INTRODUCTION
Breast cancer is a prevalent form of cancer commonly diagnosed in females around the world. According to the 2020 report from the World Health Organization (WHO), approximately 2.3 million women are diagnosed with breast cancer each year, resulting in 685,000 deaths globally. [
1] There are several subtypes of breast cancer, each with different treatment responses and prognoses [
2]. Based on the gene expression profile, triple-negative breast cancer (TNBC), which is unable to express any receptors (ER, PR, and HER2), belongs to the basal-like subgroup. The patient survival rate, immune cell infiltration, and gene expression profile have been reported to differ significantly among all the subgroups of breast cancer. This indicates that these molecular subgroups can guide any treatment regimen, gene expression profile, and prognostic value [
3].
Most cases of breast cancer develop due to the accumulation of mutations in ductal epithelial cells. These mutations can be influenced by various factors in the tumor microenvironment (TME), such as immune cells, adipocytes, fibroblasts, and the secretion of different chemokines and cytokines [
5,
6]. Tumor-infiltrating immune cells elevate the expression of various immune checkpoint modulators, including Programmed Cell Death Protein 1 (PD-1), Cytotoxic T-lymphocyte–associated Antigen 4 (CTLA-4), T-cell Immunoglobulin (Ig), Mucin Domain-containing.
Protein-3 (TIM-3), and Lymphocyte-activation Gene 3 (LAG-3) [
8,
9]. With the advancement of treatment procedures, numerous clinical trials are being conducted. It has been reported that immune checkpoint inhibitor treatment stimulates a potent immune response against tumors and greatly enhances the outcomes for cancer patients [
10,
11,
12,
13].
The immune checkpoint (IC) receptors, such as CTLA-4 and PD-1, are currently undergoing extensive research. Numerous clinical trials are currently being conducted in different phases to investigate the potential use of these ICs. However, in a phase II clinical trial for lung cancer using ipilimumab, an anti-CTLA-4 monoclonal antibody, the effectiveness was not significant [
14]. The PD-1/PD-L1 IC inhibitor was also investigated for various treatment regimens. However, as a monotherapy, this PD-1/PD-L1 IC inhibitor did not show much promise, with a success rate of only 5-20% [
15].
The recently discovered immune checkpoint inhibitor with promising therapeutic potential is the T-cell immunoglobulin and mucin domain-containing molecule-3 (TIM-3), which is also referred to as hepatitis A virus cellular receptor 2 (HAVCR2). TIM-3 is a membrane-bound molecule expressed in CD4+ and CD8+ T cells, regulatory T cells, NK cells, macrophages, and mast cells [
15,
16]. Recent evidence suggests that the resistance to anti-CTLA-4 or anti PD-1/PD-L1 inhibitors is modulated by the upregulation of TIM-3 [
17].In previous studies, the role of TIM3 has only been demonstrated in Triple negative breast cancer [
18]. However, the exact expression and role of TIM3 in various subtypes of breast cancer remain unclear.
In this present study, the expression of TIM3 in various subtypes of breast cancer and its role in survivability have been investigated. Additionally, the correlation between TIM3 expression and the infiltration of different types of immune cells has been addressed.
METHODS
Analysis of the expression of TIM-3 using transcriptional datasets
To investigate the expression of TIM3 in breast tumors, two web platforms were utilized: GEPIA and UALCAN [
19,
20]. mRNA expression data sets were obtained from the GEPIA web server for the BRCA data set (n = 1085), as well as from the University of Alabama at Birmingham Cancer Data Analysis Portal (UALCAN) (n = 1097). Both datasets, GEPIA (n = 291) and ULCAN (n = 114), included comparisons of normal breast tissue samples. Additionally, expression levels of specific subtypes including HER2 positive, Luminal, and Triple negative (TNBC) were also investigated using the UALCAN webserver.In addition, the proteomics data from the UALCAN web server were obtained and analyzed using HAVCR2 as a reference gene within the "Breast Cancer" CPTAC dataset. The Z-value of the HAVCR2 protein expression was analyzed using primary tumor samples (n=125) and normal samples (n=18) in overall breast cancer samples. Furthermore, the protein expression of HAVCR2 was also analyzed in the major subtypes, including luminal (n=64), HER2 positive (n=10), and TNBC (n=16).
Analysis of the infiltrating cell types specific expression of TIM-3 using transcriptional datasets
As TIM-3 can also be expressed in immune cells, we examined the abundance of immune cells infiltrating the tumor in relation to TIM-3 expression. Moreover, we utilized the TIMER2.0 tool to evaluate the infiltration of different immune cells in all subtypes of breast cancer patients [
21]. Specifically, we focused on four key components of the immune system: macrophage subtypes (M0, M1, M2), CD4+ T-cells, CD8+ T-cells, and B-cells. These investigations were conducted using the TCGA-BRCA gene cohort. The CIBERSORT algorithm, which is available in the TIMER2.0 webtool, was used to identify cell types by estimating the relative subsets of RNA transcripts. Spearman’s correlation and scatter plots were utilized to examine the relationship between HAVCR2 expression and tumor purity, which refers to the proportion of cancer cells in a sample. Additionally, the association between gene expression and immune cell type was investigated.
Estimation of the expression of TIM-3 in human breast tumor tissue using immunohistochemistry (IHC)
To assess the findings of the publicly accessible dataset regarding the circumstances of local breast cancer patients, tumor samples were collected from breast cancer patients. These patients had undergone surgery at Nil Ratan Sircar Medical College and Hospital (NRSMC&H) in Kolkata between 2019 and 2022, as well as at Burdwan Medical College and Hospital (BMCH), and were diagnosed with invasive ductal carcinoma (IDC). Patients who had received chemotherapy (CT) and radiation therapy (RT) were excluded from the study. A total of 41 patients were included, with histopathological confirmation of invasive ductal carcinoma (IDC) by a pathologist following the TILs working group guidelines (2014) [
27]. The clinicopathological data, such as tumor grades and the scoring of estrogen receptor (ER) and progesterone receptor (PR), as well as the HER-2 scoring status, were gathered from the case records of the respective hospitals. Samples from patients with incomplete molecular subtyping data from both hospitals were verified through immunohistochemistry (IHC) of ER, PR, and HER2 in the laboratory. Formalin-fixed paraffin-embedded samples were collected for immunohistochemistry and histopathological analysis. The expression of TIM-3 was confirmed by immunohistochemistry in all the samples from the patients. To initiate the immunohistochemistry processing, 4µm tissue sections were deparaffinized using xylene and subsequently rehydrated by passing through various concentrations of ethanol. Antigen retrieval was carried out by heating the samples in tris-buffer with EDTA (pH 9.0) for 20 minutes, followed by cooling in the same buffer. Endogenous peroxidase activity was inhibited using a peroxide solution. The tissue sections were then subjected to incubation with antibodies against ER (Leica, RTU-ER-6F11), PR (Leica-Novocastra RTU-PGR-312), HER2 (Cell Marque# 237R-18), and TIM-3(Invitrogen, PA5-115342) for one hour in a humid chamber. Subsequently, the sections were incubated with an HRP-conjugated secondary antibody (ScyTek- ABZ008) for 30 minutes in the same humid chamber. The slides were developed using diaminobenzidine and counterstained with Hematoxylin. Positive and negative controls were employed to validate and confirm the protein expression observed in this staining. A pathologist analysed and confirmed all the IHC expressions. ER and TIM-3 immunoreactivity were scored semi-quantitatively, considering both the distribution and staining intensity. The scores for proportion (PS) and intensity (IS) were combined to calculate the Total score (TS). The TS ranges from 0 to 8, depending on the proportion and intensity of staining. A score of 0 indicates no expression, scores ranging from 1 to 5 are classified as faint or low expression, and scores from 6 to 8 indicate high expression.
Analysis of the role of TIM-3in the survival analysis of breast carcinoma patients
The survival rate of breast cancer patients in relation to TIM-3 expression levels was analysed using the UALCAN portal and the GEPIA web server. The overall survival of breast carcinoma patients was analysed, and plots were generated using GEPIA, with HAVCR-2 as the reference gene in the BRCA dataset. To analyse the subtype-specific correlation of TIM-3 in breast carcinoma patients, the UALCAN portal was utilized. Here, the data from the TCGA database was compiled using the breast invasive carcinoma dataset and HAVCR2 as a reference gene. Survival plots were generated for subtype-specific BRCA patients, considering the subtypes of HER2 positive, Luminal, and Triple negative (TNBC) patients.
Statistical Analysis
IBM SPSS Statistics version 26 and GraphPad Prism version 7.0 (GraphPad Software) software were used to conduct statistical analyses. The t-test was used to determine the statistical significance between groups with different levels of gene expression. To assess the clinicopathological characteristics between the Low and High lymphocyte infiltrating groups, Fisher’s exact test and the chi-squared test were performed for categorical variables. Kaplan-Meier curves were constructed in order to compare the survival rates among patients with varying levels of total immune cell infiltration. Spearman’s rank correlation test was performed to assess the correlation of the gene. A threshold of p<0.05 was set to determine statistical significance.
RESULTS
A significant higher expression of TIM-3mRNA is present in breast tumor tissue compared to normal tissue
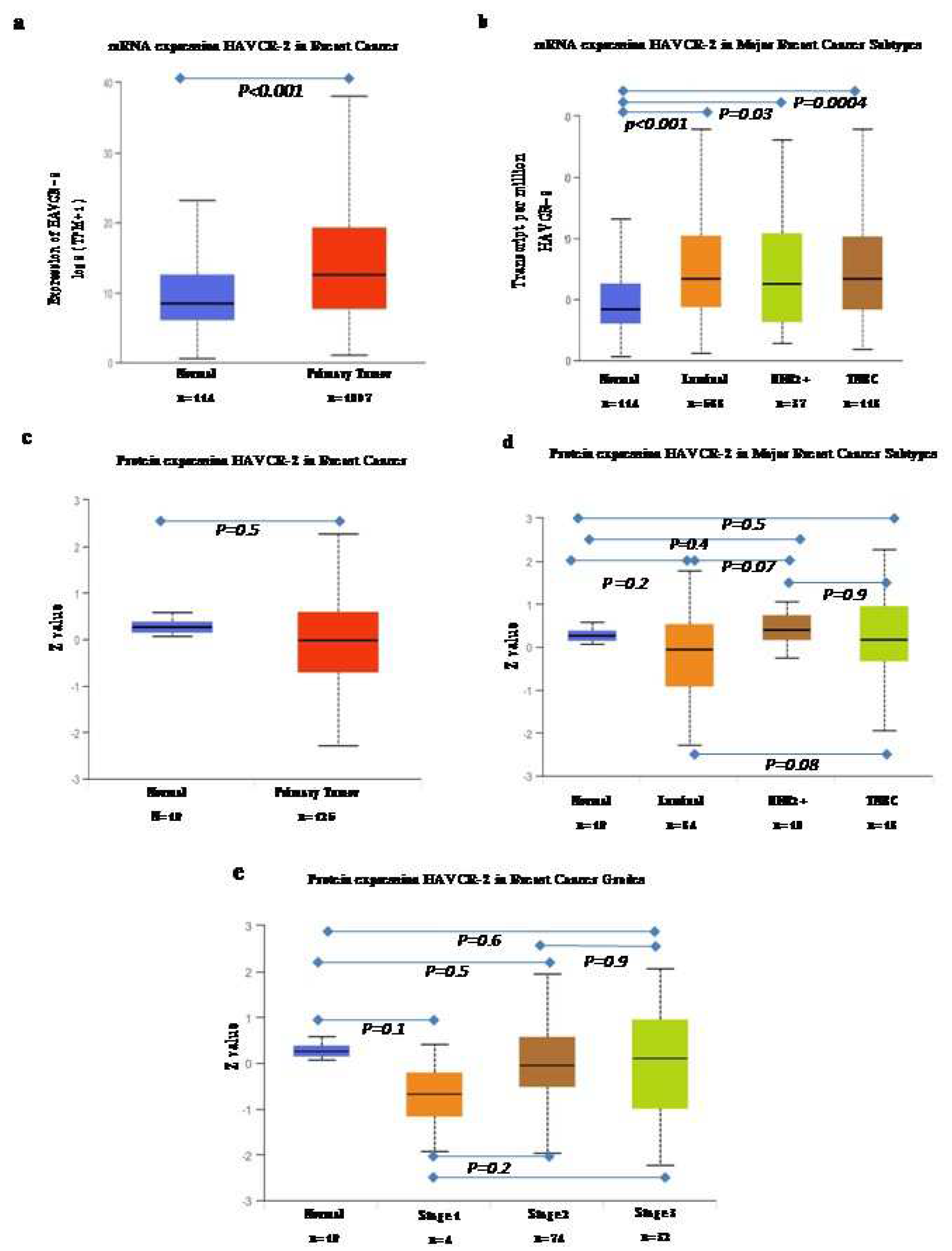
The UALCAN web server was used for the analysis of mRNA and proteomics data (
Figure 1). The analysis of mRNA data, comparing them to normal subjects, revealed significantly high expression of TIM-3 in all breast cancer subtypes compared to the normal samples (
Figure 1a). Among the subtypes, the luminal subtype exhibited the highest expression, followed by TNBC and the HER2 positive subtype. However, when we analyzed the proteomics data, we did not find any significant differences in overall pan breast cancer or its subtypes. However, what we did observe was a gradual increase in the expression of TIM3 (median value) with an increase in stage.
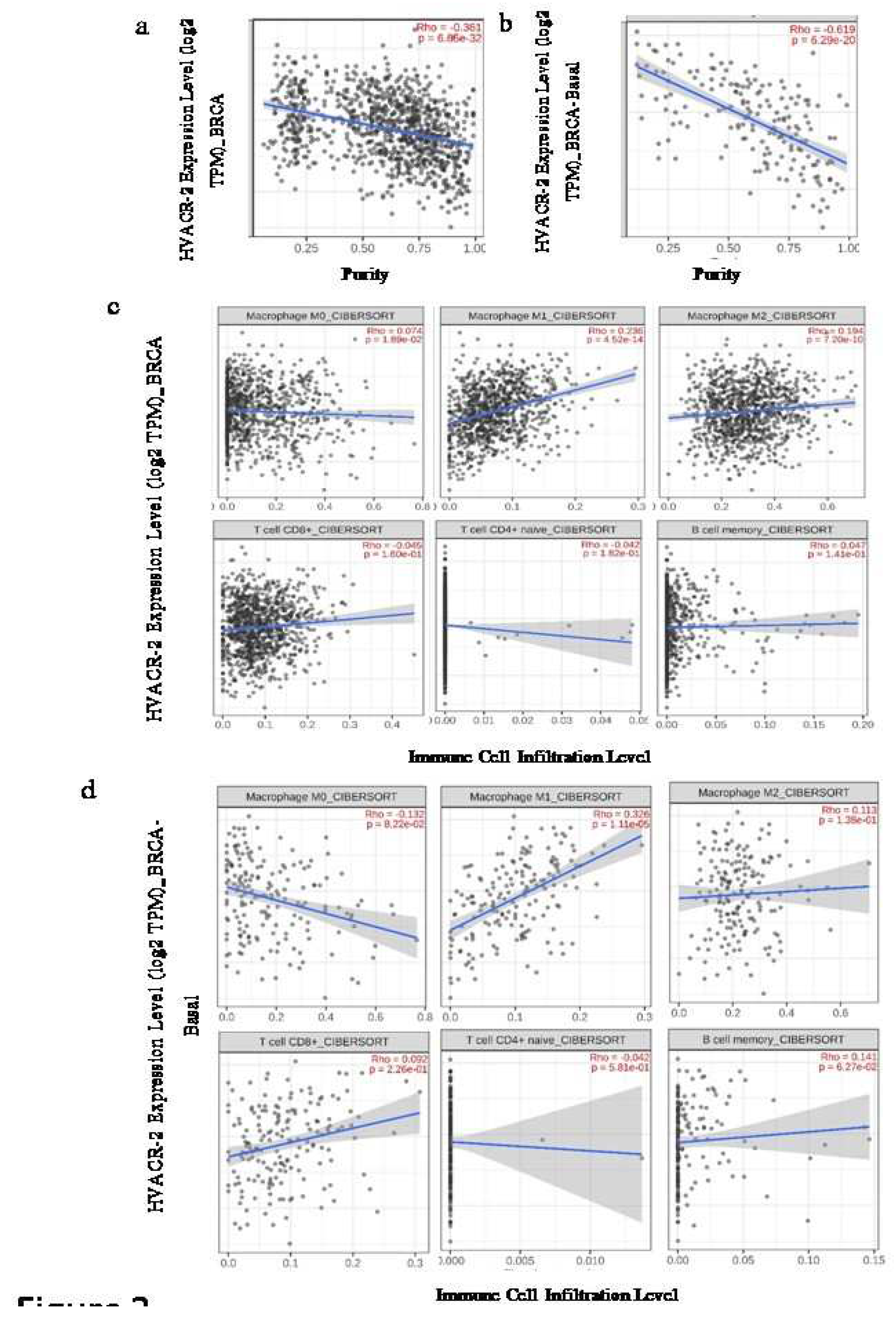
Expression of TIM-3 is correlated differently with the immune cell infiltrates
The Spearman’s rank correlation between the expression of TIM-3 and the relative abundance of various immune cells from bulk RNA sequencing data of the TCGA database was analysed using the CIBERSORT algorithm. In the Luminal A subtype, there was noticeable expression of TIM-3 that exhibited a negative correlation with CD8+ T cell infiltration. However, no significant association like this was observed in other subtypes. The infiltration of B-cells showed a significant negative correlation with the overall expression of TIM-3, except in BRCA-Basal. This correlation notably and significantly contributed to the HER-2, Luminal-A, and Luminal-B subgroups, as shown in
Table 1 (
Figure 2). The subtypes of macrophages (M0, M1, M2) showed varying rates of infiltration specific to each breast cancer subtype. On the other hand, the total BRCA sample (n=1100) demonstrated a positive correlation in all macrophage subtypes (M0, M1, M2). Nonetheless, the pattern alters when the samples are divided based on their breast cancer subtypes. Among all the groups, Luminal-A (n=568) exhibited a stable expression pattern and showed a positive correlation with TIM-3 expression in all subtypes of macrophages. However, in the Basal subtype (n=191), only M1 infiltration demonstrated a significant positive correlation with gene expression. In contrast, the HER2 positive group (n=82) exhibited a significant positive correlation between the expression of TIM-3 and the infiltration of M2 type macrophages. Finally, the Luminal-B (n=219) perceived a positive correlation with infiltration of the M0 subtype and a negative correlation with infiltration of the M2-subtype in regard of TIM-3 expression (
Table 1).
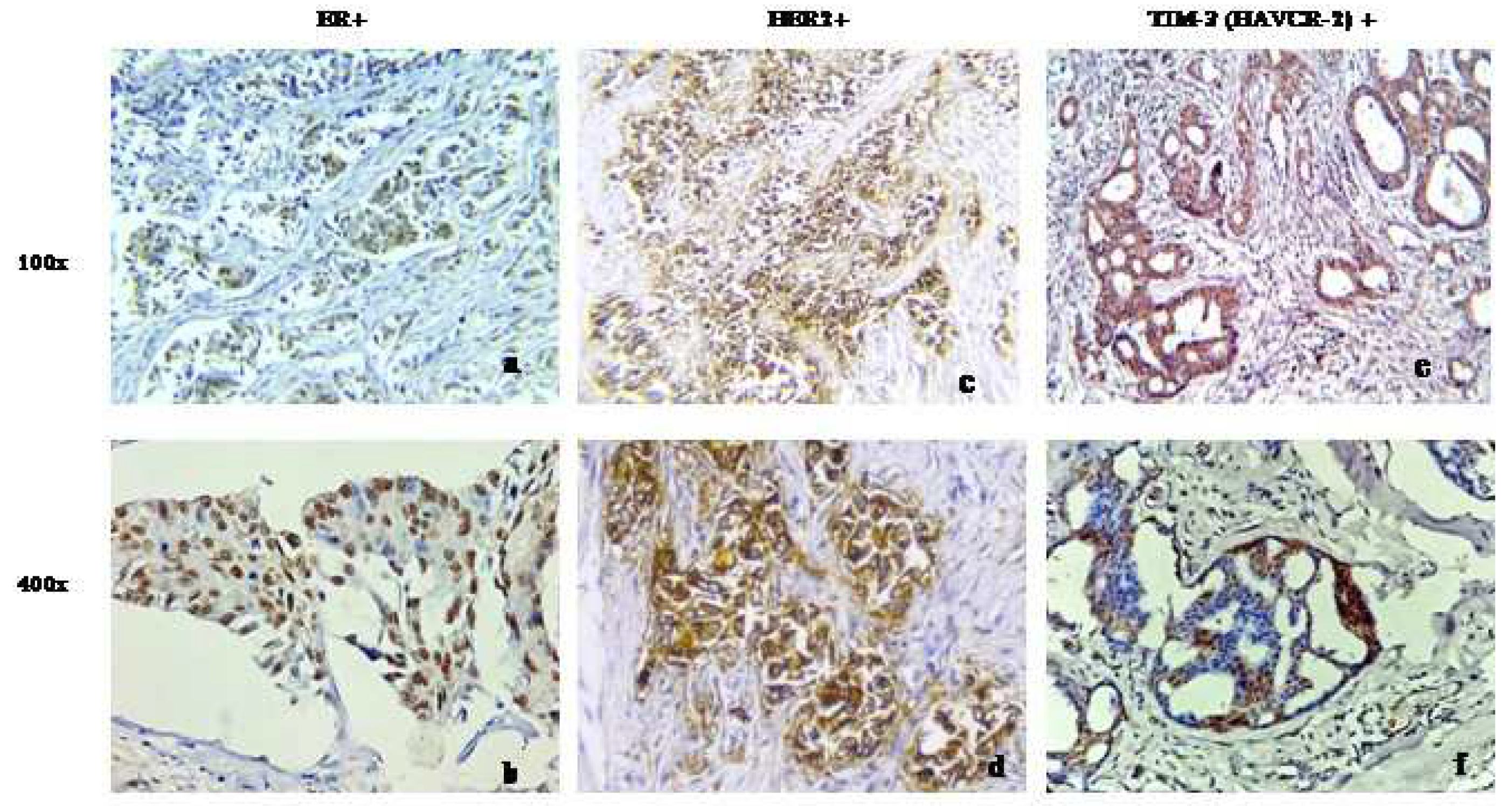
The immunohistochemical analysis of our patient cohort conducted in-house revealed an increased expression of TIM-3in patients with advanced grade
To validate the findings of the international cohort, we studied TIM-3 expression in our in-house breast tumor samples. Consistent with the aforementioned findings, our patient cohort exhibited a distinct nuclear and cytoplasmic expression of TIM-3 in the breast cancer cells of all samples (
Figure 3). Additionally, to our surprise, it was discovered that there was a significant correlation between TIM-3 expression and the tumor grade of the tested sample. In lower grades (Grade 1 and 2), a higher number of patients exhibited a higher expression of TIM-3, whereas in Grade 3, the number of patients increased in the lower expression group. However, no significant associations were found in other clinicopathological parameters that were considered in our primary breast tumor samples (
Table 2).
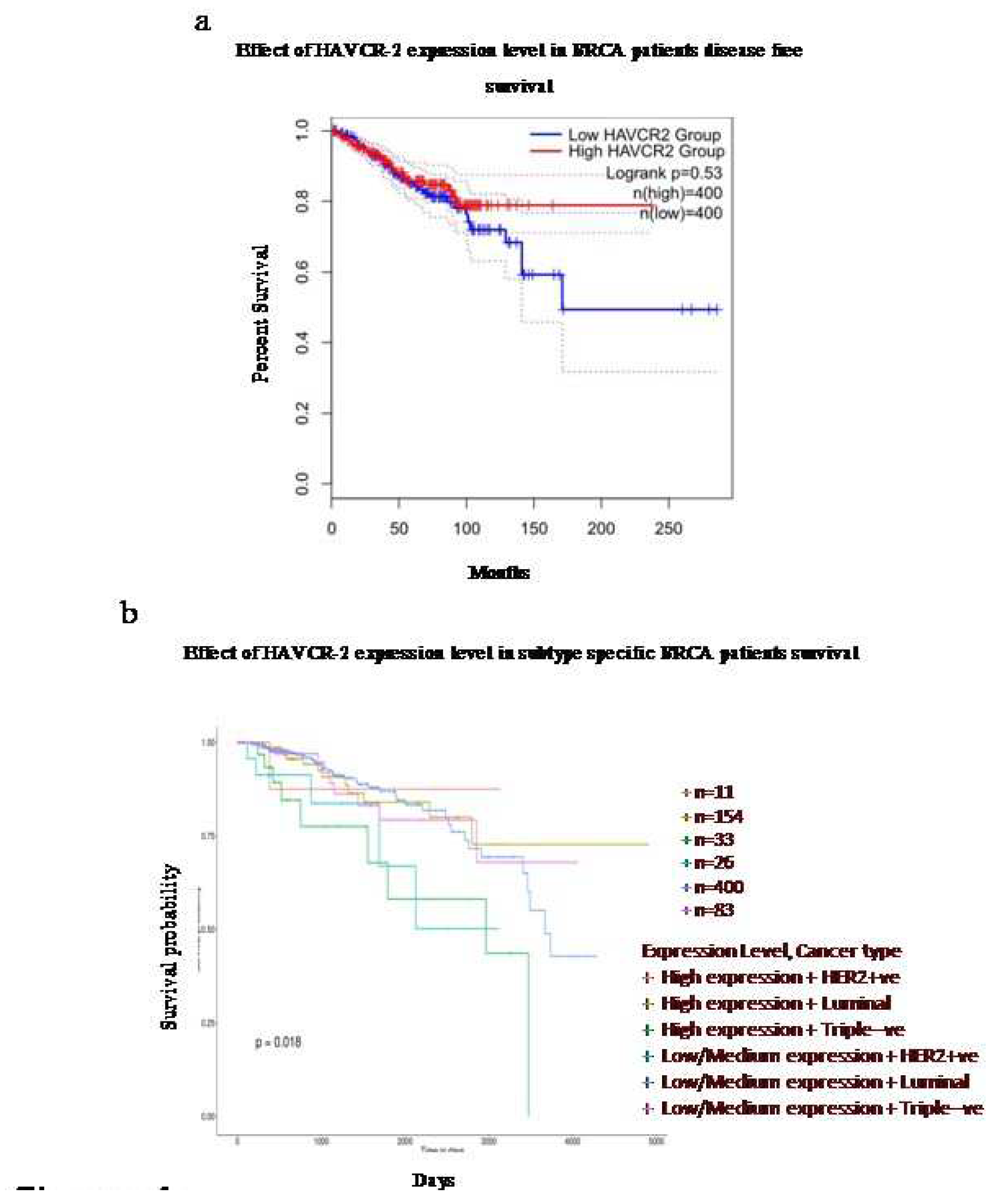
The expression of TIM-3is associated with the survival efficacy of breast cancer patients in a subtype-specific manner
The survival analysis using the Kaplan-Meier Plotter demonstrated a variable correlation depending on the breast cancer subtypes. There was an overall increase in disease-free survival with high expression of TIM3 compared to low expression (
Figure 4a). However, upon analysing different subclasses, it was further observed that the expression pattern of TIM-3 was significantly associated with patient survival in a subtype-specific manner. High expression of TIM-3 in HER-2 positive and Luminal subtypes indicated better survival effectiveness. On the other hand, in TNBC, high expression of TIM-3 predicted the lowest survival effectiveness.
DISCUSSION
Previous evidences suggest that elevated levels of TIM-3 play a significant role in the immune evasion of malignant tumor cells and enhance their invasiveness. In the present work, a significantly high mRNA expression of TIM-3was observed in primary breast tumor tissue, which is highly specific to the subtype of breast cancer.In our study, we discovered that the infiltration of major immune cells such as macrophages (M0, M1, M2), T-cells, and B-cells is negatively correlated with the expression of TIM-3. This finding suggests that as the TIM-3expression increases, the number of immune cells decreases, favouring an immune suppressive tumor microenvironment. The investigation conducted by Cong et al. in 2020revealed that overexpression of TIM-3contributed to increased aggressiveness in breast cancer cells. They demonstrated that TIM-3facilitated resistance to paclitaxel by activating the NF-κB/STAT3 signalling pathway. Additionally, previous studies have reported that TIM-3 upregulates the expression of cyclin D1 (CCND1), matrix metalloproteinase-1 (MMP1), TWIST, and vascular endothelial growth factor (VEGF), while simultaneously downregulating E-cadherin [
22]. Earlier, Cheng et al. demonstrated a correlation between the expression of TIM-3 and the inhibition of breast cancer cell apoptosis, suggesting a potential role in the recurrence and metastasis of breast cancer [
22]. Surprisingly, proteomics data did not show any remarkable increase in the tumor tissue compared to normal tissue. This may be due to post transcriptional modification in tumor tissue and greater involvement of molecular interplay in immune modulation of the TIM3 signal system and gene regulation.
Additionally, the analysis of BRCA data and our immunohistochemical study of breast tumor tissue samples revealed a high to moderate level of TIM-3 expression, especially in the ductal epithelial cells. Moreover, a significant correlation between TIM-3 expression and tumor grade was observed. Therefore, based on this finding, it can be inferred that higher TIM-3 expression is associated with a higher tumor grade.Consequently, this discovery can contribute to our understanding of cancer progression and its strong correlation with TIM-3 expression.
In renal cell carcinoma (RCC), the expression of TIM-3 in cancerous tissues is associated with an increased infiltration of tumor-associated macrophages. Therefore, it serves as a valuable biomarker for predicting clinical prognosis [
23]. According to Giraldo
et al. patients with high infiltration of exhausted TILs and Tregs (regulatory T cells) with higher TIM-3expression level presented a poor prognostic outcome in clear cell renal cell carcinoma [
24]. Therefore, we analysed the survivability in relation to the expression of TIM-3in breast cancer molecular subtypes. On the contrary, the survival data revealed that a higher expression of TIM-3 is associated with higher disease-free survival in pan breast cancer as well as higher survival rate in HER-2 positive and Luminal subtypes of breast cancer.
Conversely, in our study, it was found that lower or middle expression of TIM-3 in TNBC leads to better survival, according to the BRCA transcriptome data. This could be attributed to the relatively mild expression of TIM3 in TNBC subtypes, which may result from a statistical fallacy caused by the small sample size of TNBC in the BRCA data and high expression of TIM3.According to Yoshikawa et al. (2022), their study using IHC demonstrated that the presence of double positivity for galectin-9 and TIM-3 correlated with a favorable prognosis in TNBC [
25]. To date, no targeted treatments have been developed for TNBCs.There have been very few studies showing promising outcomes in other subtypes of breast cancer. In the past, attempts were made to use pembrolizumab (a monoclonal antibody against PD-1) or atezolizumab (a monoclonal antibody against PD-L1) in combination with chemotherapy for TNBC treatment. However, the overall success rate among TNBC patients was low. Only the subtypes that tested positive for PD-L1 showed an extension in progression-free survival [
26,
27]. A significant finding of the current study is that higher expression of TIM-3is associated with lower overall survival in TNBC, while improved overall survival is observed in HER-2 positive and Luminal subtypes. Moreover, a notable strength of this study is its ability to highlight the negative correlation between major immune cell infiltrates and TIM-3expression levels in breast cancer patients. TIM-3is currently considered an emerging area of immunotherapy, and TIM-3antibodies are undergoing phase 1/2 clinical trials in advanced solid tumors and lymphomas (NCT03489343) [
28,
29]. Our findings support the notion that TIM-3 could serve as a promising target in forthcoming immune checkpoint inhibitor-based therapies for TNBC patients, as it exhibits a harmful impact in this specific subgroup. Conversely, as TIM-3 expression is linked to decreased immune cell infiltration, inhibiting its expression may bolster the overall immune system and heighten surveillance. Therefore, with the advancement of TIM-3-mediated immunotherapy, the results of this study can be utilized to target TIM-3 for the treatment of breast cancer.
In this present work, we have demonstrated the dubious role of TIM 3 breast cancer, which is firmly opposed with other cancer. This study indicates thatexpression of TIM3 can supress tumour progression, which is indicative of a more favourable prognosis. Therefore, TIM3 plays a significant role in the progression of breast cancer. However, the current understanding of TIM3-mediated immune suppression is questionable. In the breast cancer TIM3-GAL9 system, instead of suppression, it may invoke an immune effect in the TME through an alternative pathway. Therefore, further study is crucial to understand how TIM3 controls breast cancer progression. Although the immune system usually has the capability to recognize and eliminate tumor cells, the TME disrupts this process by creating an immunosuppressive microenvironment.This impairment occurs through the manipulation of immune checkpoint modulators, which affects the function of immune cells. It has been reported that these immune checkpoints inhibit or suppress anti-tumor immune responses, making targeting immune checkpoints a modern option for cancer therapy. Therefore, targeting TIM3 with an agonist can enhance the treatment outcome.
Author Contributions
AG: BD, and BMcollected the data and perform the study and contributed in manuscript writing. AB4 and MPK assisted with patient consent and archival tissue block collection, and performed pathological interpretation. AB1* conceived and designed and supervise study, interpreted the data, edited and finalizemanuscript.
Funding
DBT, GoI [Grant No. BT/PR20699/MED/30/1748/2016] Intramural finding of National Institute of Biomedical genomics, Kalyani, W.B. [ Code no. 60122].
Ethics Approval and Consent to Participate
This study was approved by Institutional Ethics Committee Nilratan Sircar Medical College & Hospital, Kolkata (NRSMC/IEC/97/2022) and Institutional Clinical Ethics Committee, The University of Burdwan, Burdwan ( EC/BU/2021/8 and 9). Archival formalin fixed tissue blocks of breast cancer patients were collected from Nilratan Sircar Medical College & Hospital and Burdwan Medical College and Hospital with appropriate consent.
Consent for Publication
Manuscript does not contain any personal or medical information about an identifiable and has been sufficiently anonymised.
Availability of Data and Material
Data are available on reasonable request
Acknowledgements
Authors pledge to acknowledge the support of Nilratan Sircar Medical College & Hospital, Kolkata and Burdwan Medical College and Hospital, Burdwan. The authors wish to acknowledge Burdwan University authorities and Director, NIBMG for providing the necessary facilities and funds. Authors are also thankful to the patients included in the study. AG expresses gratitude to University Grants Commission (UGC), BD acknowledges Council of Scientific and Industrial Research(CSIR) and BM concedes Department of Biotechnology (DBT) for providing fellowships.
Competing interests
Authors confirm, no competing interest in the present study.
References
- WHO report: https://www.who.int/news-room/fact-sheets/detail/breast-cancer (accessed on 30-12-22).
- Karen S Johnson, Emily F Conant, Mary Scott Soo, Molecular Subtypes of Breast Cancer: A Review for Breast Radiologists, Journal of Breast Imaging, Volume 3, Issue 1, January/February 2021, 12–24. [CrossRef]
- Yin L, Duan JJ, Bian XW, Yu SC. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020;22(1):61. Published 2020 Jun 9. [CrossRef]
- Goff SL, Danforth DN. The Role of Immune Cells in Breast Tissue and Immunotherapy for the Treatment of Breast Cancer. Clin Breast Cancer 2021; 21:e63-e73. [CrossRef]
- Azizi E, Carr AJ, Plitas G, et al. Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell 2018; 174. [CrossRef]
- Ruffell B, Au A, Rugo HS, et al. Leukocyte composition of human breast cancer. PNAS 2012; 109: 2796–2801. [CrossRef]
- Yang M, Li Z, Ren M, et al. Stromal Infiltration of Tumor-Associated Macrophages Conferring Poor Prognosis of Patients with Basal-Like Breast Carcinoma. J Cancer2018;9:2308-2316. [CrossRef]
- Alemohammad H, Najafzadeh B, Asadzadeh Z, et al. The importance of immune checkpoints in immune monitoring: A future paradigm shift in the treatment of cancer. Biomed Pharmacother. 2022;146:112516. [CrossRef]
- Rupp T, Genest L, Babin D, et al. Anti-CTLA-4 and anti-PD-1 immunotherapies repress tumor progression in preclinical breast and colon model with independent regulatory T cells response. Transl Oncol. 2022;20:101405. [CrossRef]
- Mittendorf EA, Zhang H, Barrios CH, et al. Neoadjuvant atezolizumab in combination with sequential nab-paclitaxel and anthracycline-based chemotherapy versus placebo and chemotherapy in patients with early-stage triple-negative breast cancer (IMpassion031): a randomised, double-blind, phase 3 trial. Lancet. 2020;396:1090-1100. [CrossRef]
- Schmid P, Adams S, Rugo HS et al. IMpassion130 Trial Investigators. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N Engl J Med 2018; 379:2108-2121. [CrossRef]
- Cortes J, Cescon DW, Rugo HS, et al. Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for previously untreated locally recurrent inoperable or metastatic triple-negative breast cancer (KEYNOTE-355): a randomised, placebo-controlled, double-blind, phase 3 clinical trial. Lancet2020;396:1817-1828. [CrossRef]
- Venur VA, Ahluwalia MS. Novel therapeutic agents in the management of brain metastases. CurrOpin Oncol 2017; 29:395-399. [CrossRef]
- Bedognetti D, Maccalli C, Bader SB, Marincola FM, Seliger B. Checkpoint Inhibitors and Their Application in Breast Cancer. Breast Care (Basel). 2016;11(2):108-115. [CrossRef]
- He Y, Cao J, Zhao C, et al. TIM-3, a promising target for cancer immunotherapy. Onco Targets Ther. 2018;11:7005-7009.
- Harding JJ, Moreno V, Bang YJ, et al. Blocking TIM-3 in Treatment-refractory Advanced Solid Tumors: A Phase Ia/b Study of LY3321367 with or without an Anti-PD-L1 Antibody. Clin Cancer Res. 2021;27(8):2168-2178. [CrossRef]
- Tian T, Li Z. Targeting TIM-3 in Cancer with Resistance to PD-1/PD-L1 Blockade. Front Oncol. 2021;11:731175. [CrossRef]
- Cabioglu, N., Onder, S., Oner, G. et al. TIM3 expression on TILs is associated with poor response to neoadjuvant chemotherapy in patients with locally advanced triple-negative breast cancer. BMC Cancer. 2021;21, 357. [CrossRef]
- Tang, Z. et al. (2017)GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res, 10.1093/nar/gkx247. [CrossRef]
- Chandrashekar DS, Karthikeyan SK, Korla PK, Patel H, Shovon AR, Athar M, Netto GJ, Qin ZS, Kumar S, Manne U, Creighton CJ, Varambally S. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia. 2022; 25:18-27. [CrossRef]
- Taiwen Li,Jingxin Fu, Zexian Zeng, David Cohen, Jing Li, Qianming Chen, Bo Li, and X. Shirley Liu. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Research 2020 [DOI] [PubMed]. [CrossRef]
- Cong Y, Cui Y, Zhu S,et al.TIM-3 promotes cell aggressiveness and paclitaxel resistance through NF-κB/STAT3 signalling pathway in breast cancer cells. Chin J Cancer Res. 2020; 32:564-579. [CrossRef]
- Komohara Y, Morita T, Annan DA, et al. The Coordinated Actions of TIM-3 on Cancer and Myeloid Cells in the Regulation of Tumorigenicity and Clinical Prognosis in Clear Cell Renal Cell Carcinomas. Cancer Immunol Res. 2015; 3:999–1007. [CrossRef]
- Giraldo NA, Becht E, Vano Y, et al. Tumor-Infiltrating and Peripheral Blood T-cell Immunophenotypes Predict Early Relapse in Localized Clear Cell Renal Cell Carcinoma. Clin Cancer Res 2017; 23: 4416-4428. [CrossRef]
- Yoshikawa K, Ishida M, Yanai H, et al. Prognostic significance of the expression levels of T-cell immunoglobulin mucin-3 and its ligand galectin-9 for relapse-free survival in triple-negative breast cancer. Oncol Lett. 2022;23:197. [CrossRef]
- Cortes J, Cescon DW, Rugo HS, et al. Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for previously untreated locally recurrent inoperable or metastatic triple-negative breast cancer (KEYNOTE-355): a randomised, placebo-controlled, double-blind, phase 3 clinical trial. Lancet2020;396:1817-1828. [CrossRef]
- Venur VA, Ahluwalia MS. Novel therapeutic agents in the management of brain metastases. CurrOpin Oncol 2017; 29:395-399. [CrossRef]
- Lindsted T, Gad M, Grandal MV, Frolich C, Bhatia VK, Gjetting T, et al. Preclinical characterization of Sym023 a human anti-TIM3 antibody with a novel mechanism of action. Cancer Res 2018;78:5629. [CrossRef]
- Acharya N, Sabatos-Peyton C, Anderson AC. TIM-3 finds its place in the cancer immunotherapy landscape. J Immunother Cancer 2020;8:e00091. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).