Submitted:
06 February 2024
Posted:
06 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Material and methods
2.1. Patients and Data Collection
2.2. Cell sorting and scanning electron microscopy
2.3. RNA separation, library preparation and next generation sequencing
2.4. Bioinformatics and statistical analysis
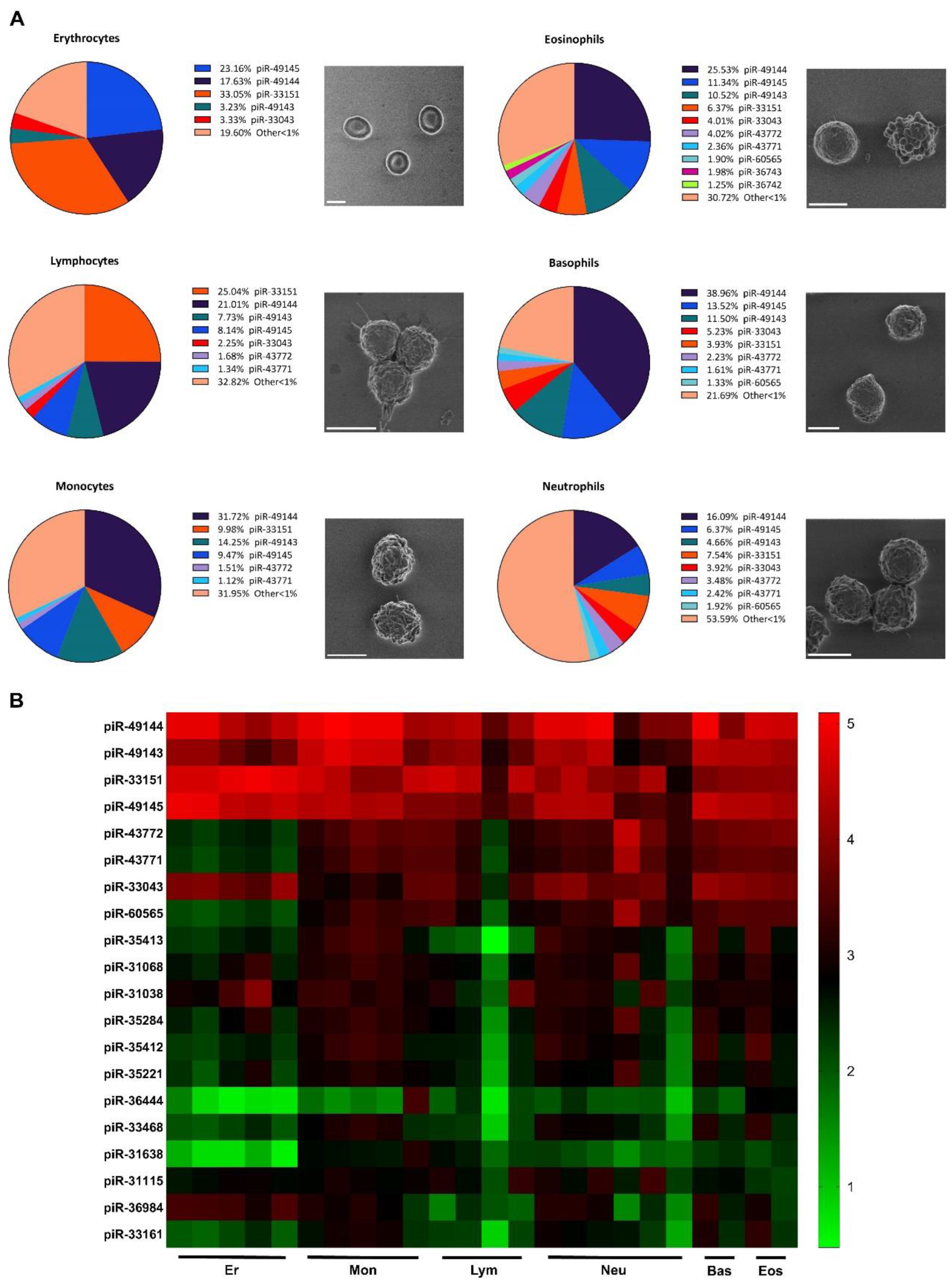
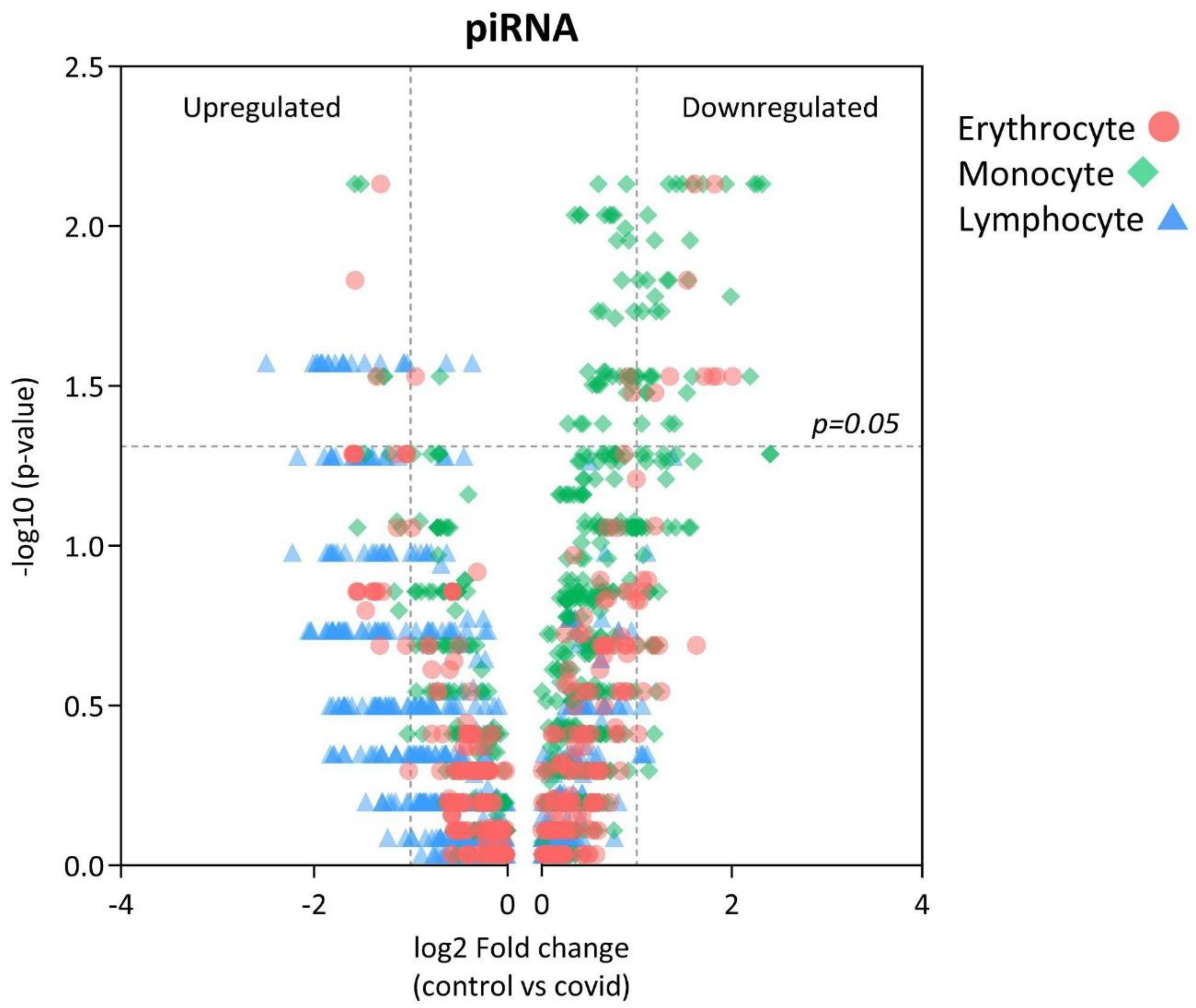
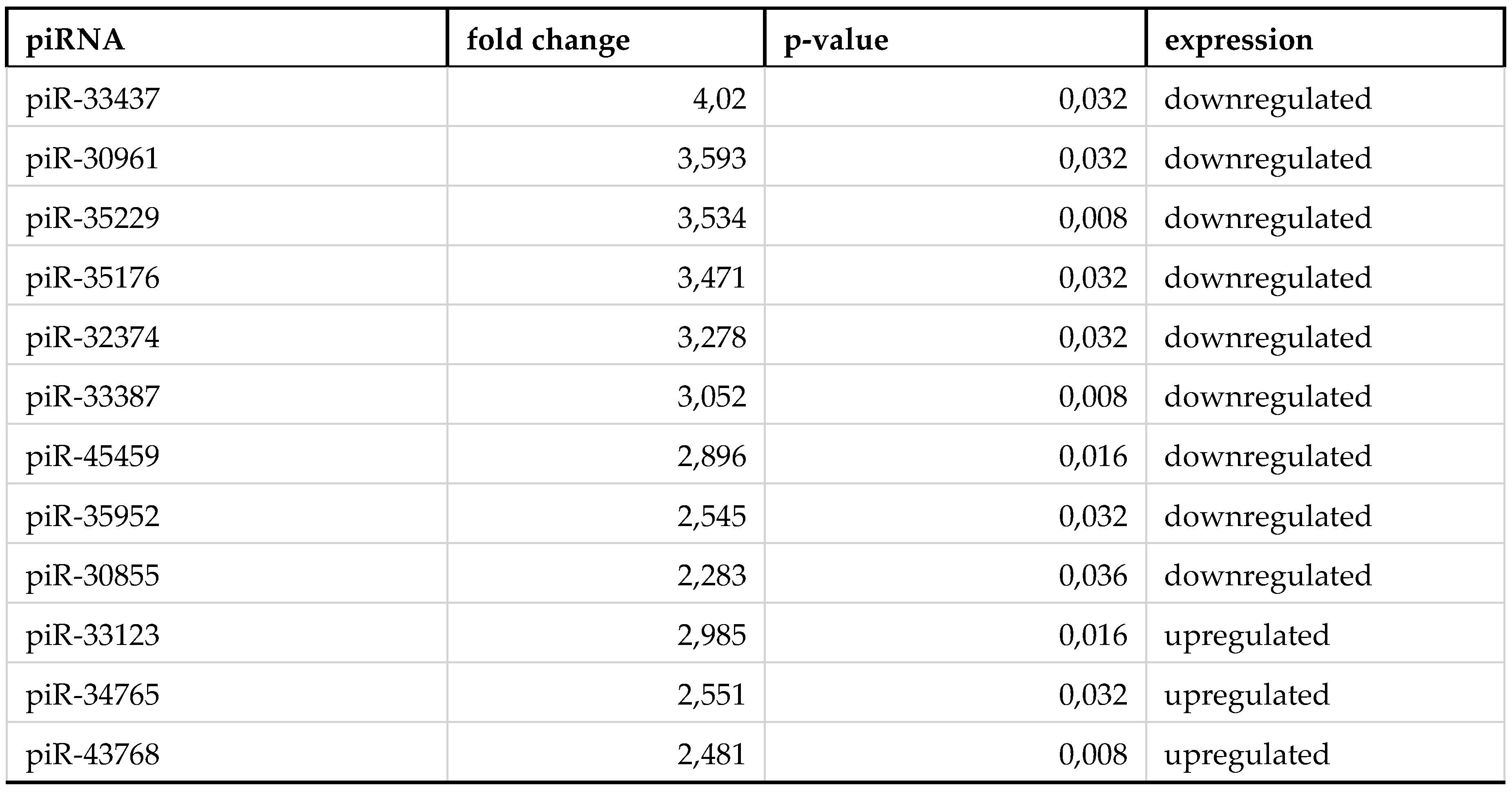
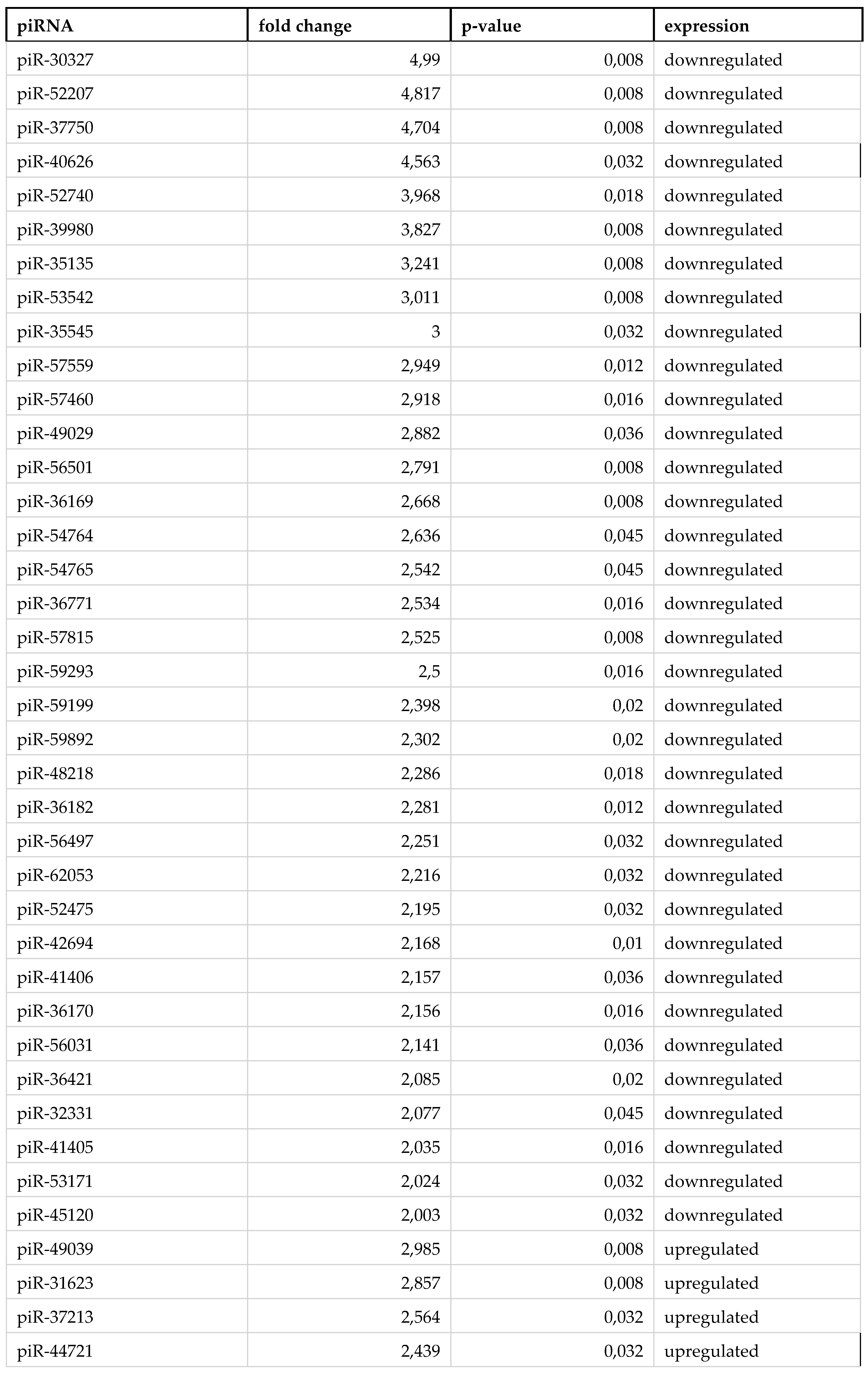
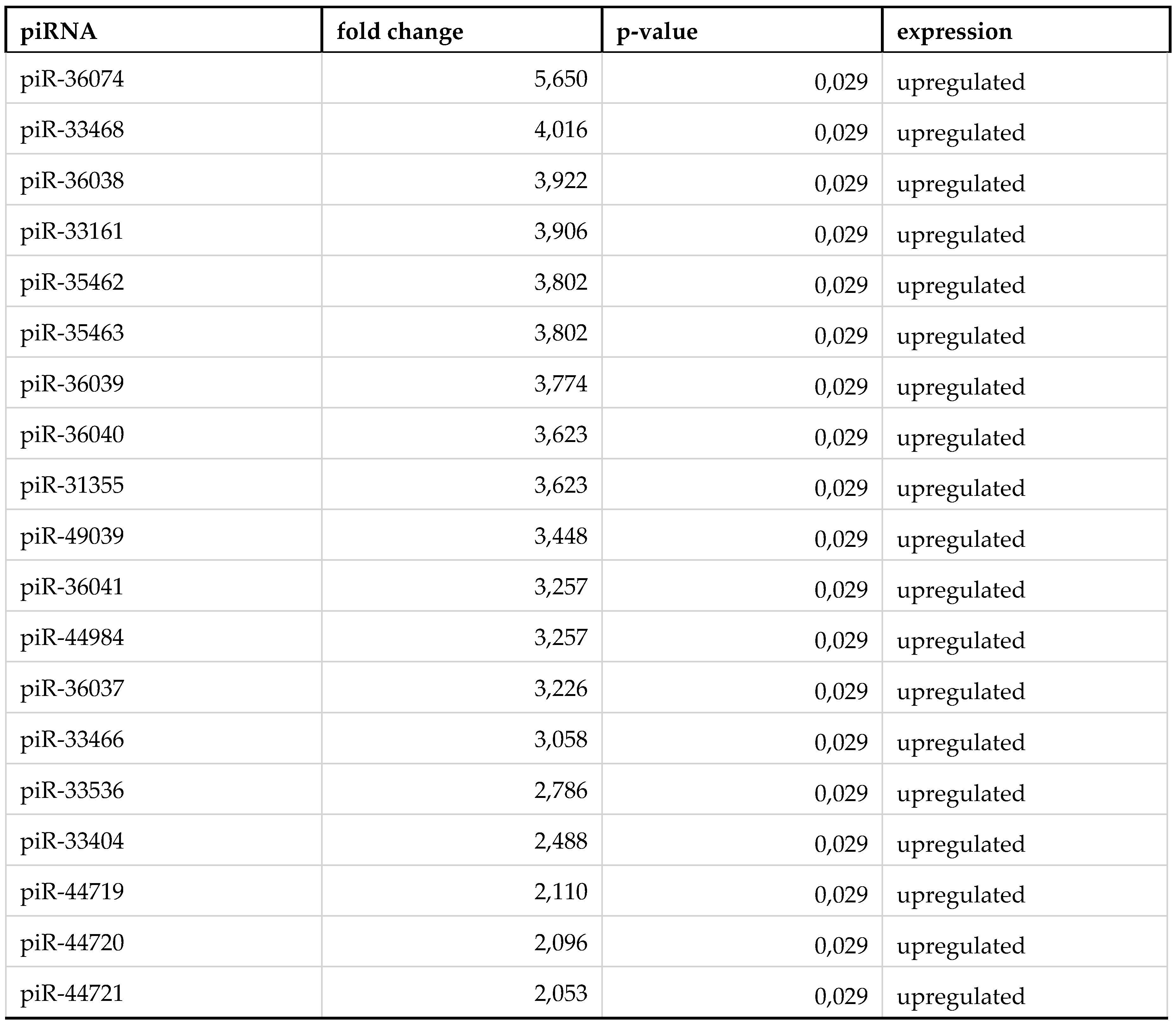
3. Results
4. Discussion
Conclusion
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Hombach, S.; Kretz, M. Non-Coding RNAs: Classification, Biology and Functioning. Adv Exp Med Biol 2016, 937, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene Regulation by Long Non-Coding RNAs and Its Biological Functions. Nat Rev Mol Cell Biol 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Palazzo, A.F.; Lee, E.S. Non-Coding RNA: What Is Functional and What Is Junk? Frontiers in Genetics 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Yoshitake, K.; Asakawa, S. A Review of Discovery Profiling of PIWI-Interacting RNAs and Their Diverse Functions in Metazoans. International Journal of Molecular Sciences 2021, 22, 11166. [Google Scholar] [CrossRef] [PubMed]
- Darricarrère, N.; Liu, N.; Watanabe, T.; Lin, H. Function of Piwi, a Nuclear Piwi/Argonaute Protein, Is Independent of Its Slicer Activity. Proc Natl Acad Sci U S A 2013, 110, 1297–1302. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Lin, H. Roles of piRNAs in Transposon and Pseudogene Regulation of Germline mRNAs and lncRNAs. Genome Biology 2021, 22, 27. [Google Scholar] [CrossRef]
- Litwack, G. Chapter 10 - Nucleic Acids and Molecular Genetics. In Human Biochemistry; Litwack, G., Ed.; Academic Press: Boston, 2018; pp. 257–317. ISBN 978-0-12-383864-3. [Google Scholar]
- Xiao, Y.; Ke, A. PIWI Takes a Giant Step. Cell 2016, 167, 310–312. [Google Scholar] [CrossRef]
- Tóth, K.F.; Pezic, D.; Stuwe, E.; Webster, A. The piRNA Pathway Guards the Germline Genome Against Transposable Elements. Adv Exp Med Biol 2016, 886, 51–77. [Google Scholar] [CrossRef]
- Zhang, T.; Wong, G. Dysregulation of Human Somatic piRNA Expression in Parkinson’s Disease Subtypes and Stages. Int J Mol Sci 2022, 23, 2469. [Google Scholar] [CrossRef]
- Rayford, K.J.; Cooley, A.; Strode, A.W.; Osi, I.; Arun, A.; Lima, M.F.; Misra, S.; Pratap, S.; Nde, P.N. Trypanosoma Cruzi Dysregulates Expression Profile of piRNAs in Primary Human Cardiac Fibroblasts during Early Infection Phase. Frontiers in Cellular and Infection Microbiology 2023, 13. [Google Scholar] [CrossRef]
- Chattopadhyay, T.; Gupta, P.; Nayak, R.; Mallick, B. Genome-Wide Profiling of Dysregulated piRNAs and Their Target Genes Implicated in Oncogenicity of Tongue Squamous Cell Carcinoma. Gene 2023, 849, 146919. [Google Scholar] [CrossRef]
- Cheng, Y.; Wang, Q.; Jiang, W.; Bian, Y.; zhou, Y.; Gou, A.; Zhang, W.; Fu, K.; Shi, W. Emerging Roles of piRNAs in Cancer: Challenges and Prospects. Aging (Albany NY) 2019, 11, 9932–9946. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Dou, M.; Song, X.; Dong, Y.; Liu, S.; Liu, H.; Tao, J.; Li, W.; Yin, X.; Xu, W. The Emerging Role of the piRNA/Piwi Complex in Cancer. Molecular Cancer 2019, 18, 123. [Google Scholar] [CrossRef] [PubMed]
- Mai, D.; Ding, P.; Tan, L.; Zhang, J.; Pan, Z.; Bai, R.; Li, C.; Li, M.; Zhou, Y.; Tan, W.; et al. PIWI-Interacting RNA-54265 Is Oncogenic and a Potential Therapeutic Target in Colorectal Adenocarcinoma. Theranostics 2018, 8, 5213–5230. [Google Scholar] [CrossRef] [PubMed]
- Kawase, M.; Ichiyanagi, K. The Expression Dynamics of piRNAs Derived From Male Germline piRNA Clusters and Retrotransposons. Frontiers in Cell and Developmental Biology 2022, 10. [Google Scholar] [CrossRef]
- Gainetdinov, I.V.; Skvortsova, Y.V.; Kondratieva, S.A.; Klimov, A.; Tryakin, A.A.; Azhikina, T.L. Assessment of piRNA Biogenesis and Function in Testicular Germ Cell Tumors and Their Precursor Germ Cell Neoplasia in Situ. BMC Cancer 2018, 18, 20. [Google Scholar] [CrossRef] [PubMed]
- Horjales, S.; Li Calzi, M.; Francia, M.E.; Cayota, A.; Garcia-Silva, M.R. piRNA Pathway Evolution beyond Gonad Context: Perspectives from Apicomplexa and Trypanosomatids. Frontiers in Genetics 2023, 14. [Google Scholar] [CrossRef] [PubMed]
- Ren, R.; Tan, H.; Huang, Z.; Wang, Y.; Yang, B. Differential Expression and Correlation of Immunoregulation Related piRNA in Rheumatoid Arthritis. Frontiers in Immunology 2023, 14. [Google Scholar] [CrossRef]
- Akimniyazova, A.; Yurikova, O.; Pyrkova, A.; Rakhmetullina, A.; Niyazova, T.; Ryskulova, A.-G.; Ivashchenko, A. In Silico Study of piRNA Interactions with the SARS-CoV-2 Genome. Int J Mol Sci 2022, 23, 9919. [Google Scholar] [CrossRef]
- Lucas, C.; Wong, P.; Klein, J.; Castro, T.B.R.; Silva, J.; Sundaram, M.; Ellingson, M.K.; Mao, T.; Oh, J.E.; Israelow, B.; et al. Longitudinal Analyses Reveal Immunological Misfiring in Severe COVID-19. Nature 2020, 584, 463–469. [Google Scholar] [CrossRef]
- Kondratov, K.A.; Artamonov, A.A.; Mikhailovskii, V.Y.; Velmiskina, A.A.; Mosenko, S.V.; Grigoryev, E.A.; Anisenkova, A.Y.; Nikitin, Y.V.; Apalko, S.V.; Sushentseva, N.N.; et al. SARS-CoV-2 Impact on Red Blood Cell Morphology. Biomedicines 2023, 11, 2902. [Google Scholar] [CrossRef] [PubMed]
- Velmiskina, A.A.; Nikitin, Yu.V.; Mikhailovskii, V.Yu.; Mosenko, S.V.; Anisenkova, A.Yu.; Apalko, S.V.; Sushentseva, N.N.; Scherbak, S.G.; Ivanov, A.M.; Galaktionov, N.K.; et al. Analysis of the Morphology of Monocytes and Lymphocytes from COVID-19 Patients Using Low-Voltage Scanning Electronic Microscopy. Bull Exp Biol Med 2023, 176, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Sun, Q.; Zhang, B.; Zhao, W.; Shen, C. The Regulation of lncRNAs and miRNAs in SARS-CoV-2 Infection. Front Cell Dev Biol 2023, 11, 1229393. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Chen, J.; Yin, X.; zhou, J. Current Understanding on Long Non-Coding RNAs in Immune Response to COVID-19. Virus Res 2022, 323, 198956. [Google Scholar] [CrossRef]
- Arman, K.; Dalloul, Z.; Bozgeyik, E. Emerging Role of microRNAs and Long Non-Coding RNAs in COVID-19 with Implications to Therapeutics. Gene 2023, 861, 147232. [Google Scholar] [CrossRef]
- Fedorov, A.; Kondratov, K.; Kishenko, V.; Mikhailovskii, V.; Kudryavtsev, I.; Belyakova, M.; Sidorkevich, S.; Vavilova, T.; Kostareva, A.; Sirotkina, O.; et al. Application of High-Sensitivity Flow Cytometry in Combination with Low-Voltage Scanning Electron Microscopy for Characterization of Nanosized Objects during Platelet Concentrate Storage. Platelets 2020, 31, 226–235. [Google Scholar] [CrossRef]
- Fehlmann, T.; Kern, F.; Laham, O.; Backes, C.; Solomon, J.; Hirsch, P.; Volz, C.; Müller, R.; Keller, A. miRMaster 2.0: Multi-Species Non-Coding RNA Sequencing Analyses at Scale. Nucleic Acids Research 2021, 49, W397–W408. [Google Scholar] [CrossRef]
- Martin, F.J.; Amode, M.R.; Aneja, A.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Becker, A.; Bennett, R.; Berry, A.; Bhai, J.; et al. Ensembl 2023. Nucleic Acids Research 2023, 51, D933–D941. [Google Scholar] [CrossRef]
- The RNAcentral Consortium RNAcentral: A Hub of Information for Non-Coding RNA Sequences. Nucleic Acids Research 2019, 47, D221–D229. [CrossRef] [PubMed]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference Sequence (RefSeq) Database at NCBI: Current Status, Taxonomic Expansion, and Functional Annotation. Nucleic Acids Res 2016, 44, D733–745. [Google Scholar] [CrossRef] [PubMed]
- Leinonen, R.; Akhtar, R.; Birney, E.; Bower, L.; Cerdeno-Tárraga, A.; Cheng, Y.; Cleland, I.; Faruque, N.; Goodgame, N.; Gibson, R.; et al. The European Nucleotide Archive. Nucleic Acids Res 2011, 39, D28–D31. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Current Protocols in Bioinformatics 2016, 54, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Shi, Y.; Zhou, H.; Zhang, P.; Song, T.; Ying, Z.; Yu, H.; Li, Y.; Zhao, Y.; Zeng, X.; et al. piRBase: Integrating piRNA Annotation in All Aspects. Nucleic Acids Research 2022, 50, D265–D272. [Google Scholar] [CrossRef] [PubMed]
- RStudio Team (2020). RStudio: Integrated Development for R. 2020. http://www.rstudio.com/.
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer-Verlag New York, 2016; ISBN 978-3-319-24277-4. https://ggplot2.tidyverse.org.
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. Journal of Open Source Software 2019, 4, 1686. [Google Scholar] [CrossRef]
- Grillone, K.; Riillo, C.; Scionti, F.; Rocca, R.; Tradigo, G.; Guzzi, P.H.; Alcaro, S.; Di Martino, M.T.; Tagliaferri, P.; Tassone, P. Non-Coding RNAs in Cancer: Platforms and Strategies for Investigating the Genomic “Dark Matter”. Journal of Experimental & Clinical Cancer Research 2020, 39, 117. [Google Scholar] [CrossRef]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA Expression Profiles Classify Human Cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Sun, L.; Yu, Y.; Niu, B.; Wang, D. Red Blood Cells as Potential Repositories of MicroRNAs in the Circulatory System. Front Genet 2020, 11, 442. [Google Scholar] [CrossRef]
- Yuan, S.; Wu, Q.; Wang, Z.; Che, Y.; Zheng, S.; Chen, Y.; Zhong, X.; Shi, F. miR-223: An Immune Regulator in Infectious Disorders. Front Immunol 2021, 12, 781815. [Google Scholar] [CrossRef]
- Zhang, Z.-W.; Cheng, J.; Xu, F.; Chen, Y.-E.; Du, J.-B.; Yuan, M.; Zhu, F.; Xu, X.-C.; Yuan, S. Red Blood Cell Extrudes Nucleus and Mitochondria against Oxidative Stress. IUBMB Life 2011, 63, 560–565. [Google Scholar] [CrossRef]
- Pretini, V.; Koenen, M.H.; Kaestner, L.; Fens, M.H.A.M.; Schiffelers, R.M.; Bartels, M.; Van Wijk, R. Red Blood Cells: Chasing Interactions. Front Physiol 2019, 10, 945. [Google Scholar] [CrossRef]
- Li, Z.; Gao, J.; Xiang, X.; Deng, J.; Gao, D.; Sheng, X. Viral Long Non-Coding RNA Regulates Virus Life-Cycle and Pathogenicity. Mol Biol Rep 2022, 49, 6693–6700. [Google Scholar] [CrossRef]
- Sato, K.; Takayama, K.; Inoue, S. Role of piRNA Biogenesis and Its Neuronal Function in the Development of Neurodegenerative Diseases. Frontiers in Aging Neuroscience 2023, 15. [Google Scholar] [CrossRef]
- Zuo, L.; Wang, Z.; Tan, Y.; Chen, X.; Luo, X. piRNAs and Their Functions in the Brain. Int J Hum Genet 2016, 16, 53–60. [Google Scholar] [CrossRef]
- Ali, S.D.; Tayara, H.; Chong, K.T. Identification of piRNA Disease Associations Using Deep Learning. Comput Struct Biotechnol J 2022, 20, 1208–1217. [Google Scholar] [CrossRef] [PubMed]
- Halic, M.; Moazed, D. Transposon Silencing by piRNAs. Cell 2009, 138, 1058–1060. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Lu, J. Silencing of Transposable Elements by piRNAs in Drosophila: An Evolutionary Perspective. Genomics, Proteomics & Bioinformatics 2017, 15, 164–176. [Google Scholar] [CrossRef]
- Roy, M.; Viginier, B.; Saint-Michel, É.; Arnaud, F.; Ratinier, M.; Fablet, M. Viral Infection Impacts Transposable Element Transcript Amounts in Drosophila. Proc Natl Acad Sci U S A 2020, 117, 12249–12257. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.S.; Nanda, H.; Aliferis, C.; Langlois, R.A. Characterization of Influenza A Virus Induced Transposons Reveals a Subgroup of Transposons Likely Possessing the Regulatory Role as eRNAs. Sci Rep 2022, 12, 2188. [Google Scholar] [CrossRef] [PubMed]
- Macchietto, M.G.; Langlois, R.A.; Shen, S.S. Virus-Induced Transposable Element Expression up-Regulation in Human and Mouse Host Cells. Life Sci Alliance 2020, 3, e201900536. [Google Scholar] [CrossRef]
- Ivancevic, A.; Chuong, E.B. Transposable Elements Teach T Cells New Tricks. Proceedings of the National Academy of Sciences 2020, 117, 9145–9147. [Google Scholar] [CrossRef]
- Marston, J.L.; Greenig, M.; Singh, M.; Bendall, M.L.; Duarte, R.R.R.; Feschotte, C.; Iñiguez, L.P.; Nixon, D.F. SARS-CoV-2 Infection Mediates Differential Expression of Human Endogenous Retroviruses and Long Interspersed Nuclear Elements. JCI Insight 6, e147170. [CrossRef]
- Li, J.; Hong, X.; Jiang, M.; Kho, A.T.; Tiwari, A.; Wang, A.L.; Chase, R.P.; Celedón, J.C.; Weiss, S.T.; McGeachie, M.J.; et al. A Novel Piwi-Interacting RNA Associates with Type 2–High Asthma Phenotypes. Journal of Allergy and Clinical Immunology 2023. [Google Scholar] [CrossRef] [PubMed]
- Girard, A.; Sachidanandam, R.; Hannon, G.J.; Carmell, M.A. A Germline-Specific Class of Small RNAs Binds Mammalian Piwi Proteins. Nature 2006, 442, 199–202. [Google Scholar] [CrossRef] [PubMed]
- Roovers, E.F.; Rosenkranz, D.; Mahdipour, M.; Han, C.-T.; He, N.; Chuva de Sousa Lopes, S.M.; van der Westerlaken, L.A.J.; Zischler, H.; Butter, F.; Roelen, B.A.J.; et al. Piwi Proteins and piRNAs in Mammalian Oocytes and Early Embryos. Cell Rep 2015, 10, 2069–2082. [Google Scholar] [CrossRef] [PubMed]
- Roy, J.; Mallick, B. Investigating Piwi-Interacting RNA Regulome in Human Neuroblastoma. Genes Chromosomes Cancer 2018, 57, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Della Bella, E.; Menzel, U.; Basoli, V.; Tourbier, C.; Alini, M.; Stoddart, M.J. Differential Regulation of circRNA, miRNA, and piRNA during Early Osteogenic and Chondrogenic Differentiation of Human Mesenchymal Stromal Cells. Cells 2020, 9, 398. [Google Scholar] [CrossRef]
- Chu, H.; Hui, G.; Yuan, L.; Shi, D.; Wang, Y.; Du, M.; Zhong, D.; Ma, L.; Tong, N.; Qin, C.; et al. Identification of Novel piRNAs in Bladder Cancer. Cancer Lett 2015, 356, 561–567. [Google Scholar] [CrossRef] [PubMed]
- Vinasco-Sandoval, T.; Moreira, F.C.; F. Vidal, A.; Pinto, P.; Ribeiro-dos-Santos, A.M.; Cruz, R.L.S.; Fonseca Cabral, G.; Anaissi, A.K.M.; Lopes, K. de P.; Ribeiro-dos-Santos, A.; et al. Global Analyses of Expressed Piwi-Interacting RNAs in Gastric Cancer. International Journal of Molecular Sciences 2020, 21, 7656. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A Web Server for Functional Enrichment Analysis and Functional Annotation of Gene Lists (2021 Update). Nucleic Acids Res 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Chahar, H.S.; Corsello, T.; Kudlicki, A.S.; Komaravelli, N.; Casola, A. Respiratory Syncytial Virus Infection Changes Cargo Composition of Exosome Released from Airway Epithelial Cells. Sci Rep 2018, 8, 387. [Google Scholar] [CrossRef]
- Lin, L.; Li, Q.; Wang, Y.; Shi, Y. Syncytia Formation during SARS-CoV-2 Lung Infection: A Disastrous Unity to Eliminate Lymphocytes. Cell Death Differ 2021, 28, 2019–2021. [Google Scholar] [CrossRef]
- Moras, M.; Lefevre, S.D.; Ostuni, M.A. From Erythroblasts to Mature Red Blood Cells: Organelle Clearance in Mammals. Front Physiol 2017, 8, 1076. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





