are merged at the end of the ISM workflow. Thus, we examine the approximation error for an embedding dimension in the neighborhood of the chosen rank. The rank for PCA, MVMDS, GFA and MOFA+ is set by inspecting the screeplot of the variance ratio.
The analysis of the Signature 915 data also examines the biological relevance of the distance between clusters in each latent multi-view space.
Detailed analysis steps are provided in workflow 3.
|
Workflow 3 Analysis steps |
| |
Input: 2D map projection of the data transformation in the latent space. |
| |
Output: Cluster purity index. |
| |
1: Perform k-means with k equal to the number of known classes; |
| |
2: For each cluster, identify the main class related to the cluster, i.e., the class corresponding to the majority of observations in the cluster; |
| |
3: Merge contiguous clusters that refer to the same class or ignore them if not contiguous; |
| |
4: for each cluster do
|
| |
5: |
proportion of the main class in relation to all elements in the cluster;
proportion of the main class in cluster c in relation to all elements of the same class; |
| |
6: |
If then
|
| |
7: |
|
Disregard cluster as the main class does not constitute an absolute majority in relation to all elements of the same class; |
| |
8: |
else |
|
| |
9: |
|
purity corrected for cluster representativity for the main class; |
| |
10: end for
|
| |
11: Calculate the global purity = sum of corrected purities over all retained clusters; |