Submitted:
26 January 2024
Posted:
26 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Discovery and Characterization of Proteins with Unknown Biocatalytic Functions
3. Discovery and Characterization of Unknown Metabolic Pathways
4. Discussion
5. Outlook
Funding
Conflicts of Interest
References
- Smoukov, S.K.; Seckbach, J.; Gordon, R. (Eds.) Conflicting Models for the Origin of Life. John Wiley & Sons, Inc., Hoboken, NJ 07030, and Scrivener Publishing LLC, Beverly, MA 01915, USA, 2023.
- Preiner, M.; Asche, S.; Becker, S.; Betts, H.C.; Boniface, A.; Camprubi, E.; Chandru, K.; Erastova, V.; Garg, S.G.; Khawaja, N.; Kostyrka, G.; Machné, R.; Moggioli, G.; Muchowska, K.B.; Neukirchen, S.; Peter, B.; Pichlhöfer, E.; Radványi, A.; Rossetto, D.; Salditt, A.; Schmelling, N.M.; Sousa, F.L.; Tria, F.D.K.; Vörös, D.; Xavier, J.C. The Future of Origin of Life Research: Bridging Decades-Old Divisions. Life 2020, 10, 20. [Google Scholar] [CrossRef]
- Oparin, A.I. 1957. The origin of life on the earth, 3rd ed.; Oliver & Boyd, Edinburgh & London, Country, UK, 1957.
- Vincent, L.; Colón-Santos, S.; Cleaves, H.J.; Baum, D.A.; Maurer, S.E. The prebiotic kitchen: A guide to composing prebiotic soup recipes to test origins of life hypotheses. Life, 2021, 11(11), 1221.
- Dodd, M.S.; Papineau, D.; Grenne, T.; Slack, J.F.; Rittner, M.; Pirajno, F.; O’Neil, J.; Little, C.T.S. Evidence for early life in Earth’s oldest hydrothermal vent precipitates. Nature 2017, 543, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Higgs, P.G. When Is a Reaction Network a Metabolism? Criteria for Simple Metabolisms That Support Growth and Division of Protocells. Life 2021, 11(9), 966. [Google Scholar] [CrossRef] [PubMed]
- Muchowska, K.B.; Varma, S.J.; Moran, J. Nonenzymatic Metabolic Reactions and Life’s Origins. Chem. Rev. 2020, 120, 15, 7708–7744. [Google Scholar] [CrossRef] [PubMed]
- Akbaria, A.; Palsson, B.O. Metabolic homeostasis and growth in abiotic cells. Proc. Natl. Acad. Sci. 2023, 120(19), e2300687120. [Google Scholar] [CrossRef] [PubMed]
- Aitken, H.R.M.; Wright, T.H.; Radakovic, A.; Szostak, J.W. Small-Molecule Organocatalysis Facilitates In Situ Nucleotide Activation and RNA Copying. J. Am. Chem. Soc, 2023; Articles ASAP. [Google Scholar] [CrossRef]
- Walsh, C.T.; Tang, Y. Natural Product Biosynthesis: Chemical Logic and Enzymatic Machinery, Royal Society of Chemistry, London, 2017.
- Shaffer, J.P.; Nothias, LF.; Thompson, L.R.; Sanders, J.G.; Salido, R.A.; Couvillion, S.P.; Brejnrod, A.D.; Lejzerowicz, F.; Haiminen, N.; Huang, S.; Lutz, H.L.; Zhu, Q.; Martino, C.; Morton, J.T.; Karthikeyan, S.; Nothias-Esposito, M.; Dührkop, K.; Böcker, S.; Kim, H.W.; Aksenov, A.A.; Bittremieux, W.; Minich, J.J.; Marotz, C.; Bryant, M.M.; Sanders, K.; Schwartz, T.; Humphrey, G.; Vásquez-Baeza, Y.; Tripathi, A.; Parida, L.; Carrieri, A.P.; Beck, K.L.; Das, P.; González, A.; McDonald, D.; Ladau, J.; Karst, S.M.; Albertsen, M.; Ackermann, G.; DeReus, J.; Thomas, T.; Petras, D.; Shade, A.; Stegen, J.; Song, S.J.; Metz, T.O.; Swafford, A.D.; Dorrestein, P.C.; Jansson, J.K.; Gilbert, J.A.; Knight, R.; Earth Microbiome Project 500 (EMP500) Consortium. Standardized multi-omics of Earth’s microbiomes reveals microbial and metabolite diversity. Nat. Microbiol. 2022, 7, 2128–2150. [Google Scholar] [CrossRef] [PubMed]
- Weng, J.K.; Lynch, J.H.; Matos, J.O.; Dudareva, N. Adaptive mechanisms of plant specialized metabolism connecting chemistry to function. Nat. Chem. Biol. 2021, 17 (10), 1037-1045. [CrossRef]
- Torres, J.P.; Eric, W. Schmidt, E.W. The biosynthetic diversity of the animal world. J. Biol. Chem 2019, 294(46), 17684–17692. [Google Scholar]
- Shendure, J.; Balasubramanian, S.; Church, G.M.; Gilbert, W.; Rogers, J.; Schloss, J.A.; Waterston, R.H. DNA sequencing at 40: past, present and future. Nature 2017, 550(7676), 345–353. [Google Scholar] [CrossRef] [PubMed]
- Stark, R.; Grzelak, M.; Hadfield, J. RNA sequencing: the teenage years. Nat. Rev. Genet. 2019, 20, 631–656. [Google Scholar] [CrossRef]
- Hoose, A.; Vellacott, R.; Storch, M.; Freemont, P.S.; Ryadnov, M.G. DNA synthesis technologies to close the gene writing gap. Nat. Rev. Chem. 2023, 7, 144–161. [Google Scholar] [CrossRef]
- Nelissen, F.H.T.; Leunissen, E.H.P.; van de Laar, L.; Tessari, M.; Heus, H.A.; Wijmenga, S.S. Fast production of homogeneous recombinant RNA—towards large-scale production of RNA. Nucleic Acids Res. 2012, 40(13), e102. [Google Scholar] [CrossRef]
- Flamme, M.; McKenzie, L.K.; Sarac, I.; Hollenstein, M. Chemical methods for the modification of RNA. Methods 2019, 161, 64–82. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium, UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; Žídek, A.; Green, T.; Tunyasuvunakool, K.; Petersen, S.; Jumper, J.; Clancy, F.; Green, R.; Vora, A.; Lutfi, M.; Figurnov, M.; Cowie, A.; Hobbs, N.; Kohli, P.; Kleywegt, G.; Birney, E.; Hassabis, D.; Velankar, S. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50(D1), D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.C.; Hu, Z., Rachlin, J.; Anton, B.P.; Kasif, S.; Roberts, R.J.; Steffen, M. COMBREX-DB: an experiment centered database of protein function: knowledge, predictions and knowledge gaps. Nucleic Acids Res. 2016, 44, D330–D335. [CrossRef]
- Oberg, N.; Zallot, R.; Gerlt, J.A. 2023. EFI-EST, EFI-GNT, and EFI-CGFP: Enzyme Function Initiative (EFI) Web Resource for Genomic Enzymology Tools. J. Mol. Biol. 2023, 435(14), 168018. [Google Scholar] [CrossRef] [PubMed]
- Sévin, D.C.; Fuhrer, T.; Zamboni, N.; Sauer, U. Nontargeted in vitro metabolomics for high-throughput identification of novel enzymes in Escherichia coli. Nat. Methods 2017, 14(2), 187–194. [Google Scholar] [CrossRef] [PubMed]
- Davidi, D.; Noor, E.; Liebermeister, W.; Bar-Even, A.; Flamholz, A.; Tummler, K.; Barenholz, U.; Goldenfeld, M.; Shlomi, T.; Milo, R. Global characterization of in vivo enzyme catalytic rates and their correspondence to in vitro kcat measurements. Proc. Natl. Acad. Sci. 2016, 113(12), 3401–3406. [Google Scholar] [CrossRef]
- Crick, F. Central Dogma of Molecular Biology. Nature 1970, 227, 561–563. [Google Scholar] [CrossRef]
- Schreiber, S. L. Small molecules: The missing link in the central dogma. Nat. Chem. Biol. 2005, 1(2), 64–66. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 2009, 324(5930), 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- McKnight, S.L. Back to the future: molecular biology meets metabolism. In: Cold Spring Harbor symposia on quantitative biology 2011, 76, 403-411. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY 11724, USA.
- Stincone, A.; Prigione, A.; Cramer, T.; Wamelink, M.M.C.; Campbell, K.; Cheung, E.; Olin-Sandoval, V.; Grüning, N.M.; Krüger, A.; Alam, M.T.; Keller, M.A.; Breitenbach, M.; Brindle, K.M.; Rabinowitz, J.D.; Ralser, M. The return of metabolism: biochemistry and physiology of the pentose phosphate pathway. Biol. Rev. 2015, 90, 927–963. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, N.N.; Zhu, J.; Thompson, C.B. The hallmarks of cancer metabolism: Still emerging. Cell Metabolism 2022, 34(3), 355–377. [Google Scholar] [CrossRef] [PubMed]
- Kavita, K.; Breaker, R.R. Discovering riboswitches: the past and the future. Trends Biochem. Sci. 2023, 48(2), P119–141. [Google Scholar] [CrossRef] [PubMed]
- Breaker, R.R. The Biochemical Landscape of Riboswitch Ligands. Biochemistry 2022, 61, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Jacob, F.; Monod, J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961, 3(3), 318–356. [Google Scholar] [CrossRef]
- Rutledge, P.; Challis, G. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol, 2015, 13, 509–523. [Google Scholar] [CrossRef]
- Diether, M.; Nikolaev, Y.; Allain, F.H.; Sauer, U. Systematic mapping of protein-metabolite interactions in central metabolism of Escherichia coli. Mol. Syst. Biol. 2019, 15(8), e9008. [Google Scholar] [CrossRef]
- Medina-Carmona, E.; Gutierrez-Rus, L.I.; Manssour-Triedo, F.; Newton, M.S.; Gamiz-Arco, G.; Mota, A.J.; Reiné, P.; Cuerva, J.M.; Ortega-Muñoz, M.; Andrés-León, E.; Ortega-Roldan, J.L.; Seelig, B.; Ibarra-Molero, B.; Sanchez-Ruiz, J.M. Cell Survival Enabled by Leakage of a Labile Metabolic Intermediate. Mol. Biol. Evol. 2023, 40(3), msad032. [Google Scholar] [CrossRef]
- Noda-Garcia, L.; Liebermeister, W.; Tawfik, D.S. Metabolite–Enzyme Coevolution: From Single Enzymes to Metabolic Pathways and Networks. Annu. Rev. Biochem. 2018, 87, 187–216. [Google Scholar] [CrossRef] [PubMed]
- Bathe, U.; Leong, B.J.; McCarty, D.R.; Henry, C.S.; Abraham, P.E.; Wilson, M.A.; Hanson, A.D. The moderately (d) efficient enzyme: catalysis-related damage in vivo and its repair. Biochemistry 2021, 60(47), 3555–3565. [Google Scholar] [CrossRef] [PubMed]
- Hult, K.; Berglund, P. Enzyme promiscuity: mechanism and applications. Trends Biotechnol. 2007, 25(5), 231–238. [Google Scholar] [CrossRef] [PubMed]
- Linster, C.L.; Van Schaftingen, E.; Hanson; A.D. Metabolite damage and its repair or pre-emption. Nat. Chem. Biol. 2013, 9, 72–80. [CrossRef] [PubMed]
- Griffith, C.M.; Walvekar, A.S.; Linster, C.L. Approaches for completing metabolic networks through metabolite damage and repair discovery. Curr. Opin. Syst. Biol. 2021, 28, 100379. [Google Scholar] [CrossRef] [PubMed]
- Haas, D.; Thamm, A.M.; Sun, J.; Huang, L.; Sun, L.; Beaudoin, G.A.; Wise, K.S.; Lerma-Ortiz, C.; Bruner, S.D.; Breuer, M.; Luthey-Schulten, Z.; Lin, J.; Wilson, M.A.; Brown, G.; Yakunin, A.F.; Kurilyak, I.; Folz, J.; Fiehn, O.; Glass, J.I.; Hanson, A.D.; Henry, C.S.; de Crécy-Lagard; V. Metabolite damage and damage control in a minimal genome. Mbio 2022, 13(4), e01630-22. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Ramesh, V.; Locasale, J.W. The evolving metabolic landscape of chromatin biology and epigenetics. Nat. Rev. Genet. 2020, 21(12), 737–753. [Google Scholar] [CrossRef]
- Goga, A.; Stoffel, M. Therapeutic RNA-silencing oligonucleotides in metabolic diseases. Nat. Rev. Drug Discov. 2022, 21(6), 417–439. [Google Scholar] [CrossRef]
- Boccaletto, P.; Stefaniak, F.; Ray, A.; Cappannini, A.; Mukherjee, S.; Purta, E.; Kurkowska, M.; Shirvanizadeh, N.; Destefanis, E.; Groza, P.; Avşar, G.; Romitelli, A.; Pir, P.; Dassi, E.; Conticello, S.G.; Aguilo, F.; Bujnicki, J.M. MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic Acids Res. 2022, 50(D1), D231–D235. [Google Scholar] [CrossRef]
- Tarazona, O.A.; Pourquie, O. Exploring the influence of cell metabolism on cell fate through protein post-translational modifications. Dev. Cell 2020, 54(2), 282–292. [Google Scholar] [CrossRef]
- McDonald, A.G.; Tipton, K.F. Enzyme nomenclature and classification: The state of the art. FEBS J. 2023, 290(9), 2214–2231. [Google Scholar] [CrossRef] [PubMed]
- Alcántara, A.R.; Dominguez de Maria, P.; Littlechild, J.A.; Schürmann, M.; Sheldon, R.A.; Wohlgemuth, R. Biocatalysis as key to sustainable industrial chemistry. ChemSusChem 2022, 15(9), e202102709. [Google Scholar] [CrossRef] [PubMed]
- He, H., Bian, G., Herbst-Gervasoni, C.J., Mori, T., Shinsky, S.A., Hou, A., Mu, X., Huang, M., Cheng, S., Deng, Z.; Christianson, D.W.; Abe, I.; Liu, T. Discovery of the cryptic function of terpene cyclases as aromatic prenyltransferases. Nat. Commun. 2020, 11(1), 3958. [CrossRef]
- Walsh, C.T.; Moore, B.S. Enzymatic cascade reactions in biosynthesis. Angew. Chem. Int. Ed. 2019, 58(21), 6846–6879. [Google Scholar] [CrossRef]
- Hertweck, C. The biosynthetic logic of polyketide diversity. Angew. Chem. Int. Ed. 2009, 48, 4688–4716. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Du, L. Iterative polyketide biosynthesis by modular polyketide synthases in bacteria. Appl. Microbiol. Biotechnol. 2016, 100, 541–557. [Google Scholar] [CrossRef] [PubMed]
- Nivina, A.; Yuet, K.P.; Hsu, J.; Khosla, C. Evolution and diversity of assembly-line polyketide synthases: focus review. Chem. Rev. 2019, 119(24), 12524–12547. [Google Scholar] [CrossRef] [PubMed]
- Steffen, W.; Richardson, K.; Rockström, J.; Cornell, S.E.; Fetzer, I.; Bennett, E.M.; Biggs, R.; Carpenter, S.R.; De Vries, W.; De Wit, C.A.; Folke, C. Planetary boundaries: Guiding human development on a changing planet. Science 2015, 347(6223), 1259855. [Google Scholar] [CrossRef] [PubMed]
- Hoskisson, P.A.; Seipke, R.F. Cryptic or silent? The known unknowns, unknown knowns, and unknown unknowns of secondary metabolism. MBio 2020, 11(5), 10–1128. [Google Scholar] [CrossRef]
- Kim, G.B.; Kim, J.Y.; Lee, J.A.; Charles, J. Norsigian, C.J.; Palsson, B.O.; Lee, S.Y. Functional annotation of enzyme-encoding genes using deep learning with transformer layers. Nat. Commun. 2023, 14, 7370. [Google Scholar] [CrossRef]
- Chang, Y.C.; Hu, Z., Rachlin, J.; Anton, B.P.; Kasif, S.; Roberts, R.J.; Steffen, M. COMBREX-DB: an experiment centered database of protein function: knowledge, predictions and knowledge gaps. Nucleic Acids Res. 2016, 44(D1), D330-D335. [CrossRef]
- Price, M.N.; Wetmore, K.M.; Waters, R.J.; Callaghan, M.; Ray, J.; Liu, H.; Kuehl, J.V.; Melnyk, R.A.; Lamson, J.S.; Suh, Y.; Carlson, H.K.; Esquivel, Z.; Sadeeshkumar, H.; Chakraborty, R.; Zane, G.M.; Rubin, B.E.; Wall, J.D.; Visel, A.; Bristow, J.; Blow, M.J.; Arkin, A.P.; Deutschbauer, A.M. Mutant phenotypes for thousands of bacterial genes of unknown function. Nature 2018, 557(7706), 503–509. [Google Scholar] [CrossRef]
- Rhee, K.Y.; Jansen, R.S.; Grundner, C. Activity-based annotation: the emergence of systems biochemistry. Trends Biochem. Sci. 2022, 47(9), 785–794. [Google Scholar] [CrossRef] [PubMed]
- Swainston, N.; Baici, A.; Bakker, B.M.; Cornish-Bowden, A.; Fitzpatrick, P.F.; Halling, P.; Leyh, T.S.; O’Donovan, C.; Raushel, F.M.; Reschel, U.; Rohwer, J.M.; Schnell, S.; Schomburg, D.; Tipton, K.F.; Tsai, M.D.; Westerhoff, H.V.; Wittig, U.; Wohlgemuth, R.; Kettner, K. STRENDA DB: enabling the validation and sharing of enzyme kinetics data. FEBS J. 2018, 1–12. [Google Scholar] [CrossRef]
- Punta, M.; Coggill, P.C.; Eberhardt, R.Y.; Mistry, J.; Tate, J.; Boursnell, C.; Pang, N.; Forslund, K.; Ceric, G.; Clements, J.; Heger, A.; Holm, L.; Sonnhammer, E.L.; Eddy, S.R.; Bateman, A.; Finn, R.D. The Pfam protein families database. Nucleic Acids Res. 2012, 40, D290–301. [Google Scholar] [CrossRef] [PubMed]
- Trudeau, D.L.; Tawfik, D.S. Protein engineers turned evolutionists—the quest for the optimal starting point. Curr. Opin. Biotechnol. 2019, 60, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.; Milpetz, F.; Bork, P.; Ponting, C.P. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. 1998, 95(11), 5857–5864. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Penny Coggill, P.; Robert D. Finn, R.D. DUFs: families in search of function. Acta Cryst. 2010, F66, 1148–1152. [CrossRef]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Lázaro Pinto, B.; Salazar, G.A.; Bileschi, M.I.; Bork, P.; Bridge, A.; Colwell, L.; Gough, J.; Haft, D.H.; Letunić, I.; Marchler-Bauer, A.; Mi, H.; Natale, D.A.; Orengo, C.A.; Pandurangan, A.P.; Rivoire, C.; Sigrist, C.A.; Sillitoe, I.; Thanki, N.; Thomas, P.D.; Tosatto, S.C.E.; Wu, C.H.; Bateman, A. InterPro in 2022, Nucleic Acids Res. , 51(D1), Pages D418–D427. [CrossRef]
- Gerlt, J.A.; Allen, K.N.; Almo, S.C.; Armstrong, R.N.; Babbitt, P.C.; Cronan, J.E.; Dunaway-Mariano, D.; Imker, H.J.; Jacobson, M.P.; Minor, W.; Poulter, C.D. The enzyme function initiative. Biochemistry 2011, 50(46), 9950–9962. [Google Scholar] [CrossRef] [PubMed]
- Gerlt, J.A.; Bouvier, J.T.; Davidson, D.B.; Imker, H.J.; Sadkhin, B.; Slater, D.R.; Whalen, K.L. Enzyme function initiative-enzyme similarity tool (EFI-EST): a web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta 2015, 1854(8), 1019–1037. [Google Scholar] [CrossRef] [PubMed]
- Zallot, R.; Oberg, N.; Gerlt, J.A. The EFI Web Resource for Genomic Enzymology Tools: Leveraging Protein, Genome, and Metagenome Databases to Discover Novel Enzymes and Metabolic Pathways. Biochemistry 2019, 58(41), 4169–4182. [Google Scholar] [CrossRef] [PubMed]
- Oberg, N.; Zallot, R.; Gerlt, J.A. EFI-EST, EFI-GNT, and EFI-CGFP: Enzyme Function Initiative (EFI) Web Resource for Genomic Enzymology Tools. J. Mol. Biol. 2023, 435, 168018. [Google Scholar] [CrossRef]
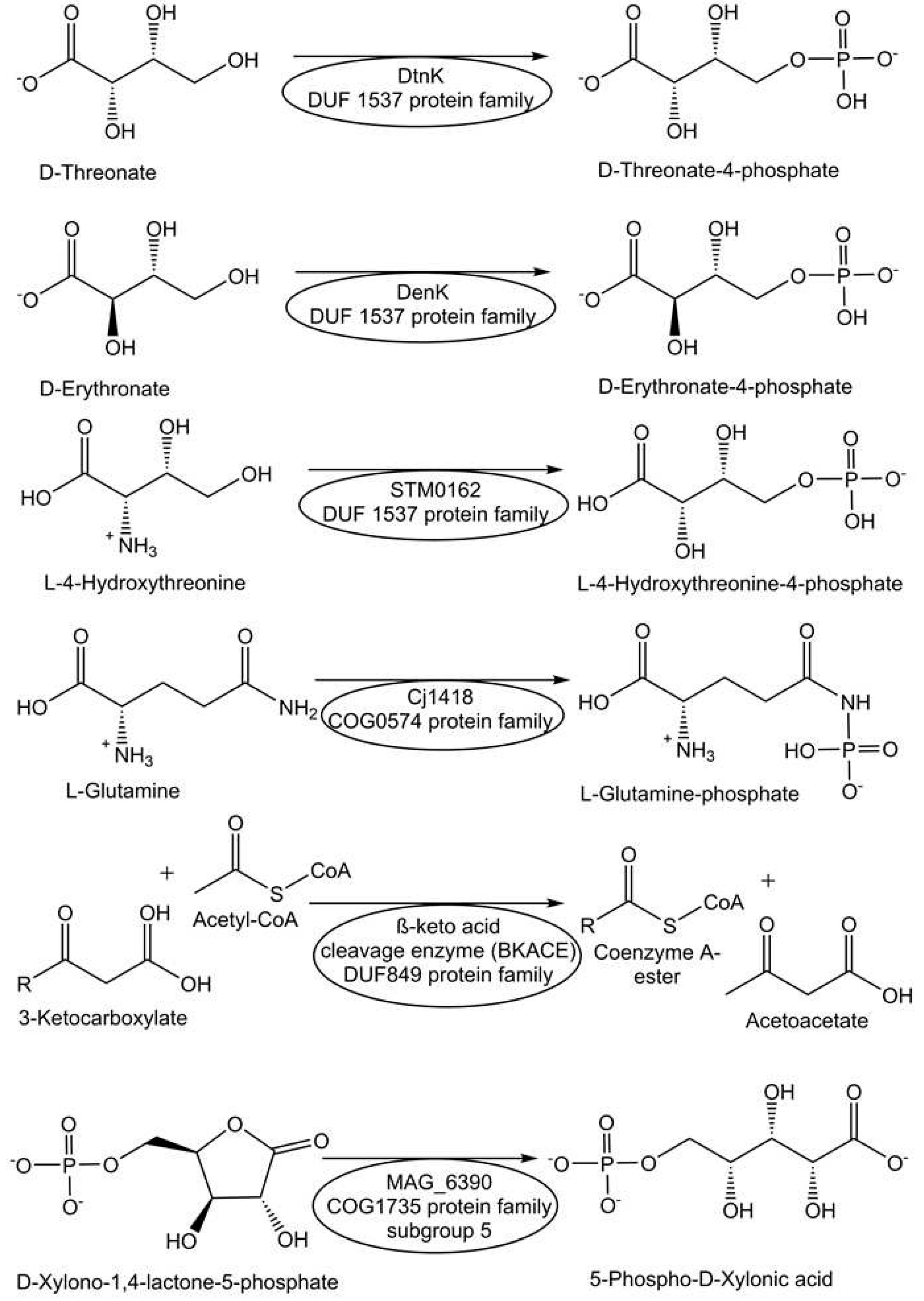
- Zhang, X.; Carter, M.S.; Vetting, M.W.; San Francisco, B.; Zhao, S.; Al-Obaidi, N.F.; Solbiati, J.O.; Thiaville, J.J.; de Crécy-Lagard, V.; Jacobson, M.P.; Almo, S.C. Assignment of function to a domain of unknown function: DUF1537 is a new kinase family in catabolic pathways for acid sugars. Proc. Natl. Acad. Sci. 2016, 113(29), E4161–E4169. [Google Scholar] [CrossRef]
- Taylor, Z.W.; Brown, H.A.; Narindoshvili, T.; Wenzel, C.Q.; Szymanski, C.M.; Holden, H.M.; Raushel, F.M. Discovery of a glutamine kinase required for the biosynthesis of the O-methyl phosphoramidate modifications found in the capsular polysaccharides of Campylobacter jejuni. J. Am. Chem. Soc. 2017, 139(28), 9463–9466. [Google Scholar] [CrossRef]
- Bastard, K.; Smith, A.A.T.; Vergne-Vaxelaire, C.; Perret, A.; Zaparucha, A.; De Melo-Minardi, R.; Mariage, A.; Boutard, M.; Debard, A.; Lechaplais, C.; Pelle, C. Revealing the hidden functional diversity of an enzyme family. Nat. Chem. Biol. 2014, 10(1), 42–49. [Google Scholar] [CrossRef] [PubMed]
- Korczynska, M.; Xiang, D.F.; Zhang, Z.; Xu, C.; Narindoshvili, T.; Kamat, S.S.; Williams, H.J.; Chang, S.S.; Kolb, P.; Hillerich, B.; Sauder, J.M.; Burley, S.K.; Almo, S.C.; Swaminathan, S.; Shoichet, B.K.; Raushel, F.M. Functional annotation and structural characterization of a novel lactonase hydrolyzing D-xylono-1, 4-lactone-5-phosphate and L-arabino-1, 4-lactone-5-phosphate. Biochemistry, 2014, 53(28), 4727-4738. [CrossRef]
- Malpartida, F.; Hopwood, D.A. Molecular cloning of the whole biosynthetic pathway of a Streptomyces antibiotic and its expression in a heterologous host. Nature 1984, 309(5967), 462–464. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Huo, Y.X. Using genome and transcriptome analysis to elucidate biosynthetic pathways. Curr. Opin. Biotechnol. 2022, 75, 102708. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.T.; Fischbach, M.A. Natural Products Version 2.0: Connecting Genes to Molecules. J. Am. Chem. Soc. 2010, 132, 2469–249392469. [Google Scholar] [CrossRef] [PubMed]
- MohammadiPeyhani, H.; Hafner, J.; Sveshnikova, A.; Viterbo, V.; Hatzimanikatis, V. Expanding biochemical knowledge and illuminating metabolic dark matter with ATLASx. Nat. Commun. 2022, 13, 1560. [Google Scholar] [CrossRef] [PubMed]
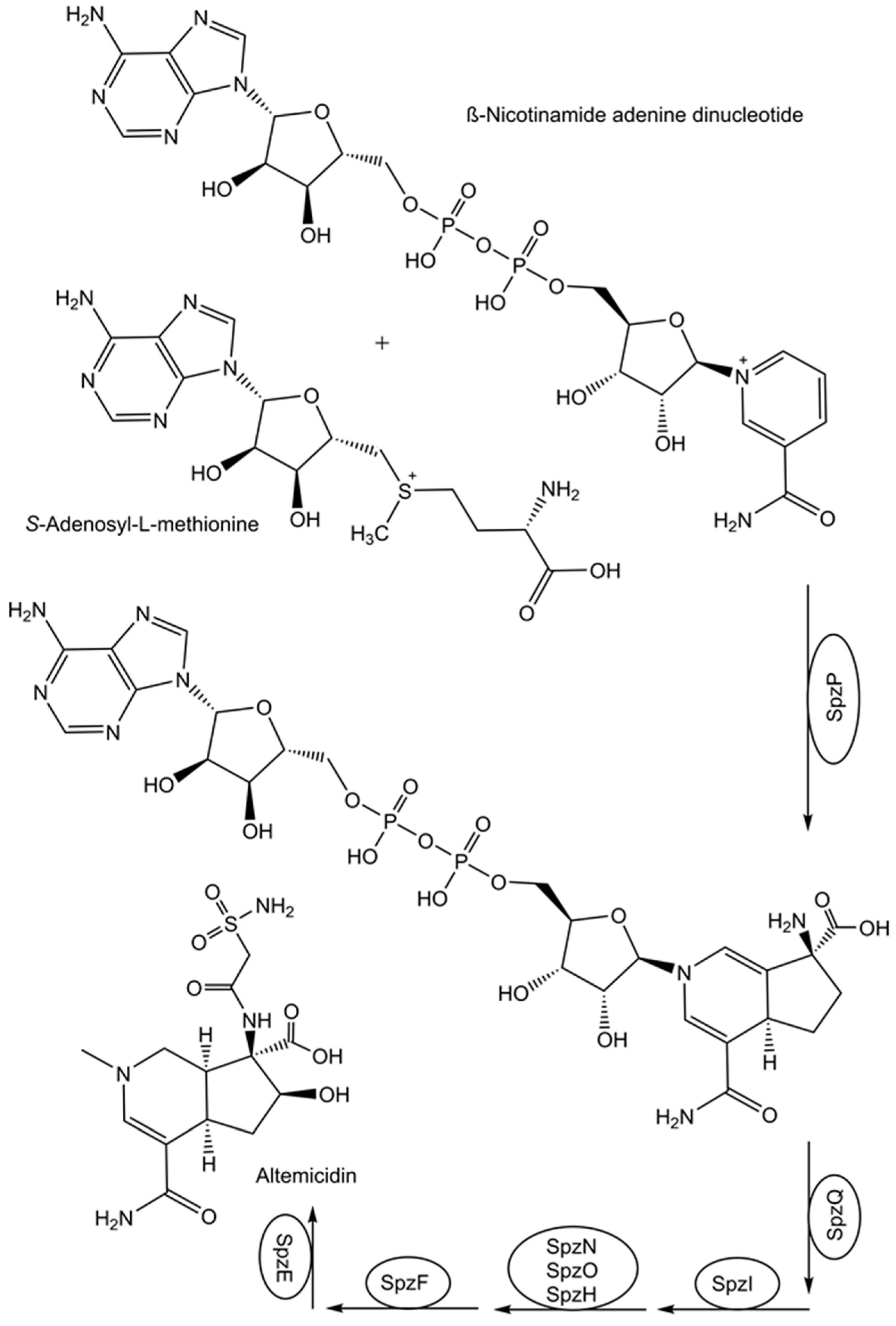
- Barra, L.; Awakawa, T.; Shirai, K.; Hu, Z.; Bashiri, G.; Abe, I. β-NAD as a building block in natural product biosynthesis. Nature 2021, 600(7890), 754–758. [Google Scholar] [CrossRef] [PubMed]
- Caputi, L.; Franke, J.; Farrow, S.C.; Chung, K.; Payne, R.M.; Nguyen, T.D.; Dang, T.T.T.; Soares Teto Carqueijeiro, I.; Koudounas, K.; Dugé de Bernonville, T.; Ameyaw, B. , Jones, D.M.; Curcino Vieira, I.J.; Courdavault, V.; O’Connor, S.E. Missing enzymes in the biosynthesis of the anticancer drug vinblastine in Madagascar periwinkle. Science 2018, 360(6394), 1235-1239.
- Qu, Y.; Easson, M.E.A.M.; Simionescu, R.; Hajicek, J.; Thamm, A.M.K.; Salim, V.; De Luca, V. Solution of the multistep pathway for assembly of corynanthean, strychnos, iboga, and aspidosperma monoterpenoid indole alkaloids from 19E-geissoschizine. Proc. Natl. Acad. Sci. 2018, 115(12), 3180–3185. [Google Scholar] [CrossRef]
- Qu, Y.; Easson, M.L.; Froese, J.; Simionescu, R.; Hudlicky, T.; De Luca, V. Completion of the seven-step pathway from taber-sonine to the anticancer drug precursor vindoline and its assembly in yeast. Proc. Natl. Acad. Sci. 2015, 112(19), 6224–6229. [Google Scholar] [CrossRef]
- Qu, Y.; Safonova, O.; De Luca, V. Completion of the canonical pathway for assembly of anticancer drugs vincristine/vinblastine in Catharanthus roseus. Plant J. 2019, 97(2), 257–266. [Google Scholar] [CrossRef]
- Zhang, J.; Hansen, L.G.; Gudich, O.; Viehrig, K.; Lassen, L.M.M.; Schrübbers, L.; Adhikari, K.B.; Rubaszka, P.; Carrasquer-Alvarez, E.; Chen, L.; D’Ambrosio, V.; Lehka, B.; Haidar, A.K.; Nallapareddy, S.; Giannakou, K.; Laloux, M.; Arsovska, D.; Jørgensen, M.A.K.; Chan, L.J.G.; Kristensen, M.; Christensen, H.B.; Sudarsan, S.; Stander, E.A.; Baidoo, E.; Petzold, C.J.; Wulff, T.; O’Connor, S.E.; Courdavault, V.; Jensen; M.K.; Keasling, J.D. A microbial supply chain for production of the anti-cancer drug vinblastine. Nature 2022, 609, 341–347. [CrossRef]
- Nunoura, T.; Chikaraishi, Y.; Izaki, R.; Suwa, T.; Sato, T.; Harada, T.; Mori, K.; Kato, Y.; Miyazaki, M.; Shimamura, S.; Yanagawa, K.; Shuto, A.; Ohkouchi, N.; Fujita, N.; Takaki, Y.; Atomi, H.; Takai, K. A primordial and reversible TCA cycle in a facultatively chemolithoautotrophic thermophile. Science 2018, 359, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Pascal Andreu, V.; Augustijn, H.E.; Chen, L.; Zhernakova, A.; Fu, J.; Fischbach, M.A.; Dodd, D.; Medema, M.H. gutSMASH predicts specialized primary metabolic pathways from the human gut microbiota. Nat. Biotechnol. 2023, 41, 1416–1423. [Google Scholar] [CrossRef] [PubMed]
- Shimosaka, T.; Makarova, K.S.; Koonin, E.V.; Atomi, H. Identification of dephospho-coenzyme A (dephospho-CoA) kinase in Thermococcus kodakarensis and elucidation of the entire CoA biosynthesis pathway in Archaea. Mbio 2019, 10(4), 10–1128. [Google Scholar] [CrossRef]
- Medema, M.H.; de Rond, T.; Moore, B.S. Mining genomes to illuminate the specialized chemistry of life. Nat. Rev. Genet. 2021, 22(9), 553–571. [Google Scholar] [CrossRef] [PubMed]
- Robey, M.T.; Caesar, L.K.; Drott, M.T.; Keller, N.P.; Kelleher, N.L. An interpreted atlas of biosynthetic gene clusters from 1,000 fungal genomes. Proc. Natl. Acad. Sci. 2021, 118(19), e2020230118. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Medema, M.H.; Weber, T. The antiSMASH database version 4: additional genomes and BGCs, new sequence-based searches and more. Nucleic Acids Res. 2024, 52, D586–D589. [Google Scholar] [CrossRef] [PubMed]
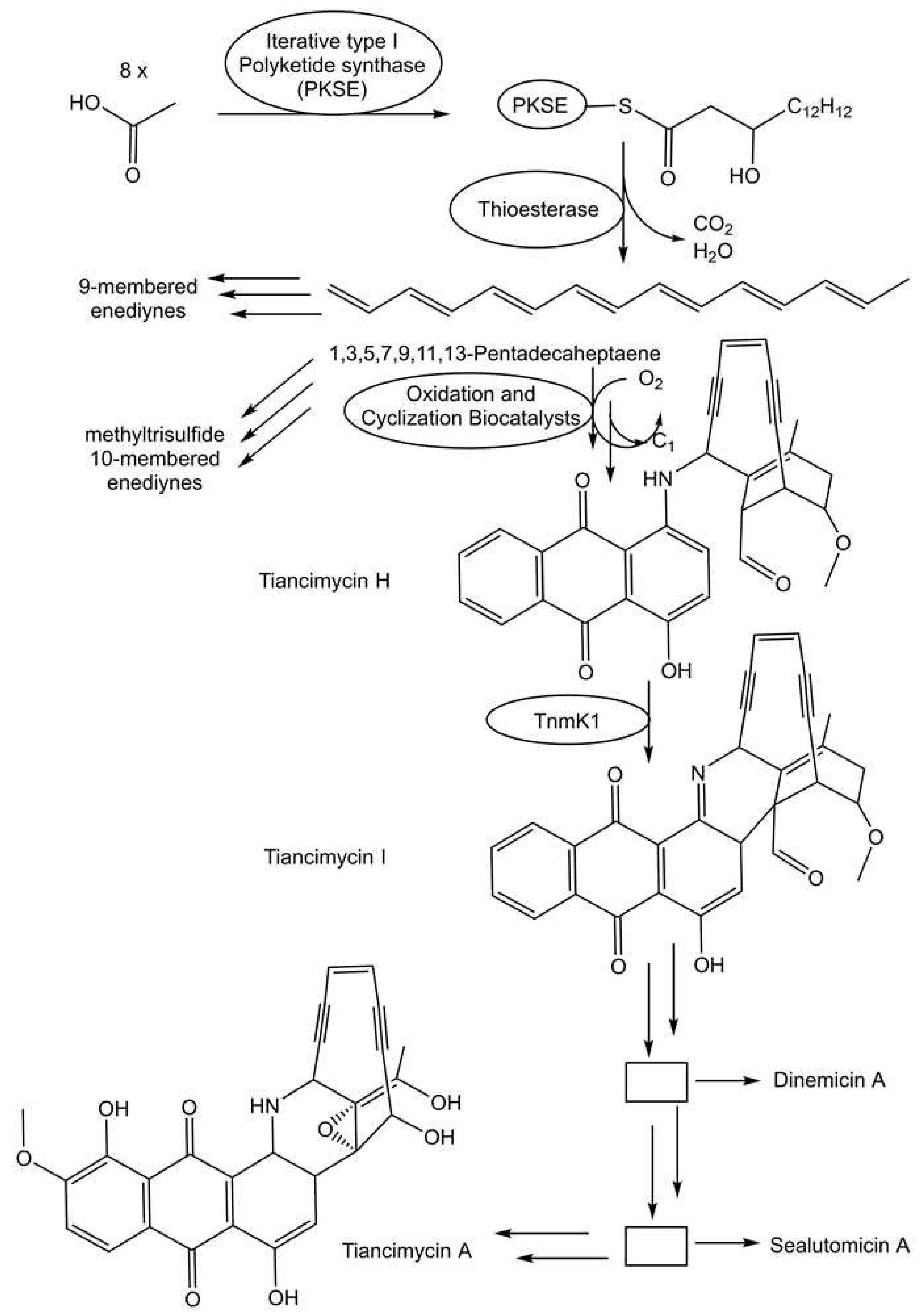
- Bhardwaj, M.; Cui, Z.; Hankore, E.D.; Moonschi, F.H.; Esfahani, H.S.; Kalkreuter, E.; Gui, C.; Yang, D.; Phillips Jr., G. N.; Thorson, J.S.; Shen, B.; Van Lanen, S.G. A discrete intermediate for the biosynthesis of both the enediyne core and the anthraquinone moiety of enediyne natural products. Proc. Natl. Acad. Sci. 2023, 120(9), e2220468120. [Google Scholar] [CrossRef]
- Scherlach, K.; Hertweck, C. Mining and unearthing hidden biosynthetic potential. Nat. Commun. 2021, 12, 3864. [Google Scholar] [CrossRef]
- Terlouw, B.R.; Blin, K.; Navarro-Muñoz, J.C.; Avalon, N.E.; Chevrette, M.G.; Egbert, S.; Lee, S.; Meijer, D.; Recchia, M.J.J.; Reitz, Z.L.; van Santen, J.A.; Selem-Mojica, N., Tørring, T.; Zaroubi, L.; Alanjary, M.; Aleti, G.; Aguilar, C.; Al-Salihi, S.A.A.; Augustijn, H.E.; Avelar-Rivas, J.A.; Avitia-Domínguez, L.A.; Barona-Gómez, F.; Bernaldo-Agüero, J.; Bielinski, V.A.; Biermann, F.; Booth, T.J.; Carrion Bravo, V.J.; Castelo-Branco, R.; Chagas, F.O.; Cruz-Morales, P.; Du, C.; Duncan, K.R.; Gavriilidou, A.; Gayrard, D.; Gutiérrez-García, K.; Haslinger, K.; Helfrich, E.J.N.; van der Hooft, J.J.J.; Jati, A.P.; Kalkreuter, E.; Kalyvas, N.; Kang, K.B.; Kautsar, S.; Kim, W.; Kunjapur, A.M.; Li, Y.X.; Lin, G.M.; Loureiro, C.; Louwen, J.J.R.; Louwen, N.L.L.; Lund, G.; Parra, J.; Philmus, B.; Pourmohsenin, B.; Pronk, L.J.U.; Rego, A.; Rex, D.A.B.; Robinson, S.; Rosas-Becerra, L.R.; Roxborough, E.T.; Schorn, M.A.; Scobie, D.J:, Singh, K.S.; Sokolova, N.; Tang, X.; Udwary, D.; Vigneshwari, A.; Vind, K.; Vromans, S.P.J.M.; Waschulin, V.; Williams, S.E.; Winter, J.M.; Witte, T.E.; Xie, H.; Yang, D.; Yu, J.; Zdouc, M.; Zhong, Z.; Collemare, J.; Linington, R.G.; Weber, T.; Medema, M.H. MIBiG 3.0: a community-driven effort to annotate experimentally validated biosynthetic gene clusters, Nucleic Acids Res. 2023, 51(D1), D603–D610. [CrossRef]
- Medema, M.H.; Blin, K.; Cimermancic, P.; de Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids oRes. 2011, 39, W339–W346. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.; van Wezel, G.P., Medema, M.H.; Weber, T. antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023, 51(W1), W46–W50. [CrossRef]
- Ren, H.; Shi, C.; Zhao, H. Computational tools for discovering and engineering natural product biosynthetic pathways. Iscience 2020, 23(1), 100795. [Google Scholar] [CrossRef]
- Kountz, D.J.; Balskus, E.P. Leveraging Microbial Genomes and Genomic Context for Chemical Discovery. Acc. Chem. Res. 2021, 54, 2788–2797. [Google Scholar] [CrossRef] [PubMed]
- Stark, R.; Grzelak, M.; Hadfield, J. RNA sequencing: the teenage years. Nat. Rev. Genetics 2019, 20, 631–656. [Google Scholar] [CrossRef] [PubMed]
- Doroghazi, J.R.; Albright, J.C.; Goering, A.W.; Ju, K.S.; Haines, R.R.; Tchalukov, K.A.; Labeda, D.P.; Kelleher, N.L.; Metcalf, W.W. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat. Chem. Biol. 2014, 10(11), 963–968. [Google Scholar] [CrossRef] [PubMed]
- Avalon, N.E.; Murray, A.E.; Baker, B.J. Integrated Metabolomic–Genomic Workflows Accelerate Microbial Natural Product Discovery. Anal. Chem. 2022, 94, 35, 11959–11966. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, B.; Timári, I.; Somogyi, Á.; Li, D.W.; Adcox, H.E.; Gunn, J.S.; Bruschweiler-Li, L.; Brüschweiler, R. Accurate and efficient determination of unknown metabolites in metabolomics by NMR-based molecular motif identification. Anal. Chem. 2019, 91(24), 15686–15693. [Google Scholar] [CrossRef] [PubMed]
- Sidda, J.D.; Song, L.; Poon, V.; Al-Bassam, M.; Lazos, O.; Buttner, M.J.; Challis, G.L.; Corre, C. Discovery of a family of γ-aminobutyrate ureas via rational derepression of a silent bacterial gene cluster. Chem. Sci. 2014, 5, 86–89. [Google Scholar] [CrossRef]
- Alberti, F.; Leng, D.J.; Wilkening, I.; Song, L.; Tosin, M.; Corre, C. Triggering the expression of a silent gene cluster from genetically intractable bacteria results in scleric acid discovery. Chem. Sci. 2019, 10, 453–463. [Google Scholar] [CrossRef]
- Mao, D.; Okada, B.K.; Wu, Y.; Xu, F.; Seyedsayamdost, M.R. Recent advances in activating silent biosynthetic gene clusters in bacteria. Curr. Opin. Microbiol. 2018, 45, 156–163. [Google Scholar] [CrossRef]
- Covington, B.C.; Xu, F.; Seyedsayamdost, M.R. A natural product chemist's guide to unlocking silent biosynthetic gene clusters. Ann. Rev. Biochem. 2021, 90, 763–788. [Google Scholar] [CrossRef]
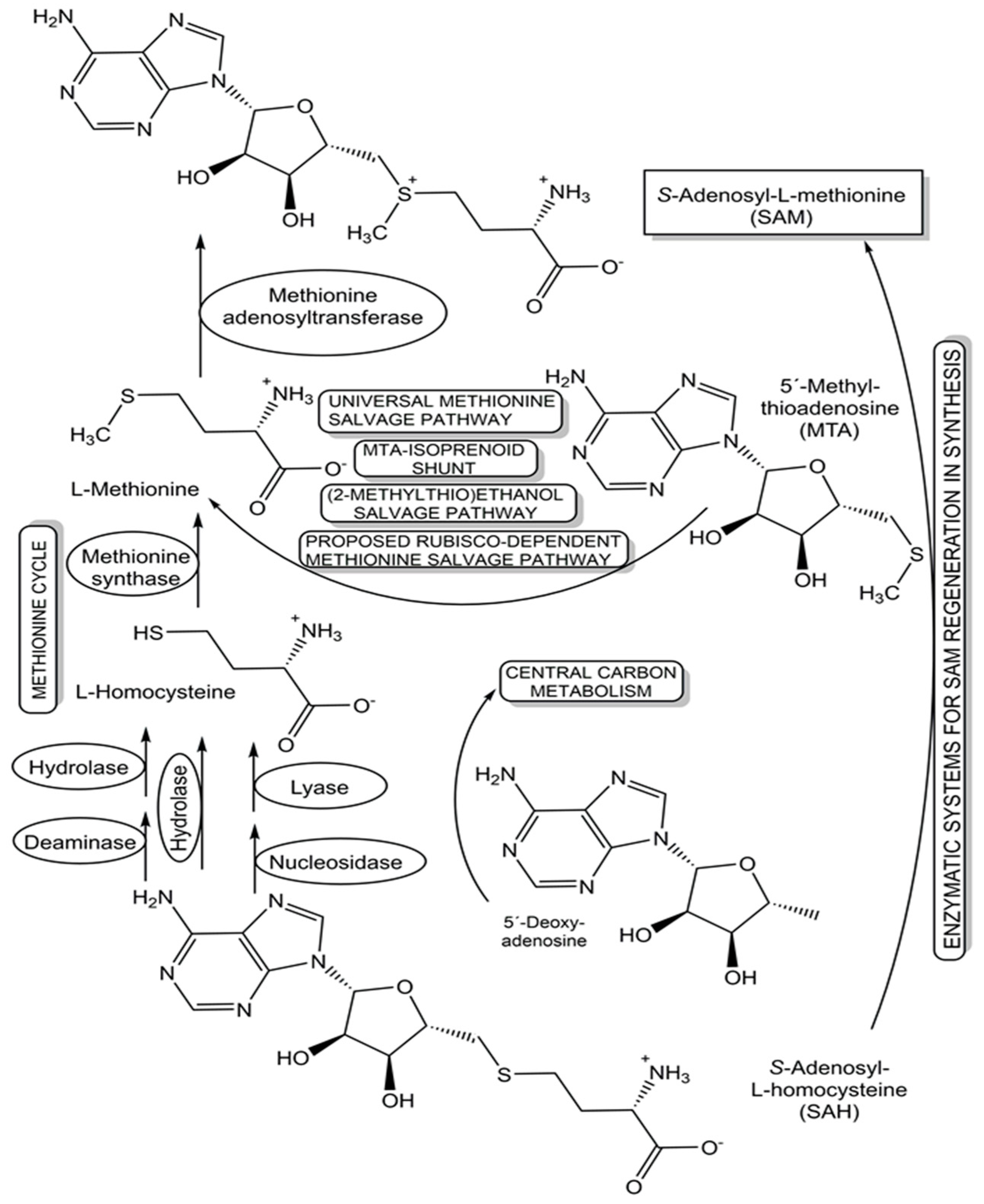
- Hartl, J.; Kiefer, P.; Meyer, F.; Vorholt; J. A. Longevity of major coenzymes allows minimal de novo synthesis in microorganisms. Nat. Microbiol. 2017, 2, 17073. [Google Scholar] [CrossRef]
- Katsyuba, E.; Mottis, A.; Zietak, M.; De Franco, F.; van der Velpen, V.; Gariani, K.; Ryu, D.; Cialabrini, L.; Matilainen, O.; Liscio, P.; Giacchè, N.; Stokar-Regenscheit, N.; Legouis, D.; de Seigneux, S.; Ivanisevic, J.; Raffaelli, N.; Schoonjans, K.; Pellicciari, R.; Auwerx, J. . De novo NAD+ synthesis enhances mitochondrial function and improves health. Nature 2018, 563(7731), 354–359. Nature 2018, 563(7731), 354–359. [Google Scholar] [CrossRef] [PubMed]
- Preiss, J.; Handler, P. Biosynthesis of Diphosphopyridine Nucleotide. I. Identification of Intermediates. J. Biol. Chem. 1958, 233(2), 488–492. [Google Scholar] [CrossRef] [PubMed]
- Preiss, J.; Handler, P. Biosynthesis of Diphosphopyridine Nucleotide. II. Enzymatic Aspects. J. Biol. Chem. 1958, 233(2), 493–500. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.G.; Liu, J.; Makarov, M.V.; Wu, X.; Lih, F.B.; Deterding, L.J.; Lim, C.; Xu, X.; Randall, T.A.; Lee, E.; Li, W.; Fan, W.; Li, J.L.; Sokolsky, M.; Kabanov, A.V.; Li, L.; Migaud, M.E.; Locasale, J.W.; Li, X. Bacteria Boost Mammalian Host NAD Metabolism by Engaging the Deamidated Biosynthesis Pathway. Cell Metabolism 2020, 31(3), 564–579. [Google Scholar] [CrossRef]
- Bogan, K.L.; Brenner, C. Nicotinic acid, nicotinamide, and nicotinamide riboside: a molecular evaluation of NAD+ precursor vitamins in human nutrition. Annu. Rev. Nutr. 2008, 28, 115–130. [Google Scholar] [CrossRef] [PubMed]
- Bieganowski, P.; Brenner, C. Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans. Cell 2004, 117(4), 495–502. [Google Scholar] [CrossRef] [PubMed]
- Belenky, P.; Christensen, K.C.; Gazzaniga, F.; Pletnev, A.A.; Brenner, C. Nicotinamide riboside and nicotinic acid riboside salvage in fungi and mammals: Quantitative basis for Urh1 and purine nucleoside phosphorylase function in NAD+ metabolism. J. Biol. Chem. 2009, 284(1), 158–164. [Google Scholar] [CrossRef]
- Giroud-Gerbetant, J.; Joffraud, M.; Giner, M.P.; Cercillieux, A.; Bartova, S.; Makarov, M.V.; Zapata-Pérez, R.; Sánchez-García, J.L.; Houtkooper, R.H.; Migaud, M.E.; Moco, S. A reduced form of nicotinamide riboside defines a new path for NAD+ biosynthesis and acts as an orally bioavailable NAD+ precursor. Mol. Metab. 2019, 30, 192–202. [Google Scholar] [CrossRef]
- Yang, Y.; Mohammed, F.S.; Zhang, N.; Sauve, A.A. Dihydronicotinamide riboside is a potent NAD+ concentration enhancer in vitro and in vivo, J. Biol. Chem. 2019, 294(23), 9295–9307. [Google Scholar] [CrossRef]
- Yoshino, J.; Baur, J.A.; Imai, S.I. NAD+ intermediates: the biology and therapeutic potential of NMN and NR. Cell Metab. 2018, 27(3), 513–528. [Google Scholar] [CrossRef]
- Katsyuba, E.; Romani, M.; Hofer, D.; Auwerx, J. NAD+ homeostasis in health and disease. Nature Metabolism 2020, 2, 9–31. [Google Scholar] [CrossRef] [PubMed]
- Covarrubias, A.J.; Perrone, R.; Grozio, A.; Verdin, E. NAD+ metabolism and its roles in cellular processes during ageing. Nat. Rev. Mol. Cell Biol. 2021, 22(2), 119–141. [Google Scholar] [CrossRef] [PubMed]
- Gude, S.; Pherribo, G.J.; Taga, M.A. A Salvaging Strategy Enables Stable Metabolite Provisioning among Free-Living Bacteria. mSystems 2022, 7(4), 1-14. [CrossRef]
- North, J.A.; Wildenthal, J.A.; Erb, T.J.; Evans, B.S.; Byerly, K.M. , Gerlt, J.A.; Fred R. Tabita, F.R. A bifunctional salvage pathway for two distinct S-adenosylmethionine by-products that is widespread in bacteria, including pathogenic Escherichia coli. Mol. Microbiol, 2020; 113, 923–937. [Google Scholar] [CrossRef]
- Gericke, L.; Mhaindarkar, D.; Karst, L.C.; Jahn, S.; Kuge, M.; Mohr, M.K.F.; Gagsteiger, J.; Cornelissen, N.V.; Wen, X.; Mordhorst, S.; Jessen, H.J.; Rentmeister, A.; Seebeck, F.P.; Layer, G.; Loenarz, C.; Andexer, J.A. Biomimetic S-Adenosylmethionine Regeneration Starting from Multiple Byproducts Enables Biocatalytic Alkylation with Radical SAM Enzymes. ChemBioChem 2023, 24, e202300133. [Google Scholar] [CrossRef] [PubMed]
- Jeffryes, J.G.; Lerma-Ortiz, C.; Liu, F.; Golubev, A.; Niehaus, T.D.; Elbadawi-Sidhu, M.; Fiehn, O.; Hanson, A.D.; Tyo, K.E.; Henry, C.S. Chemical-damage MINE: A database of curated and predicted spontaneous metabolic reactions. Metab. Eng. 2022, 69, 302–312. [Google Scholar] [CrossRef] [PubMed]
- Bommer, G.T.; Van Schaftingen, E.; Veiga-da-Cunha, M. Metabolite repair enzymes control metabolic damage in glycolysis. Trends Biochem. Sci. 2020, 45(3), 228–243. [Google Scholar] [CrossRef] [PubMed]
- Niehaus, T.D.; Katie, B. Hillmann, K.B. Enzyme promiscuity, metabolite damage, and metabolite damage control systems of the tricarboxylic acid cycle. FEBS J. 2020, 287, 1343–1358. [Google Scholar] [CrossRef] [PubMed]
- Leister, D.; Sharma, A.; Kerber, N.; Nägele, T.; Reiter, B.; Pasch, V.; Beeh, S.; Jahns, P.; Barbato, R.; Pribil, M.; Rühle, T. An ancient metabolite damage-repair system sustains photosynthesis in plants. Nat. Commun. 2023, 14, 3023. [Google Scholar] [CrossRef]
- Thiaville, J.J.; Flood, J.; Yurgel, S.; Prunetti, L.; Elbadawi-Sidhu, M.; Hutinet, G.; Forouhar, F.; Zhang, X.; Ganesan, V.; Reddy, P.; Fiehn, O.; Gerlt, J.A.; Hunt, J.F.; Copley, S.D.; de Crécy-Lagard, V. Members of a novel kinase family (DUF1537) can recycle toxic intermediates into an essential metabolite. ACS Chem. Biol. 2016, 11(8), 2304–2311. [Google Scholar] [CrossRef]
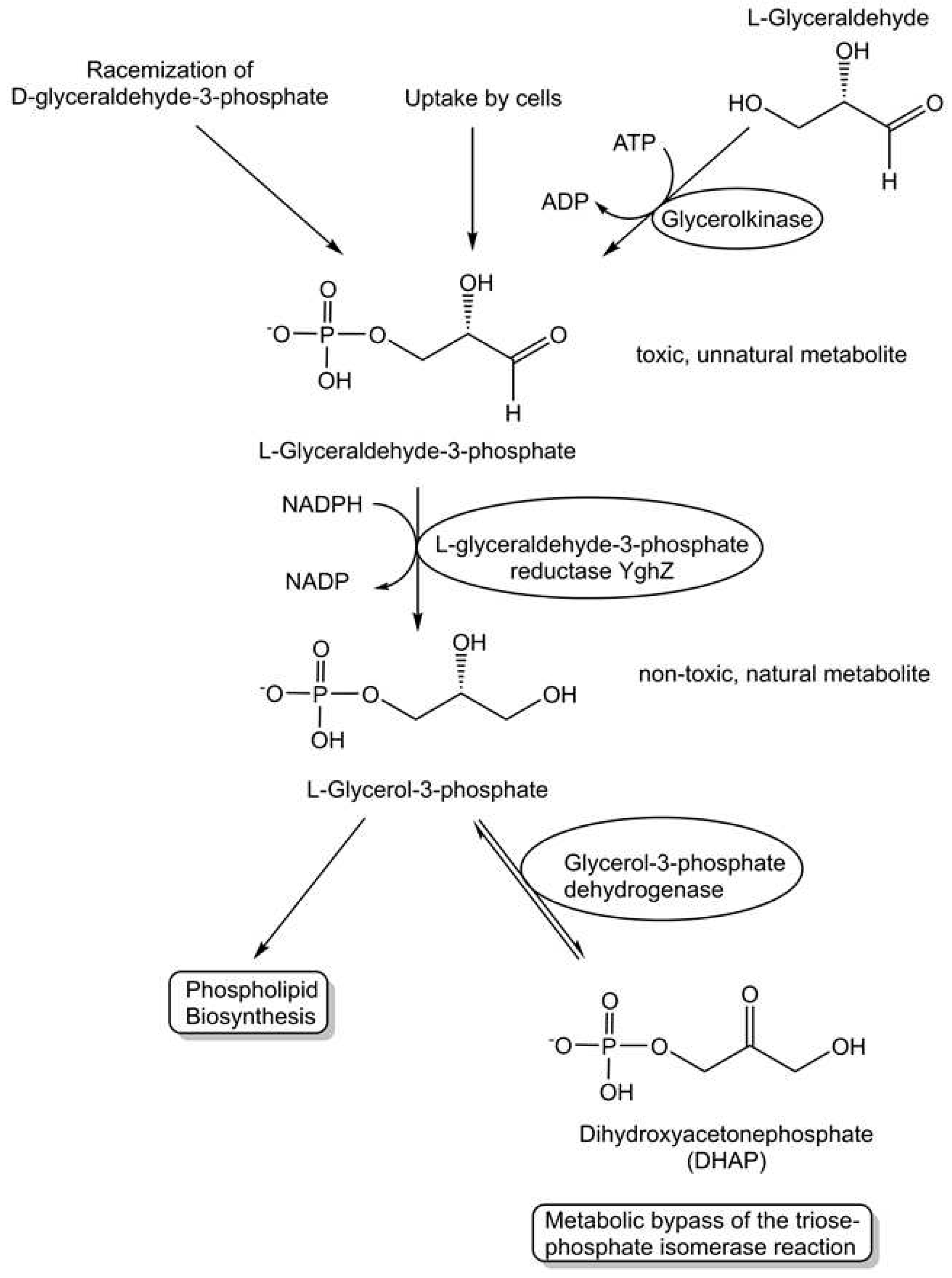
- Hall, A.; Knowles, J.R. The Uncatalyzed Rates of Enolization of Dihydroxyacetone Phosphate and of Glyceraldehyde 3-Phosphate in Neutral Aqueous Solution. The Quantitative Assessment of the Effectiveness of an Enzyme Catalyst. Biochemistry 1975, 14(19), 4348–4351. [Google Scholar] [CrossRef]
- Kalyananda, M.K.G.S.; Engel, R.; Tropp, B.E. Metabolism of L-Glyceraldehyde 3-Phosphate in Escherichia coli. J. Bact. 1987, 169(6), 2488–2493. [Google Scholar] [CrossRef]
- Desai, K.D.; Miller, B.G. A Metabolic Bypass of the Triosephosphate Isomerase Reaction. Biochemistry 2008, 47, 7983–7985. [Google Scholar] [CrossRef] [PubMed]
- Lewin, H.A.; Richards, S.; Lieberman Aiden, E.; Allende, M.L.; Archibald, J.M.; Bálint, M.; Barker, K.B.; Baumgartner, B.; Belov, K.; Bertorelle, G.; Blaxter, M.L. The earth BioGenome project 2020: Starting the clock. Proc. Natl. Acad. Sci. 2022, 119(4), e2115635118. [Google Scholar] [CrossRef] [PubMed]
- Ebenezer, T.E.; Muigai, A.W.T.; Nouala, S.; Badaoui, B.; Blaxter, M.; Buddie, A. G.; Jarvis, E.D.; Korlach, J.; Kuja, J.O.; Lewin, H.A.; Majewska, R.; Mapholi, N.; Maslamoney, S.; Mbo’o-Tchouawou, M.; Osuji, J.O.; Seehausen, O.; Shorinola, O.; Tiambo, C.K.; Mulder, N.; Ziyomo, C.; Djikeng, A. Africa: sequence 100,000 species to safeguard biodiversity. Nature 2022, 603(7901), 388–392. [Google Scholar] [CrossRef] [PubMed]
- Nayfach, S.; Roux, S.; Seshadri, R.; Udwary, D.; Varghese, N.; Schulz, F.; Wu, D.; Paez-Espino, D.; Chen, I.M.; Huntemann, M.; Palaniappan, K.; Ladau, J.; Mukherjee, S.; Reddy, T.B.K.; Nielsen, T.; Kirton, E.; Faria, J.P.; Edirisinghe, J.N.; Henry, C.S.; Jungbluth, S.P.; Chivian, D.; Dehal, P.; Wood-Charlson, E.M.; Arkin, A.P.; Tringe, S.G.; Visel, A.; IMG/M Data Consortium, Woyke, T.; Mouncey, N.J.; Ivanova, N.N.; Kyrpides, N.C.; Eloe-Fadrosh, E.A. A genomic catalog of Earth’s microbiomes. Nat. Biotechnol. 2021, 39, 520. [CrossRef]
- de Crécy-Lagard, V.; Amorin de Hegedus, R.; Arighi, C.; Babor, J.; Bateman, A.; Blaby, I.; Blaby-Haas, C.; Bridge, A.J.; Burley, S.K.; Cleveland, S.; Colwell, L.J.; Conesa, A.; Dallago, C.; Danchin, A.; de Waard, A.; Deutschbauer, A.; Dias, R.; Ding, Y.; Fang, G.; Friedberg, I.; Gerlt, J.; Goldford, J.; Gorelik, M.; Gyori, B.M.; Henry, C.; Hutinet, G.; Jaroch, M.; Karp, P.D.; Kondratova, L.; Lu, Z.; Marchler-Bauer, A.; Martin, M.J.; McWhite, C.; Moghe, G.D.; Monaghan, P.; Morgat, A.; Mungall, C.J.; Natale, D.A.; Nelson, W.C.; O’Donoghue, S.; Orengo, C.; O’Toole, K.H.; Radivojac, P.; Reed, C.; Roberts, R.J.; Rodionov, D.; Rodionova, I.A.; Rudolf, J.D.; Saleh, L.; Sheynkman, G.; Thibaud-Nissen, F.; Thomas, P.D.; Uetz, P.; Vallenet, D.; Watson Carter, E.; Weigele, P.R.; Wood, V.; Wood-Charlson, E.M.; Xu, J. A roadmap for the functional annotation of protein families: a community perspective. Database 2022, 2022, baac062. [Google Scholar] [CrossRef] [PubMed]
- Harmange Magnani, C.S.; Hernández-Meléndez, J.R.; Tantillo, D.J.; Maimone, T.J. Total Synthesis of Altemicidin: A Surprise Ending for a Monoterpene Alkaloid. JACS Au 2023, 3, 2883–2893. [Google Scholar] [CrossRef] [PubMed]
- Hetzler, B. E.; Trauner, D.; Lawrence, A. L. Natural product anticipation through synthesis. Nat. Rev. Chem. 2022, 6, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Caesar, L.K.; Montaser, R.; Keller, N.P.; Kelleher, N.L. Metabolomics and Genomics in Natural Products Research: Complementary Tools for Targeting New Chemical Entities. Nat. Prod. Rep. 2021, 38, 2041–2065. [Google Scholar] [CrossRef]
- Caesar, L.K.; Butun, F.A.; Robey, M.T.; Ayon, N.J.; Gupta, R.; Dainko, D.; Bok, J.W.; Nickles, G.; Stankey, R.J.; Johnson, D.; Mead, D.; Cank, K.B.; Earp, C.E.; Raja, H.A.; Oberlies, N.H.; Keller, N.P.; Kelleher, N.L. Correlative metabologenomics of 110 fungi reveals metabolite–gene cluster pairs. Nat. Chem. Biol. 2023, 19(7), 846–854. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




