Introduction
Muscadine (
Vitis rotundifolia Michx) is a grape crop native to the south-eastern and south-central regions of the United States [
1]. To date, this crop is being mainly produced commercially in Florida, Georgia, Alabama, Louisiana, South Carolina, and North Carolina [
2]. Many years of breeding efforts have created multiple superior cultivars for fresh market, wine, and unfermented juice industries [
3].
Anthocyanins are main active nutraceuticals in muscadine berries, which provide antioxidative and other health values [
4,
5,
6,
7]. To date, anthocyanins have been analyzed in intensive studies in a number of muscadine cultivars. The first anthocyanin molecule isolated from muscadine berries of the Hunt cultivar was named muscadinin (3, 5-diglycosidyl-3’-O-methyldelphinidin, namely petunidin 3, 5-diglucoside) by W.L. Brown in 1940 [
1]. Main anthocyanins were then identified from berries and other tissues [
8,
9,
10,
11,
12,
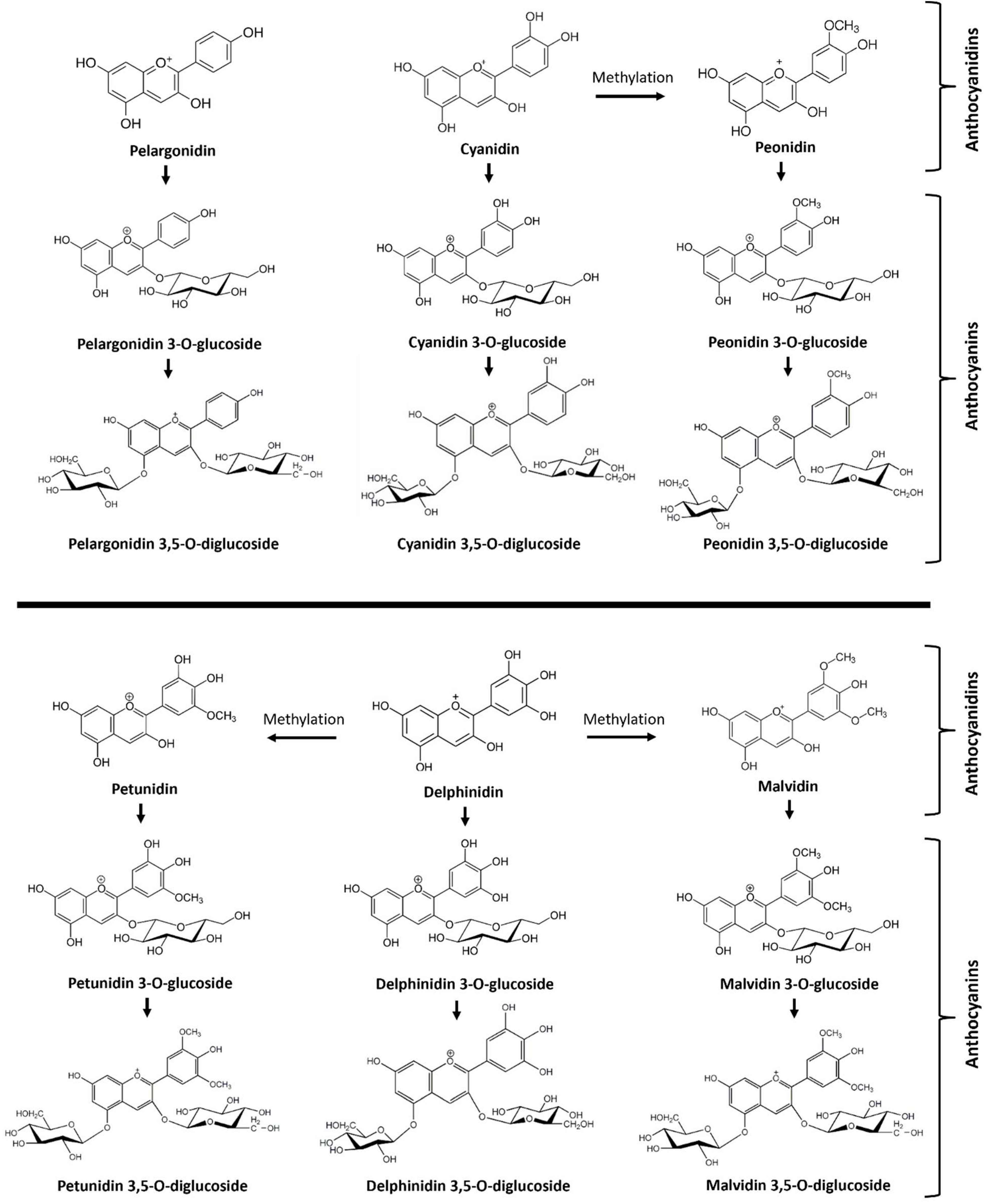
13]. The most common muscadine anthocyanidins are delphinidin, malvidin, petunidin, cyanidin, pelargonidin, and peonidin (
Figure 1).
The most common anthocyanins are non-acylated 3,5 diglucosides of delphinidin, malvidin, petunidin, cyanidin, pelargonidin, and peonidin, which have been elucidated from fresh fruits of Noble, Tarheel, and other cultivars [
9,
11,
12,
14,
15]. The contents of these six anthocyanins in muscadine berries varies widely among cultivars. More importantly, the abundance of each of six anthocyanins has been demonstrated to control wine and juice color. For example, high quality wine color was found to relate to high amounts and percentages of malvidin 3,5-diglucoside but low amounts and percentages of delphinidin 3,5-diglucoside and cyanidin 3,5-diglucoside [
9]. Wine production research has showed that muscadine varieties with high amounts of cyanidin 3,5-diglucoside produce the poorest wine color and color stability [
9]. Given that malvidin 3,5-diglucoside is structurally featured with two methyl groups in the B-ring, there is a general assumption that the degree of methylation of each aglycone is associated with pigment stability of muscadine wine [
16].
However, maintaining pigment stability remains a challenging problem with muscadine wine and juice products [
12,
17]. To address this problem, both intraspecific and interspecific muscadine hybrids have been generated to attempt to produce new genotypes with more color stable anthocyanin pigment ratios [
12,
18,
19,
20]. These studies have achieved progress in not only improving anthocyanin ratios of methylated to non-methylated ones, but also producing additional muscadine anthocyanins. For example, 25 anthocyanins including five common anthocyanidin-3,5-diglucosides and new anthocyanins were identified in 14 black muscadine hybrids [
18]. Although pelargonidin and its monoglucoside and diglucoside have not been reported from common commercial verities such as Noble and Nesbitt, these metabolites were observed in hybrids. These results demonstrated that conventional breeding methods can produce genotypes with improved anthocyanin ratios and new anthocyanins for color quality improvement of wine and juice products.
FLH 13-11 is an interspecific muscadine F1 hybrid that resulted from the cross of ‘Marsh’ x ‘Magoon’ made by the grape breeding program of the University of Florida, located at Leesburg, Florida. ‘Marsh’ is a wild selection of Vitis munsoniana and ‘Magoon’ is a V. rotundifolia cultivar that resulted from the cross ‘Thomas’ x ‘Burgaw’. FLH 13-11 muscadine variety is being cropped at the Castle Hayne research station in Wilmington, North Carolina. In this study, our goal was to use HPLC-qTOF-MS/MS technology to determine the anthocyanin profile and structure in berries of FLH 13-11 hybrid muscadine genotype.
Materials and Methods
Chemical agents. Peonidin 3-O-glucoside (≥97%, HPLC grade, cat# 42008), Cyanidin 3,5-
diglucoside ((≥90%, HPLC grade, cat# 74397), pelargonidin chloride (HPLC grade, cat# P1659), and cyanidin chloride (≥95%, HPLC grade, cat# 79457) were purchased from Sigma-Aldrich® (St Louis, MO, USA). Delphinidin chloride (≥95%, HPLC grade, cat# 43725) was purchased from Fluka™ Chemical (Ronkonkoma, NY, USA). Hydrochloric acid (36.5–38%) was purchased from BDH (cat#: BHH3028-2.5L, West Chester, PA, 19380, USA). Acetonitrile (LC-MS grade, cat#: 9829-03), glacial acetic acid (HPLC grade, cat#: 9515-03) and methanol (LC-MS grade, cat#: 9830-03) were purchased from Avantor® (Center Valley, PA 18034, USA). Ethyl alcohol, 200 proof (cat#: EX0276-1) was purchased from EMD (Burlington, MA 01803, USA).
Plant material. FLH 13-11 vines grows at the Castle Hayne Station in Wilmington, North Carolina. Berries were fully ripened in the first two weeks of September each year. Berries were collected on Sept. 6 in 2011 and Sept. 10 in 2012. Fruits were harvested and immediately placed on ice in a cooler and transported to the laboratory. All fresh berries were frozen in liquid nitrogen and then stored in -80°C freezers. Frozen berries were ground to fine powder in liquid nitrogen using a steel blender. Powdered samples were completely dried via lyophilization from -40°C to -20°C for 72 hours. Dried powder samples were stored in -80°C until extraction of anthocyanins described below.
Extraction and measurement of anthocyanins. One hundred milligrams of freeze-dried berry powder were suspended in 1.0 ml extraction buffer, which was composed of 0.5% HCl in methanol: dH2O (50:50, v/v) in a 2 ml Eppendorf tube at room temperature. The tube was vigorously vortexed for 45 sec, sonicated for 10 min, and then centrifuged at 10,000 rpm for 10 min. The supernatant was transferred into a new 1.5 ml tube. This step was repeated using 0.5 ml extraction buffer. The two extractions were pooled together in the 1.5 ml tube. To remove chlorophyll and non-polar lipids in the extraction, the 1.5 ml ethanol: water extraction was mixed with 0.5 ml chloroform in a 2 ml tube. The mixture was vortexed vigorously for 45 sec and centrifuged at the speed of 10,000 rpm for 5 min. The resulting upper ethanol: water-phase (about 750 µl) containing red pigment was pipetted into a new 1.5 ml tube. The bottom chloroform phase containing chlorophyll and non-polar lipids was disposed of into a waste container. This step was repeated once. The resulting upper red phase was stored at -20°C for anthocyanin analysis described below. Three replications were included varieties in this experiment.
The absorbance (ABS) of ethanol-water phase extracts was recorded at the wavelength of 530 nm on a HEλIOSγ UV-Visible spectrophotometer. The extraction buffer was used as a blank control. Ten µl extract was added to 990 µl extraction buffer to dilute anthocyanin concentrations to measure ABS value. Authentic standard peonidin 3-O-glucoside was used to establish a standard curve. Total anthocyanin content in berries was estimated as peonidin 3-O-glucoside equivalent (µg/g) according to the standard curve.
After measurement of total anthocyanin contents for each sample, 720 µl ethanol-water anthocyanin extract was dried off using a SpeedVac Concentrator connected to Refrigerated Condensation Trap for 2 hrs. The remaining pellet was dissolved in 720 µl of 0.1% HCl in methanol 100% in a 1.5 ml tube. The tube was centrifuged at 10,000 rpm for 10 min. The resulting clear supernatant was transferred to a new 1.5 ml tube and then stored at -20°C for anthocyanin analysis. Two hundred µl HCl-methanol extract for each sample was transferred to a glass insert, which was placed in a 1.5 ml glass vial for HPLC and HPLC-qTOF-MS/MS analysis described below. Fifty µl HCl-methanol extract was used for hydrolysis.
Hydrolysis of anthocyanins. Hydrolysis of anthocyanins followed our protocol reported previously [
21]. In brief, fifty µl of anthocyanin extraction was added into 450 µl of
n-butanol: HCl (95:5, v/v) solvent contained in a 1.5 ml tube. This mixture was boiled for 1 hr. After the sample was cooled down to room temperature, it was dried off with flow nitrogen gas. The remaining residue was suspended in 200 µl of 0.1% HCl-methanol. The sample was centrifuged at 12,000 rpm for 10 min. The supernatant was transferred to a new 1.5 ml tube and stored at -20°C for anthocyanidin analysis. Two hundred µl HCl-methanol extract for each sample was transferred to a glass insert, which was placed in a 1.5 ml glass vial for HPLC and LC-MS/MS analysis described below.
High performance liquid chromatography- diode array detectors analysis. Anthocyanins profiling was carried out using high performance liquid chromatography-diode array detector (HPLC-DAD) on 2,010 eV LC instrument (Shimadzu, Japan) as reported previously [
21,
22]. Both anthocyanidins and anthocyanins were separated on an analytical column of Eclipse XDB-C18 (250 mm x 4.6 mm, 5 µm, Agilent) as also reported previously [
22]. The mobile phase solvents were composed of 1% acetic acid in water (solvent A: 1% HPLC grade acetic acid in LC-MS grade water) and 100% acetonitrile (solvent B) (LC-MS grade). The column was equilibrated with 30 min using solvent A: B (80:20). Then a gradient solvent system was developed to separate metabolites. It was composed of ratios of solvent A to B: 80:20 (0-5 min), 80:20 to 70:30 (5-10 min), 70:30 to 65:35 (10-20 min), 65:35 to 60:40 (20-30 min), 60:40 to 55:45 (30-40 min), 55:45 to 50:50 (40-45 min), 50:50 to 48:52 (45-50 min), 48:52 to 45:55 (50-55 min), 45:55 to 40:60 (55-58 min), 40:60 to 10:90 (58-58.5 min), 10:90 to 80:20 (58.5-60 min). After these gradient steps, the column was equilibrated and washed with solvent A: B (80:20)10 min. The flow rate was 0.4 ml/min and the injection volume was 20 µl. The UV spectrum was recorded from 190 to 800 nm. Pelargonidin chloride, cyanidin chloride, delphinidin chloride, and peonidin 3-glucoside were used as authentic standard controls.
HPLC- quadrupole time-of-flight-tandem mass spectrometer (HPLC-qTOF-MS/MS) analysis. HPLC-TOF-MS/MS analysis was performed on an Agilent Technologies (Santa Clara, CA, USA) 6210 time-of-flight LC-MS/MS as reported previously [
23]. The mobile phase solvents were composed of 1% acetic acid in water (solvent A: 1% HPLC grade acetic acid in LC-MS grade water) and 100% acetonitrile (solvent B) (LC-MS grade), which formed another gradient solvent system to separate anthocyanins and anthocyanidins for LC/MS/MS assay. A gradient solvent system was composed of gradient ratios of solvent A to B: 85:15 (0-10 min), 85:15 to 80:20 (10-20 min), 80:20 to 75:25 (20-30 min), 75:25 to 65:35 (30-35 min), 65:35 to 60:40 (35-40 min), 60:40 to 50:50 (40-55 min), 50:50 to 10:90 (55-60 min), 10:90 to 90:10 (60-70 min). After the last gradient step, the column was equilibrated and washed 10 min with solvents A: B (85:15). The flow rate was 0.4 ml/min. The injection volume of samples was 5.0 µl. The drying gas flow was set to 12 l/min, and the nebulizer pressure was set to 50 psi. As our recent report using an optimized protocol for anthocyanin ionization [
23], a negative mode was used for ionization. Mass spectrum was scanned from 100 to 3000 m/z. The acquisition rate was three spectra per second. Other parameters include fragmentor: 150 v, skimmer: 65 v, OCT 1 RF Vpp: 750 v, and collision energy: 30. In addition, the UV spectrum was recorded from 190 to 600 nm. Pelargonidin chloride, cyanidin chloride, delphinidin chloride, and peonidin 3-glucoside were used as authentic standard controls.
Structure annotation. Anthocyanidin and anthocyanin structure annotation was performed using Agilent MassHunter Software for 6200 Series TOF and 6500 Series G-TOF version B.05.00. To identify anthocyanidins released from hydrolysis of anthocyanin extracts, retention time, extracted ion chromatogram (EIC), and mass to charge (m/z) ratio for each peak was analyzed to compare with available standards. For those peaks without standards, their EICs and m/z ratios were used for annotation. For each anthocyanin peak detected at 530 nm by HPLC-DAD in two different instruments, their EIC, m/z ratio, finger fragments from CID, and maximum UV spectrum were integrated for structure annotation. Anthocyanin structures reported in the literature were utilized as our references for annotation. If anthocyanin peaks couldn’t match a reported structure, their EIC, m/z, CID fragments, and UV spectrum were provided to show molecular features.
Statistical analysis. Student`s t test was used to statistically compare contents of total anthocyanins. Standard deviation was calculated to reflect variation of contents between biological replicates.
Results and Discussion
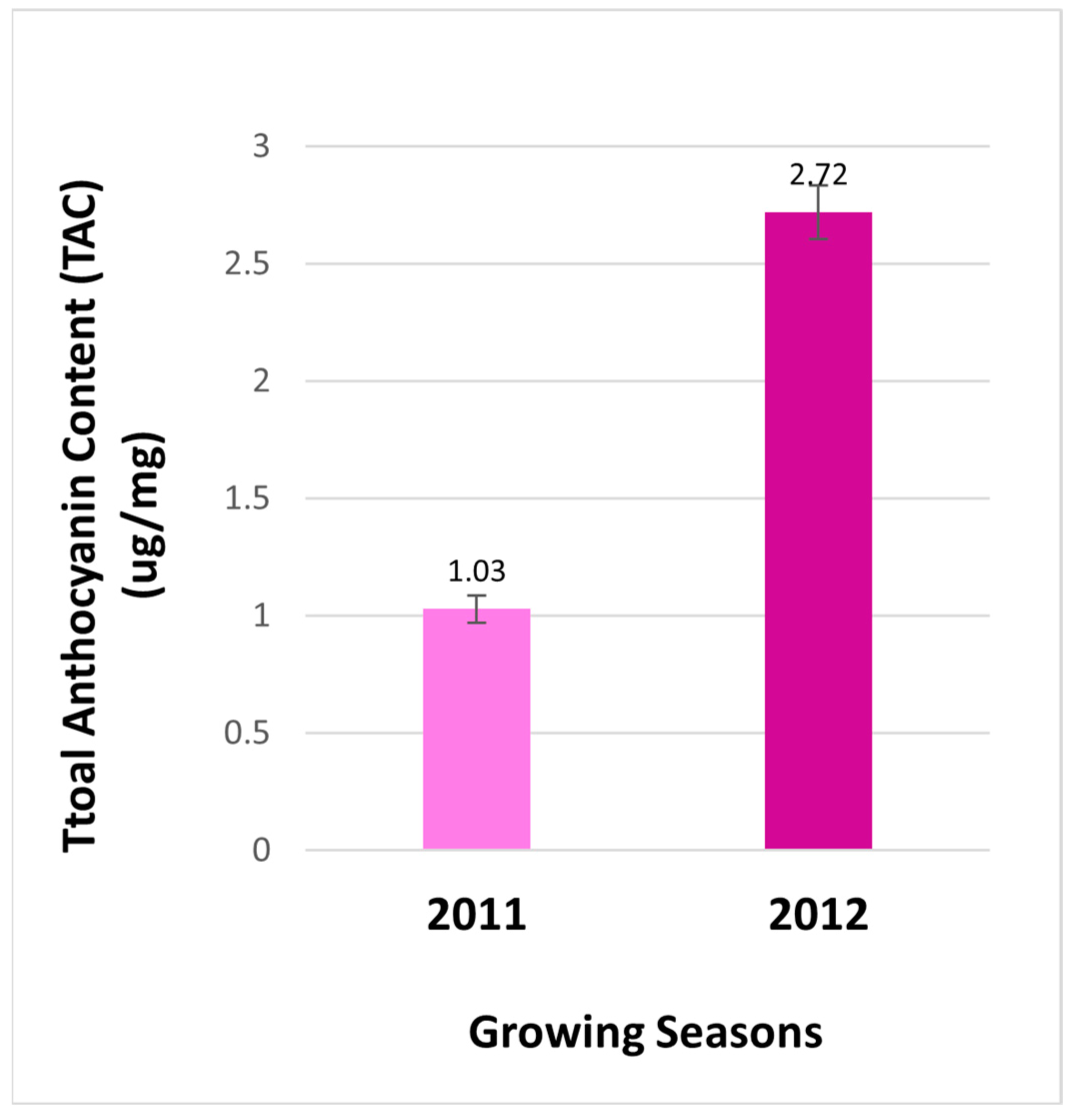
Total anthocyanin contents in berries. Anthocyanins were measured to compare effects of two cropping seasons on the total anthocyanin content (TAC) in berries from the field. The resulting data revealed that the contents of anthocyanin were 2.64 folds higher in berries of 2012 than those of in 2011 (
Figure 2). This result indicates that the production of total anthocyanins is regulated by two different growth seasons. This result is a common phenomenon, given that the biosynthesis of anthocyanins is highly regulated by different environmental conditions. Light conditions and temperatures are two main environmental factors that have been demonstrated to tightly control anthocyanin biosynthesis in plant tissues [
21,
24,
25,
26,
27].
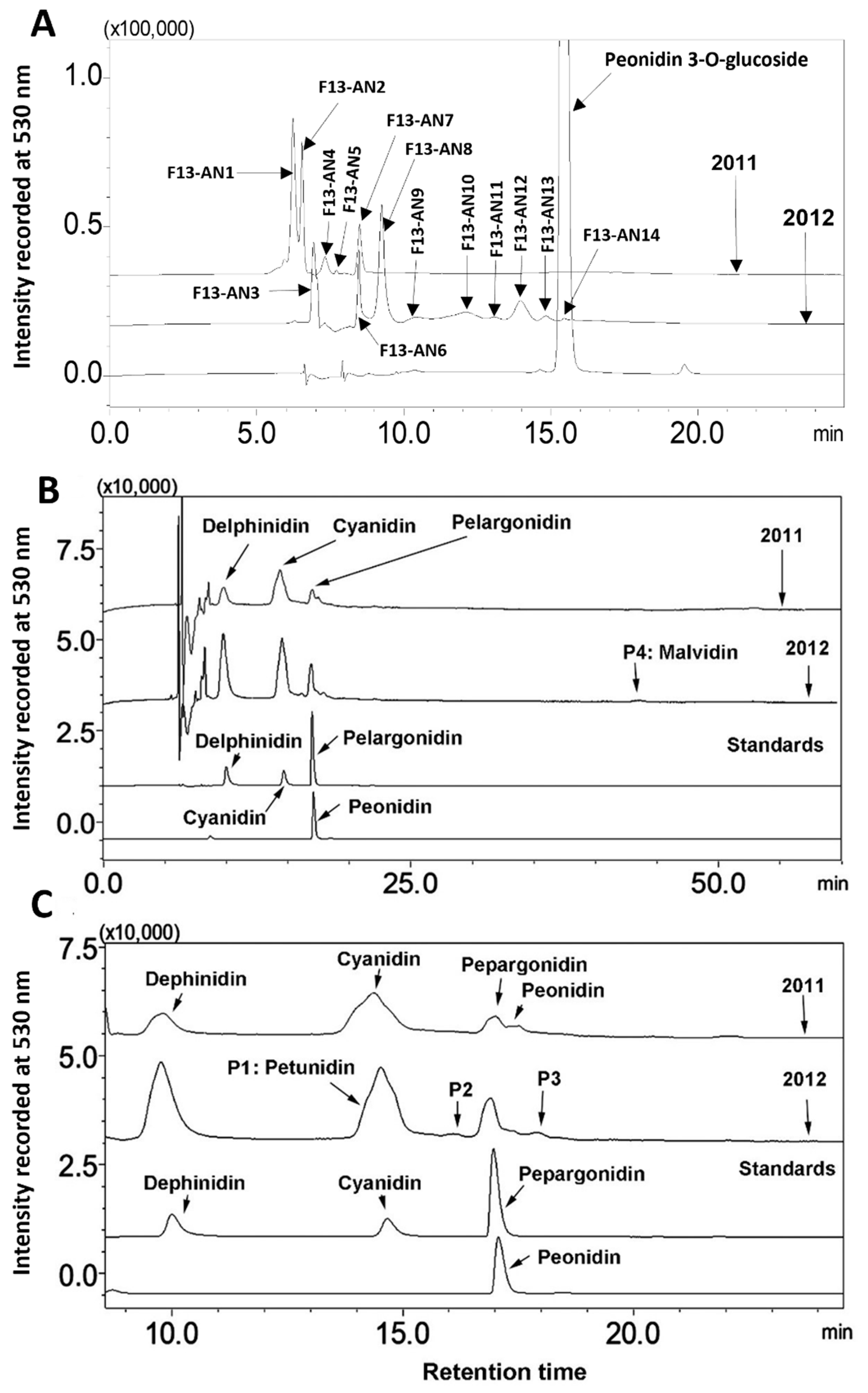
Alteration of anthocyanin profiles. Anthocyanin profiles were analyzed by HPLC-DAD based profiling. Chromatographic peaks were recorded at 530 nm to compare anthocyanin profiles in berries between two growth seasons. At least three experimental replicates were performed to understand anthocyanin profiles in berries from the two seasons. The resulting peak profiles showed that anthocyanin profiles were altered during two growing seasons (
Figure 3A).
Regardless of years, 14 anthocyanin peaks were detected from extracts of two seasons’ berries. Based on retention times, these peaks were labelled as from F13-AN1 to F13-AN14 (
Figure 3A). F13-AN1, 2, 4, 5, 6, and 7 were detected in berries harvested in 2011. Nine peaks, F13-AN3, 6, and 8-14 were detected in berries harvested in 2012. F13-AN6 was the only one detected in both years. These results revealed that the anthocyanin profiles in muscadine berries can be dramatically altered by two cropping seasons. This alteration most likely resulted from differences in weather in the two years. It is well understood that environmental factors can not only control anthocyanin production as discussed above but also can dramatically alter anthocyanin molecule complexity in the same plant. For example,
Arabidopsis thaliana was reported to produce two anthocyanin molecules under regular growing condition [
28]. However, light condition changes can enhance the formation of nearly 30 anthocyanins in this model plant [
21].
Anthocyanidin profiles. The different chromophores of anthocyanidins are the structural bases of anthocyanin hues. Butanol: HCl based boiling was performed to completely hydrolyze anthocyanin extracts to release all anthocyanidins. HPLC-DAD based profiling and HPLC-qTOF-MS/MS were performed to analyze anthocyanidins. The resulting data showed that anthocyanidin profiles were the same in berries from two years (
Figure 3B). These data showed that although berries produced different anthocyanin profiles in two cropping seasons (
Figure 3A), they biosynthesized the same anthocyanidins. Based on four authentic standards, delphinidin, cyanidin, pelargonidin, and peonidin, which were co-eluted as positive controls, hydrolysis of anthocyanins produced four main peaks with the same retention time as these three core anthocyanidins (
Figure 3C). In addition, four additional peaks were detected and labelled as P1, 2, 3, and 4 (
Figure 3B,C).
HPLC-qTOF-MS analysis using the negative mode of ionization further showed that the primary mass-to-charge ratio values of pelargonidin (C
15H
11O
5+, molecular weight, MW, 271.24), cyanidin (C
15H
12O
6+, MW, 287.24), delphinidin (C
15H
13O
7+, MW, 303.24), and peonidin (C
16H
13O
6+, MW, 301.24) standards, 269.265, 285.223, 301.213 [M-2H], and 299. 213 respectively. Two ions were reduced from these standards in the negative mode. This ionization result was likely associated the flavylium cation form of anthocyanidins in the acidic condition. In addition, a second main m/z values for each standard were created by ESI. The m/z value (18) for each standard was added. Therefore, the 2
nd m/z values for the four standards were 287.265, 303.223, 319.213, and 317.213 [M+18-2H]
-. Based on these MS features, berries produced all these four anthocyanidins. In addition, there were four peaks detected at 530 nm, labelled as P1 through P4, (
Figure 3B,C). P1 was shown as a shoulder together with cyanidin because two were closely co-eluted. HPLC-qTOF-MS analysis revealed that the m/z values of P1were 315.1347 [M-2H] and 333.1347 [M+18-2H]. The m/z values for P4 were 331.2968 [M-2H] and 349.2968 [M+18-2H]. Based on these mass spectra and retention times, P1and P4 were annotated to be petunidin and malvidin, respectively. The m/z ratios of P2 and P3 were 365.1982 and 381.1314. [M-H], respectively. Their structures remain to be elucidated in the future.
HPLC-qTOF-MS/MS based characterization of anthocyanins. HPLC-qTOF-MS/MS was performed to annotate anthocyanins detected in extracts of berries. Fourteen anthocyanins (
Figure 3A) were ionized using the negative mode and each anthocyanin ion was formed in the ion source. The resulting ions (primary ions) were separated by mass-to-charge ratio in the first stage of mass spectrometry (MS1). Each particular mass-to-charge ratio from each anthocyanin peak was selected to create fragment ions by collision induced dissociation (CID). The resulting fragment ions (secondary ions) were separated and detected in a second stage of mass spectrometry (MS2). Primary and secondary ions generated for each anthocyanin peak were analyzed to annotate a structure.
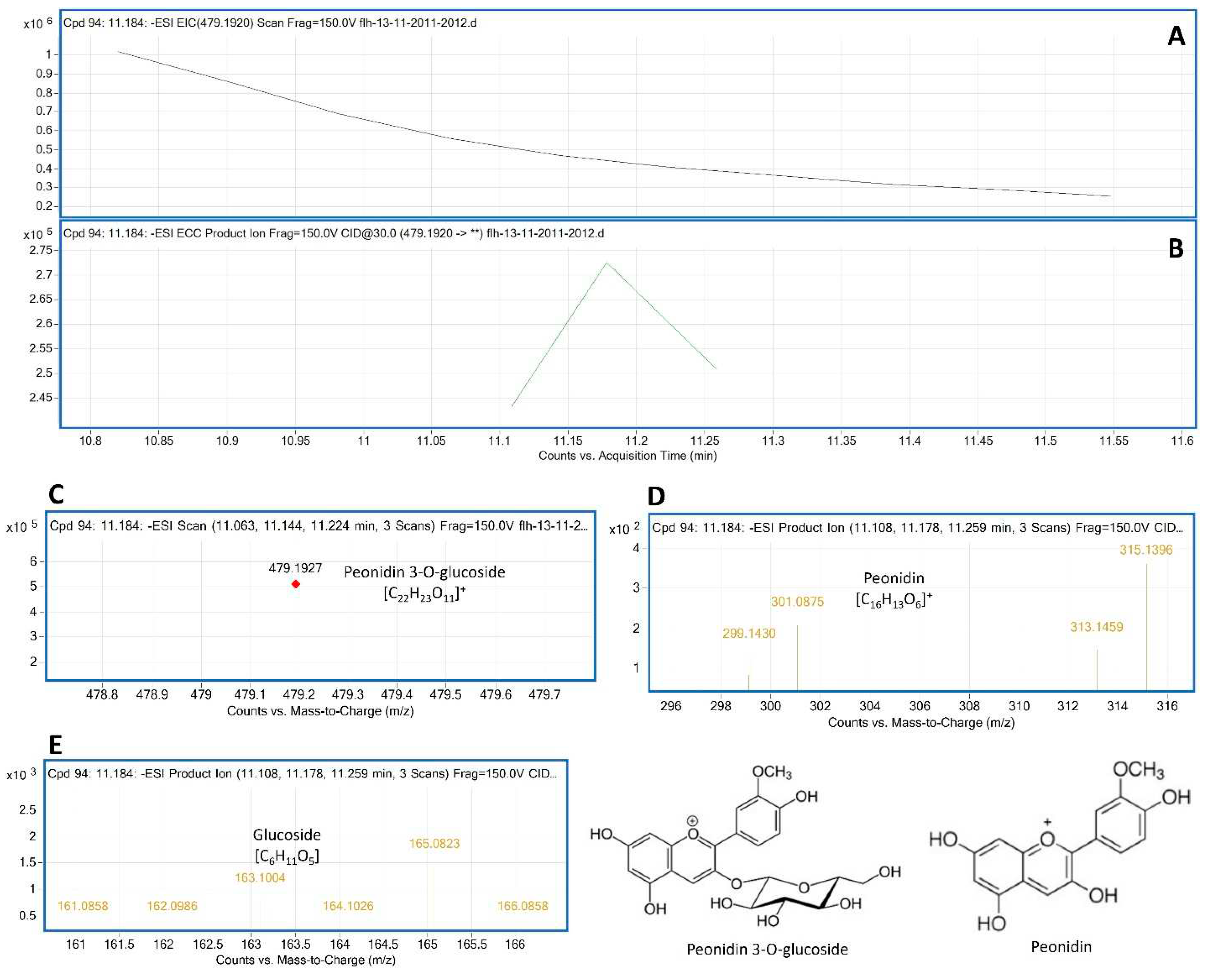
To annotate anthocyanin peaks, peonidin 3-glycoside (Pn-3-G) standard was used as reference to understand main ion and CID features generated from our instrument. The molecular weight (MW) of Pn-3-G is 463.415. After ESI, its EIC was searched from total ion chromatographs (TIC). The resulting EIC and enhanced charge capacity (ECC) ion products showed that two primary m/z ratio values were 479.2307 and 461.1927 [M-2H] (
Figure 4A–C). The m/z ratio of 479.1927 was derived from 463.415+18-2H [M+18-2H]. CID was performed to demonstrate the core structure in 479.1927 and 461.1927 [m/z]¯. CID analysis revealed that two typical fragments 299.1927 and 163.0682 (
Figure 4D,E) were generated from 479.2307 and 461.1927 [m/z]¯.
These ion fragments resulted from the homolytic dissociation of peonidin (299.1927) and the glucose group (163.0682) in both 461.1927 and 479.1927 [m/z]¯. In addition, fragments relating to peonidin observed from CID included 300.1927 and 301.1927. Fragments relating to glucose observed from CID consisted of 161.06 to 163.0682 and 165.0841 to 168.1036 (
Figure 4E). These CID fragmentation profiles resulted from heterolytic fragmentation that has been commonly observed in MS/MS analysis [
29,
30,
31,
32]. All these data showed that in addition to an expected m/z ratio and homolytic fragmentations from CID of Pn-3-G, other m/z ratios and heterolytic dissociation fragmentation were generated from this anthocyanin molecule. Based on these observations, anthocyanin peaks from berries were annotated in the following descriptions.
The peak F13-AN14 had the same retention time as Pn-3-G (
Figure 3A). MS/MS generated its EIC m/z value and profiles of CID fragmentation. The resulting data that its m/z values and EIC were the same as those of Pn-3-G (
Figure 5A–C). Its CID fragment profiles were also highly identical to those of Pn-3-G (
Figure 5D,E). Based on these features, F13-AN14 was identified to be Pn-3-G (
Figure 5F).
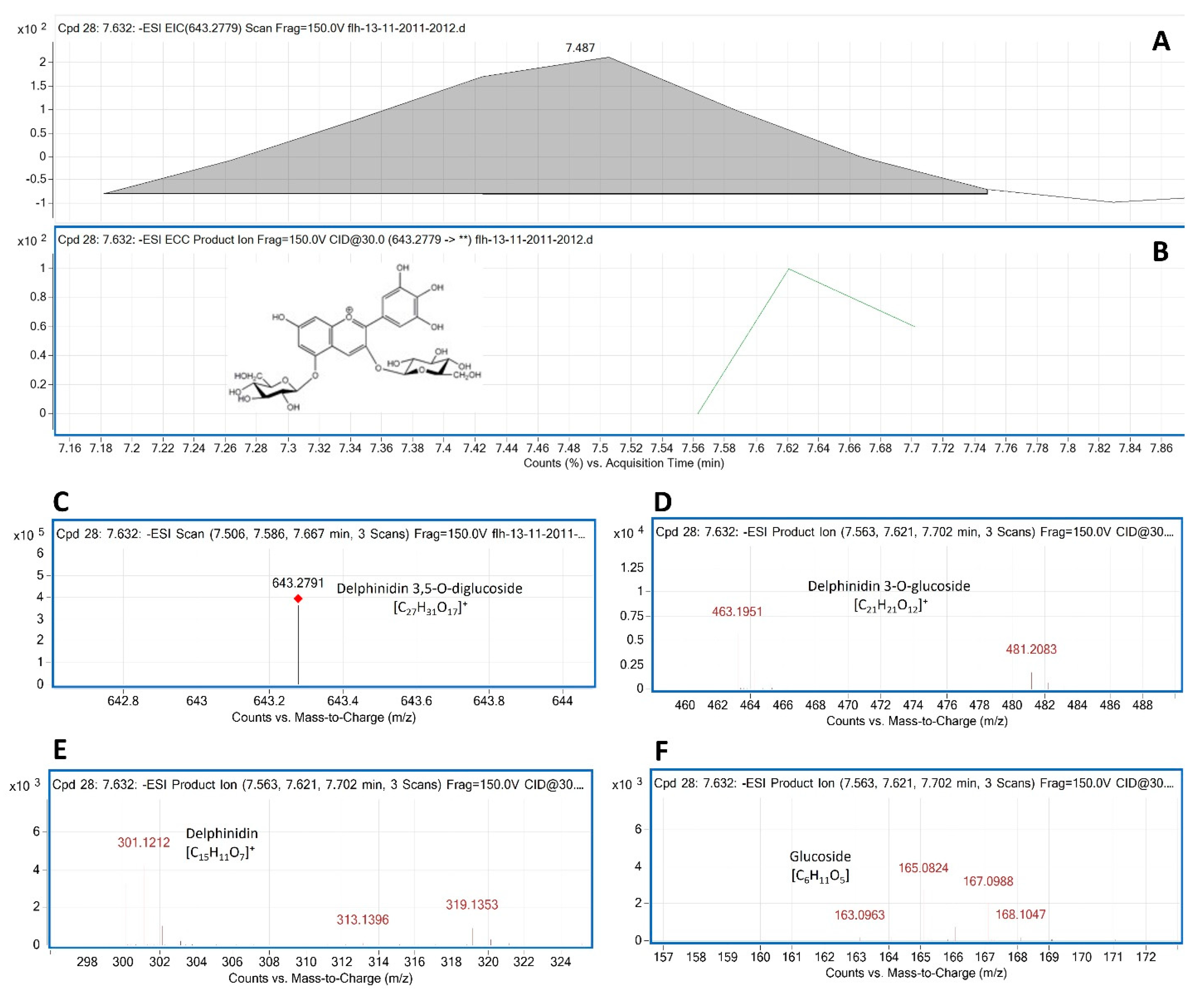
Delphinidin 3,5- diglucoside (Del-3,5-dG) is a common anthocyanin molecule in berries of muscadine. In our samples, F13-AN6 detected by HPLC-DAD was annotated to be Del-3,5-dG by LC-qTOF-MS/MS analysis. The MW of Del-3,5-dG is 627.5280. MS search from TIC obtained two primary m/z values for this peak, 643.2822 [M+18-2H] and 625.2822 [M-2H], which were further demonstrated with its extracted EIC and EIC-ECC ion products (
Figure 6A–C). CID of 625.2822 and 643.2822 generated five groups of secondary ion fragments (
Table 1), group 1: 481.2083 [463.1951+18] and group 2: 463.1951 (
Figure 6D), group 3: 301.0886 and 303.1268 and group 4: 317.1254 and 319.1353 (
Figure 6E), and group 5: 163.0963, 165.0832, 166.0824, 167.0988, and 168.1047 (
Figure 6F).
Based on these ion fragment features, the second group that resulted from the dissociation of the first glucose group (163.0686) from 625.2822 was relating to Del-3-glucoside. The third group that resulted from another dissociation of the second glucose group from 463.1951 was relating to delphinidin aglycone. The third group that resulted from the dissociation from 625.2822 and 464.2128-467.2174 was relating to glucose. Based on these features, this F13-AN13 peak was annotated to be Del-3,5-dG.
Cyanidin 3,5- diglucoside (Cy-3, 5-dG) is another common anthocyanin molecule formed in berries of different muscadine cultivars. Based on MS/MS data, F13-AN2 (
Figure 3A) was annotated to Cy-3, 5-dG (
Figure S1). After ESI, two primary m/z values of this peak were 609.529 [M-2H]¯ and 627.529 [M+18-2H]¯ (
Figure S1A–C). CID of 609.529 and 627.529 generated fragments (
Figure S1D–F), which were characterized by five groups of secondary m/z values (
Table 1). Fragments 447.1979, 285.1244 and 163.0677 were relating to Cy-3-G, cyanidin aglycone, and glucose.
Accordingly, based on MS1 and MS2 generated from MS/MS analysis, 11 additional peaks were annotated to either a known anthocyanin or characterized to be a specific anthocyanidin-related anthocyanin molecule (
Table 1,
Figures S2–S12). Based on anthocyanin molecules reported in the muscadine literature [
9,
11,
14,
15,
18,
20], the common five 3,5-diglucosides of anthocyanidins were found from anthocyanin extracts (
Table 1). Furthermore, this analysis identified four monoglucosides of anthocyanidins. Three were peonidin 3- glucoside, pelargonidin 3- glucoside like, and malvidin 3-glucoside like anthocyanins that only detected in 2012, while one was delphinidin-3 glucoside like anthocyanin that was only detected in 2011. These results indicate that anthocyanin profiles in berries of FLH 13-11 is closely associated with the cropping seasons.
Figure 1.
Overview of anthocyanidins and anthocyanins identified in berries of different muscadine varieties. Six anthocyanidins include pelargonidin, cyanidin, delphinidin, peonidin, petunidin, and malvidin. Anthocyanins include six monoglucosides of anthocyanidins and six diglucosides of anthocyanidins.
Figure 1.
Overview of anthocyanidins and anthocyanins identified in berries of different muscadine varieties. Six anthocyanidins include pelargonidin, cyanidin, delphinidin, peonidin, petunidin, and malvidin. Anthocyanins include six monoglucosides of anthocyanidins and six diglucosides of anthocyanidins.
Figure 2.
The total anthocyanin content of extract from berries of FLH 13-11 collected in 2011 and 2012 growing seasons.
Figure 2.
The total anthocyanin content of extract from berries of FLH 13-11 collected in 2011 and 2012 growing seasons.
Figure 3.
Comparison of anthocyanin and anthocyanidin profiles in berries of FLH 13-11 harvested in the 2011 and 2012 cropping seasons. A, comparison of anthocyanin profiles in berries from 2011 and 2012; B, anthocyanidin profiles released from the hydrolysis of anthocyanins in 2011 and 2012; C, four anthocyanidin standards.
Figure 3.
Comparison of anthocyanin and anthocyanidin profiles in berries of FLH 13-11 harvested in the 2011 and 2012 cropping seasons. A, comparison of anthocyanin profiles in berries from 2011 and 2012; B, anthocyanidin profiles released from the hydrolysis of anthocyanins in 2011 and 2012; C, four anthocyanidin standards.
Figure 4.
Extracted ion chromatogram (EIC) of primary mass spectrum (MS1) and m/z features of secondary ion fragments (MS2) derived from LC-MC/MS of peonidin 3-gluoside (Pn-3-G, molecular weight: 463.415) standard. A: EIC of primary ion 479.2305 [M+18-2H]¯, B: enhanced charge capacity (ECC) ion product for 479.2305, C: a MS profile showing two extracted m/z values, 461.1927 [M-2H]¯ and 479.2307 [M+18-2H]¯, D-E: fragments from CID of 479.2307 and 461.1927 showing 299.1435 [m/z]¯ and 301.1927 [m/z]¯ relating to peonidin aglycone (D) and 161.0858-165.082, 166.0858 relating to glucose (E).
Figure 4.
Extracted ion chromatogram (EIC) of primary mass spectrum (MS1) and m/z features of secondary ion fragments (MS2) derived from LC-MC/MS of peonidin 3-gluoside (Pn-3-G, molecular weight: 463.415) standard. A: EIC of primary ion 479.2305 [M+18-2H]¯, B: enhanced charge capacity (ECC) ion product for 479.2305, C: a MS profile showing two extracted m/z values, 461.1927 [M-2H]¯ and 479.2307 [M+18-2H]¯, D-E: fragments from CID of 479.2307 and 461.1927 showing 299.1435 [m/z]¯ and 301.1927 [m/z]¯ relating to peonidin aglycone (D) and 161.0858-165.082, 166.0858 relating to glucose (E).
Figure 5.
Extracted ion chromatogram (EIC) of primary mass spectrum and m/z features of secondary ion fragments derived from LC-MC/MS of peak F13-AN14. These data show this peak being peonidin 3-glucoside. A: EIC of primary ion 479.2305 [m/z]¯, [M+18-2H], B: enhanced charge capacity (ECC) ion product for 479.2305, C: a MS profile showing one extracted m/z value, 479.2307 [m/z]¯, D-E: fragments from CID of 479.2307 showing 299.1435 [m/z] and 301.1927 [m/z] relating to peonidin aglycone (D), and 161.0858-165.0823, 166.0858 [m/z] relating to glucose (E), F: structures of peonidin 3-glucoside and peonidin.
Figure 5.
Extracted ion chromatogram (EIC) of primary mass spectrum and m/z features of secondary ion fragments derived from LC-MC/MS of peak F13-AN14. These data show this peak being peonidin 3-glucoside. A: EIC of primary ion 479.2305 [m/z]¯, [M+18-2H], B: enhanced charge capacity (ECC) ion product for 479.2305, C: a MS profile showing one extracted m/z value, 479.2307 [m/z]¯, D-E: fragments from CID of 479.2307 showing 299.1435 [m/z] and 301.1927 [m/z] relating to peonidin aglycone (D), and 161.0858-165.0823, 166.0858 [m/z] relating to glucose (E), F: structures of peonidin 3-glucoside and peonidin.
Figure 6.
Extracted ion chromatogram (EIC) of primary mass spectrum and m/z features of secondary ion fragments derived from LC-MC/MS of peak F13-AN6. These data annotate this peak to be delphinidin 3, 5-digluoside (Del-3,5-dG, molecular weight: 427.528). A: EIC of primary ion 643.2791 [m/z]¯, [M+18-2H], B: enhanced charge capacity (ECC) ion product for 643.2791 [m/z]¯, C: a MS profile showing an extracted m/z value, 643.2791 [m/z]¯, [M+18-2H], D-F: fragments from CID of 643.2791 showing 463.1951 and 464.1953 [m/z]¯ relating to Del-3-G (D), 300.111, 301.1212 and 302.1213 [m/z]¯ relating to delphinidin aglycone (E), and 163.0963-168.1047 relating to glucose (F).
Figure 6.
Extracted ion chromatogram (EIC) of primary mass spectrum and m/z features of secondary ion fragments derived from LC-MC/MS of peak F13-AN6. These data annotate this peak to be delphinidin 3, 5-digluoside (Del-3,5-dG, molecular weight: 427.528). A: EIC of primary ion 643.2791 [m/z]¯, [M+18-2H], B: enhanced charge capacity (ECC) ion product for 643.2791 [m/z]¯, C: a MS profile showing an extracted m/z value, 643.2791 [m/z]¯, [M+18-2H], D-F: fragments from CID of 643.2791 showing 463.1951 and 464.1953 [m/z]¯ relating to Del-3-G (D), 300.111, 301.1212 and 302.1213 [m/z]¯ relating to delphinidin aglycone (E), and 163.0963-168.1047 relating to glucose (F).
Table 1.
Mass spectrum characterization and annotation of 14 anthocyanin peaks by HPLC-qTOF-MS/MS analysis.
Table 1.
Mass spectrum characterization and annotation of 14 anthocyanin peaks by HPLC-qTOF-MS/MS analysis.
| Peak # |
λ (nm) and
Rt (min) |
MW and
MS1 [m/z]⁻ |
MS2 [m/z] fragments from CID |
Anthocyanin annotation |
Seasons
(Figures) |
| F13-AN1 |
λ: 525
Rt: 6.258 |
MW: Unknown
MS1: 643.2778 |
-
1.
481.2036; -
2.
463.1935,465.1953; -
3.
301.1211-304.986; -
4.
319.1345; -
5.
163.0615; 165.0822; 167.1998 |
Delphinidin 3,5-O-diglucoside like anthocyanin |
2011
Figure S4
|
| F13-AN2 |
λ: 522
Rt: 6.269 |
MW: Unknown
MS1: 679.2582 |
-
1.
447.1949, 455.0124, 463.1942; -
2.
283.1062-285.1186; -
3.
301.1217-302.1217; -
4.
163. 0826, 165.0836-166.0834 |
Cyanidin 3,5-O-diglucoside like anthocyanin |
2011
Figure S5
|
| F13-AN3 |
λ: 523
Rt: 7.163 |
MW: 465.3870
MS: 481.1751 |
-
1.
301.0852-302.0852; -
2.
317.3565; -
3.
165.0864, 167.0622, 168.0651 |
Delphinidin 3-O-glucoside like anthocyanin |
2012
Figure S6
|
| F13-AN4 |
λ: 525
Rt: 7.238 |
MW: Unknown
MS: 793.3089 |
-
1.
477.2128, 481.2091, 482.2091, 495.2120; -
2.
315.1352, 319.14; -
3.
329.1790, 331.1528; -
4.
165.0829 |
Petunidin 3,5-O-diglucoside like anthocyanin |
2011
Figure S7
|
| F13-AN5 |
λ: 529
Rt: 7.310 |
MW: Unknown
MS: 741.2607 |
-
1.
481.2082, 482.2082; -
2.
463.1966, 464.2045; -
3.
297.0398, 299.0396, 301.1168; -
4.
317.1776, 319.1397; -
5.
162.1067, 165.0836, 167.0892 |
Delphinidin 3,5-O-diglucoside like anthocyanin |
2011
Figure S8
|
| F13-AN6 |
λ: 523
Rt: 7.632 |
MW: 627.5280
MS1: 643.2791 |
-
1.
481.2083; -
2.
463.1951; -
3.
301.0886; 303.1268; -
4.
317.1254,319.1353; -
5.
163.0963, 165.0832, 166.0824, 167.0988,168.1047 |
Delphinidin 3,5-O-diglucoside |
2011, 2012
Figure 6
|
| F13-AN7 |
λ: 517
Rt: 8.285 |
MW: 611.5290
MS1: 627.2824 |
-
1.
465.2126; 466.2126; -
2.
447.1979, 448.1979; -
3.
285.1244; 287.1318; -
4.
301.0886, 303.1381; -
5.
163.0677, 164.0716, 165.0832, 166.0919 |
Cyanidin 3,5-O-diglucoside |
2011
Figure S1
|
| F13-AN8 |
λ: 520
Rt: 8.371 |
MW: 641.2820
MS1: 657.2963 |
-
1.
495.2253, 496.2263; -
2.
477.2127, 478.2127; -
3.
327.1626, 329.1787, 331.1731, 333.1531; -
4.
314. 1392, 315.1392, 317.1399; -
5.
163.0843, 165.0822, 167.0933, 169.0802 |
Petunidin 3,5-O-diglucoside |
2012
Figure S2
|
| F13-AN9 |
λ: 514
Rt: 9.988 |
MW: Unknown
MS1: 493.2101 |
-
1.
330.1279, 331.1153, 332.1253; -
2.
315.1028, 316.1028, 317.1058; -
3.
298.1063, 299.1063, 301.110; -
4.
163.0836, 165.0836, 166.0824, 167.0988 |
Peonidin-3-O-glucoside like anthocyanin |
2012
Figure S9
|
| F13-AN10 |
λ: 523
Rt:10.462 |
MW: 625.5560
MS1: 641.2635 |
-
1.
461.1782, 479.1915; -
2.
315.1414, 317.1224; -
3.
299.1090, 301.1226; -
4.
163.1029, 165.0835, 166.0863 |
Peonidin 3,5-O-diglucoside |
2012
Figure S3
|
| F13-AN11 |
λ: 523
Rt: 0.693 |
MW: Unknown
MS1: 633.1995 |
-
1.
481.1651; -
2.
463.1594, 464.1745, 465.1745, 466.1779; -
3.
300.0765, 301.0857, 302.0890, 303.0909; -
4.
161.0926, 163.1045-165.0810, 167.0624 |
Delphinidin 3,5-O-diglucoside like anthocyanin |
2012
Figure S10
|
| F13-AN12 |
λ: 518
Rt:11.028 |
MW: Unknown
MS1: 453.1748 |
-
1.
271.1062, 272.1154, 273.1232, 274.1285; -
2.
163.1029, 165.0829, 166.0863 |
Pelargonidin 3-O-glucoside like anthocyanin |
2012
Figure S11
|
| F13-AN13 |
λ: 524
Rt: 1.039 |
MW: 655.587
MS1: 673.2911 |
-
1.
493.2083, 494.2080, 495.2101; -
2.
330.1328, 331.1448, 332.1328; -
3.
161.1010, 163.0708,165.0829, 166.0869 |
Malvidin 3,5-O-diglucoside |
2012
Figure S12
|
| F13-AN14 |
λ: 529
Rt: 1.184 |
MW: 463.4150
MS1: 479.1927 |
-
1.
313.1459, 315.1396 -
2.
299.1430, 301.0875; -
3.
161.0858, 163.0709, 165.082, 166.0858 |
Peonidin 3-O-glucoside |
2012
Figure 5
|