Submitted:
26 January 2024
Posted:
29 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Experimental Data
2.2. Primary Independent Characteristics of Individual Genes and Functional Pathways
2.3. Derived Characteristics of Individual Genes
2.3. Quantification of Transcriptomic Changes
2.3.1. Significant Regulation of the Average Expression Value
2.3.2. Weighted Individual (Gene) Regulation (WIR) and Weighted Pathway Regulation (WPR)
2.3.3. Regulation of the Expression Control and Expression Coordination
2.4. Functional Pathways
- i)
-
carbohydrate metabolism:
- ii)
-
Energy metabolism
- -
- (OXP) mmu00190 Oxidative phosphorylation [35]
- iii)
-
Lipid metabolism
- iv)
-
Nucleotide metabolism
- v)
-
Amino acid metabolism
- vi)
-
Glycan biosynthesis and metabolism
- -
- (NGL) mmu00510 N-Glycan biosynthesis [47].
- vii)
-
Xenobiotics biodegradation and metabolism
3. Results
3.1. The Global Picture
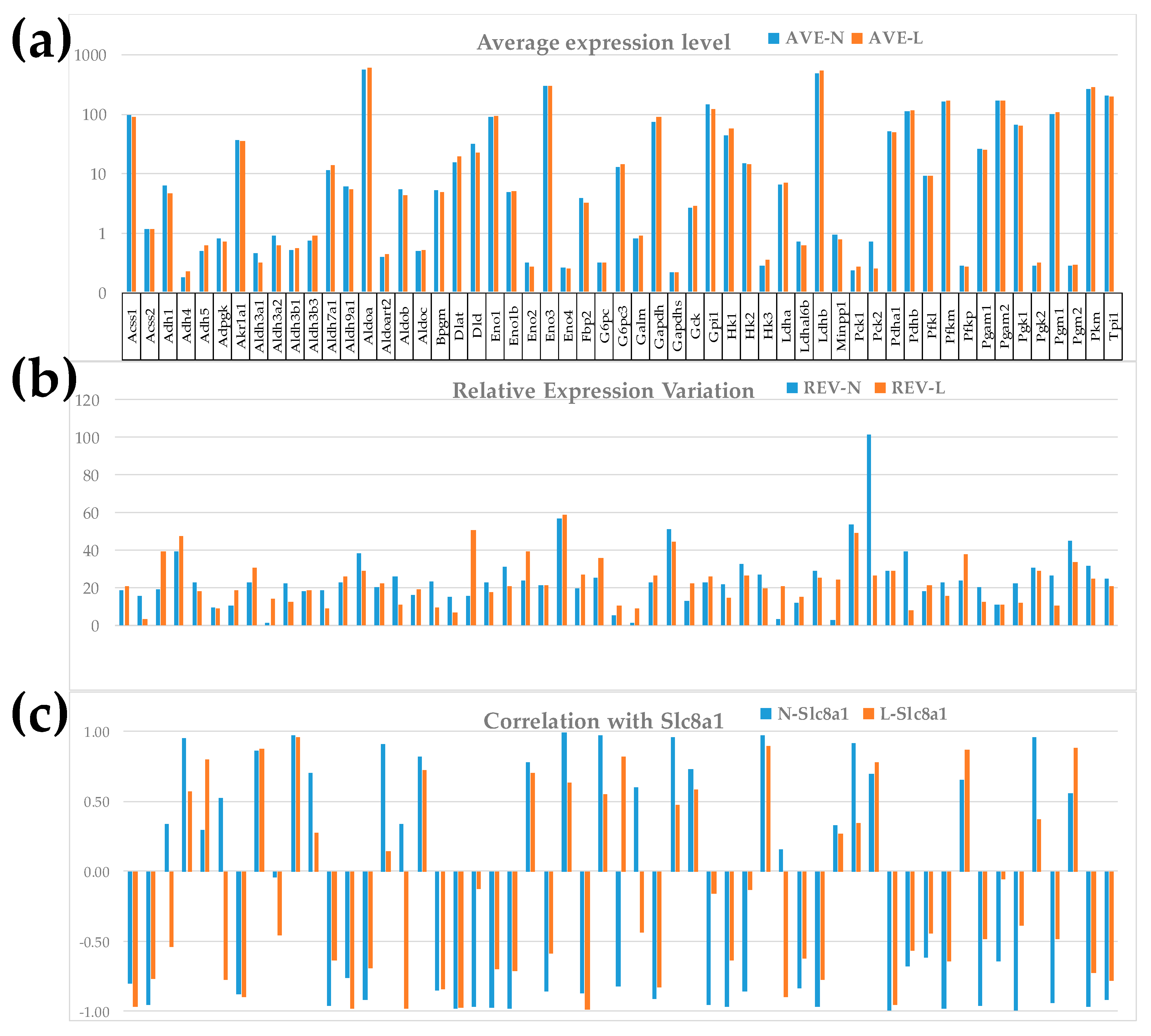
3.2. Independence of the Three Types of Primary Expression Characteristics of Individual Genes
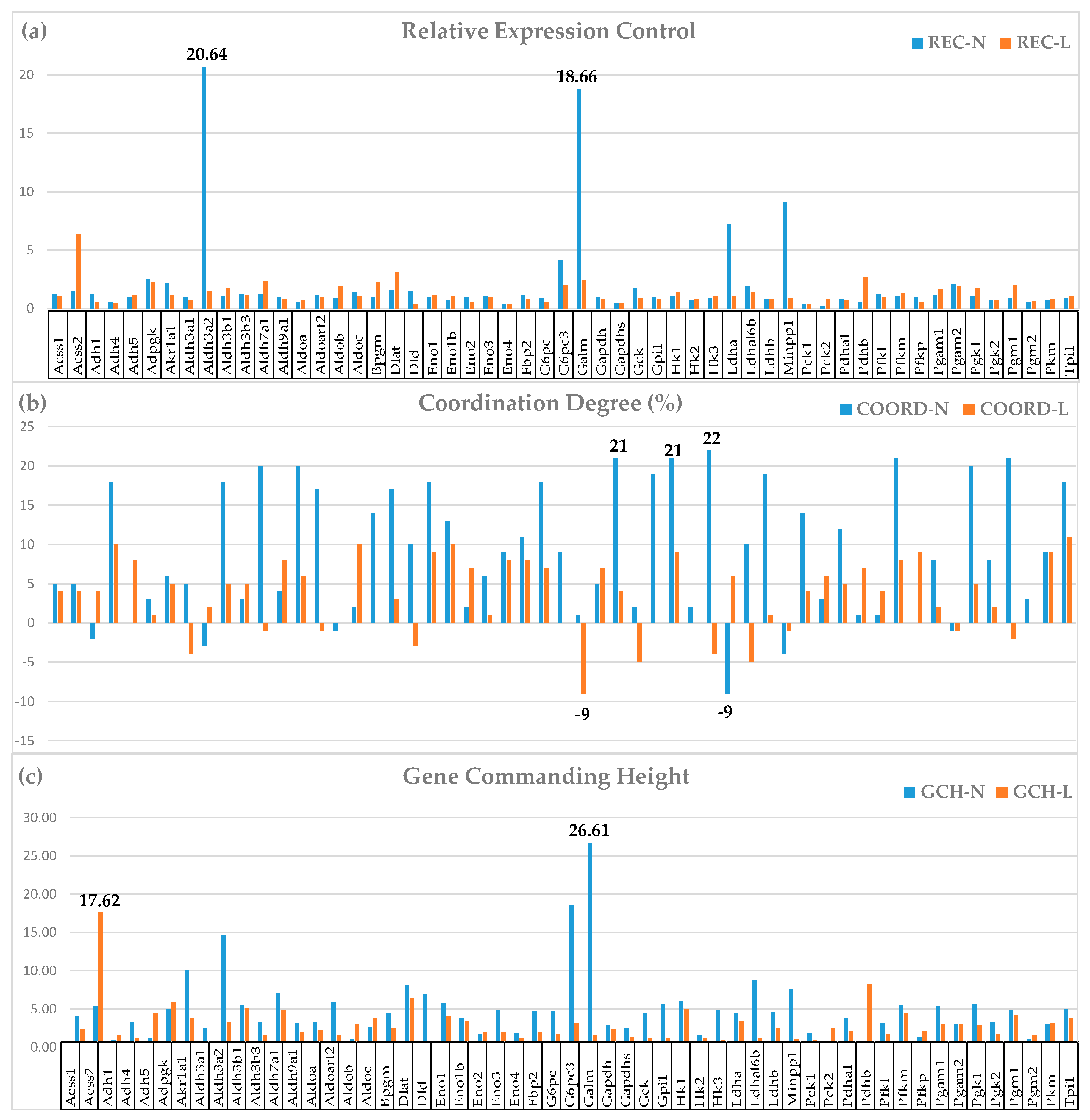
3.3. Important Derived Characteristics of the Individual Genes
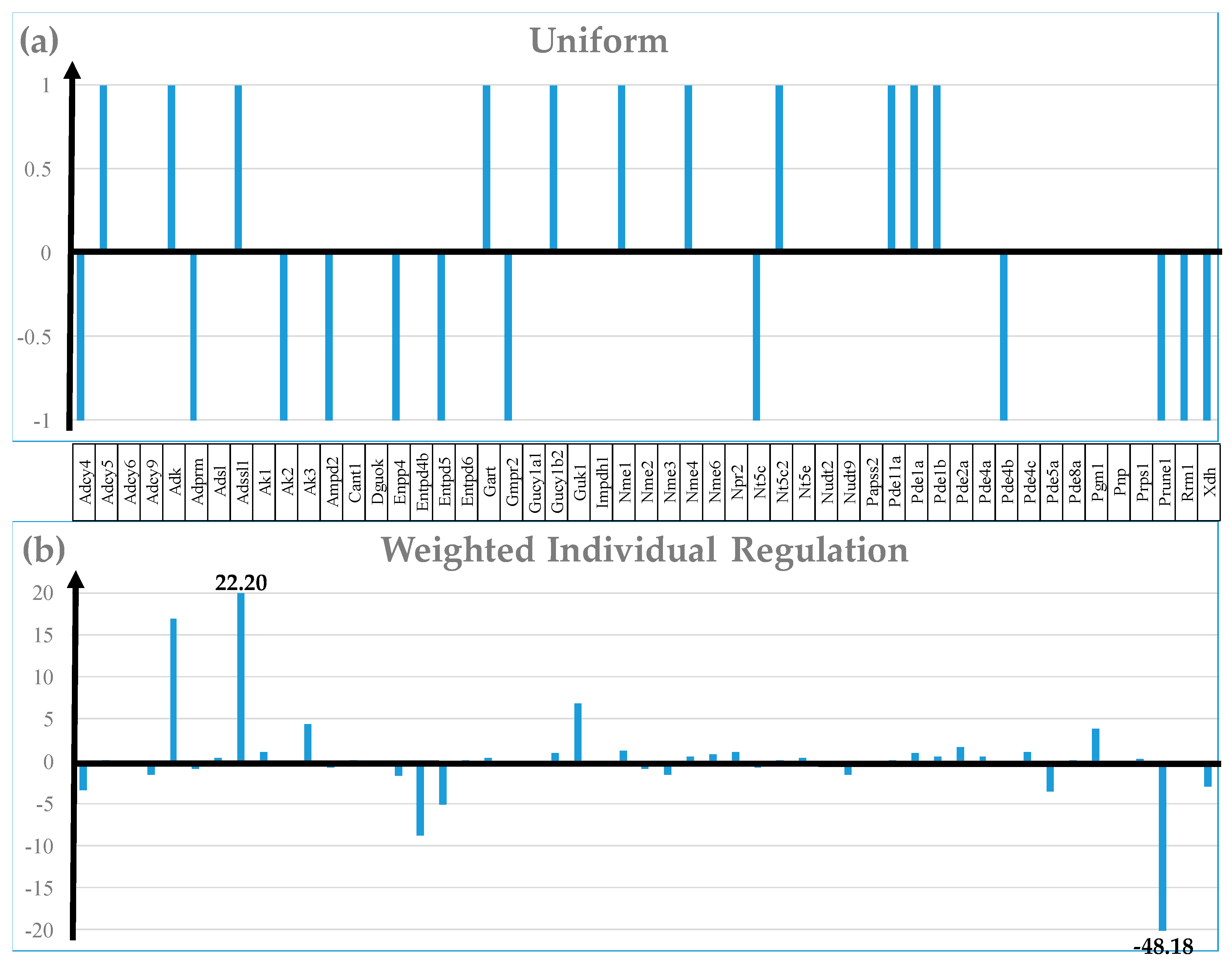
3.4. Measures of Transcriptomic Regulation
3.5. Correcting the False Hits of the Traditional Significant Regulation Analysis
3.5. Overall Regulation of Expression Level and Transcription Control within Selected Metabolic, Circulatory System, and Cardiac Chronic Diseases’ Pathways
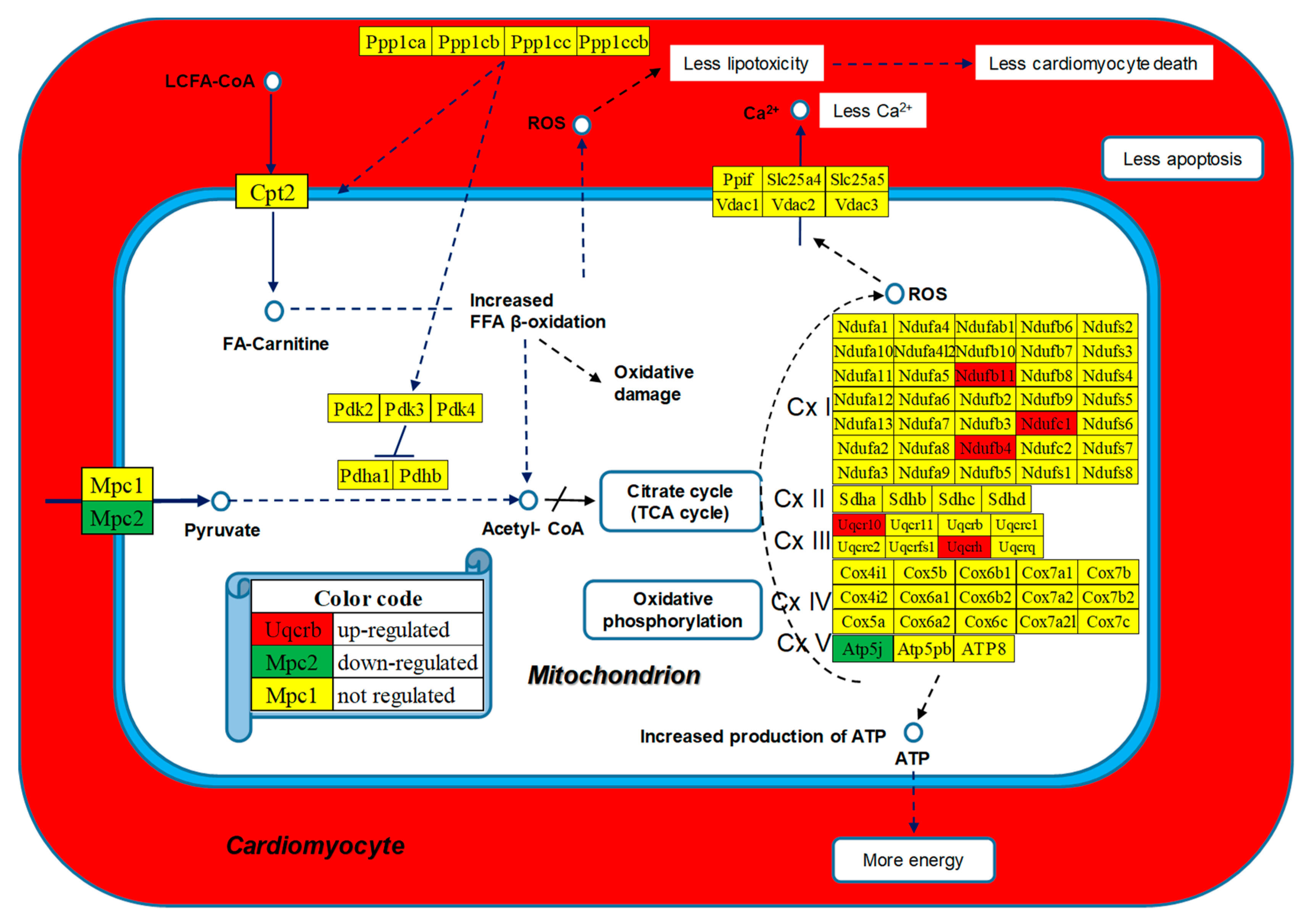
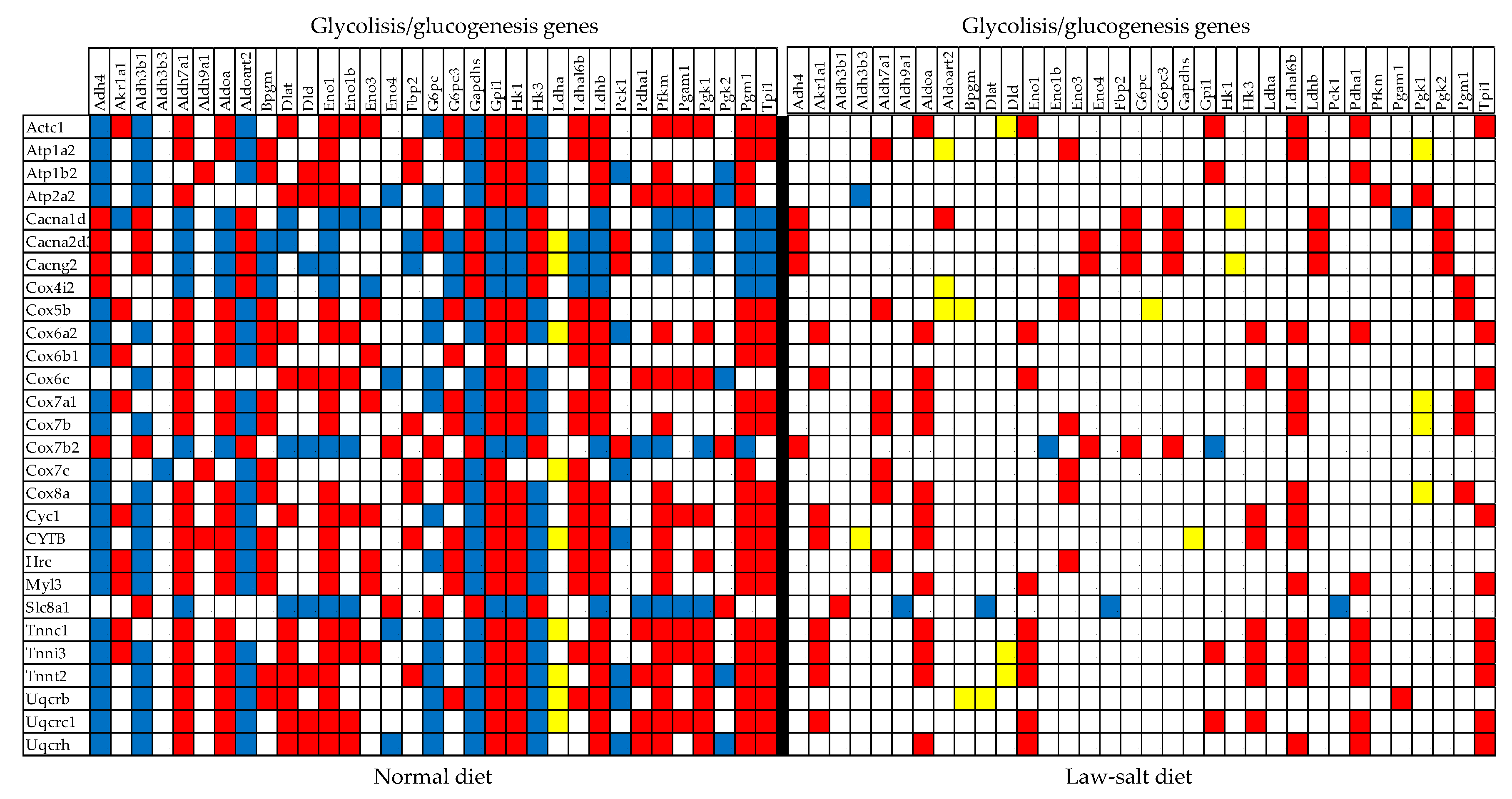
3.6. Regulated Genes within Selected Metabolic Pathways
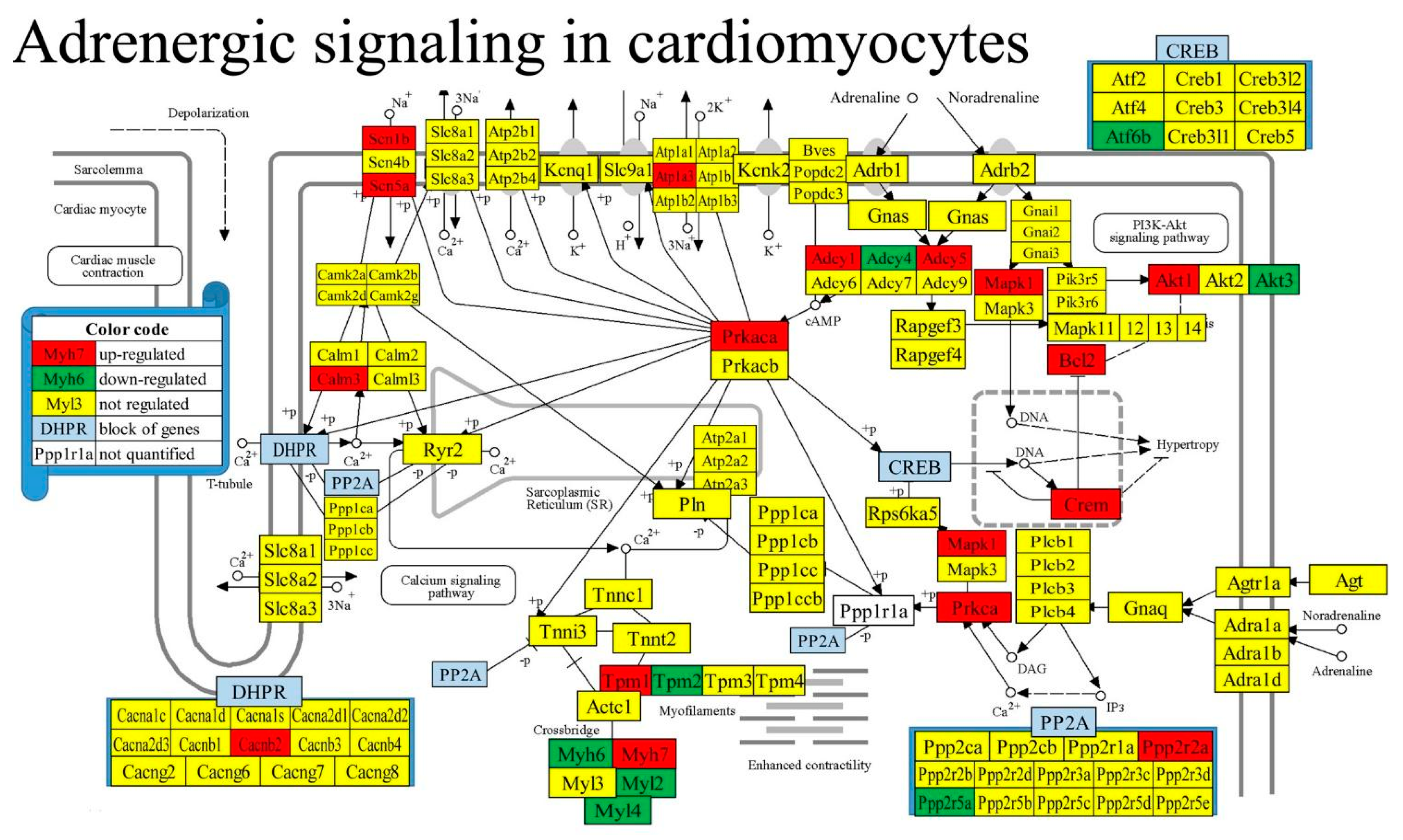
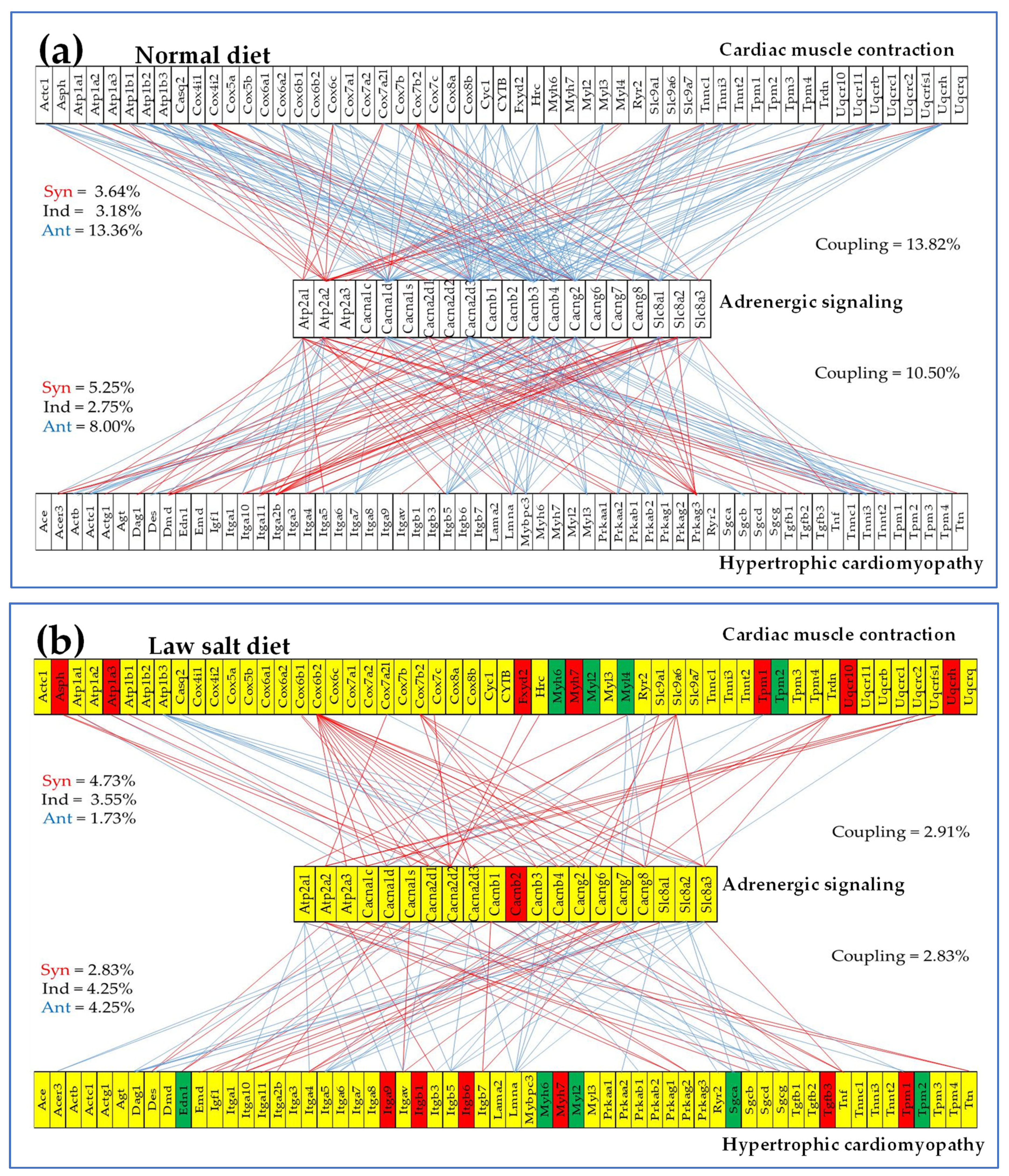
3.7. Regulation of Selected Signaling Pathways
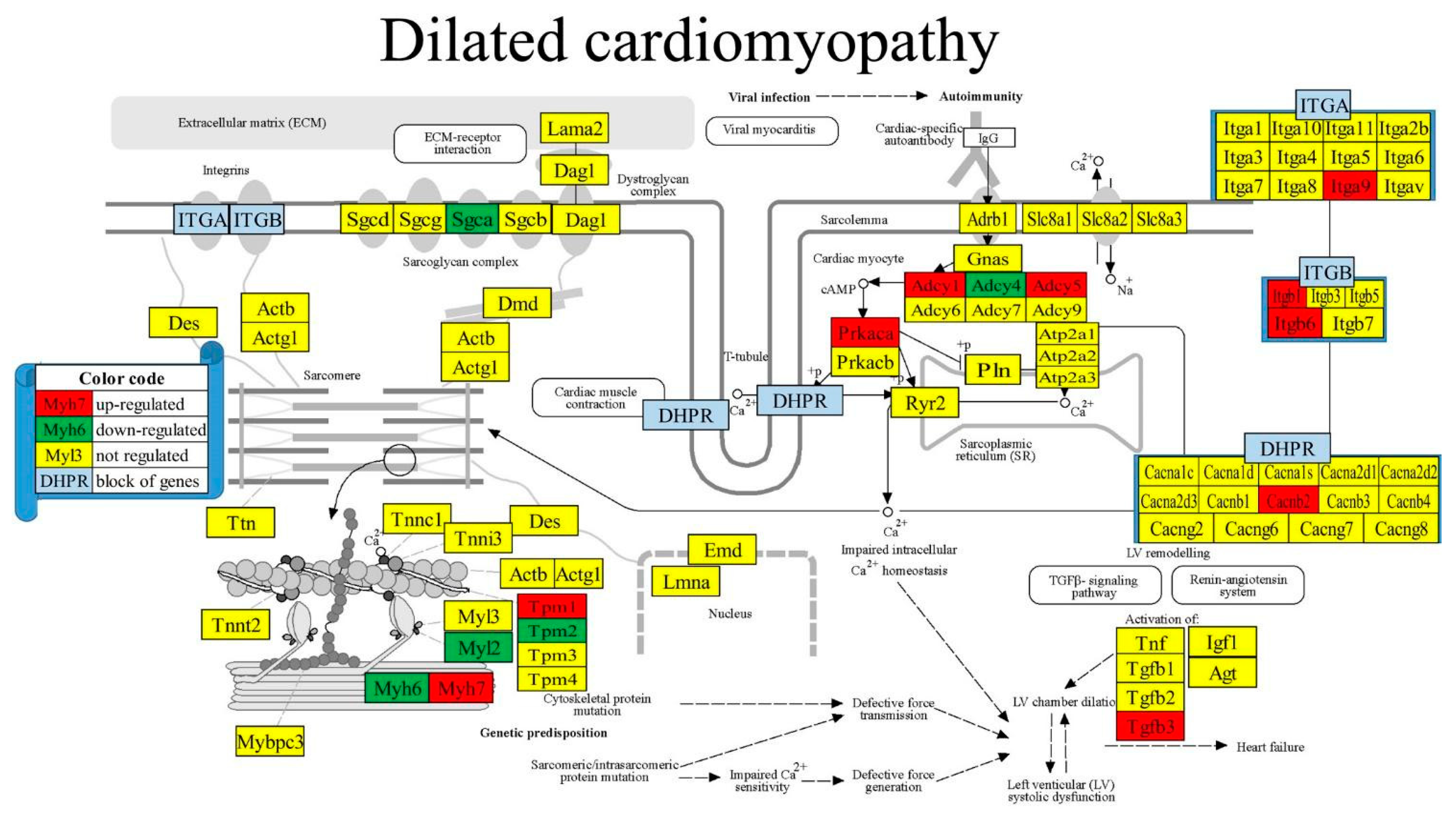
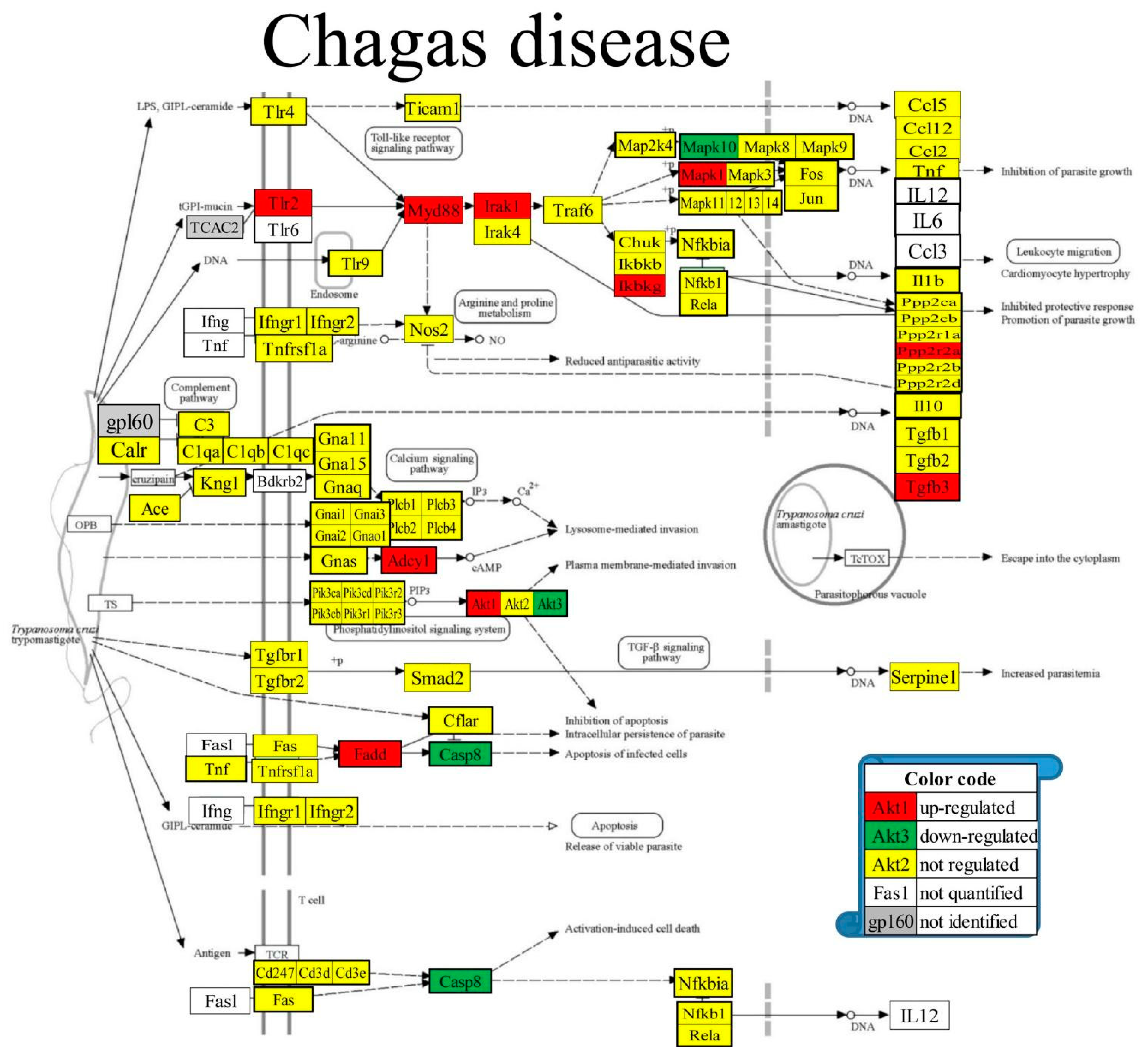
3.8. Regulated Genes within Pathways of Selected Cardiac Diseases
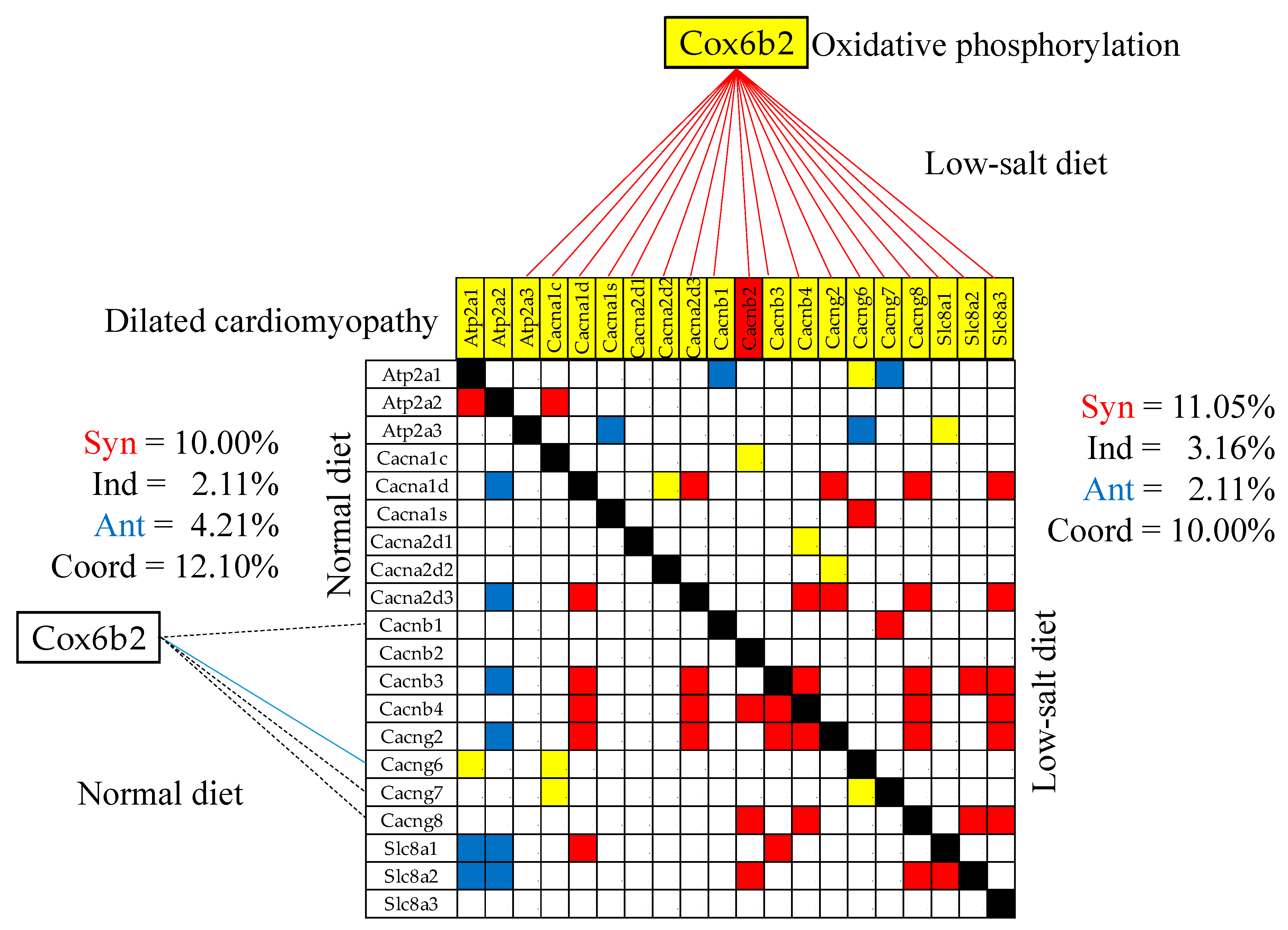
3.9. Remodeling of the Gene Networks
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Appendix A: Independent Primary Expression Characteristics of Individual Gene and Functional Pathways
Appendix B: Derived Characteristics of Individual Genes and Their Averages over Functional Pathways
Appendix C: Measures of Transcriptomic Regulation
References
- Aliasgharzadeh, S.; Tabrizi, J.S.; Nikniaz, L.; Ebrahimi-Mameghani, M.; Lotfi Yagin, N. Effect of salt reduction interventions in lowering blood pressure: A comprehensive systematic review and meta-analysis of controlled clinical trials. PLoS One. 2022, 17(12):e0277929. [CrossRef]
- Ma, H.; Wang, X.; Li, X.; Heianza, Y.; Qi, L. Adding Salt to Foods and Risk of Cardiovascular Disease. J Am Coll Cardiol. 2022; 80(23):2157-2167. [CrossRef]
- Padilla-Moseley, J.; Blanco-Metzler, A.; L’Abbé, M.R.; Arcand, J. A Program Evaluation of a Dietary Sodium Reduction Research Consortium of Five Low- and Middle-Income Countries in Latin America. Nutrients. 2022; 14(20):4311. [CrossRef]
- Shanmugam, R.; Worsley, A. Current levels of salt knowledge: a review of the literature. Nutrients. 2014; 6:5534–59. [CrossRef]
- Kotchen, T.A.; Cowley, A.W. Jr.; Frohlich, E.D. Salt in health and disease-a delicate balance. N Engl J Med. 2013; 368:1229–37. [CrossRef]
- World Health Organization. Guideline: Sodium Intake for Adults and Children. World Health Organization, Geneva. 2012. https://pubmed.ncbi.nlm.nih.gov/23658998.
- Aksentijević, D.; Shattock, M.J. With a grain of salt: Sodium elevation and metabolic remodeling in heart failure. J Mol Cell Cardiol. 2021; 161:106-115. [CrossRef]
- Ertuglu, L.A.; Kirabo, A. Dendritic Cell Epithelial Sodium Channel in Inflammation, Salt-Sensitive Hypertension, and Kidney Damage. Kidney360. 2022; 3(9):1620-1629. [CrossRef]
- Xiao, H.; Lu, H.; Xue, Y.; Jia, Z.; Dai, M.; He, K. et al. Deleterious effect in endothelin receptor-mediated coronary artery smooth muscle contractility in high-salt diet rats. Nutr Metab Cardiovasc Dis. 2022; S0939-4753(22)00422-7. [CrossRef]
- Hunter, R.W.; Dhaun, N.; Bailey, M.A. The impact of excessive salt intake on human health. Nat Rev Nephrol. 2022; 18(5):321-335. [CrossRef]
- Jaques, D.A.; Wuerzner, G.; Ponte, B. Sodium Intake as a Cardiovascular Risk Factor: A Narrative Review. Nutrients. 2021; 13(9):3177. [CrossRef]
- Dumančić, D.; Stupin, A.; Kožul, M.; Šerić, V.; Kibel, A.; Goswami, N. et al. Increased cerebral vascular resistance underlies preserved cerebral blood flow in response to orthostasis in humans on a high-salt diet. Eur J Appl Physiol. 2023. 123(4):923-933. [CrossRef]
- Carll, A.P.; Haykal-Coates, N.; Winsett, D.W.; Hazari, M.S.; Nyska, A.; Richards, J.H. et al. Dietary salt exacerbates isoproterenol-induced cardiomyopathy in rats. Toxicol Pathol. 2011; 39(6):925-37. [CrossRef]
- Xu, H.; Qing, T.; Shen, Y.; Huang, J.; Liu, Y.; Li, J. et al. RNA-seq analyses the effect of a high-salt diet on hypertension. Gene. 2018; 677:245-250. [CrossRef]
- Yim, J.; Cho, H.; Rabkin, S.W. Gene expression and gene associations during the development of heart failure with preserved ejection fraction in the Dahl salt-sensitive model of hypertension. Clin Exp Hypertens. 2018; 40(2):155-166. [CrossRef]
- Chen, X.; Wu, H.; Huang, S. Excessive Sodium Intake Leads to Cardiovascular Disease by Promoting Sex-Specific Dysfunction of Murine Heart. Front Nutr. 2022; 9:830738. [CrossRef]
- Corona, G.; Giuliani, C.; Parenti, G.; Norello, D.; Verbalis, J.G.; Forti, G. et al. Moderate Hyponatremia Is Associated with Increased Risk of Mortality: Evidence from a Meta-Analysis. PLOS ONE. 2013; 8:e80451. [CrossRef]
- Iacobas, D.A.; Xi, L. Theory and Applications of the (Cardio) Genomic Fabric Approach to Post-Ischemic and Hypoxia-Induced Heart Failure. J Pers Med. 2022; 12(8):1246. [CrossRef]
- Adesse, D.; Goldenberg, R.C.; Fortes, F.S.; Jasmin, Iacobas, D.A.; Iacobas, S. et al. Gap junctions and Chagas disease. Adv Parasitol. 2011; 76:63-81. [CrossRef]
- Adesse, D.; Iacobas, D.A.; Iacobas, S.; Garzoni, L.R.; Meirelles Mde, N.; Tanowitz, H.B. et al. Transcriptomic signatures of alterations in a myoblast cell line infected with four distinct strains of Trypanosoma cruzi. Am J Trop Med Hyg. 2010; 82(5):846-54. [CrossRef]
- Boudina, S.; Abel, E.D. Diabetic cardiomyopathy, causes, and effects. Rev Endocr Metab Disord. 2010; 11(1):31-9. [CrossRef]
- Xu, C.; Liu, C.; Xiong, J.; Yu, J. Cardiovascular aspects of the (pro)renin receptor: Function and significance. FASEB J. 2022; 36(4):e22237. [CrossRef]
- Luk, A.; Ahn, E.; Soor, G.S.; Butany, J. Dilated cardiomyopathy: a review. J Clin Pathol 2009; 62:219-25. [CrossRef]
- Fatkin, D.; Graham, R.M. Molecular mechanisms of inherited cardiomyopathies. Physiol Rev 2002; 82:945-80. [CrossRef]
- Akhtar, H.; Al Sudani, H.; Hussein, M.; Farhan, M.U.N.; Elkholy, K. Effects of Renin-Angiotensin-Aldosterone System Inhibition on Left Ventricular Hypertrophy, Diastolic Function, and Functional Status in Patients With Hypertrophic Cardiomyopathy: A Systematic Review. Cureus. 2022; 14(7):e26642. [CrossRef]
- Kyoto Encyclopedia of Genes and Genomes. Wiring diagrams of molecular interactions, reactions and relations. Available on line at: https://www.genome.jp/kegg/pathway.html. Accessed on Jan 7th, 2024.
- Transcriptomic effects of law salt diet on the mouse left ventricle. Available online at: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE72561. Accessed 1/27/2023.
- P-Value from Pearson (R) Calculator. Available online at: https://www.socscistatistics.com/pvalues/pearsondistribution.aspx (accessed 12/27/2023).
- Iacobas, S.; Ede, N.; Iacobas, D.A. The Gene Master Regulators (GMR) Approach Provides Legitimate Targets for Personalized, Time-Sensitive Cancer Gene Therapy. Genes (Basel). 2019; 10(8):560. [CrossRef]
- Iacobas, S.; Iacobas, D.A. A Personalized Genomics Approach of the Prostate Cancer. Cells. 2021; 10(7):1644. [CrossRef]
- Fructose and manose metabolism. Available online at: https://www.genome.jp/pathway/mmu00051. Accessed 1/11/2024.
- Galactose metabolism. Available online at: https://www.genome.jp/kegg-bin/show_pathway?mmu00052. Accessed 1/11/2024.
- Glycolysis/Glucogenesis. Available online at: https://www.genome.jp/pathway/mmu00010. Accessed 1/11/2024.
- Inositol phosphate metabolism. Available online at: https://www.genome.jp/kegg-bin/show_pathway?mmu00562. Accessed 1/11/2024.
- Oxidative phosphorylation. Available online at: https://www.genome.jp/kegg-bin/show_pathway?mmu00190. Accessed 1/11/2024.
- Fatty acid biosynthesis. Available online at: https://www.genome.jp/kegg-bin/show_pathway?mmu00061. Accessed 1/11/2024.
- Glycerolipid metabolism. Available online at: https://www.genome.jp/kegg-bin/show_pathway?mmu00561. Accessed 1/11/2024.
- Glycerophospholipid metabolism. Available online at: https://www.genome.jp/kegg-bin/show_pathway?mmu00564. Accessed 1/11/2024.
- Steroid biosynthesis. Available online at: https://www.genome.jp/kegg-bin/show_pathway?mmu00100. Accessed 1/11/2024.
- Steroid hormone biosynthesis. Available online at: https://www.genome.jp/kegg-bin/show_pathway?mmu00140. Accessed 1/11/2024.
- Purine metabolism. Available online at: https://www.genome.jp/kegg-bin/show_pathway?mmu00230. Accessed 1/11/2024.
- Pyrimidine metabolism. Available online at: https://www.genome.jp/kegg-bin/show_pathway?mmu00240. Accessed 1/11/2024.
- Cysteine and methionine metabolism. Available on line at: https://www.genome.jp/kegg-bin/show_pathway?mmu00270. Accessed 1/11/2024.
- Glutathione metabolism. Available on line at: https://www.genome.jp/kegg-bin/show_pathway?mmu00480. Accessed 1/11/2024.
- Thyrosine metabolism. Available on line at: https://www.genome.jp/kegg-bin/show_pathway?mmu00350. Accessed 1/11/2024.
- Valine, leucine and isoleucine degradation. Available on line at: https://www.genome.jp/kegg-bin/show_pathway?mmu00280. Accessed 1/11/2024.
- N-Glycan biosynthesis. Available on line at: https://www.genome.jp/kegg-bin/show_pathway?mmu00510. Accessed 1/11/2024.
- Drug metabolism – cytochrome P450. Available on line at: https://www.genome.jp/kegg-bin/show_pathway?mmu00982. Accessed 1/11/2024.
- Drug metabolism – other enzymes. Available on line at: https://www.genome.jp/kegg-bin/show_pathway?mmu00983. Accessed 1/11/2024.
- Adrenergic signaling in cardiomyocytes. Available on line at: https://www.genome.jp/kegg-bin/show_pathway?mmu04261. Accessed 1/11/2024.
- Cardiac muscle contraction. Available on line at: https://www.genome.jp/kegg-bin/show_pathway?mmu04260. Accessed 1/11/2024.
- Chagas disease. Available on line at: https://www.genome.jp/kegg-bin/show_pathway?mmu05142. Accessed 1/11/2024.
- Diabetic cardiomyopathy. Available on line at: https://www.genome.jp/kegg-bin/show_pathway?mmu05415. Accessed 1/11/2024.
- Dilated cardiomyopathy. Available on line at: https://www.genome.jp/kegg-bin/show_pathway?mmu05414. Accessed 1/11/2024.
- Hypertrophic cardiomyopathy. Available on line at: https://www.genome.jp/kegg-bin/show_pathway?mmu05410. Accessed 1/11/2024.
- MAPK signaling pathway. Available on line at https://www.genome.jp/kegg-bin/show_pathway?hsa04010. Accessed 1/11/2024.
- PIK3-Akt signaling pathway. Available on line at https://www.genome.jp/kegg-bin/show_pathway?hsa04151. Accessed 1/11/2024.
- Rap1 signaling pathway. Available on line at https://www.genome.jp/kegg-bin/show_pathway?hsa04015. Accessed 1/11/2024.
- Ras signaling pathway. Available on line at https://www.genome.jp/kegg-bin/show_pathway?hsa04014. Accessed 1/11/2024.
- Chemokine signaling pathway. Available on line at https://www.genome.jp/kegg-bin/show_pathway?hsa04062. Accessed 1/11/2023.
- Calcium signaling pathway. Available on line at https://www.genome.jp/kegg-bin/show_pathway?hsa04020. Accessed 1/11/2024.
- CAMP signaling pathway. Available on line at https://www.genome.jp/kegg-bin/show_pathway?hsa04024. Accessed 1/11/2024.
- CGMP-PKG signaling pathway. Available on line at https://www.genome.jp/kegg-bin/show_pathway?hsa04022. Accessed 1/11/2024.
- MTOR signaling pathway. Available on line at https://www.genome.jp/kegg-bin/show_pathway?hsa04150. Accessed 1/11/2024.
- Wnt signaling pathway. Available on line at https://www.genome.jp/kegg-bin/show_pathway?hsa04310. Accessed 1/11/2024.
- Central carbon metabolism in cancer. Available on line at: https://www.genome.jp/kegg-bin/show_pathway?mmu05230. Accessed 1/11/2024.
- Choline metabolism in cancer. Available online at: https://www.genome.jp/kegg-bin/show_pathway?mmu05231. Accessed 1/11/2024.
- Bayne, E.F.; Rossler, K.J.; Gregorich, Z.R.; Aballo, T.J.; Roberts, D.S.; Chapman, E.A. et al. Top-down proteomics of myosin light chain isoforms define chamber-specific expression in the human heart. J Mol Cell Cardiol. 2023; 181:89-97. [CrossRef]
- Iacobas, S.; Amuzescu, B.; Iacobas, D.A. Transcriptomic uniqueness and commonality of the ion channels and transporters in the four heart chambers. Sci Rep. 2021; 11(1):2743. [CrossRef]
- Sun, J.H.; Liu, X.K.; Xing, X.W.; Yang, Y.; Xuan, H.H.; Fu, B.B. Value of Cardiac Troponin, Myoglobin Combined with Heart-type Fatty Acid-binding Protein Detection in Diagnosis of Early Acute Myocardial Infarction. Pak J Med Sci. 2023; 39(6):1690-1694. [CrossRef]
- Yu, M.; Tcheandjieu, C.; Georges, A.; Xiao, K.; Tejeda, H.; Dina, C. et al. Computational estimates of annular diameter reveal genetic determinants of mitral valve function and disease. JCI Insight. 2022; 7(3):e146580. [CrossRef]
- Ning, Z.; Huang, Y.; Lu, H.; Zhou, Y, Tu T, Ouyang F et al. Novel Drug Targets for Atrial Fibrillation Identified Through Mendelian Randomization Analysis of the Blood Proteome. Cardiovasc Drugs Ther. 2023. [CrossRef]
- Iacobas, D.A.; Iacobas, S.; Chachua, T.; Goletiani, C.; Sidyelyeva, G.; Velíšková, J. et al. Prenatal corticosteroids modify glutamatergic and GABAergic synapse genomic fabric: insights from a novel animal model of infantile spasms. J Neuroendocrinol. 2013; 25(11):964-79. [CrossRef]
- Zhang, Y.; Zhao, G.; Yu, L.; Wang, X.; Meng, Y.; Mao, J. et al. Heat-shock protein 90α protects NME1 against degradation and suppresses metastasis of breast cancer. Br J Cancer. 2023; 129(10):1679-1691. [CrossRef]
- Jarrar, Y.; Zihlif, M.; Al Bawab, A.Q.; Sharab, A. Effects of Intermittent Hypoxia on Expression of Glucose Metabolism Genes in MCF7 Breast Cancer Cell Line. Curr Cancer Drug Targets. 2020; 20(3):216-222. [CrossRef]
- Iacobas, D.A.; Iacobas, S.; Lee, P.R.; Cohen, J.E.; Fields, R.D. Coordinated Activity of Transcriptional Networks Responding to the Pattern of Action Potential Firing in Neurons. Genes (Basel). 2019; 10(10):754. [CrossRef]
- Iacobas, D.A.; Obiomon, E.A.; Iacobas, S. Genomic Fabrics of the Excretory System’s Functional Pathways Remodeled in Clear Cell Renal Cell Carcinoma. Current Issues in Molecular Biology. 2023; 45(12):9471-9499. [CrossRef]
- Guo, G.L.; Sun, L.Q.; Sun, M.H.; Xu, H.M. LncRNA SLC8A1-AS1 protects against myocardial damage through activation of the cGMP-PKG signaling pathway by inhibiting SLC8A1 in mice models of myocardial infarction. J Cell Physiol. 2019; 234(6):9019-9032. [CrossRef]
- Sommer, N.; Hüttemann, M.; Pak, O.; Scheibe, S.; Knoepp, F.; Sinkler, C.; Malczyk, M. et al. Mitochondrial Complex IV Subunit 4 Isoform 2 Is Essential for Acute Pulmonary Oxygen Sensing. Circ Res. 2017; 121(4):424-438. [CrossRef]
- Bayer, A.L.; Alcaide, P. MyD88: At the heart of inflammatory signaling and cardiovascular disease. J Mol Cell Cardiol. 2021; 161:75-85. [CrossRef]
- Geering, K. Function of FXYD proteins, regulators of Na, K-ATPase. J Bioenerg Biomembr. 2005; 37(6):387-92. [CrossRef]
- Nordin, H.; Nakagawa, R.; Wallin, M.; Pernow, J.; Kass, D.A.; Ståhlberg, M. Regional protein expression changes within the left ventricle in a mouse model of dyssynchronous and resynchronized heart failure. ESC Heart Fail. 2020; 7(6):4438-4442. [CrossRef]
- Yusuf, A.M.; Qaisar, R.; Al-Tamimi, A.O.; Jayakumar, M.N.; Woodgett, J.R.; Koch, W.J. et al. Cardiomyocyte-GSK-3β deficiency induces cardiac progenitor cell proliferation in the ischemic heart through paracrine mechanisms. J Cell Physiol. 2022; 37(3):1804-1817. [CrossRef]
- Munasinghe, P.E.; Saw, E.L.; Reily-Bell, M.; Tonkin, D.; Kakinuma, Y.; Fronius, M. et al. Non-neuronal cholinergic system delays cardiac remodeling in type 1 diabetes. Heliyon. 2023; 9(6):e17434. [CrossRef]
- Zhang, W.; Jiang, H.; Huang, P.; Wu, G.; Wang, Q.; Luan, X. et al. Dracorhodin targeting CMPK2 attenuates inflammation: A novel approach to sepsis therapy. Clin Transl Med. 2023; 13(10):e1449. [CrossRef]
- Gérus, M.; Bonnart, C.; Caizergues-Ferrer, M.; Henry, Y.; Henras, A.K. Evolutionarily conserved function of RRP36 in early cleavages of the pre-rRNA and production of the 40S ribosomal subunit. Mol Cell Biol. 2010; 30(5):1130-44. [CrossRef]
- Drouard, G.; Hagenbeek, F.A.; Whipp, A.; Pool, R. et al., BIOS Consortium; BBMRI-NL Metabolomics Consortium et al.. Longitudinal multi-omics study reveals common etiology underlying association between plasma proteome and BMI trajectories in adolescent and young adult twins. BMC Med. 2023; 21(1):508. [CrossRef]
- Bassaganyas, L.; Popa, S.J.; Horlbeck, M.; Puri, C.; Stewart, S.E.; Campelo, F. et al. New factors for protein transport identified by a genome-wide CRISPRi screen in mammalian cells. J Cell Biol. 2019; 218(11):3861-3879. [CrossRef]
- Yu, Y.; Chen, G.; Jiang, C.; Guo, T.; Tang, H.; Yuan, Z. et al. USP31 serves as a potential biomarker for predicting prognosis and immune responses for clear cell renal cell carcinoma via single-cell and bulk RNA-sequencing. J Gene Med. 2023: e3594. [CrossRef]
- Iacobas, D.A.; Mgbemena, V.E.; Iacobas, S.; Menezes, K.M.; Wang, H.; Saganti, P.B. Genomic Fabric Remodeling in Metastatic Clear Cell Renal Cell Carcinoma (ccRCC): A New Paradigm and Proposal for a Personalized Gene Therapy Approach. Cancers (Basel). 2020; 12(12):3678. [CrossRef]
- Kristofova, M.; Ori, A.; Wang, Z.Q. Multifaceted Microcephaly-Related Gene MCPH1. Cells. 2022; 11(2):275. [CrossRef]
- Martínez-Moreno, R.; Carreras, D.; Sarquella-Brugada, G.; Pérez, G.J.; Selga, E.; Scornik, F.S. et al. Loss of sodium current caused by a Brugada syndrome-associated variant is determined by patient-specific genetic background. Heart Rhythm. 2023: S1547-5271(23)02966-1. [CrossRef]
- Balestrini, S.; Mikati, M.A.; Álvarez-García-Rovés, R.; Carboni, M.; Hunanyan, A.S.; Kherallah, B. et al. Cardiac phenotype in ATP1A3-related syndromes: A multicenter cohort study. Neurology. 2020; 95(21):e2866-e2879. [CrossRef]
- Slack, R.J.; Macdonald, S.J.F.; Roper, J.A.; Jenkins, R.G.; Hatley, R.J.D. Emerging therapeutic opportunities for integrin inhibitors. Nat Rev Drug Discov. 2022; 21(1):60-78. [CrossRef]
- Song, Z.; Tian, X.; Shi, Q. Fas, Caspase-8, and Caspase-9 pathway-mediated bile acid-induced fetal cardiomyocyte apoptosis in intrahepatic cholestasis pregnant rat models. J Obstet Gynaecol Res. 2021; 47(7):2298-2306. [CrossRef]
- Pahlavani, H.A. Exercise-induced signaling pathways to counteracting cardiac apoptotic processes. Front Cell Dev Biol. 2022; 10:950927. [CrossRef]
- Iacobas, D.A.; Iacobas, S.; Tanowitz, H.B.; Campos de Carvalho, A.; Spray, D.C. Functional genomic fabrics are remodeled in a mouse model of Chagasic cardiomyopathy and restored following cell therapy. Microbes Infect. 2018; 20(3):185-195. [CrossRef]
- Nisimura, L.M.; Coelho, L.L.; de Melo, T.G.; Vieira, P.C.; Victorino, P.H.; Garzoni, L.R. et al. Trypanosoma cruzi Promotes Transcriptomic Remodeling of the JAK/STAT Signaling and Cell Cycle Pathways in Myoblasts. Front Cell Infect Microbiol. 2020; 10:255. [CrossRef]
- Kolobarić, N.; Mihalj, M.; Kozina, N.; Matić, A.; Mihaljević, Z.; Jukić, I. et al. Tff3-/- Knock-Out Mice with Altered Lipid Metabolism Exhibit a Lower Level of Inflammation following the Dietary Intake of Sodium Chloride for One Week. Int J Mol Sci. 2023; 24(8):7315. [CrossRef]
- Bochi, A.P.G.; Ferreira, G.D.S.; Del Bianco, V.; Pinto, P.R.; Rodrigues, L.G.; Trevisani, M.D.S. et al. Aerobic Exercise Training Reduces Atherogenesis Induced by Low-Sodium Diet in LDL Receptor Knockout Mice. Antioxidants (Basel). 2022; 11(10):2023. [CrossRef]
- Launonen, H.; Pang, Z.; Linden, J.; Siltari, A.; Korpela, R.; Vapaatalo, H. Evidence for local aldosterone synthesis in the large intestine of the mouse. J Physiol Pharmacol. 2021; 72(5). [CrossRef]
| GENE | DESCRIPTION | X | P | CUT | WIR |
|---|---|---|---|---|---|
| False down-regulated genes | |||||
| Ifitm5 | interferon induced transmembrane protein 5 | -2.350 | 0.030 | 2.427 | -0.428 |
| Hinfp | histone H4 transcription factor | -2.164 | 0.039 | 2.639 | -0.263 |
| Prdm11 | PR domain containing 11 | -2.000 | 0.026 | 2.170 | -0.376 |
| Myl7 | myosin, light polypeptide 7, regulatory | -1.887 | 0.022 | 2.468 | -4.566 |
| Trim71 | tripartite motif-containing 71 | -1.852 | 0.036 | 2.285 | 0.173 |
| Usf1 | upstream transcription factor 1 | -1.837 | 0.023 | 1.928 | -0.341 |
| Chkb | choline kinase beta | -1.829 | 0.025 | 2.633 | -5.056 |
| Cntnap5c | contactin associated protein-like 5C | -1.824 | 0.025 | 1.922 | -5.270 |
| Dnajb1 | DnaJ heat shock protein family | -1.812 | 0.034 | 2.129 | -9.529 |
| Csrnp2 | cysteine-serine-rich nuclear protein 2 | -1.797 | 0.032 | 2.176 | -0.228 |
| Missed down-regulated genes | |||||
| Gsk3b | glycogen synthase kinase 3 beta | -1.490 | 0.017 | 1.341 | -4.025 |
| Aldh3a2 | aldehyde dehydrogenase family 3, subfamily A2 | -1.462 | 0.007 | 1.198 | -0.422 |
| Mapk10 | mitogen-activated protein kinase 10 | -1.455 | 0.028 | 1.306 | -2.712 |
| Myl2 | myosin, light polypeptide 2, regulatory, cardiac, slow | -1.431 | 0.007 | 1.329 | -0.868 |
| Tpm2 | tropomyosin 2, beta | -1.421 | 0.027 | 1.359 | -1.751 |
| Atp5j | ATP synthase H+ transporting mitochondrial F0 complex subunit F | -1.401 | 0.013 | 1.272 | -0.171 |
| Gmpr2 | guanosine monophosphate reductase 2 | -1.371 | 0.009 | 1.238 | -0.350 |
| Enpp4 | ectonucleotide pyrophosphatase/phosphodiesterase 4 | -1.362 | 0.028 | 1.316 | -1.748 |
| Chat | choline acetyltransferase | -1.353 | 0.024 | 1.292 | -0.253 |
| Dbt | dihydrolipoamide branched chain transacylase E2 | -1.323 | 0.024 | 1.274 | -1.046 |
| Missed up-regulated genes | |||||
| Lpin3 | lipin 3 | 1.372 | 0.004 | 1.146 | 0.366 |
| Pde1a | phosphodiesterase 1A, calmodulin-dependent | 1.374 | 0.008 | 1.219 | 0.974 |
| Gpam | glycerol-3-phosphate acyltransferase, mitochondrial | 1.374 | 0.019 | 1.214 | 0.427 |
| B4galt1 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 | 1.391 | 0.005 | 1.334 | 3.184 |
| Ncf4 | neutrophil cytosolic factor 4 | 1.397 | 0.046 | 1.320 | 0.273 |
| Bcl2 | B cell leukemia/lymphoma 2 | 1.401 | 0.005 | 1.164 | 0.392 |
| Ndufc1 | NADH: ubiquinone oxidoreductase subunit C1 | 1.410 | 0.018 | 1.303 | 58.827 |
| Ikbkg | inhibitor of kappaB kinase gamma | 1.424 | 0.005 | 1.233 | 0.260 |
| Atp6v1b2 | ATPase, H+ transporting, lysosomal V1 subunit B2 | 1.438 | 0.045 | 1.381 | 0.265 |
| Gucy1b2 | guanylate cyclase 1, soluble, beta 2 | 1.490 | 0.034 | 1.426 | 0.943 |
| False up-regulated genes | |||||
| Kif3c | kinesin family member 3C | 1.706 | 0.009 | 1.832 | 3.179 |
| Nt5el | 5’ nucleotidase, ecto-like | 1.720 | 0.028 | 2.153 | 0.097 |
| Zfp362 | zinc finger protein 362 | 1.758 | 0.024 | 1.852 | 0.637 |
| Ctsg | cathepsin G | 1.887 | 0.018 | 1.890 | 0.192 |
| Tmem231 | transmembrane protein 231 | 1.912 | 0.027 | 2.196 | 0.128 |
| Adam12 | a disintegrin and metallopeptidase domain 12 | 1.966 | 0.036 | 2.313 | 0.423 |
| Ftcd | formiminotransferase cyclodeaminase | 1.979 | 0.033 | 2.214 | 0.163 |
| Ap1m1 | adaptor-related protein complex AP-1, mu subunit 1 | 2.063 | 0.006 | 2.079 | 11.060 |
| Lrrc71 | leucine rich repeat containing 71 | 2.153 | 0.034 | 2.559 | 0.138 |
| Gclc | glutamate-cysteine ligase, catalytic subunit | 2.330 | 0.028 | 2.456 | 1.332 |
| mmu | PATH | Description | GENES | D% | U% | WPR | ΔREC (%) |
|---|---|---|---|---|---|---|---|
| 04261 | ASC | Adrenergic signaling in cardiomyocytes | 130/156 | 6.15 | 13.08 | 19.97 | -3.71 |
| 04260 | CMC | Cardiac muscle contraction | 75/87 | 5.33 | 10.67 | 45.30 | -1.38 |
| 05142 | CHA | Chagas disease | 85/103 | 3.61 | 12.05 | 3.31 | -6.71 |
| 05415 | DIA | Diabetic cardiomyopathy | 184/211 | 3.80 | 7.07 | 29.55 | 0.40 |
| 05414 | DIL | Dilated cardiomyopathy | 81/94 | 6.17 | 12.35 | 7.05 | -0.45 |
| 00061 | FAB | Fatty acids biosynthesis | 18/19 | 0.00 | 5.56 | 2.49 | 17.10 |
| 00561 | GLM | Glycerolipid metabolism | 52/63 | 3.85 | 15.38 | 4.63 | -5.88 |
| 00564 | GPL | Glycerophospholipid metabolism | 83/98 | 4.82 | 9.64 | 1.54 | 2.27 |
| 00010 | GLY | Glycolisis/glucogenesis | 55/64 | 3.64 | 1.82 | 5.51 | 6.18 |
| 05410 | HCM | Hypertrophic cardiomyopathy | 78/91 | 6.41 | 8.97 | 6.73 | 2.23 |
| 00510 | NGL | N-Glycan biosynthesis | 50/53 | 4.00 | 4.00 | 14.18 | 14.63 |
| 00190 | OXP | Oxidative phosphorylation | 110/135 | 1.82 | 6.36 | 37.42 | 12.39 |
| 00230 | PUM | Purine metabolism | 114/134 | 10.53 | 11.40 | 5.42 | 4.19 |
| 00240 | PYR | Pyrimidine metabolism | 47/56 | 8.51 | 10.64 | 1.64 | -5.83 |
| 00100 | STB | Steroid biosynthesis | 17/20 | 0.00 | 5.88 | 0.69 | -11.37 |
| 00140 | SHB | Steroid hormone biosynthesis | 42/93 | 7.14 | 9.52 | 8.27 | -18.74 |
| 00280 | VLI | Valine, leucine and isoleucine degradation | 48/57 | 6.25 | 2.08 | 9.28 | 5.72 |
| ALL | All quantified genes | 19,605 | 3.65 | 5.96 | 15.67 | 0.30 |
| PATHWAY | R | GENES |
|---|---|---|
| Purine metabolism | D | Adcy4; Adprm; Ak2; Ampd2; Enpp4; Entpd5; Gmpr2; Nt5c; Pde4b; Prune1; Rrm1; Xdh |
| U | Adcy1; Adcy5; Adk; Adssl1; Gart; Gucy1b2; Nme1; Nme4; Nt5c2; Pde11a; Pde1a; Pde1b; Prps2 | |
| Choline metabolism in cancer | D | Akt3; Gpcpd1; Mapk10; Pdgfd; Pdgfra; Pdgfrb; Rac2 |
| U | Akt1; Egfr; Hif1a; Kras; Mapk1; Pdpk1; Pip5k1a; Plpp1; Plpp2; Plpp3; Prkca; Prkcb; Rac1; Slc44a1 | |
| Drug metabolism - other enzymes | D | Ces1d; Gsta3; Gstt1; Gstt2; Rrm1; Xdh |
| U | Cmpk1; Gsta4; Gstm1; Gstm6; Gstm7; Gstp1; Gusb; Nat2; Nme1; Nme4; Upp1 | |
| Glycerophospholipid metabolism | D | Adprm; Chat; Gpcpd1; Selenoi |
| U | Etnk2; Gpam; Lpin3; Mboat1; Pla1a; Plpp1; Plpp2; Plpp3 | |
| Glutathione metabolism | D | Gsta3; Gstt1; Gstt2; Rrm1 |
| U | Chac1; Gsta4; Gstm1; Gstm6; Gstm7; Gstp1; Odc1; Srm | |
| Central carbon metabolism in cancer | D | Akt3; Fgfr3; Pdgfra; Pdgfrb; Slc1a5 |
| U | Akt1; Egfr; Hif1a; Kras; Mapk1; Sco2 | |
| Drug metabolism - cytochrome P450 | D | Fmo1; Gsta3; Gstt1; Gstt2 |
| U | Fmo5; Gsta4; Gstm1; Gstm6; Gstm7; Gstp1 | |
| Glycerolipid metabolism | D | Aldh3a2; Mgll |
| U | Akr1b8; Aldh1b1; Gpam; Lpin3; Mboat1; Plpp1; Plpp2; Plpp3 | |
| Pyrimidine metabolism | D | Cmpk2; Entpd5; Nt5c; Rrm1 |
| U | Cmpk1; Nme1; Nme4; Nt5c2; Upp1 | |
| Cysteine & methionine metabolism | D | Agxt2; Amd2; Mpst |
| U | Adi1; Apip; Mtap; Srm; Tst | |
| Inositol phosphate metabolism | D | Inpp1; Isyna1 |
| U | Pi4k2a; Pik3c2b; Pip5k1a; Plcd3; Synj2 | |
| Fructose and mannose metabolism | D | Pfkfb1 |
| U | Akr1b8; Gmds; Khk; Pfkfb3; Pfkfb4 | |
| Galactose metabolism | U | Akr1b8; B4galt1; Gaa; Ugp2 |
| Tyrosine metabolism | U | Comt; Dct; Mif; Th |
| MAPK | PI3K-Akt | Rap1 | Ras | Chemokine | |||||
|---|---|---|---|---|---|---|---|---|---|
| 36U | 14D | 28U | 17D | 28U | 13D | 27U | 11D | 21U | 10D |
| Akt1 | Akt3 | Akt1 | Akt3 | Adcy1 | Adcy4 | Abl2 | Akt3 | Adcy1 | Adcy4 |
| Cacnb2 | Cacna1g | Bcl2 | Atf6b | Adcy5 | Adora2a | Akt1 | Fgfr3 | Adcy5 | Akt3 |
| Crk | Fgfr3 | Cdkn1a | Ddit4 | Adora2b | Akt3 | Calm3 | Igf2 | Akt1 | Cxcl11 |
| Csf1 | Hspa1a | Col4a1 | Epor | Akt1 | Fgfr3 | Csf1 | Mapk10 | Ccl21b | Cxcl14 |
| Dusp6 | Igf2 | Col4a2 | Fgfr3 | Calm3 | Map2k6 | Efna3 | Pdgfd | Ccl6 | Dock2 |
| Dusp8 | Map2k6 | Col4a5 | Foxo3 | Crk | P2ry1 | Egfr | Pdgfra | Ccr7 | Foxo3 |
| Efna3 | Map3k11 | Csf1 | Gsk3b | Csf1 | Pdgfd | Ets1 | Pdgfrb | Crk | Gsk3b |
| Egfr | Map3k2 | Efna3 | Igf2 | Efna3 | Pdgfra | Exoc2 | Rac2 | Cx3cr1 | Rac2 |
| Fgf18 | Mapk10 | Egfr | Mlst8 | Egfr | Pdgfrb | Fgf18 | Rapgef5 | Gnb3 | Rhoa |
| Gadd45b | Max | Eif4e | Pck2 | Enah | Prkd2 | Gnb3 | Rgl1 | Gng7 | Stat2 |
| Gna12 | Pdgfd | Fgf18 | Pdgfd | Fgf18 | Rac2 | Gng7 | Rhoa | Grk3 | |
| Ikbkg | Pdgfra | Gnb3 | Pdgfra | Itgal | Rapgef5 | Ikbkg | Ikbkg | ||
| Irak1 | Pdgfrb | Gng7 | Pdgfrb | Itgb1 | Rhoa | Kras | Kras | ||
| Kras | Rac2 | Ikbkg | Ppp2r5a | Itgb2 | Mapk1 | Mapk1 | |||
| Lamtor3 | Il4ra | Sgk1 | Kras | Mras | Prkaca | ||||
| Map3k3 | Itga9 | Thbs2 | Krit1 | Nf1 | Prkcb | ||||
| Map3k7 | Itgb1 | Tnxb | Mapk1 | Ngf | Prkcd | ||||
| Mapk1 | Itgb6 | Mras | Pla1a | Ptk2b | |||||
| Mapt | Kras | Ngf | Prkaca | Rac1 | |||||
| Mknk2 | Mapk1 | Pard6a | Prkca | Stat5b | |||||
| Mras | Ngf | Pfn1 | Prkcb | Tiam1 | |||||
| Myd88 | Pdpk1 | Prkca | Rab5a | ||||||
| Nf1 | Ppp2r2a | Prkcb | Rab5b | ||||||
| Ngf | Prkca | Rac1 | Rac1 | ||||||
| Ppp3ca | Rac1 | Rap1gap | Ralgapa2 | ||||||
| Prkaca | Thbs1 | Sipa1l2 | Stk4 | ||||||
| Prkca | Thbs4 | Thbs1 | Tiam1 | ||||||
| Prkcb | Tlr2 | Tiam1 | |||||||
| Ptpn5 | |||||||||
| Rac1 | |||||||||
| Relb | |||||||||
| Srf | |||||||||
| Stk3 | |||||||||
| Stk4 | |||||||||
| Tgfb3 | |||||||||
| Traf2 | |||||||||
| Calcium | cAMP | cGMP-PKG | mTOR | Wnt | |||||
|---|---|---|---|---|---|---|---|---|---|
| 15U | 14D | 14U | 11D | 15U | 10D | 16U | 9D | 13U | 12D |
| Adcy1 | Adcy4 | Adcy1 | Adcy4 | Adcy1 | Adcy4 | Akt1 | Akt3 | Crebbp | Fzd4 |
| Adora2b | Adora2a | Adcy5 | Adora2a | Adcy5 | Akt3 | Atp6v1b2 | Castor2 | Csnk2a1 | Gpc4 |
| Asph | Cacna1g | Akt1 | Akt3 | Adra2b | Atf6b | Clip1 | Ddit4 | Dvl1 | Gsk3b |
| Calm3 | Fgfr3 | Atp1a3 | Edn1 | Akt1 | Itpr2 | Dvl1 | Fzd4 | Map3k7 | Mapk10 |
| Egfr | Grm1 | Calm3 | Mapk10 | Atp1a3 | Itpr3 | Eif4e | Gsk3b | Notum | Porcn |
| Fgf18 | Itpr2 | Crebbp | Myl9 | Calm3 | Myh6 | Kras | Mlst8 | Ppp3ca | Prickle1 |
| Ngf | Itpr3 | Fxyd2 | Pde4b | Fxyd2 | Myl9 | Lamtor3 | Rhoa | Prkaca | Rac2 |
| Pde1a | Mst1r | Hcn2 | Ppp1r12a | Gna12 | Mylk4 | Lpin3 | Rictor | Prkca | Rhoa |
| Pde1b | Mylk4 | Mapk1 | Ppp1r1b | Gtf2ird1 | Ppp1r12a | Mapk1 | Sgk1 | Prkcb | Sfrp5 |
| Plcd3 | P2rx1 | Prkaca | Rac2 | Gucy1b2 | Rhoa | Pdpk1 | Rac1 | Sox17 | |
| Ppp3ca | Pdgfd | Rac1 | Rhoa | Mapk1 | Prkca | Smad3 | Tle2 | ||
| Prkaca | Pdgfra | Sst | Myh7 | Prkcb | Wnt1 | Tle3 | |||
| Prkca | Pdgfrb | Sstr5 | Nppb | Stradb | Wnt5b | ||||
| Prkcb | Phkg1 | Tiam1 | Ppp3ca | Wdr59 | |||||
| Ptk2b | Srf | Wnt1 | |||||||
| Wnt5b | |||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





