Submitted:
02 February 2024
Posted:
05 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Virus Concentration
2.3. RNA Extraction and Virus Detection
2.4. Estimation of SARS-CoV-2 Genome Copy Number in Wastewater
2.5. Genome Sequencing and Variant Typing of SARS-CoV-2
2.6. Environmental Samples and SARS-CoV-2 Detection in Antarctic Wildlife
2.7. RNA extraction from Wildlife Samples
2.8. RT-qPCR and RT-PCR Analysis
3. Results
3.1. SARS-CoV-2 RNA Detection in WWTPs
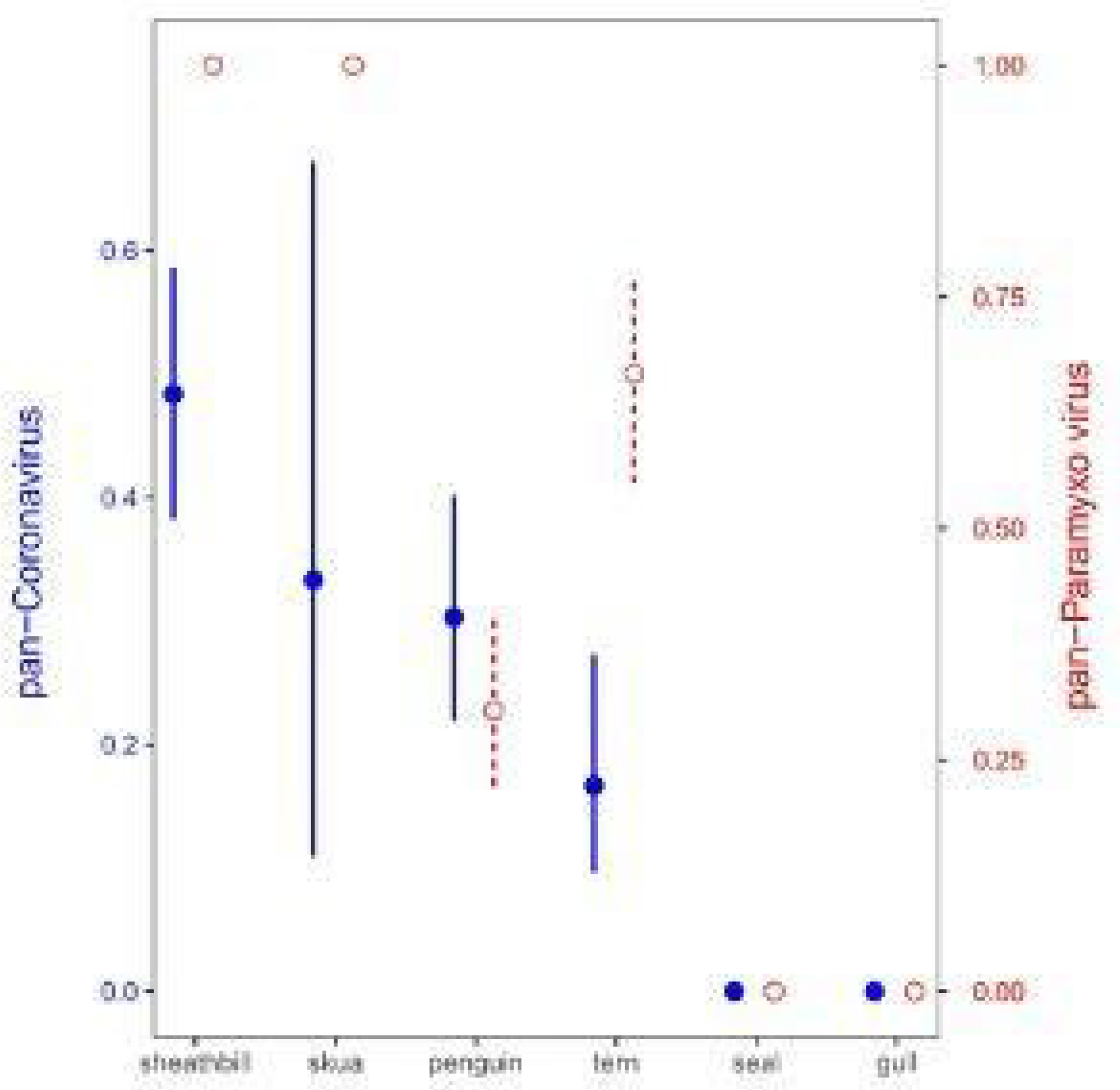
3.2. Environmental Animal Viral Detections by Real-Time RT-PCR Assays
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of interest
References
- Aronson, R.B.; Thatje, S.; McClintock, J.B.; Hughes, K.A. Anthropogenic impacts on marine ecosystems in Antarctica. Ann N Y Acad Sci. 2011, 1223, 82–107. [Google Scholar] [CrossRef]
- Chwedorzewska, K.J.; Korczak-Abshire, M.; Znój, A. Is Antarctica under threat of alien species invasion? Glob Chang Biol. 2020, 26, 1942–1943. [Google Scholar] [CrossRef]
- Lalonde, M.M.L.; Marcus, J.M. A global molecular phylogeny yields insights into the dispersal and invasion history of Junonia, a butterfly genus with remarkable dispersal abilities. Proc R Soc. 2022, 289, 20212801. [Google Scholar]
- Leihy, R.I.; Peake, L.; Clarke, D.A.; Chown, S.L.; McGeoch, M.A. Introduced and invasive alien species of Antarctica and the Southern Ocean islands. Sci Data. 2023, 10, 200. [Google Scholar] [CrossRef]
- Marcelino, V.R.; Wille, M.; Hurt, A.C.; González-Acuña, D.; Klaassen, M.; Schlub, T.E.; Eden, J.S.; Shi, M.; Iredell, J.R.; Sorrell, T.C.; Holmes, E.C. Meta-transcriptomics reveals a diverse antibiotic resistance gene pool in avian microbiomes. BMC Biol. 2019, 17, 1–11. [Google Scholar] [CrossRef]
- Barbosa, A.; Varsani, A.; Morandini, V.; Grimaldi, W.; Vanstreels, R.E.T.; Diaz, J.I.; Boulinier, T.; Dewar, M.; González-Acuña, D.; Gray, R.; McMahon, C.R.; Miller, G.; Power, M.; Gamble, A.; Wille, M. Risk assessment of SARS-CoV-2 in Antarctic wildlife. Sci Total Environ. 2021, 755, 143352. [Google Scholar] [CrossRef]
- Hughes, K.A.; Convey, P. Implications of the COVID-19 pandemic for Antarctica. Antarct Sci 2020, 32, 426–439. [Google Scholar] [CrossRef]
- Kitajima, M.; Ahmed, W.; Bibby, K.; Carducci, A.; Gerba, C.P.; Hamilton, K.A.; Haramoto, E. , Rose, J.B. SARS-CoV-2 in wastewater: State of the knowledge and research needs. Sci Total Environ. 2020, 739, 139076. [Google Scholar] [CrossRef] [PubMed]
- La Rosa, G.; Bonadonna, L.; Lucentini, L.; Kenmoe, S.; Suffredini, E. 2020. Coronavirus in water environments: Occurrence, persistence and concentration methods-A scoping review. Water Res. 2020, 179, 115899. [Google Scholar] [CrossRef] [PubMed]
- Olivares-Pacheco, J.; Adell, A.D.; Hepp, M.I.; Reis, A.S.; Echeverría, C.; Ibacache-Quiroga, C.; Assmann, P.; Gaggero, A. Detección y cuantificación de SARS-CoV-2 en plantas de tratamiento de aguas residuales de diferentes ciudades de Chile: hacia la implementación de una vigilancia centinela permanente. REVINF. 2022, 39, 690–698. [Google Scholar] [CrossRef]
- Tandukar, S.; Sthapit, N.; Thakali, O.; Malla, B.; Sherchan, S.P.; Shakya, B.M.; Shrestha, L.P.; Sherchand, J.B.; Joshi, D.R.; Lama, B.; Haramoto, E. Detection of SARS-CoV-2 RNA in wastewater, river water, and hospital wastewater of Nepal. Sci Total Environ. 2022, 824, 153816. [Google Scholar] [CrossRef]
- Hamouda, M.; Mustafa, F.; Maraqa, M.; Rizvi, T.; Aly Hassan, A. . Wastewater surveillance for SARS-CoV-2: Lessons learnt from recent studies to define future applications. Sci Total Environ. 2021, 759, 143493. [Google Scholar] [CrossRef]
- Yang, K., Guo, J.; Møhlenberg, M.; Zhou, H. SARS-CoV-2 surveillance in medical and industrial wastewater-a global perspective: a narrative review. Environ Sci Pollut Res Int. 2023, 30, 63323–63334. [CrossRef]
- Carducci, A.; Federigi, I.; Balestri, E.; Lardicci, C.; Castelli, A.; Maltagliati, F.; Zhao, H.; Menicagli, V.; Valente, R.; De Battisti, D.; Verani, M. Virus contamination and infectivity in beach environment: Focus on sand and stranded material. Mar Pollut Bull. 2022, 185, 114342. [Google Scholar] [CrossRef]
- Contrant, M.; Bigault, L.; Andraud, M.; Desdouits, M.; Rocq, S.; Guyader, F.S.; Le, Blanchard, Y. Porcine epidemic diarrhea virus, surrogate for coronavirus decay measurement in French coastal waters and contribution to coronavirus risk evaluation. Microbiol Spectr. 2023, 11, e01844–23.
- Mahlknecht, J. Presence and persistence of SARS-CoV-2 in aquatic environments: A mini-review. Curr Opin Environ Sci Health. 2022, 100385. [Google Scholar] [CrossRef]
- Novoa, B.; Ríos-Castro, R.; Otero-Muras, I.; Gouveia, S.; Cabo, A.; Saco, A.; Rey-Campos, M.; Pájaro, M.; Fajar, N.; Aranguren, R.; Romero, A.; Panebianco, A.; Valdés, L.; Payo, P.; Alonso, A.A.; Figueras, A.; Cameselle, C. Wastewater and marine bioindicators surveillance to anticipate COVID-19 prevalence and to explore SARS-CoV-2 diversity by next generation sequencing: One-year study. Sci Total Environ. 2022, 833, 155140. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, Y.; Thongchankaew-Seo, U.; Yamazaki, W. Very low likelihood that cultivated oysters are a vehicle for SARS-CoV-2: 2021–2022 seasonal survey at supermarkets in Kyoto, Japan. Heliyon. 2022, 8. [Google Scholar] [CrossRef] [PubMed]
- Atoui, A.; Cordevant, C.; Chesnot, T.; Gassilloud, B. SARS-CoV-2 in the environment: Contamination routes, detection methods, persistence and removal in wastewater treatment plants. Sci Total Environ. 2023, 881, 163453. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ash, K.T.; Joyner, D.C.; Williams, D.E.; Alamilla, I.; McKay, P.J.; Iler, C.; Green, B.M.; Kara-Murdoch, F.; Swift, C.M.; Hazen, T.C. Decay of enveloped SARS-CoV-2 and non-enveloped PMMoV RNA in raw sewage from university dormitories. Front Microbiol. 2023, 14, 1144026. [Google Scholar] [CrossRef] [PubMed]
- Gröndahl, F.; Sidenmark, J.; Thomsen, A. Survey of waste water disposal practices at Antarctic research stations. Polar Res. 2016, 28, 298–306. [Google Scholar] [CrossRef]
- Masindi, V.; Foteinis, S.; Nduli, K.; Akinwekomi, V. Systematic assessment of SARS-CoV-2 virus in wastewater, rivers and drinking water –A catchment-wide appraisal. Sci Total Environ. 2021, 800, 149298. [Google Scholar] [CrossRef]
- Calgua, B.; Fumian, T.; Rusinol, M.; Rodriguez-Manzano, J.; Mbayed, V.A.; Bofill-Mas, S.; Miagostovich, M.; Girones, R. Detection and quantification of classic and emerging viruses by skimmed-milk flocculation and PCR in river water from two geographical areas. Water Res. 2013, 47, 2797–2810. [Google Scholar] [CrossRef]
- Guerrero-Latorre, L.; Ballesteros, I.; Villacrés-Granda, I.; Granda, M.G.; Freire-Paspuel, B., Ríos-Touma, B. 2020. SARS-CoV-2 in river water: Implications in low sanitation countries. Sci Total Environ. 2020, 743, 140832.
- Melgaço, F.G.; Corrêa, A.A.; Ganime, A.C.; Brandão, M.L.L.; Medeiros, V. de M.; Rosas, C. de O.; Lopes, S.M. dos R.; Miagostovich, M.P. Evaluation of skimmed milk flocculation method for virus recovery from tomatoes. Braz J Microbiol. 2018, 49, 34–39. [Google Scholar] [CrossRef]
- Rio, D.C.; Ares, M.; Hannon, G.J.; Nilsen, T.W. Purification of RNA Using TRIzol (TRI Reagent). Cold Spring Harb Protoc. 2010, pdb.prot5439. [Google Scholar] [CrossRef]
- Randazzo, W.; Truchado, P.; Cuevas-Ferrando, E.; Simón, P.; Allende, A.; Sánchez, G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020, 181, 115942. [Google Scholar] [CrossRef] [PubMed]
- Ai, Y.; Davis, A.; Jones, D.; Lemeshow, S.; Tu, HH; He, FF; Ru, PP; Pan, X.; Bohrerova, Z.; Lee, J. Wastewater SARS-CoV-2 monitoring as a community-level COVID-19 trend tracker and variants in Ohio, United States. Sci Total Environ. 2021, 801, 149757. [CrossRef] [PubMed]
- Giraud-Billoud, M.; Cuervo, P., Altamirano, J.C.; Pizarro, M.; Aranibar, J.N.; Catapano, A.; Cuello, H.; Masachessi, G.; Vega, I.A.. Monitoring of SARS-CoV-2 RNA in wastewater as an epidemiological surveillance tool in Mendoza, Argentina. Sci Total Environ. 2021, 796, 148887. [CrossRef] [PubMed]
- Rosiles-González, G.; Carrillo-Jovel, V.H.; Alzate-Gaviria, L.; Betancourt, W.Q.; Gerba, C.P.; Moreno-Valenzuela, O.A.; Tapia-Tussell, R.; Hernández-Zepeda, C. Environmental Surveillance of SARS-CoV-2 RNA in Wastewater and Groundwater in Quintana Roo, Mexico. Food Environ Virol. 2021, 13, 457–469. [Google Scholar] [CrossRef] [PubMed]
- Scott, L.C.; Aubee, A.; Babahaji, L.; Vigil, K.; Tims, S.; Aw, T.G. 2021. Targeted wastewater surveillance of SARS-CoV-2 on a university campus for COVID-19 outbreak detection and mitigation. Environ Res. 2021, 200, 111374. [Google Scholar] [CrossRef] [PubMed]
- Barriga, G.P.; Boric-Bargetto, D.; Cortez-San Martin, M.; Neira, V.; van Bakel, H.; Thompsom, M.; Tapia, R.; Toro-Ascuy, D.; Moreno, L.; Vasquez, Y.; Sallaberry, M.; Torres-Pérez, F.; González-Acuña, D.; Medina, R.A. Avian influenza virus H5 strain with North American and Eurasian lineage genes in an Antarctic penguin. Emerg Infect Dis. 2016, 22, 2221. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. 1987... Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Vijgen L.; Moës E.; Keyaerts E,; Li S.; Van Ranst M. A pancoronavirus RT-PCR assay for detection of all known coronaviruses. SARS-and other coronaviruses: laboratory protocols. 2008, 3, 3–12.
- van Boheemen, S.; Bestebroer, T.M.; Verhagen, J.H.; Osterhaus, A.D.; Pas, S.D.; Herfst, S.; Fouchier, R. A family-wide RT-PCR assay for detection of paramyxoviruses and application to a large-scale surveillance study. PLoS One. 2012, 7, e34961. [Google Scholar] [CrossRef] [PubMed]
- Hale, V.L.; Dennis, P.M.; McBride, D.S.; Nolting, J.M.; Madden, C.; Huey, D.; Ehrlich, M.; Grieser, J.; Winston, J.; Lombardi, D.; Gibson, S.; Saif, L.; Killian, M.L.; Lantz, K.; Tell, R.M.; Torchetti, M.; Robbe-Austerman, S.; Nelson, M.I.; Faith, S.A.; Bowman, A.S. SARS-CoV-2 infection in free-ranging white-tailed deer. Nature. 2022, 602, 481–486. [Google Scholar] [CrossRef]
- Sharun, K.; Dhama, K.; Pawde, A.M.; Gortázar, C.; Tiwari, R.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J.; De La Fuente, J.; Michalak, I.; Attia, Y.A.; Gort Azar, C.; De La Fuente, J.E.; Pandit, P.; Dayal, D.; Chikitsa, U.P.; Vishwavidyalaya, V.; Go, E. ; Sansthan, A SARS-CoV-2 in animals: potential for unknown reservoir hosts and public health implications. Vet Q. 2021, 41, 181–201. [Google Scholar] [CrossRef]
- Polo, D., Lois, M.; Fernández-Núñez, M.T.; Romalde, J.L. Detection of SARS-CoV-2 RNA in bivalve mollusks and marine sediments. Sci Total Environ. 2021, 786, 147534. [CrossRef]
- Casanova, L.; Rutala, W.A.; Weber, D.J.; Sobsey, M.D. Survival of surrogate coronaviruses in water. Water Res. 2009, 43, 1893–1898. [Google Scholar] [CrossRef]
- Paul, D.; Kolar, P.; Hall, S.G. A review of the impact of environmental factors on the fate and transport of coronaviruses in aqueous environments. NPJ Clean Water. 2021, 4, 7. [Google Scholar] [CrossRef]
- Kumar, M.; Taki, K.; Gahlot, R.; Sharma, A.; Dhangar, K. A. chronicle of SARS-CoV-2: Part-I - Epidemiology, diagnosis, prognosis, transmission and treatment. Sci Total Environ. 2020, 734, 139278. [Google Scholar] [CrossRef]
- Shutler, J.D.; Zaraska, K.; Holding, T.; Machnik, M.; Uppuluri, K.; Ashton, I.G.C.; Migdał, Ł.; Dahiya, R.S. Rapid assessment of SARS-CoV-2 transmission risk for fecally contaminated river water. ACS ES&T Water. 2021, 1, 949–957. [Google Scholar]
- Sala-Comorera, L.; Reynolds, L.J.; Martin, N.A.; O’Sullivan, J.J.; Meijer, W.G.; Fletcher, N.F. Decay of infectious SARS-CoV-2 and surrogates in aquatic environments. Water Res. 2021, 201, 117090. [Google Scholar] [CrossRef]
- Allinson, M.; Kadokami, K.; Shiraishi, F.; Nakajima, D.; Zhang, J.; Knight, A.; Gray, S.R.; Scales, P.J.; Allinson, G. Wastewater recycling in Antarctica: Performance assessment of an advanced water treatment plant in removing trace organic chemicals. J Environ Manage. 2018, 224, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Hemnani, M.; Rodrigues, D.; Santos, N.; Santos-Silva, S.; Figueiredo, M.E., Henriques, P.; Ferreira-e-Silva, J.; Rebelo, H.; Poeta, P.; Thompson, G.; Mesquita, J.R. 2022. Molecular detection and characterization of coronaviruses in migratory ducks from Portugal show the circulation of Gammacoronavirus and Deltacoronavirus. Animals. 2022, 12, 3283.
- Monchatre-Leroy, E.; Boué, F.; Boucher, J.M.; Renault, C.; Moutou, F.; Gouilh, M.A.; Umhang, G. Identification of Alpha and Beta Coronavirus in wildlife species in France: bats, rodents, rabbits, and hedgehogs. Viruses. 2017, 9, 364. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M.; Talukder, A.; Chowdhury, M.M.H.; Talukder, R.; Akter, R. Coronaviruses in wild birds – A potential and suitable vector for global distribution. Vet Med Sci. 2021, 7, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Wille, M.; Harvey, E.; Shi, M.; Gonzalez-Acuña, D.; Holmes, E.C.; Hurt, A.C. Sustained RNA virome diversity in Antarctic penguins and their ticks. ISME J. 2020, 14, 1768–1782. [Google Scholar] [CrossRef]
- Zamora, G.; Aguilar Pierlé, S.; Loncopan, J.; Araos, L.; Verdugo, F.; Rojas-Fuentes, C.; Krüger, L.; Gaggero, A.; Barriga, G.P. Scavengers as Prospective Sentinels of Viral Diversity: the Snowy Sheathbill Virome as a Potential Tool for Monitoring Virus Circulation, Lessons from Two Antarctic Expeditions. Microbiol Spectr. 2023, 11, e03302-22. [Google Scholar] [CrossRef]
- Damas, J.; Hughes, G.M.; Keough, K.C.; Painter, C.A.; Persky, N.S.; Corbo, M.; Hiller, M.; Koepfli, K.P.; Pfenning, A.R.; Zhao, H.; Genereux, D.P.; Swofford, R.; Pollard, K.S.; Ryder, O.A.; Nweeia, M.T.; Lindblad-Toh, K.; Teeling, E.C.; Karlsson, E.K.; Lewin, H.A. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc Natl Acad Sci. 2020, 117, 22311–22322. [Google Scholar] [CrossRef]
- León, M.R. De; Hughes, K.A.; Morelli, E.; Convey, P. International response under the Antarctic treaty system to the establishment of a non-native fly in Antarctica. Environ Manage. 2021, 67, 1043–1059. [Google Scholar] [CrossRef] [PubMed]
- Balaraman, V.; Drolet, B.S.; Mitzel, D.N.; Wilson, W.C.; Owens, J.; Gaudreault, N.N.; Meekins, D.A.; Bold, D.; Trujillo, J.D.; Noronha, L.E.; Richt, J.A.; Nayduch, D. Mechanical transmission of SARS-CoV-2 by house flies. Parasites Vectors. 2021, 14, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Cashman, J.S.; Cozier, G.E.; Harrison, C.; Isaac, R.E.; Acharya, K.R. Crystal structures of angiotensin-converting enzyme from Anopheles gambiae in its native form and with a bound inhibitor. Biochem J. 2019, 476, 3505–3520. [Google Scholar] [CrossRef] [PubMed]
- Higgs, S.; Huang, Y.J.S.; Hettenbach, S.M.; Vanlandingham, D.L. SARS-CoV-2 and Arthropods: A Review. Viruses. 2022, 14, 985. [Google Scholar] [CrossRef]

| Date | Source | Frei | Escudero | O’Higgins | Lineages detected | |
|---|---|---|---|---|---|---|
| Dec 21st 2020 | I | n/s | (+) | n/s | B.1.1.451 | |
| E | n/s | (+) | n/s | B.1.1 | ||
| Feb 10th 2021 | I | (+) | n/d | n/s | B.1.1 | |
| E | (+) | n/d | n/s | B.1.1 | ||
| Feb 17th 2021 | I | n/d | n/d | n/s | n/s | |
| E | n/d | (+) | n/s | No sequence | ||
| Feb 23rd 2021 | I | n/s | n/s | (+) | B.1.1 | |
| E | n/s | n/s | (+) | B.1.1 | ||
| Feb 24th 2021 | I | n/d | n/s | n/s | n/s | |
| E | n/d | n/s | n/s | n/s | ||
| Feb 25th 2021 | I | n/s | n/d | n/s | n/s | |
| E | n/s | (+) | n/s | No sequence | ||
| Mar 2nd 2021 | I | n/s | n/s | (+) | B.1.1 | |
| E | n/s | n/s | (+) | B.1.1 | ||
| Mar 3rd 2021 | I | n/s | n/d | n/s | n/s | |
| E | n/s | n/d | n/s | n/s | ||
| Mar 9th 2021 | I | n/s | n/s | (+) | B.1.1.409 | |
| E | n/s | n/s | (+) | No sequence |
| Environmental pool | Pan-Coronavirus | SARS-CoV-2 | pan-Paramyxo virus | Influenza A |
|---|---|---|---|---|
| 1 | (-) | (-) | (-) | (-) |
| 2 | (-) | (-) | (-) | (-) |
| 3 | (-) | (-) | (-) | (-) |
| 4 | (-) | (-) | (-) | (-) |
| 5 | (-) | (-) | (+) | (-) |
| 6 | (+) | (-) | (+) | (-) |
| 7 | (-) | (-) | (+) | (-) |
| 8 | (-) | (-) | (+) | (-) |
| 9 | (+) | (-) | (+) | (-) |
| 10 | (-) | (-) | (-) | (-) |
| 11 | (-) | (-) | (-) | (-) |
| 12 | (+) | (-) | (-) | (-) |
| 13 | (-) | (-) | (-) | (-) |
| 14 | (-) | (-) | (-) | (-) |
| 15 | (-) | (-) | (+) | (-) |
| 16 | (-) | (-) | (+) | (-) |
| 17 | (+) | (-) | (+) | (-) |
| 18 | (+) | (-) | (+) | (-) |
| 19 | (-) | (-) | (+) | (-) |
| 20 | (+) | (-) | (+) | (-) |
| 21 | (-) | (-) | (+) | (-) |
| Negative control | (-) | (-) | (-) | (-) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





