Submitted:
29 January 2024
Posted:
30 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Selection of bacterial strains
2.2. Antibiotic susceptibility
2.3. DNA extraction and amplification of genes associated with DAP non-susceptibility in S. aureus
2.4. Visualization of amplicons and purification of PCR product
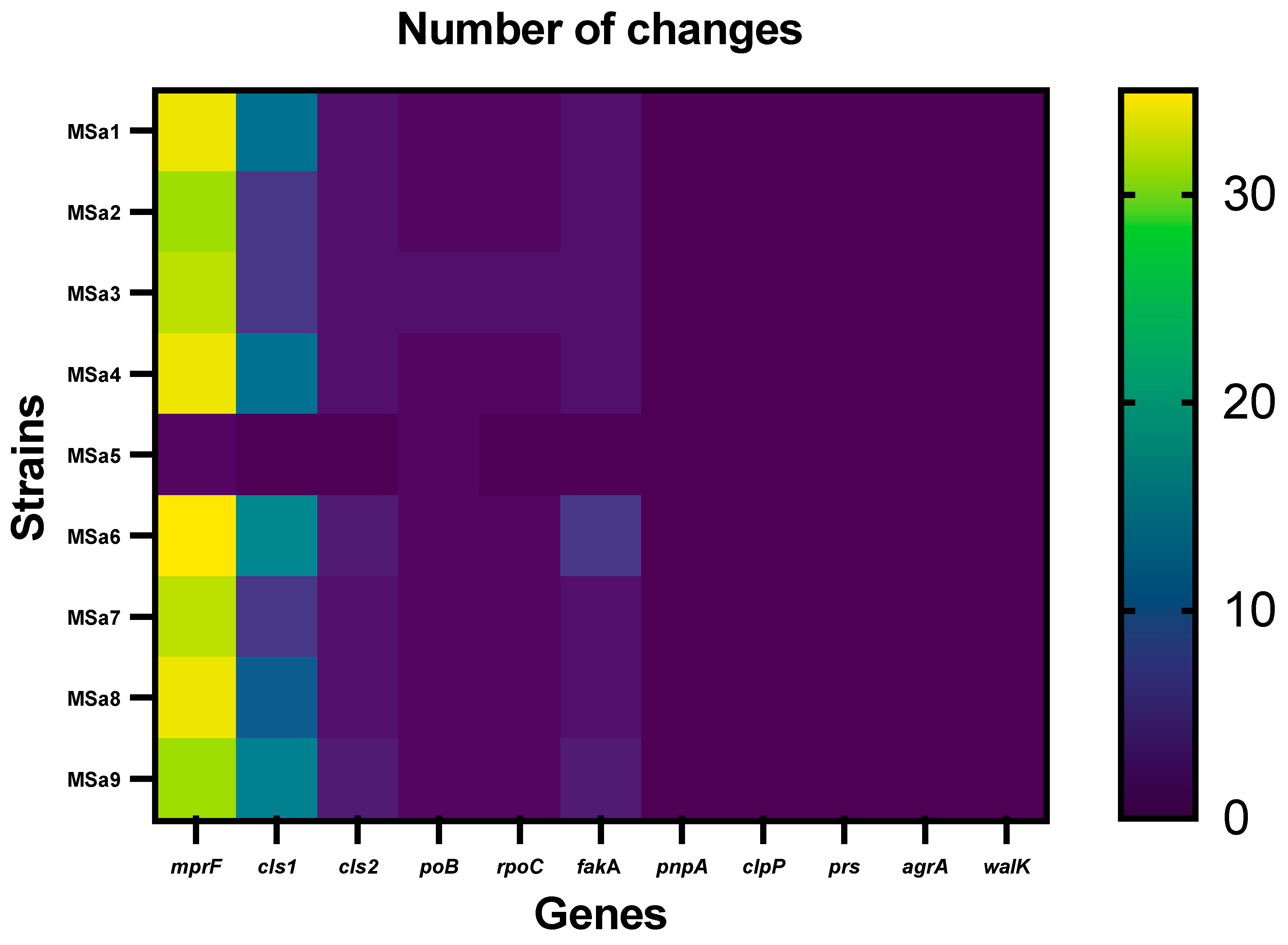
3. Results
3.1. Antibiotic susceptibility
3.2. Mutations profiles associated with non-susceptibility
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eisenstein, B.I.; Oleson, F.B. Jr.; Baltz, R.H. Daptomycin: from the mountain to the clinic, with essential help from Francis Tally, MD. Clin Infect Dis 2010, 50 Suppl 1, S10–S15. [Google Scholar] [CrossRef]
- Steenbergen, J.N.; Alder, J.; Thorne, G.M.; Tally, F.P. Daptomycin: a lipopeptide antibiotic for the treatment of serious Gram-positive infections. J Antimicrob Chemother 2005, 55, 283–8. [Google Scholar] [CrossRef]
- Anastasiou, D.M.; Thorne, G. M.; Luperechio, S.A.; Alder, J.D. In vitro activity of daptomycin against clinical isolates with reduced susceptibilities to linezolid and quinupristin/ dalfopristin. Int J Antimicrob Agents 2006, 28, 385–8. [Google Scholar] [CrossRef]
- Cotroneo, N.; Harris, R.; Perlmutter, N.; Beveridge, T.; Silverman, J.A. Daptomycin exerts bactericidal activity without lysis of Staphylococcus aureus. Antimicrob Agents Chemother 2008, 52, 2223–2225. [Google Scholar] [CrossRef]
- Bhullar, K.; Waglechner, N.; Pawlowski, A.; et al. Antibiotic resistance is prevalent in an isolated cave microbiome. PloS One 2012, 7, 1–11. e34953. [Google Scholar] [CrossRef]
- Bayer, A.S.; Schneider, T.; Sahl, H.G. Mechanisms of daptomycin resistance in Staphylococcus aureus: role of the cell membrane and cell wall. Ann N Y Acad Sci 2013, 1277, 139–58. [Google Scholar] [CrossRef]
- Friedman, L.; Alder, J.D.; Silverman, J.A. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob Agents Chemother 2006, 50, 2137–2145. [Google Scholar] [CrossRef]
- Mishra, N.N.; Rubio, A.; Nast, C.C.; Bayer, A.S. Differential Adaptations of Methicillin-Resistant Staphylococcus aureus to Serial In Vitro Passage in Daptomycin: Evolution of Daptomycin Resistance and Role of Membrane Carotenoid Content and Fluidity. Int J Microbiol 2012, 2012, 683450. [Google Scholar] [CrossRef]
- Pillai, S.K.; Gold, H.S.; Sakoulas, G.; Wennersten, C.; Moellering, R.C. Jr.; Eliopoulos, G.M. Daptomycin nonsusceptibility in Staphylococcus aureus with reduced vancomycin susceptibility is independent of alterations in MprF. Antimicrob. Agents Chemother 2007, 51, 2223–2225. [Google Scholar] [CrossRef]
- Song, Y.; Rubio, A.; Jayaswal, R.K.; Silverman, J.A.; Wilkinson, B.J. Additional routes to Staphylococcus aureus daptomycin resistance as revealed by comparative genome sequencing, transcriptional profiling, and phenotypic studies. PLoS One 2013, 8, e58469. [Google Scholar] [CrossRef]
- Peleg, A.Y.; Miyakis, S.; Ward, D.V.; et al. Whole genome characterization of the mechanisms of daptomycin resistance in clinical and laboratory derived isolates of Staphylococcus aureus. PLoS One 2012, 7, e28316. [Google Scholar] [CrossRef]
- Mehta, S.; Cuirolo, A.X.; Plata, K.B.; et al. VraSR two-component regulatory system contributes to mprF-mediated decreased susceptibility to daptomycin in in vivo-selected clinical strains of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 2012, 56, 92–102. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 27th ed.; CLSI Supplement M100S; Clinical Laboratory Standards Institute: Wayne, Philadelphia (USA), 2017. [Google Scholar]
- Fujimura, S.; Nakano, Y.; Watanabe, A. A correlation between reduced susceptibilities to vancomycin and daptomycin among the MRSA isolates selected in mutant selection window of both vancomycin and daptomycin. J Infect Chemother 2014, 20, 752–756. [Google Scholar] [CrossRef]
- Humphries, R. M, Pollett, S.; Sakoulas, G. A current perspective on daptomycin for the clinical microbiologist. Clin Microbiol Rev 2013, 26, 759–780. [Google Scholar] [CrossRef]
- Cameron, D.R.; Mortin, L.I.; Rubio, A.; et al. Impact of daptomycin resistance on Staphylococcus aureus virulence. Virulence. 2015, 6, 127–131. [Google Scholar] [CrossRef]
- Gómez-Casanova, N.; Siller Ruiz, M.; Muñoz Bellido, J.L. Mechanisms of resistance to daptomycin in Staphylococcus aureus. Rev Esp Quimioter 2017, 30, 391–396. [Google Scholar]
- Ernst, C.M.; Peschel, A. Broad spectrum antimicrobial peptide resistance by MprF-mediated aminoacylation and flipping of phospholipids. Mol Microbiol 2011, 80, 290–299. [Google Scholar] [CrossRef]
- Ernst, C.M.; Staubitz, P.; Mishra, N.; Yang, S.J.; Hornig, G.; Kalbacher, H.; Bayer, A.S.; Kraus, D.; Peschel, A. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinilation and antimicrobial peptide repulsion. PLOS Pathogens 2009, 5, e1000660. [Google Scholar] [CrossRef]
- Kristian, S.A.; Dürr, M.; Van Strijp, J.A.; Neumeister, B.; Peschel, A. MprF-mediated lysinylation of phospholipids in Staphylococcus aureus leads to protection against oxygen-independent neutrophil killing. Infect Immun 2003, 71, 546–549. [Google Scholar] [CrossRef]
- Yang, S.J.; Nast, C.C.; Mishra, N.N.; Yeaman, M.R.; Fey, P.D.; Bayer, A.S. Cell wall thickening is not a universal accompaniment of the daptomycin nonsusceptibility phenotype in Staphylococcus aureus: evidence for multiple resistance mechanisms. Antimicrob Agents Chemother 2010, 54, 3079–3085. [Google Scholar] [CrossRef]
- Bayer, A.S.; Mishra, N.N.; Sakoulas, G.; Nonejuie, P.; Nast, C.C.; Pogliano, J.; Chen, K.T.; Ellison, S.N.; Yeaman, M.R.; Yang, S.J. Heterogeneity of mprF sequences in methicillin-resistant Staphylococcus aureus clinical isolates: role in cross-resistance between daptomycin and host defense antimicrobial peptides. Antimicrob Agents Chemother 2014, 58, 7462–7. [Google Scholar] [CrossRef]
- Murthy, M.H.; Olson, M.E.; Wickert, R.W.; Fey, P.D.; Jalali, Z. Daptomycin non-susceptible methicillin-resistant Staphylococcus aureus USA 300 isolate. J Med Microbiol 2008, 57, 1036–1038. [Google Scholar] [CrossRef]
- Sabat, A.J.; Tinelli, M.; Grundmann, H.; Akkerboom, V.; Monaco, M.; del Grosso, M.; Errico, G.; Pantosti, A.; Friedrich, A.W. Daptomycin resistant Staphylococcus aureus clinical strain with novel non-synonymous mutations in the mprF and vraS genes: a new insight into daptomycin resistance. Front Microbiol 2018, 9, 2705. [Google Scholar] [CrossRef]
- Cui, L.; Isii, T.; Fukuda, M.; Ochiai, T.; Neoh, H.; Camargo, I.L.B.D.C.; Watanabe, Y.; Shoji, M.; Hiramatsu, K. An rpoB mutation confers dual heteroresistance to daptomycin and vancomycin in S. aureus. Antimicrob Agents Chemother 2010, 54, 5222–33. [Google Scholar] [CrossRef]
- Bæk, K.T.; Thøgersen, L.; Mogenssen, R.G.; Mellergaard, M.; Thomsen, L.E.; Petersen, A.; Skov, S.; Cameron, D.R.; Peleg, A.Y.; Frees, D. Stepwise decrease in daptomycin susceptibility in clinical S. aureus isolates associated with an initial mutation in rpoB and a compensatory inactivation of the clpX gene. Antimicrob Agents Chemother 2015, 59, 6983–91. [Google Scholar] [CrossRef]
- Hagiya, H.; Sugawara, Y.; Kimura, K.; Hamaguchi, S.; Nishi, I.; Hayashi, M.; Akeda, Y.; Tomono, K. Emergence of daptomycin non-susceptible coagulase-negative Staphylococci in patients with cardiovascular device infections: Two cases report investigated by whole genome analysis. Medicine (Baltimore) 2018, 97, e13487. [Google Scholar] [CrossRef]
- Dubrac, S.; Bisicchia, P.; Devine, K.M.; et al. A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol Microbiol 2008, 7, 1307–1322. [Google Scholar] [CrossRef]
- Howden, B.P.; McEvoy, C.R.; Allen, D.L.; Chua, K.; Gao, W.; Harrison, P.F.; Bell, J.; Coombs, G.; Bennett-Wood, V.; Porter, J.L.; Robins-Browne, R.; Davies, J.K.; Seemann, T.; Stinear, T.P. Evolution of multidrug resistance during Staphylococcus aureus infection involves mutation of the essential two component regulator WalKR. PLoS Pathog 2011, 7, e1002359. [Google Scholar] [CrossRef]
- Koprivnjak, T.; Zhang, D.; Ernst, C.M.; Peschel, A.; Nauseef, W.M.; Weiss, J.P. Characterization of Staphylococcus aureus cardiolipin synthases 1 and 2 and their contribution to accumulation of cardiolipin in stationary phase and within phagocytes. J Bacteriol 2011, 193, 4134–4142. [Google Scholar] [CrossRef]
- Camargo, I. L.; Neoh, H. M.; Cui, L.; Hiramatsu, K. Serial daptomycin selection generates daptomycin-nonsusceptible Staphylococcus aureus strains with a heterogeneous vancomycin-intermediate phenotype. Antimicrob. Agents Chemother 2008, 52, 4289–4299. [Google Scholar] [CrossRef]
| Strains | DAP | VAN | OXA | LZD | DLV | TLV |
| MSa1 | 4.0 (R) | 1.0 (S) | 16 (R) | 2.0 (S) | 0.064 (S) | 0.125 (S) |
| MSa2 | 1.0 (S) | 1.0 (S) | 256 (R) | 2.0 (S) | 0.047 (S) | 0.047 (S) |
| MSa3 | 4.0 (R) | 2.0 (S) | 0.125 (S) | 2.0 (S) | 0.100 (S) | 0.125 (S) |
| MSa4* | 2.0 (R) | 2.0 (S) | 2.0 (S) | 2.0 (S) | 0.064 (S) | 0.125 (S) |
| MSa5 | 4.0 (R) | 1.0 (S) | 0.064 (S) | 2.0 (S) | 0.047 (S) | 0.125 (S) |
| MSa6 | 4.0 (R) | 1.0 (S) | 4.0 (R) | 2.0 (S) | 0.032 (S) | 0.125 (S) |
| MSa7* | 2.0 (R) | 1.0 (S) | 2.0 (S) | 2.0 (S) | 0.064 (S) | 0.094 (S) |
| MSa8 | 4.0 (R) | 1.0 (S) | 0.125(S) | 2.0 (S) | 0.047 (S) | 0.125 (S) |
| MSa9 | 2.0 (R) | 1.0 (S) | 0.5 (S) | 2.0 (S) | 0.032 (S) | 0.094 (S) |
| Nº of isolate (MSax) | Amino acid mutations1 | Base changes* |
| 1, 2, 3, 4, 6, 7, 8, 9 |
V26A; N160D; A171V; L174F; Y194F; V223A; L371I; Y400F; I406L; I409T; L413F; V426A; A430V; I446V; L451I; I459L; I464V; F473L; V478I; K489R; I494L; V503I; A505M; N522K; E525D; D531N; D554N; N556T; I575L; T696E; N710E |
T1353395C; A1353696G; C1353730T-T1353731A; G1353740T; A1353799T; T1353886C-C1353887T; C1354329A; A1354417T; A1354434T-C1354436A; T1354444C-T1354445A; A1354457C; T1354495C; C1354507T; A1354554G-T1354556A; C1354569A; A1354593T; A1354608G-C1354610T; T1354637A; G1354650A; A1354683C-A1354684G-A1354685C; A1354698C; G1354725A-T1354727A; G1354731A-C1354732T-A1354733G; T1354784A; G1354793T; G1354809A; G1354878A; A1354885C-T1354886A; A1354941T- C1354943A; A1355305G-C1355304A; A1355346G-T1355348A |
| 1, 4 |
I375M; I461T |
A1354343G; T1354600C-A1354601T |
| 1, 5 |
P314T |
C1354158A-G1354160T |
| 6 |
I9V; G105A; P314L2; P721T |
A1353243G; G1353532C; C1354159T-G1354160T; C1355379 |
| 3 |
L291I |
T1354089A |
| 4, 7 |
T345I2,4,5,6 |
C1354252T |
| 8 |
V287I; S295P3; A500S |
G1354077A-G1354079A; T1354101C; G1354716T- A1354718T |
| Genes | Nº of strain (MSax) | Amino acid mutations1 | Base changes* |
| cls1 | 1, 4, 6, 8, 9 |
K147Q; K170Q; H174K; N197K |
A1306452C; A1306521C; C1306533A-T1306535A; T1306604A |
| 1, 4, 9 | F389Y; I421M; N448K | T1307179A; C1307276G; T1307357G | |
| 1, 2, 3, 4, 6, 7, 8, 9 | I238V | A1306725G-T1306727A | |
| 1, 2, 3, 4, 7, 8 | E469K | G1307418A | |
| 1, 2, 3, 4, 7, 8 | Q2R; F3Y; S4T | A1306018G; T1306021A; T1306023A | |
| 1, 2, 3, 4, 7, 8 | V18A | T1306166C | |
|
6, 9 |
V18A; G87A; V132I; A175V |
T1306166C-C1306167A; G1306273C-A1306274G; G1306407A; C1306537T-T1306538G |
|
| 9 | S43A; T44K | T1306140G; C1306144A-T1306145A | |
| 6, 9 | G20A | G1306072C-A1306073C | |
| 6 |
V300E; G308K; P309S; L310F; S313A; V455A |
T1306912A-T1306913A; G1306935A-G1306936A; C1306938T; G1306943C; T1306950G-A1306952G; T1307377C | |
| 5 | NC | NC | |
|
cls2 |
1, 2, 3, 4, 6, 7, 8, 9 |
V135I; H205R |
G2125012A; A2125223G |
| 6 | A471E | C2126021A | |
| 9 | I459L | A2125984T | |
| 5 | NC | NC |
| Genes | Nº of strain (MSax) | Amino acid mutations1 | Base changes* |
| rpoB | 1, 2, 3, 4, 5, 6, 7, 8, 9 | F737Y | T545943A |
| 3 | M513I | G545272A | |
| rpoC | 1, 2, 3, 4, 6, 7, 8, 9 | V864I | G550011A-T550013A |
| 3 | P100L | C547720T | |
| 5 | NC | NC | |
| fakA | 1, 2, 3, 4, 6, 7, 8, 9 | L214I; D497E | C1191817A; T1192668A |
| 9 | E431D | A1192470C | |
| 6 |
I144V; E277K; Y287H; A513E |
A1191607G; G1192006A; T1192036C; C1192715A | |
| 5 | NC | NC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





