1. Introduction
Human papillomavirus (HPV) is a sexually transmitted virus that can cause infections through abrasion of the skin and sexual intercourse [
1,
2] Infections by high-risk HPVs have been implicated in various cancers, such as cervical, penile, anal, and oropharynx cancers, emphasizing the oncogenic potential of this viral family [
3,
4,
5,
6].
The existing literature suggests an association between high-risk HPV and diverse cancer types, including breast and prostate cancers [
7,
8,
9,
10]. While the detection of HPV in various cancers raises questions about its potential role, direct evidence of HPV transmission from the site of initial infection to adjacent organs within the same individual remains an unexplored aspect. The metastatic spread of HPV-associated cancers to distant organs has been documented in rare cases, highlighting the need for further investigation into the mechanisms underlying viral transportation and its implications for cancer development [
11,
12].
In this study, we presented a unique case of concurrent high-risk HPV35, HPV45, and HPV59 infections in prostate and bladder cancer tissues of a single patient. To the best of our knowledge, this is the first documented case worldwide demonstrating the presence of identical HPV types in two adjacent organs of the same patient, shedding light on the potential transport of HPV from an organ of initial infection to a nearby organ. Therefore, this discovery not only expands our understanding of HPV-related oncogenesis but also provides an insight into the mechanism of HPV transmission within human body.
2. Materials and Methods
2.1. Case Presentation
An 87-year-old Caucasian male patient, a smoker, with a history of multiple health conditions including recurrent visible hematuria and extended-spectrum beta-lactamases (ESBL) in urine, was admitted to Kingston Hospital. Initial cystoscopy examination revealed suspected bladder lesions, which upon resection pathology displayed a low-grade, non-muscle-invasive bladder carcinoma with a clinical stage of (pT1B). Diagnostic imaging, including abdominal and pelvic computed tomography scans (CT), was negative for metastatic lesions. One year later, this progressed to a metastatic transitional cell carcinoma (TCC) of the bladder involving the prostate. Bladder and Prostate specimens were obtained from this patient following a written informed consent, approved by IRAS (Project ID 204705 – Leicester Central Research Ethics Committee) to investigate the presence and the expression of high-risk (HR) HPV genotypes in bladder and prostate cancer biopsies.
2.2. Detection and Genotyping of HPV DNA
Cellular DNA was extracted from the collected bladder and prostate cancer tissue samples by using Gen Elute RNA/DNA/Protein Purification Plus kit (Sigma -Aldrich, UK) in accordance with the manufacturer’s protocol. The concentration and purty of the extracted DNA were assessed using NanoVue plus spectrophotometer (GE Lifesciences, Chicago, Illionis, USA). A multiplex-PCR approach was employed for simultaneous amplification of four targeted regions of the HPV gene, focusing on four different HPV DNA types in one tube. The HPV DNA amplification was carried out in three tubes, each distinctly amplifying the following HPV types: 16/31/33/35, 18/39/45/59, and 52/56/58/66. This methodology enabled the identification of HPV infections and co-infections of 12 distinct high-risk HPV subtypes in each sample. The PCR experiment was repeated in triplicate for each sample to ensure data accuracy. Positive amplification of the β-globin gene (fragment size 723 bp) was observed in all samples analyzed, indicating the quality of the extracted DNA. For the analysis and determination of HPV genotypes, amplified PCR products were electrophoresed on a 3% (w/v) agarose gel stained with SYBR Safe (Invitrogen, Carlsbad, CA, USA). β-globin amplification in each sample served as an internal control to confirm the adequacy of the extracted DNA for amplification. Gel visualization under UV was conducted using a Gel Doc XR + System (Bio-Rad, Hercules, CA, USA).
2.3. HPV DNA sequencing
To confirm the presence of HPV genes,,PCR-amplified products from positive samples were s direct sequencing. purification using the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany), sequencing was performed with an Applied Biosystems 3730xL analyzer. The resulting data were analyzed using the NCBI BLAST software [
13,
14]. This method served as an additional means of validating the obtained results.
2.4. Western Blotting
To investigate the expresssion of HPV oncoproteins in bladder and prostate cancer tissue samples, a standard semi-dry Western blotting technique was performed. Total protein extracted using Gen Elute Protein Pruficiation kit and equal amounts of proteins were electrophoresed. The membrane was blocked with 5% bovine serum albumin (BSA) in TBS–T (10 mM Tris-HCI, pH 7.5, 150 mM NaCI, and 1% (v/v) Tween 20) at room temperature for 2 h to reduce nonspecific binding and incubated with mAb Cervimax HPV E7 antibody followed by Donkey anti-mouse (1:10,000) and actin antibody (1:10,000). The membrane was then visualized using the OdysseyClx Imaging System (Li-COR).
3. Results
3.1. Detection of HPVs 35,45 and 59 in bladder and prostate cancer
To identify high-risk HPV types in prostate and bladder cancer specimens, targeted regions of HPV genes were subjected to PCR amplification. The amplification of the β-globin gene, with a fragment size of 723 bp, yielded positive results for all analyzed samples. To ensure the reliability of the obtained data, the PCR experiment was meticulously repeated in triplicate, confirming the robustness and accuracy of the results across all samples. As a standardized protocol, each sample run in gel electrophoresis included both HPV positive controls and negative controls, ensuring the validity of the experimental setup.
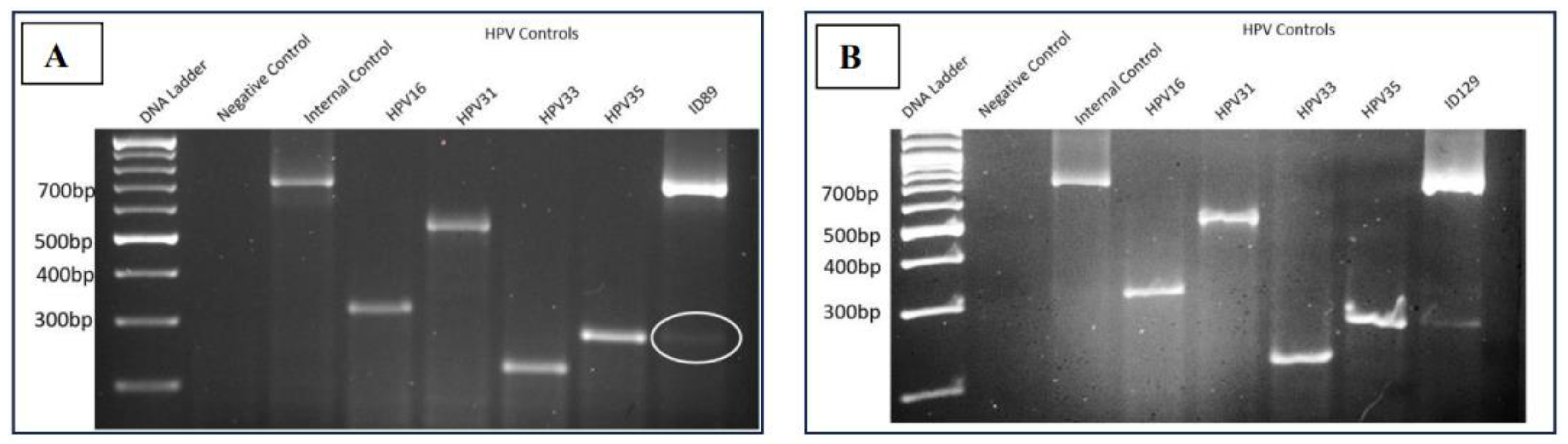
3.1.1. Presence HPV35 in Bladder and Prostate Cancer
The gel electrophoresis pattern of four high-risk HPV types (16/31/33/35) for a fresh bladder cancer sample (ID89) and prostate cancer sample (ID129) fom same patient revealed molecular weights of the amplified DNA products comparable to the internal positive control β-Globin (723bp), confirming the adequacy of the extracted DNA. Notably, samples ID89 and ID129 exhibited a DNA band analogous to the HPV35 positive control, while no DNA band was observed for the negative control, confirming the absence of contaminations (
Figure 1). This finding showed that bladder and prostate cancer samples from same patient have positive for HPV35.
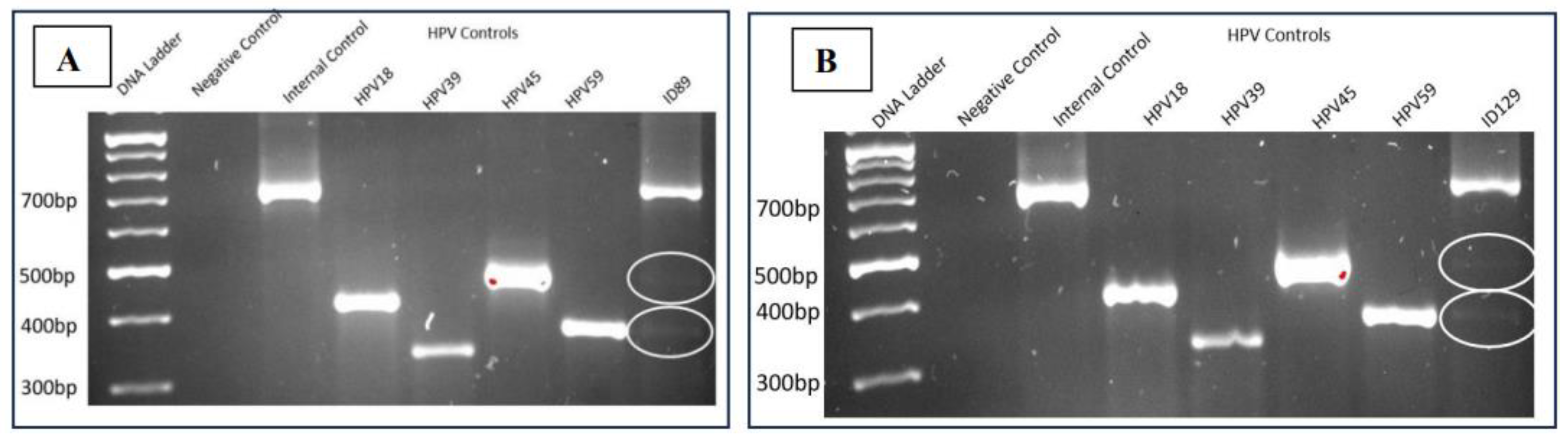
3.1.2. Presence of HPV45 and HPV59 in Bladder and Prostate Cancer
The gel electrophoresis pattern of four high-risk HPV types (18/39/45/59) for the bladder cancer sample (ID89) demonstrated molecular weights of the amplified DNA product comparable to the internal positive control β-Globin (723bp), validating the quality of the extracted DNA. Samples ID89 and ID129 exhibited DNA bands akin to the HPV45 and HPV59 positive controls, respectively. As in the previous analysis, the absence of a DNA band in the negative control confirmed the absence of contaminations. This finding also revealed that bladder and prostate cancer samples from same patient have positive for HPV45 and HPV59.
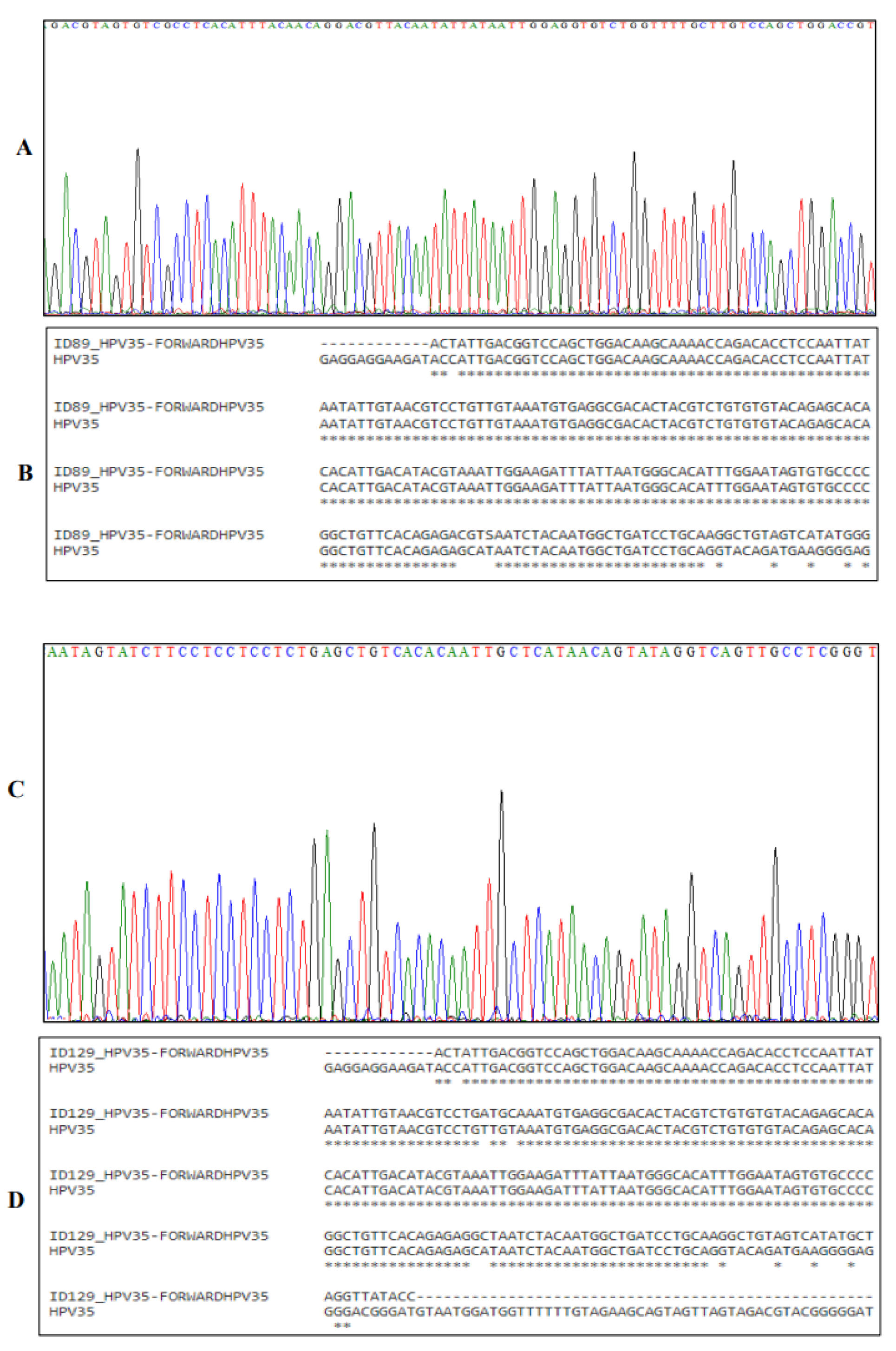
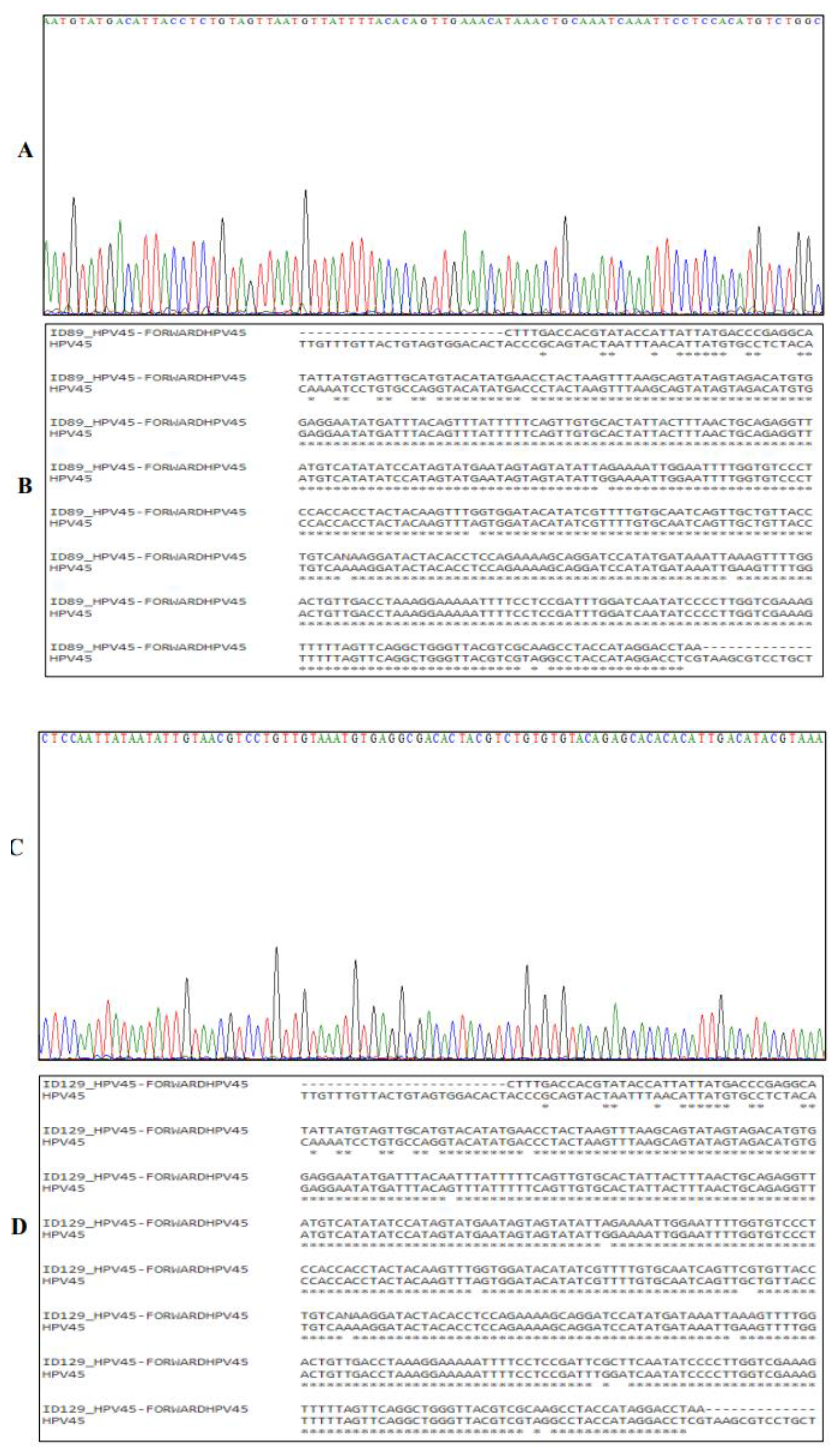
3.2. Sanger Sequencing of HPV DNA
Sanger sequencing was employed to confirm and enhance the validity of the PCR results. The four-colour chromatogram depicts the partial results of the sequencing in
Figure 3,
Figure 4 and
Figure 5. Blast analysis revealled a robust concordance of over 90% in HPV DNA sequences across all samples. This consistent data futher affirms the accuracy and reliability of our findings.
3.4. The Expression of HPV E7 Oncoprotein in Bladder and Prostate Cancer Samples
To investigate the active expresion of HPV DNA detected in both bladder and prostate cancer tissues, a Western blot analysis was conducted using an HPV E7 antibody. The Western blot analysis confirmed the expression of HPV E7 protein in both bladder and prostate cancer tissues (
Figure 6).
4. Discussion
Prostate and bladder cancers pose significant challenges to global public health, contributing substantial morbidity and mortality [
15,
16]. Despite extensive research efforts, the precise etiology of these cancers remains elusive. It has been shown previously that HPV has the capacity to cause the development of certain cancers, such as cervical, oral, and breast cancers [
17]. Hence, it is possible that HPV may also play a major role in the development of prostate and bladder cancers. Indeed, several studies have found a significant number of samples of bladder and prostate cancers expressing HPV oncoproteins [
10,
18,
19,
20]
Interestingly , our investigation focuses on a patient diagnosed with concurrent prostate and bladder cancers, providing a unique opportunity to explore the presence and expression of different HPV types. Our results revealed the presence and expression of HPV types 35, 45, and 59 in both organs, a novel finding not previously demonstrated in any study worldwide. This significant discovery suggests that HPV may not only be transmitted from person to person through sexual or skin-to-skin contact but also from one organ to another within the same individual. Specifically, our patient exhibits three distinct HPV infections concurrently in two different organs, highlighting the potential for intra-organ transmission.
Contrary to the widely accepted notion that HPV lacks a viremic phase during infection, our findings prompt consideration of alternative transmission routes. The potential transmission of HPV in peripheral blood, raises the possibility that HPV may transmit hematogenously under certain conditions [
21,
22,
23,
24,
25]. This alternative route could explain the simultaneous presence of HPV types 35, 45, and 59 in both prostate and bladder cancers in this patient. The exceptional outcome of this case report not only challenges conventional understanding but also contributes substantially to our knowledge of HPV transportation and transmission between different organs.
In conclusion, our study provides compelling evidence of the presence and expression of diverse HPV types in both prostate and bladder cancers within the same individual. This groundbreaking finding underscores the potential for HPV intra-organ transmission and necessitates further exploration of alternative transmission routes. The implications of our results offer new insights into the complex dynamics of viral transmission in cancer pathogenesis.
Author Contributions
Conceptualization, G.H.A.; Methodology, M.Y.A. and G.H.A.; validation, G.H.A.; M.Y.A.; N.A.S.; M.O.C.; formal analysis, G.H.A.; M.Y.A.; N.A.S.; M.O.C.; data curation, M.Y.A; writing—original draft preparation, M.O.C.; writing—review and editing, G.H.A.; M.Y.A.; N.A.S.; M.O.C.; supervision, G.H.A.; project administration, G.H.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
This study was conducted in accordance with the Declaration of Helsinki. Bladder and Prostate specimens were obtained from this patient following a written informed consent, approved by IRAS (Project ID 204705 – Leicester Central Research Ethics Committee) to investigate the presence and the expression of high-risk (HR) HPV genotypes in bladder and prostate cancer biopsies.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. Also, informed consent has been obtained from the patient(s) to publish this paper.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
The authors acknowledge the patients who donated tissue and assistance of the surgical team at Kingston Hospital (Kingston upon Thames, London, UK). Also, authors would like to acknowledge Kingston University London for providing laboratory facilities.
Conflicts of Interest
The authors declare no conflict of interests.
References
- Veldhuijzen, N.J.; Snijders, P.J.; Reiss, P.; Meijer, C.J.; van de Wijgert, J.H. Factors Affecting Transmission of Mucosal Human Papillomavirus. Lancet Infect Dis 2010, 10, 862–874. [Google Scholar] [CrossRef] [PubMed]
- Ryndock, E.J.; Meyers, C. A Risk for Non-Sexual Transmission of Human Papillomavirus? Expert Rev Anti Infect Ther 2014, 12, 1165–1170. [Google Scholar] [CrossRef] [PubMed]
- Gillison, M.L. Evidence for a Causal Association Between Human Papillomavirus and a Subset of Head and Neck Cancers. J Natl Cancer Inst 2000, 92, 709–720. [Google Scholar] [CrossRef] [PubMed]
- Gosens, K.C.M.; Richel, O.; Prins, J.M. Human Papillomavirus as a Cause of Anal Cancer and the Role of Screening. Curr Opin Infect Dis 2017, 30, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Diorio, G.J.; Giuliano, A.R. The Role of Human Papilloma Virus in Penile Carcinogenesis and Preneoplastic Lesions. Urologic Clinics of North America 2016, 43, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Burd, E.M. Human Papillomavirus and Cervical Cancer. Clin Microbiol Rev 2003, 16, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Sher, G.; Salman, N.A.; Kulinski, M.; Fadel, R.A.; Gupta, V.K.; Anand, A.; Gehani, S.; Abayazeed, S.; Al-Yahri, O.; Shahid, F.; et al. Prevalence and Type Distribution of High-Risk Human Papillomavirus (HPV) in Breast Cancer: A Qatar Based Study. Cancers (Basel) 2020, 12, 1528. [Google Scholar] [CrossRef] [PubMed]
- Salman, N.A.; Davies, G.; Majidy, F.; Shakir, F.; Akinrinade, H.; Perumal, D.; Ashrafi, G.H. Association of High Risk Human Papillomavirus and Breast Cancer: A UK Based Study. Sci Rep 2017, 7, 43591. [Google Scholar] [CrossRef] [PubMed]
- Medel-Flores, O.; Valenzuela-Rodríguez, V.A.; Ocadiz-Delgado, R.; Castro-Muñoz, L.J.; Hernández-Leyva, S.; Lara-Hernández, G.; Silva-Escobedo, J.-G.; Vidal, P.G.; Sánchez-Monroy, V. Association between HPV Infection and Prostate Cancer in a Mexican Population. Genet Mol Biol 2018, 41, 781–789. [Google Scholar] [CrossRef]
- Ahmed, M.Y.; Salman, N.A.; Sandhu, S.; Cakir, M.O.; Seddon, A.M.; Kuehne, C.; Ashrafi, G.H. Detection of High-Risk Human Papillomavirus in Prostate Cancer from a UK Based Population. Sci Rep 2023, 13, 7633. [Google Scholar] [CrossRef]
- Sacks, R.; Law, J.Y.; Zhu, H.; Beg, M.S.; Gerber, D.E.; Sumer, B.D.; Myers, L.L.; Truelson, J.M.; Nedzi, L.; Sher, D.; et al. Unique Patterns of Distant Metastases in HPV-Positive Head and Neck Cancer. Oncology 2020, 98, 179–185. [Google Scholar] [CrossRef]
- Brkic, F.F.; Kadletz-Wanke, L.; Kenner, L.; Füreder, T.; Jank, B.; Brunner, M.; Heiduschka, G. An Analysis of Distant Metastasis Cases from HPV-Associated Oropharyngeal Squamous Cell Carcinoma. Journal of Cranio-Maxillofacial Surgery 2021, 49, 312–316. [Google Scholar] [CrossRef]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A Greedy Algorithm for Aligning DNA Sequences. Journal of Computational Biology 2000, 7, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J Mol Biol 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Wang, L.; Lu, B.; He, M.; Wang, Y.; Wang, Z.; Du, L. Prostate Cancer Incidence and Mortality: Global Status and Temporal Trends in 89 Countries From 2000 to 2019. Front Public Health 2022, 10. [Google Scholar] [CrossRef]
- Cai, Q.; Chen, Y.; Xin, S.; Zhang, D.; Pan, J.; Xie, Z.; Xu, C.; Li, S.; Zhang, X.; Gao, Y.; et al. Temporal Trends of Bladder Cancer Incidence and Mortality from 1990 to 2016 and Projections to 2030. Transl Androl Urol 2020, 9, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Pešut, E.; Đukić, A.; Lulić, L.; Skelin, J.; Šimić, I.; Milutin Gašperov, N.; Tomaić, V.; Sabol, I.; Grce, M. Human Papillomaviruses-Associated Cancers: An Update of Current Knowledge. Viruses 2021, 13, 2234. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Xu, J.; Liu, C.; An, Y.; Xu, M.; Zhong, X.; Zeng, N.; Ma, S.; He, H.; Hu, J.; et al. The Association between Human Papillomavirus and Bladder Cancer: Evidence from Meta-analysis and Two-sample Mendelian Randomization. J Med Virol 2023, 95. [Google Scholar] [CrossRef]
- Khatami, A.; Salavatiha, Z.; Razizadeh, M.H. Bladder Cancer and Human Papillomavirus Association: A Systematic Review and Meta-Analysis. Infect Agent Cancer 2022, 17, 3. [Google Scholar] [CrossRef]
- Tsydenova, I.A.; Ibragimova, M.K.; Tsyganov, M.M.; Litviakov, N. V. Human Papillomavirus and Prostate Cancer: Systematic Review and Meta-Analysis. Sci Rep 2023, 13, 16597. [Google Scholar] [CrossRef]
- Capone, R.B.; Pai, S.I.; Koch, W.M.; Gillison, M.L.; Danish, H.N.; Westra, W.H.; Daniel, R.; Shah, K.V; Sidransky, D. Detection and Quantitation of Human Papillomavirus (HPV) DNA in the Sera of Patients with HPV-Associated Head and Neck Squamous Cell Carcinoma. Clin Cancer Res 2000, 6, 4171–4175. [Google Scholar] [PubMed]
- Dong, S.M.; Pai, S.I.; Rha, S.-H.; Hildesheim, A.; Kurman, R.J.; Schwartz, P.E.; Mortel, R.; McGowan, L.; Greenberg, M.D.; Barnes, W.A.; et al. Detection and Quantitation of Human Papillomavirus DNA in the Plasma of Patients with Cervical Carcinoma. Cancer Epidemiol Biomarkers Prev 2002, 11, 3–6. [Google Scholar] [PubMed]
- Liu, V.W.S.; Tsang, P.; Yip, A.; Ng, T.-Y.; Wong, L.-C.; Ngan, H.Y.S. Low Incidence of HPV DNA in Sera of Pretreatment Cervical Cancer Patients. Gynecol Oncol 2001, 82, 269–272. [Google Scholar] [CrossRef] [PubMed]
- Pao, C.C.; Lin, S.-S.; Lin, C.-Y.; Maa, J.-S.; Lai, C.-H.; Hsieh, T.-T. Identification of Human Papillomavirus DNA Sequences in Peripheral Blood Mononuclear Cells. Am J Clin Pathol 1991, 95, 540–546. [Google Scholar] [CrossRef]
- Bodaghi, S.; Wood, L.V.; Roby, G.; Ryder, C.; Steinberg, S.M.; Zheng, Z.-M. Could Human Papillomaviruses Be Spread through Blood? J Clin Microbiol 2005, 43, 5428–5434. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).