Introduction
Malaria is a universal public health burden, with approximately 241 million confirmed cases and a fatality of approximately 627,000 as at 2020 (WHO, 2022). These statistics underscore an alarming rise in malaria cases and deaths compared to WHO’s 2019 report. In 2019, the WHO reported 227 million confirmed cases and approximately 558,000 deaths worldwide (WHO, 2022). Unfortunately, Africa contributes a disproportionally high portion of the global malaria burden, especially sub-Saharan Africa. In 2020, the African region contributed 95% of malaria-confirmed cases and 96% of malaria deaths (Oladipo et al., 2022). Children aged five years and below accounted for approximately 80% of all malaria deaths in the African region (Oladipo et al., 2022). Four countries within sub-Saharan Africa accounted for over half of malaria deaths globally (WHO, 2022). These shockingly high morbidity and mortality rates call for an in-depth understanding of malaria’s etiology, transmission, and pathogenesis and how malaria intervention strategies can be more effective.
Mosquitoes are the primary vectors of malaria, which primarily affects individuals in subtropical and tropical areas. Obligate protozoan parasites of the genus Plasmodium cause the ailment (Sá, Costa, & Tavares, 2022). Mosquito vectors that carry malaria transmit the parasites to people during a blood meal. Once within the host, the parasites first infect the liver cells before moving on to the red blood cells. Ramos et al. (2021) explain that by altering the infected erythrocytes to make them cytoadherent, malaria parasites cause malaria pathology at the blood stage. Malaria infection, whose incubation period is 7-14 days, can cause lethargy, fatigue, muscle aches, nausea, stomach aches, fever, and shivering (Laurens, 2020; Nadeem et al., 2022). Due to the massive loss of red blood cells, the disease can cause jaundice and anemia. Furthermore, if not appropriately treated, malaria can become lethal and cause coma, mental confusion, seizures, kidney failure, and finally, death (CDC, 2022).
To date, P. falciparum, P. knowlesi, P. ovale, P. vivax, and P. malariae are the five species of Plasmodium that can infect individuals and cause malaria. P. falciparum has the highest morbidity and fatality rates and hence poses a severe threat to public well-being in locations where malaria is endemic (Ayanful-Torgby et al., 2018; Chew et al., 2022; Laurens, 2020; Nadeem et al., 2022). Even so, the devastating effects of the other four Plasmodium species on human health should not be underrated. Currently, malaria kills approximately 400,000 individuals yearly, primarily kids in sub-Saharan Africa because they have not yet developed any immunity to the illness. It affects 200-400 million people annually (Chew et al., 2022; Gross, 2019; Wang et al., 2020). Developing tools and strategies that can interrupt the transmission of a specified malaria parasite species is a way of reducing the devastating impacts of the disease on human well-being and safeguarding the well-being of the millions of people affected by the illness yearly.
The WHO recommended malaria prevention strategies and tools that have been in use for almost two decades. Some of these strategies are malaria vaccines and the utilization of anti-malarial drugs. Even though such malaria prevention strategies and tools have reduced the global burden of the illness, the upsurge in the number of malaria’s confirmed cases and deaths from 2019 to 2020 underlies the need for more effective intervention strategies (WHO, 2022). Wang et al. (2020) outline that between 2000 and 2015, anti-malarial medications, insecticide-treated mosquito nets, and other public health initiatives helped reduce malaria cases worldwide by 50–75%. The recently developed malaria vaccine is an essential tool to alleviate the enormous socioeconomic burden that malaria causes. Despite these initiatives, malaria incidences have grown since 2015 in numerous places for various reasons (Wang et al., 2020). One of the outstanding reasons include the parasites developing resistance to anti-malarial medicines.
Anti-malarial drug resistance, which increases malaria mortality and morbidity, has become a menace that casts doubt on the effectiveness of the currently available anti-malarial medications. Only P. falciparum and P. vivax have been proven to be resistant to anti-malarial drugs currently on the market (CDC, 2018). No information is available on P. ovale or P. malariae treatment resistance. P. knowlesi, a zoonotic monkey malaria that affects individuals in Southeast Asian forested environment, is completely sensitive to chloroquine and other commonly utilized medications (CDC, 2018). P. falciparum and P. vivax are resistant to chloroquine. P. falciparum is also resistant to other anti-malarial drugs, including quinine, halofrantrine, mefloquine, and sulfadoxine/pyrimethamine (CDC, 2018). Although resistance to these medicines tends to be noticeably less geographically widespread, the impacts of such multi-drug resistant malaria can be catastrophic in various parts of the globe (CDC, 2018).
Researchers have argued that Artemisinin-based Combination Therapy (ACT) is the only efficacious malaria treatment (Maiga et al., 2021; Pousibet-Puerto et al., 2016). However, Stofberg et al. (2021) explain that the extended ACTs clearance times, which are linked to the emergence of artemisinin monotherapy resistance, have been recorded most recently in Africa and the Great Mekong region, pose a danger to its efficacy. This trend in drug resistance is comparable to the medicine chloroquine’s fate, where drug resistance first appeared in the early 2000s in the Mekong region before spreading widely to many areas, including Africa and East Asia. Genotypes of parasites resistant to the artemisinin medication have recently been discovered in Rwanda, indicating extensive artemisinin compound resistance (Tintó-Font et al., 2021). It is well known that parasite-targeted gene mutations cause the development of medication resistance (Stofberg et al., 2021). Antiparasitic drug concentrations are influenced by the proper operation of mutant genes connected to the inflow and efflux pumps; for instance, molecular chaperones such as the Hsps are responsible for the proper folding of mutated chloroquine resistance transporter (CRT). All proteins can fold naturally when under stress, including pressure from drugs, thanks to these Hsps. Therefore, the Hsps are invaluable antimalarial drug targets.
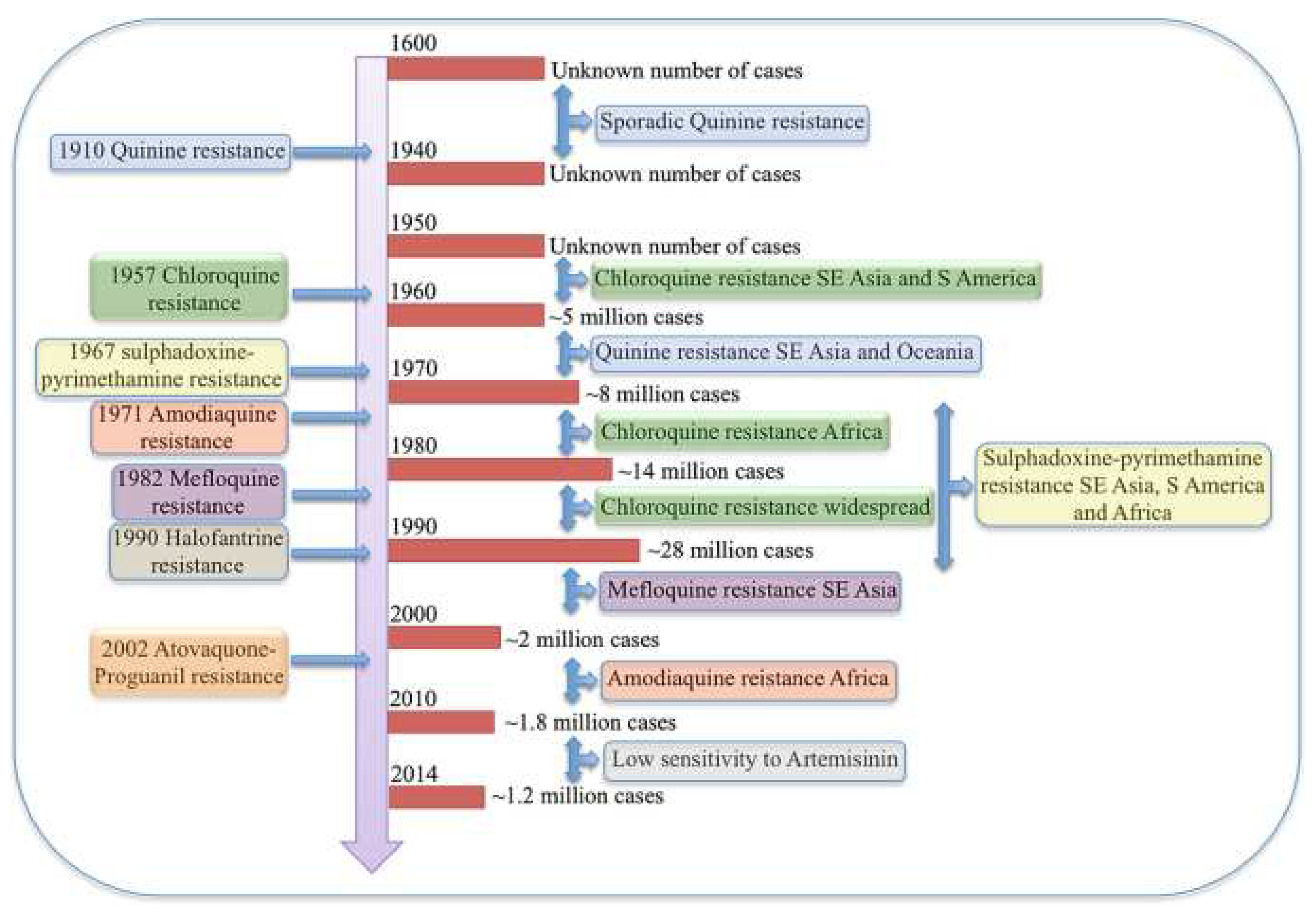
Additionally, due to their crucial function in protein quality regulation, the genes, such as the CRT gene linked to multidrug resistance, are situated in the same Hsp90 gene cluster (Zininga et al., 2015). This implies that the expression of the mutant CRT gene and the chaperone may be similarly co-regulated by cis-regulatory elements like transcription factors. Furthermore, it has been established that various molecular chaperones interact directly and indirectly with CRT through an unknown mechanism (Stofberg et al., 2021). Hsps’ chaperoning function on proteins might facilitate drug resistance during the stress response. Since the 1600s, attempts have been made to control and manage antimalaria drug resistance in vain (Chakrabarti et al., 2019). Reported malaria cases have always been in millions, as displayed in
Figure 1 (Chakrabarti et al., 2019). Hsp90 can be considered a drug target whose inhibition prevents its role in protein folding and facilitates malaria treatment by reducing the level of antimalarial drug resistance that the parasite can mount within the body. In this regard, Hsp90 can be used as a target protein to discover its inhibitors and potential antimalarial drugs against
Plasmodium malaria using computational approaches.
This timeline shows how P. falciparum has been mounting resistance to various antimalarial drugs over the years (Chakrabarti et al., 2019).
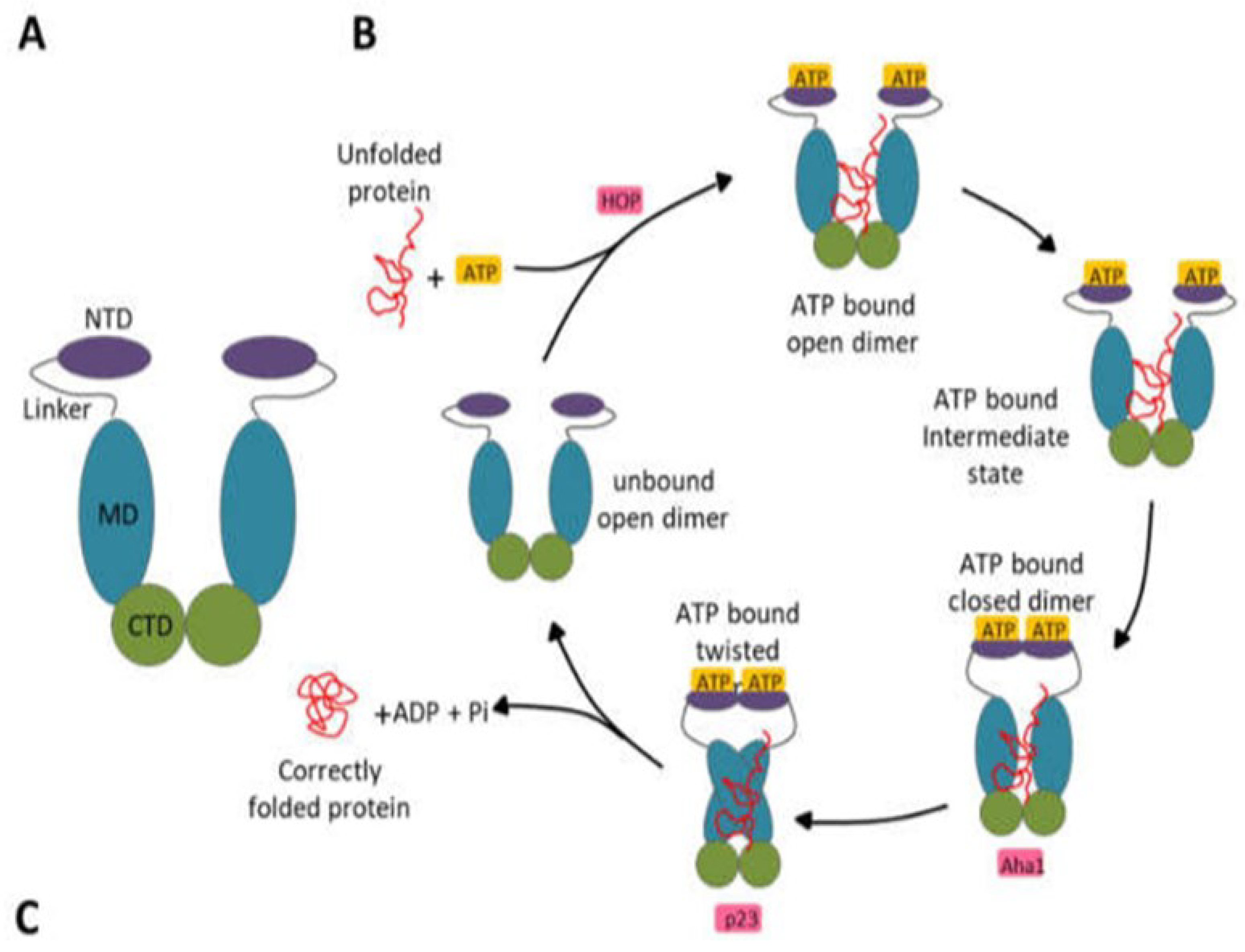
Hsp90 has been well-characterized in the existing literature. Understanding its functional cycle is essential to identify its active sites or binding pockets in which inhibitors can be docked. Spiegelberg et al. (2020) consider the Hsp90 an evolutionary conserved and widely expressed molecular chaperones group that accounts for around 2% of the cellular proteome. Almost all organisms possess Hsp90 proteins, which are necessary for eukaryotes to survive but optional for some eubacterial species to survive under normal circumstances (Honoré et al., 2017; Schopf et al., 2017). Hsp90 chaperone is essential in reducing stress and preserving cellular homeostasis. Schopf et al. (2017) explain that as a critical regulator of vital physiological functions, including protein folding, stress control, DNA repair, growth, signaling pathways, and immunological response, Hsp90 cooperates with a wide range of client proteins. Hsp90s are desirable pharmacological targets for numerous illnesses, including neurological conditions, cancer, and infectious illnesses like malaria, because of their crucial role in cell survival. The Hsp90 family members are ATP-dependent chaperones involved in numerous biological functions (Schopf et al., 2017). Adenosine triphosphate (ATP) binding and subsequent hydrolysis power the Hsp90, which occurs as a V-shaped homodimer functional cycle (
Figure 2) (Stofberg et al., 2021).
During this cycle, Hsp90 binds to the ATP-bound state and open-dimer configuration of an unfolded substrate/client protein (Mader et al., 2020). The N5, N4, and N1 helixes in the NTD undergo a conformational shift; as a result, closing over the ATP binding pocket and serve as a lid over the cavity (Li et al., 2019). Following their association and dimerization, the NTDs undergo additional conformational changes that lead to the middle domains (MDs) attachment. The catalytic loop in the MDs is moved due to these modifications. The crucial catalytic residues Gln423, Arg419, and Asn416 that hydrolyze ATP are part of the catalytic loop in P. falciparum, which extends from residues 414 to 427 (Silva et al., 2020). The release of the appropriately folded client protein is then made possible by the dissociation of the NTDs brought on by ATP hydrolysis (Radli & Rüdiger, 2018; Rashid et al., 2020). Therefore, Hsp90’s active site is found within the NTD, where ATP can bind and initiate the folding of different client proteins. Finding compounds that can bind to Hsp90’s N-terminal ATP binding site can help curtail the protein’s function of assisting the folding of other client proteins.
In early studies, Hsp90 inhibitors were primarily used to treat cancer (Koren & Blagg, 2020). Due to the cancer cells’ insatiable addiction to Hsp90 (Koren & Blagg, 2020), these inhibitors mainly targeted Hsp90 NTD’s ATP binding pocket (Stofberg et al., 2021). Therefore, Hsp90 overexpression in cancerous cells suggests it is involved in developing various oncogenic client proteins. These characteristics make Hsp90 a promising target for anti-cancer medications (Han et al., 2018). In P. falciparum, similar to other eukaryotes, all four Hsp90s may be necessary for parasite persistence and changes in erythrocytic stage transitions (Stofberg et al., 2021). It ought to be highlighted, nonetheless, that several investigations on parasite Hsp90s utilize renowned human Hsp90 inhibitors to assess the role and necessity of those particular proteins. Since it has not been meticulously established that the impact of such inhibitors on parasite growth is solely due to the protein of interest and no other target proteins, care must be used in interpreting such results. Despite this drawback, Hsp90 proteins are desirable therapeutic targets for several reasons. First, Hsp90s are ATPases with differing activity levels in various organisms (Whitesell et al., 2019), and diseased cells have higher ATP hydrolysis rates, making them more vulnerable to ATPase inhibitors (Stofberg et al., 2021).
Similarly,
P. falciparum Hsp90 is more susceptible to inhibition than the human homolog due to its increased ATPase activity (Stofberg et al., 2021). Second, in various organisms, Hsp90s interact with various co-chaperones. These different co-chaperone interactions that are species-specific could also be used for selective inhibition (Mak et al., 2021). Lastly, it has been established that the minor variations in amino acid residues among Hsp90 proteins from various organisms and cellular compartments resulted in specific structural variances (Park et al., 2020). Identifying drug targets based on distinctive Hsp90 protein conformations has become easier because of the elucidation of various Hsp90 crystal structures (Huck et al., 2019). High-resolution crystal structures have revealed structural information that has allowed the identification and screening of new Hsp90 inhibitors. Numerous crystal structures of various Hsp90 NTDs in complex with inhibitors and nucleotides have been resolved (Que et al., 2018). Additionally, Hsp90’s complete structural characterization has prompted the creation of inhibitors targeting its MD (Silva et al., 2020; Zhang et al., 2018; Mak et al., 2021) and CTD (Bopp et al., 2016).
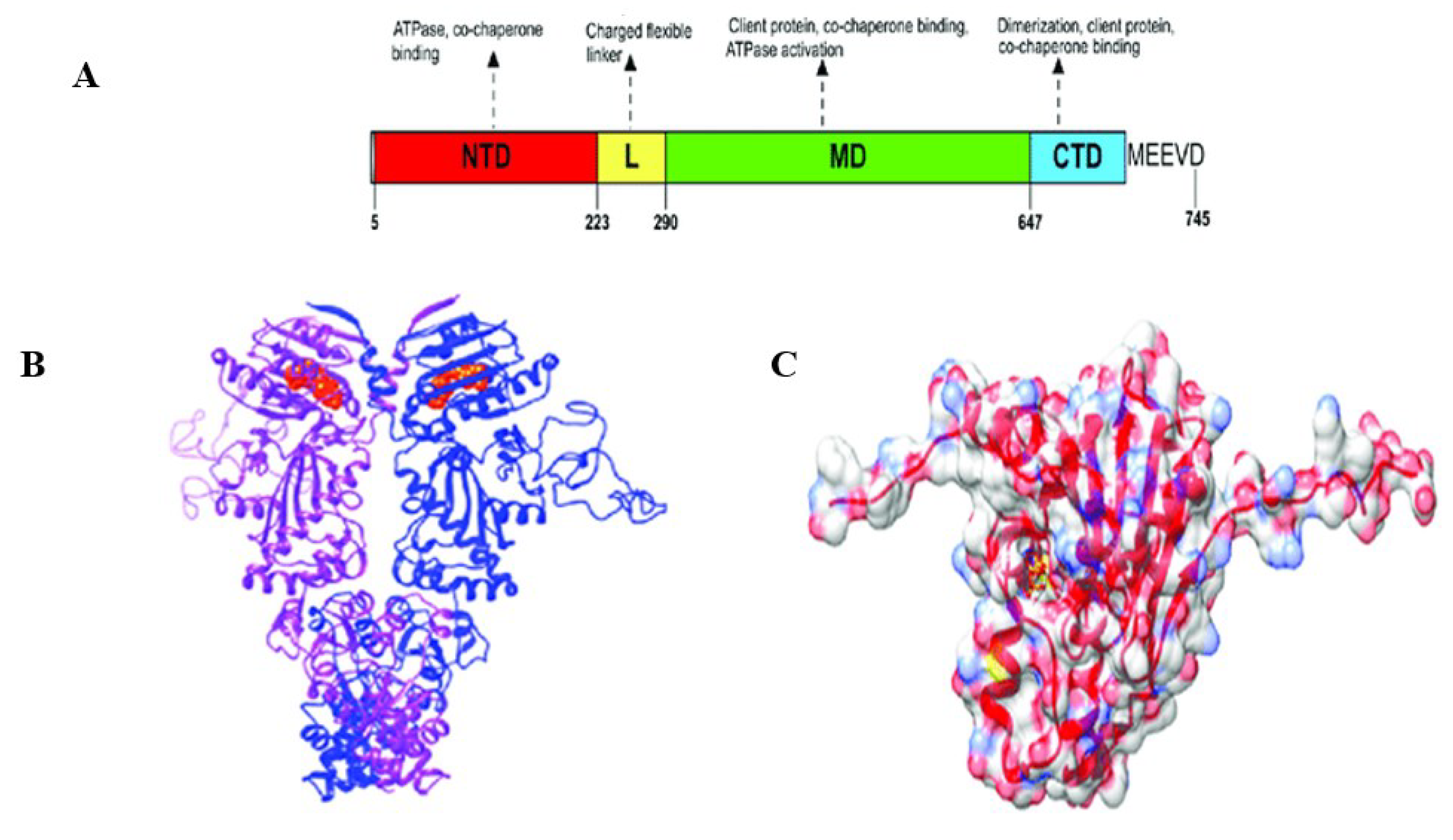
Figure 3 (Dutta et al., 2022) shows PfHsp90’s domain organization and structure. Through targeting Hsp90, some inhibitors have been demonstrated to be efficient at reducing the parasite’s development (Posfai et al., 2018; Wang et al., 2016; Zininga & Shonhai, 2019). Therefore, PfHsp90 was used as the target protein to determine novel PfHsp90 inhibitors with pharmacological properties against
Plasmodium malaria.
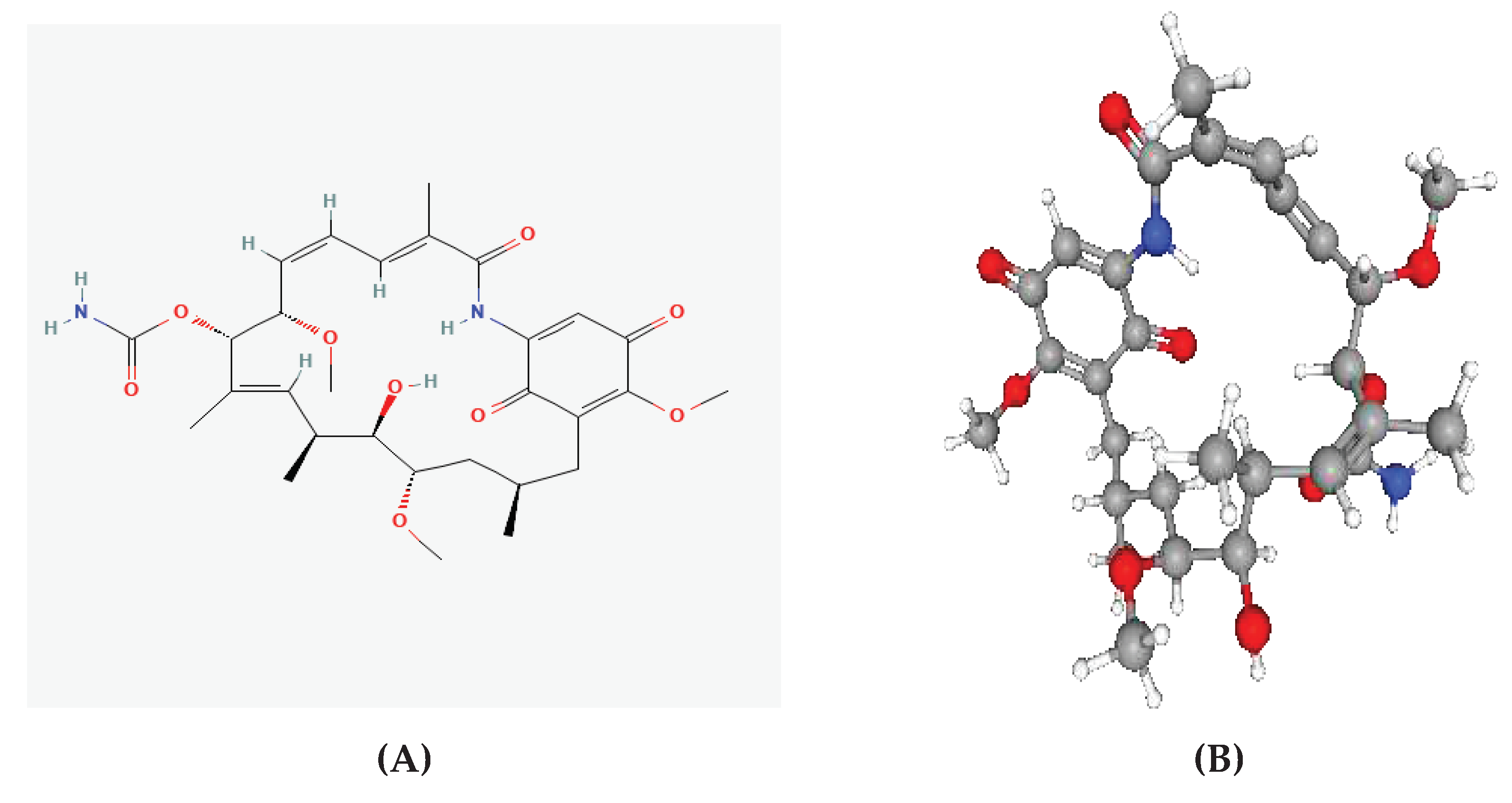
One of the first Hsp90 inhibitors discovered was geldanamycin (GDM), a benzoquinone ansamycin molecule naturally generated by
Streptomyces hygroscopicus (Stofberg et al., 2021). GDM was initially believed to be an antibiotic that inhibited kinases, but it was later discovered that it had a high degree of selectivity in its binding to Hsp90 (Stofberg et al., 2021). Some of these initial studies targeted Hsp90 in tumor cells and eventually adapted their findings to treat other illnesses, such as malaria. As a result, it was demonstrated that some cancer treatments and inhibitors have strong anti-plasmodial efficacy (Posfai et al., 2018). GDM competitively binds to the PfHsp90’s ATPase domain (Stofberg et al., 2021). The unfolded client protein is then degraded due to GDM’s ability to prevent PfHsp90-client protein interaction (Stofberg et al., 2021). In a study at an IC
50 similar to the well-known anti-malarial chloroquine (20 nM and 15 nM, respectively), GDM suppressed
in vitro parasite development (Stofberg et al., 2021). PfHsp90 may be crucial for the growth of parasites because its inhibition causes a stage evolution arrest for intra-erythrocytic parasite phases, primarily the transition from the ring to the trophozoite stages (Stofberg et al., 2021). Furthermore, GDM has been noted to be similarly effective against strains that are sensitive to and resistant to chloroquine (Stofberg et al., 2021). Compared to human HSPC2, independent research found GDM to be extra efficacious at decreasing PfHsp90’s ATPase action (Stofberg et al., 2021). This shows that GDM is more selective in abrogating PfHsp90’s enzymatic role than its human counterpart. Therefore, it was used as the reference ligand during virtual screening to determine novel PfHsp90 inhibitors with pharmacological properties against
Plasmodium malaria.
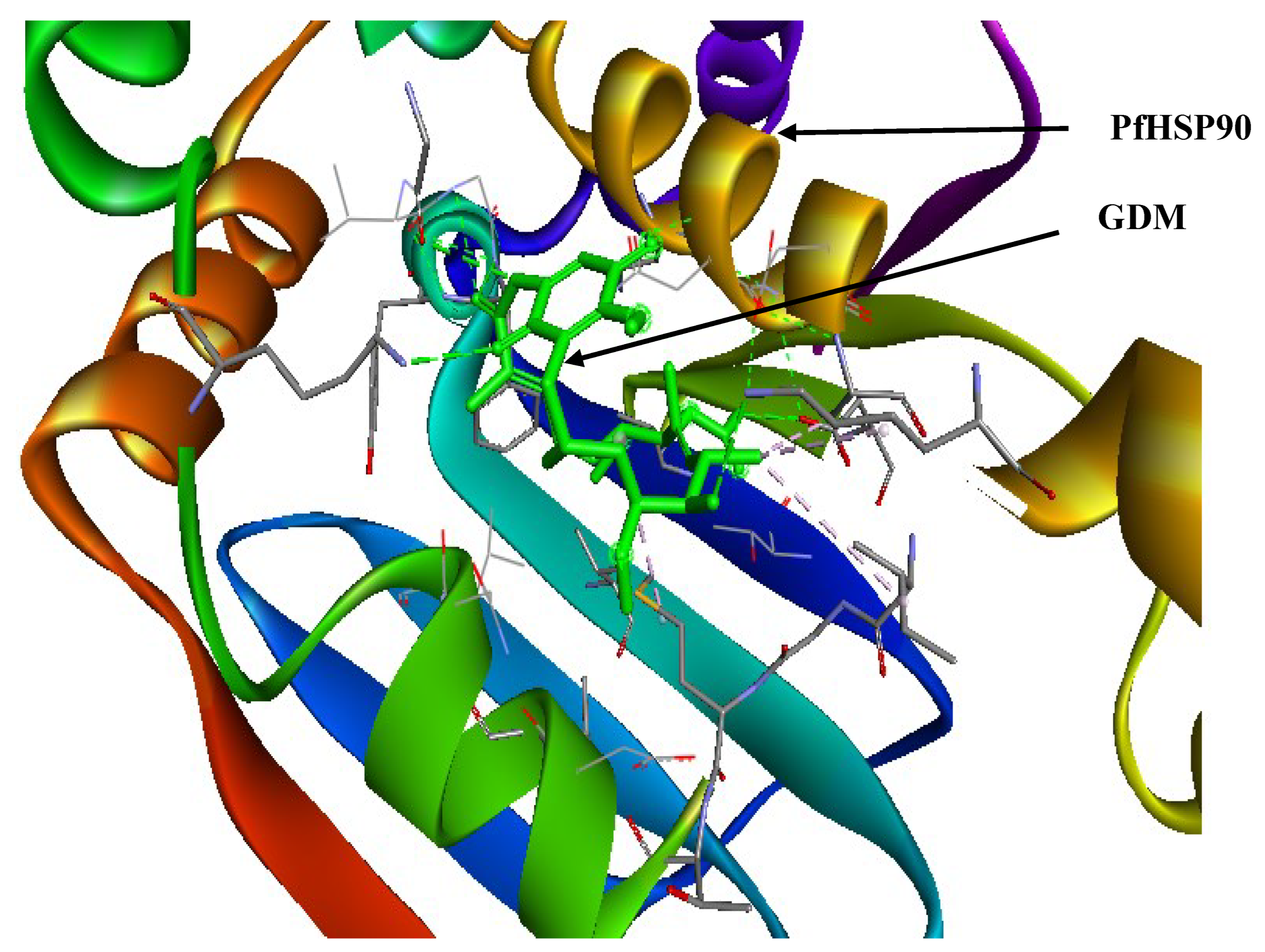
Figure 4 shows the interaction between PfHsp90 and GDM.
Several researchers have resorted to the use of in-silico means to design and develop drugs, shying away from the traditional techniques that are expensive and time-consuming. Some of the most common computational methods used in drug design and discovery include drug design visualization, hierarchical virtual screening, molecular dynamics simulations, and molecular docking (Onyango, 2023). These in-silico methods have been applied by numerous researchers to create drug candidates. For instance, Onyango et al. (2022) used silico techniques to identify prospective anti-SARS-CoV-2 main protease (Mpro) medicines. Similarly, Mengist et al. (2021) discovered 15 effective anti-viral Mpro compounds using in-silico methods, including chloroquine, cilexetil, dipyridamole, hydroxychloroquine, and candesartan. Computational techniques are crucial in drug discovery to find potential antimalarial drug candidates.
Furthermore, in-silico techniques can be used to test the anti-plasmodial potential of synthetic molecules in vivo through molecular binding. For instance, Tahghighi et al. (2020) used in-silico methods to ascertain that the synthetic derivatives of 1-(heteroaryl)-2-((5-nitroheteroaryl) methylene) hydrazine that showed anti-plasmodial activities in vitro, also possessed the same capability in vivo. From the molecular docking results in the study, the authors verified Pf lactate dehydrogenase (LDH) inhibition and the inhibitory effect on the haemozoin formation for the studied compounds (Tahghighi et al., 2020). In a similar study, Sachdeva et al. (2020) used in-silico approaches to assess the capability of repurposing approved antimalarial medicines against COVID-19. The researchers discovered that the antimalarial drug doxycycline (DOX) could be an ideal candidate for repurposing for COVID-19 because it bound effectively to the spike protein of SARS-CoV-2 (Sachdeva et al., 2020). These studies prove that in-silico methodologies are ideal alternatives to drug design, development, and discovery.
Using computational approaches to discover drug candidates often occur during the pre-clinical phase of drug design and development. After discovering potential drug candidates, undertaking further in vitro and in vivo validation of their therapeutic capacity is essential. Sachdeva et al. (2020) recommended further in vitro and in vivo studies to ascertain the actual potential of DOX against COVID-19. In most cases, the in vitro drug sensitivity assays are utilized depending on the illness and pathogen of interest. Sinha et al. (2017) undertook a systematic review pointing out the various drug sensitivity assay utilized for antimalarial drug efficacy testing targeting different stages of the parasite’s development. Some of those assays include blood stages assays (Schizont maturation, microscopic assay, radioisotopic assay, and enzymatic assay), gametocytes stage assays (oxido-reduction indicator, Alamar blue, and SMFA), liver stage assays (infrared fluorescence detection method), and HTS (fluorescence-based assay and in vitro beta-hematin formation assay) (Sinha et al., 2017).
One of the most common antimalarial drug sensitivity assays is the SYBR green assay. It is considered the GOLD standard for in vitro malaria drug sensitivity testing because of its reliability as a drug screening and surveillance tool (Cheruiyot et al., 2016). It is also described as a simple and cost-effective methodology that has been utilized to determine the 50% inhibitory concentrations (IC50) of clinical isolates (Cheruiyot et al., 2016). Researchers have used this assay in their studies, including Traoré et al. (2019) when assessing the susceptibility of P. falciparum isolates to antimalarial medicines in Mali. Similarly, Duan et al. (2022) determined susceptibilities of P. falciparum isolates to 11 antimalarial medicines using the SYBR green assay. The antimalarial drugs included PND, lumefantrine (LMF), quinine (QN), artemether (AM), DHA, artesunate (AS), pyrimethamine (PY), NQ, mefloquine (MFQ), PPQ, and chloroquine (CQ) (Duan et al., 2022). Therefore, this assay determined the inhibitory capability of the selected PfHsp90 inhibitors.
Discussion
While several licensed treatment regimens exist, malaria still causes considerable fatality because of antimalarial drug resistance, heightening the need for novel chemotherapeutic interventions (Su et al., 2019; Varo et al., 2020). Targeting a distinct parasitic pathway that differs significantly from the host’s is a valuable means to prevent the spread of the malarial parasite. Since PfHsp90 facilitates parasite growth during its asexual blood phase (Posfai et al., 2018), Plasmodium falciparum uses it to cause malaria. In this regard, inhibiting this protein may hold promise for a Plasmodium falciparum malaria cure.
Currently, there is a paradigm shift from traditional to computational approaches of novel drug design and discovery. In-silico approaches are gaining traction as options for drug discovery. Numerous studies have used in-silico approaches in the discovery of novel therapeutic compounds against malaria, which have progressed to clinical trials (Ashton et al., 2019; White et al., 2018). P. falciparum dihydroorotate dehydrogenase (PfDHODH) inhibitor (DSM265) has been reported to possess antimalarial activity, which has progressed through phase 2 of clinical trials (Alzain et al., 2022; Murphy et al., 2018; Phillips et al., 2015; Sulyok et al., 2017).
Alzain et al. (2022) discovered Z1481646084, Z24317941, and Z951873618 as potential antimalarial compounds inhibiting PfDHODH using in-silico means without subjecting them to in vitro and in vivo validation. Now that the use of in-silico methods to find novel antimalarial compounds is limited, and the existing in-silico studies focus mainly on P. falciparum DHODH and no other parasite enzymes or proteins like PfHsp90, there is need to widen the scope of antimalarial drugs discovery. This study sought to discover PfHsp90 inhibitors as antimalaria drugs.
Virtual screening process was performed using GDM as the ligand to identify the best scoring natural compounds with the ability to inhibit PfHsp90. Due to GDM’s vast stereochemical properties that could limit the number of ZINCPHARMER hits obtained, a pharmacophore model was developed using some of its pharmacophore features. The pharmacophore-based virtual screening yielded 17 hits (
Table 2). All the ZINCPHARMER hits were subjected to other in-silico processes because of their low RMSD values (below 1 Å), which suggests that all the hits are structurally similar to GDM. Alzain et al. (2022) and Oduselu et al. (2023) performed pharmacophore-based virtual screening and focused on RMSD values of their hits to determine the most suitable inhibitors against different
P. falciparum proteins. Usually, more similar compounds or proteins have small RMSD values (Oduselu et al., 2023). Therefore, RMSD values below 1 Å found in this current study suggests the suitability of all the ZINCPHARMER hits as potential PfHsp90 inhibitors. However, this is subject to confirmation via other in-silico processes like drug-likeness test, ADMET property analysis, molecular docking, and MDS.
Even though several studies perform molecular docking of lead compounds to target proteins before drug-likeness and ADMET property analysis (Alzain et al., 2022; Gao et al., 2023; Oduselu et al., 2023; Onyango et al., 2022), drug-likeness and ADMET property analysis were done before molecular docking to get rid of all lead compounds with unsuitable drug characteristics. This approach is consistent with other studies (Oduselu et al., 2023; Onyango et al., 2022). The drug-likeness test and ADMET properties analysis yielded 9 potential PfHsp90 inhibitors with drug-likeness and ADMET characteristics for further molecular docking (
Figure 8).
After determining the 9 potential PfHsp90 inhibitors, our intent to determine the stability of the complexes they form with PfHsp90 necessitated two in-silico process, molecular docking and MDS. These two in-silico approaches are common in drug design and discovery (Onyango, 2023). Regarding the binding affinity of GDM to PfHsp90 (-7.5 kcal/mol), only five of the nine PfHsp90 inhibitors had better binding affinities: ZINC72163401 (-7.7 kcal/mol), ZINC72133064 (-7.8 kcal/mol), ZINC09060002 (-8.2 kcal/mol), ZINC72358557 (-7.6 kcal/mol), and ZINC72358537 (-8.1 kcal/mol) (
Figure 9). This means that five PfHsp90 inhibitors bind more strongly to PfHsp90 than GDM.
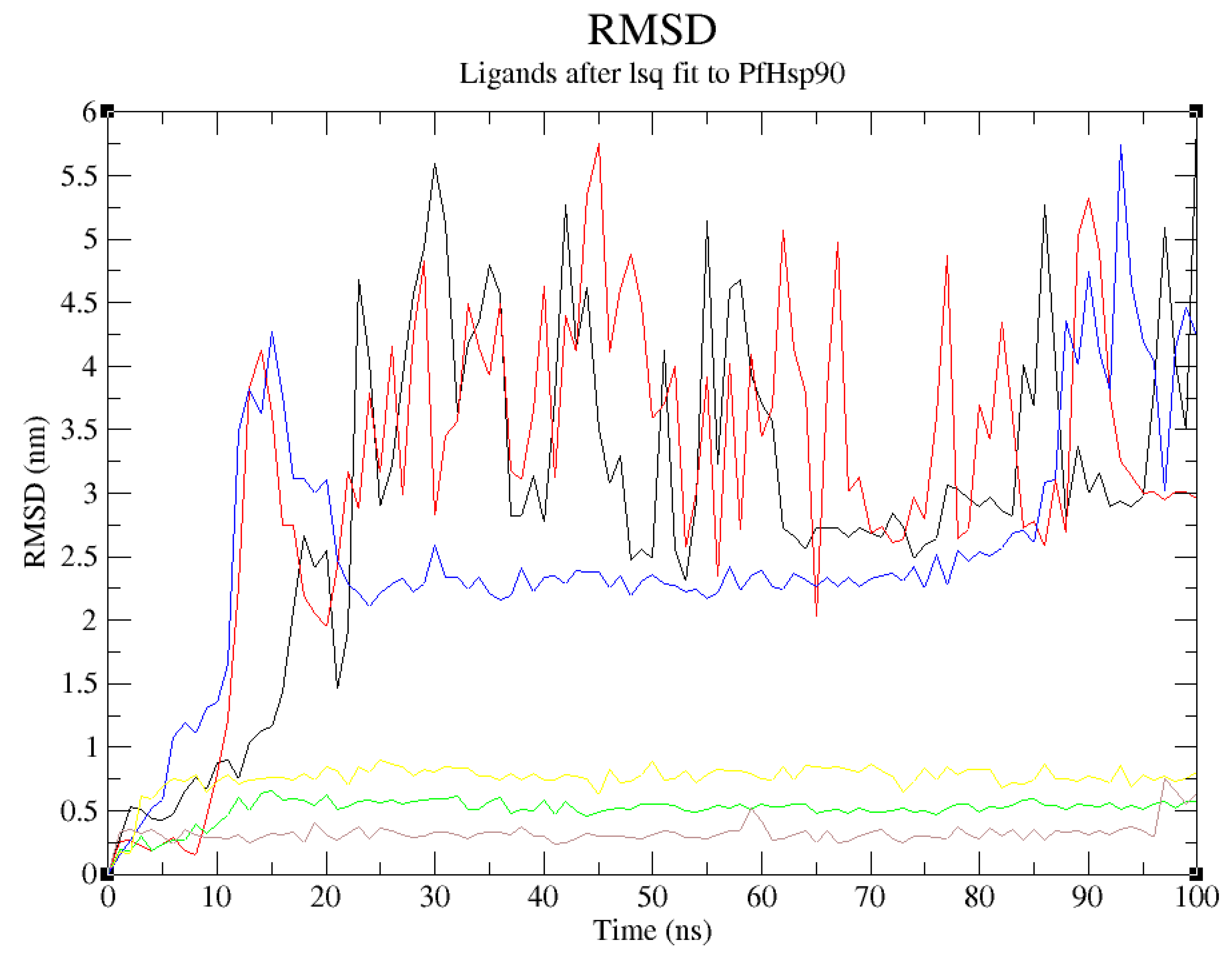
Despite the positive results of the molecular docking, which validated this study’s design rationale, additional MDS studies were performed to confirm and validate the stability of PfHsp90-ligands complexes. To identify, analyze, and provide insights for future lead optimization, six molecular dynamic simulation tests were done. The stability of the ligand when complexed with PfHsp90 and its binding pocket is shown by the ligands’ RMSD analysis. The RMSD was used to evaluate the structural changes of the six PfHsp90-ligands complexes. In this study, a stable ligand-protein interaction was indicated by the plot of ligand RMSD vs time (100 ns), which was within the average limits . This finding is true for all the lead compounds except ZINC09060002 and ZINC72133064 that did not demonstrate the required stability, evidenced by the high RMSD values reaching 4nm (
Figure 10).
ZINC72163401, ZINC72358537, and ZINC72358557, with average RMSD value of 0.25nm suggested that the conformation of the complex formed between these candidate drugs and PfHsp90 remained relatively stable throughout the 100ns simulation. Usually, low RMSD values regarding the true binding pose between a ligand and target protein suggest low binding energy or high binding affinity that facilitates stability (Zheng et al., 2022). Oduselu et al., (2023) concluded that protein-ligand complexes that deviate at distances between 1.5 and 4.0 Å, with an average RMSD of less than 3 Å remain relatively stable throughout the MD simulation. Similarly, Alzain et al. (2022) found out that the ligand and protein RMSD in their study remained between 1.125 and 2.25 Ǻ, indicating that the conformations attained from MDS were structurally stable and ideal for further in-silico assessment. These findings are consistent with those of the current study, proving that ZINC72163401, ZINC72358537, and ZINC72358557 form stable complexes with PfHsp90.
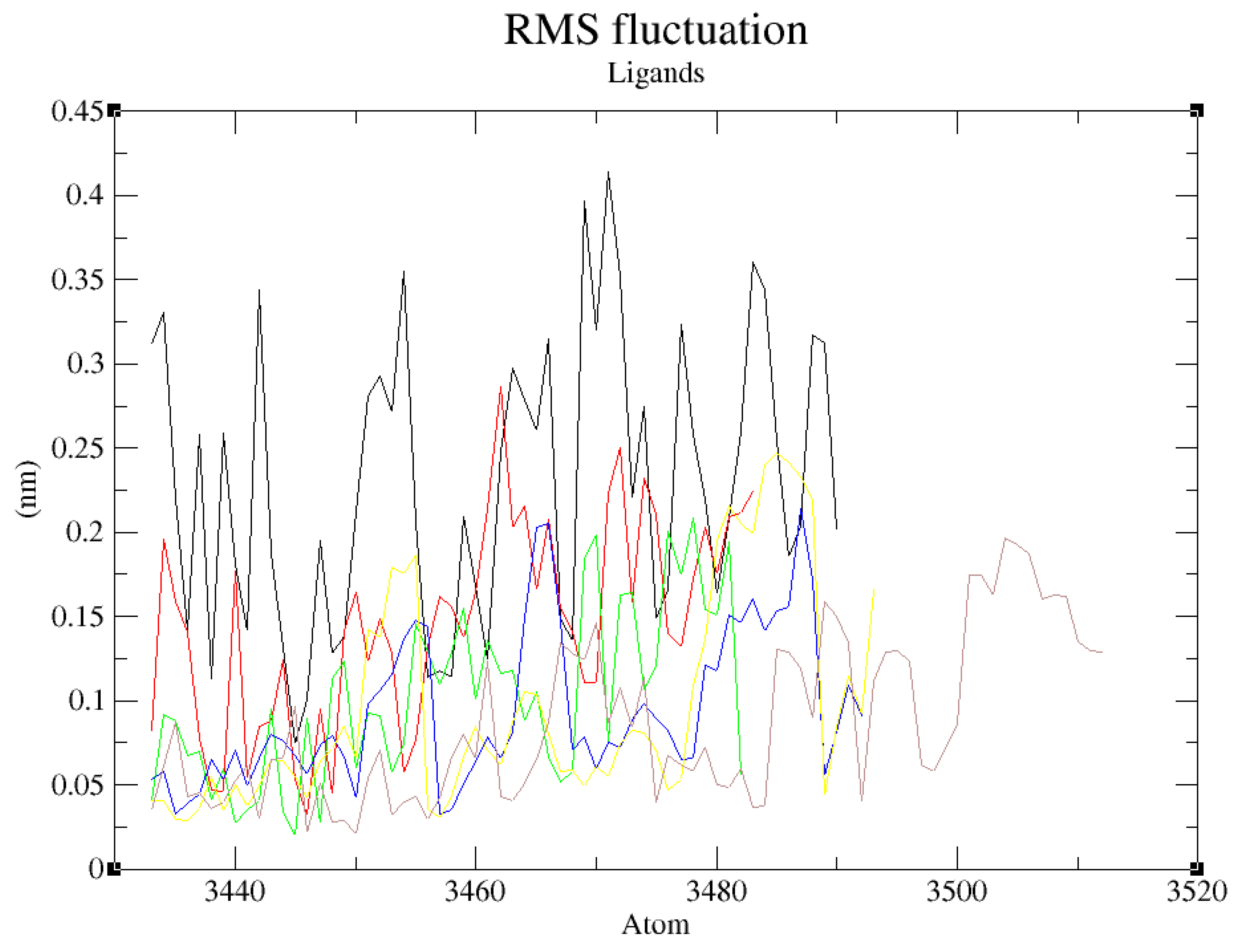
Further RMSF and hydrogen bonds analysis confirmed the stability of the complexes formed between ZINC72163401, ZINC72358557, and ZINC72358537 with PfHsp90. The fluctuations were within RMSF values of 0.2nm, which is acceptable when compared with the RMSF value of the reference ligand (GDM). There were no major fluctuations to indicate that the inhibitors’ atoms shift from their average positions during the 100ns simulation (
Figure 11). This finding is consistent with Oduselu et al.’s (2023) and Razzaghi-Asl et al.’s (2022) studies that ensured RMSF values of their protein-ligand complexes are within acceptable levels to infer their stability.
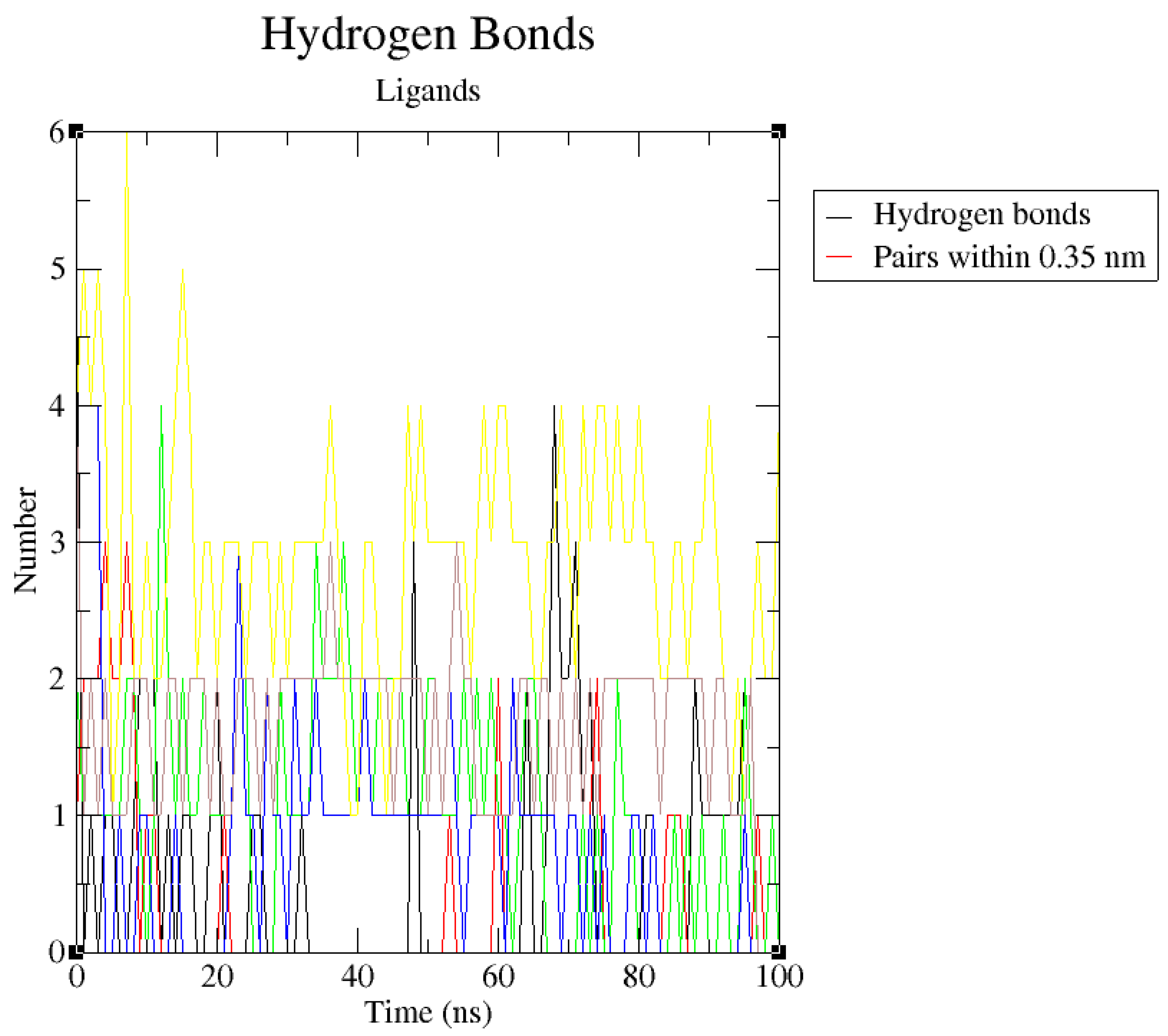
The hydrogen bond analyses (
Figure 12) demonstrated that the potential PfHsp90 inhibitors maintained stable conformation in PfHsp90’s active site during the 100ns simulation, signifying their inhibitory capability. Hydrogen bond formation between a ligand and target protein is essential in complexes’ stability because it increases the binding affinity between the two molecules (Oduselu et al., 2023). Oduselu et al. (2023) discovered that the nine hydrogen bonds between ASP 430 as the acceptor and CSMS00081585868 as the donor increased their binding affinity and inhibitory potential. This finding suggests that ZINC09060002, ZINC72163401, and ZINC72358537 that forms a maximum of 4 hydrogen bonds with PfHsp90 and ZINC72358557 that forms 6, all higher than the 3 hydrogen bonds formed between PfHsp90 and GDM, are bound more tightly to the target protein and might possess better inhibitory capability than GDM.
Since
in vitro validation of the inhibitory potential and capability of the lead compounds was also a primary objective of this study, ascertaining their stability in complex with their target proteins was not enough.
In vitro validation has become a popular approach following MDS in recent drug design and discovery processes (Onyango, 2023). Cheng et al. (2023), Kant et al. (2022), and Ornnork et al. (2020) performed
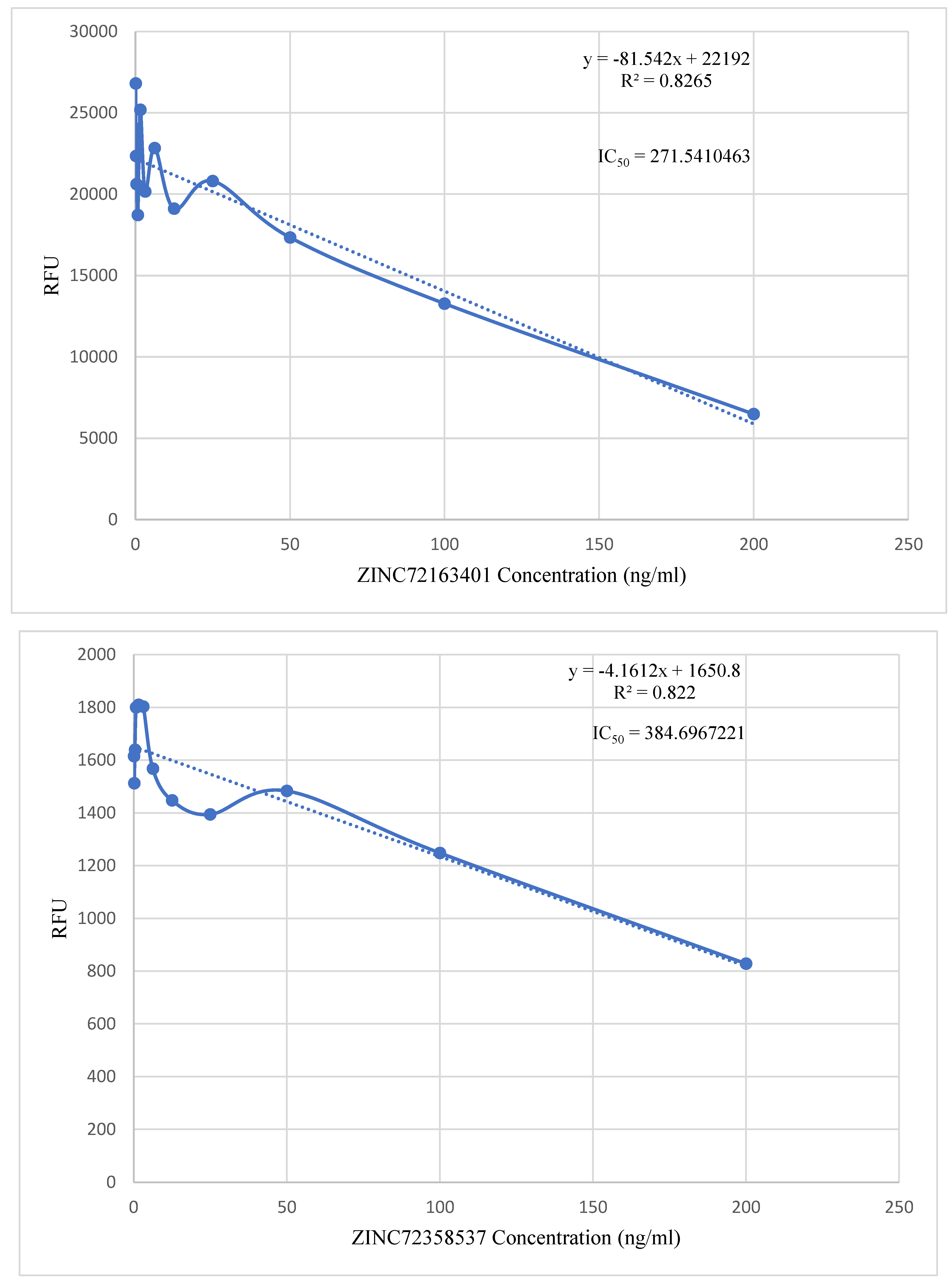
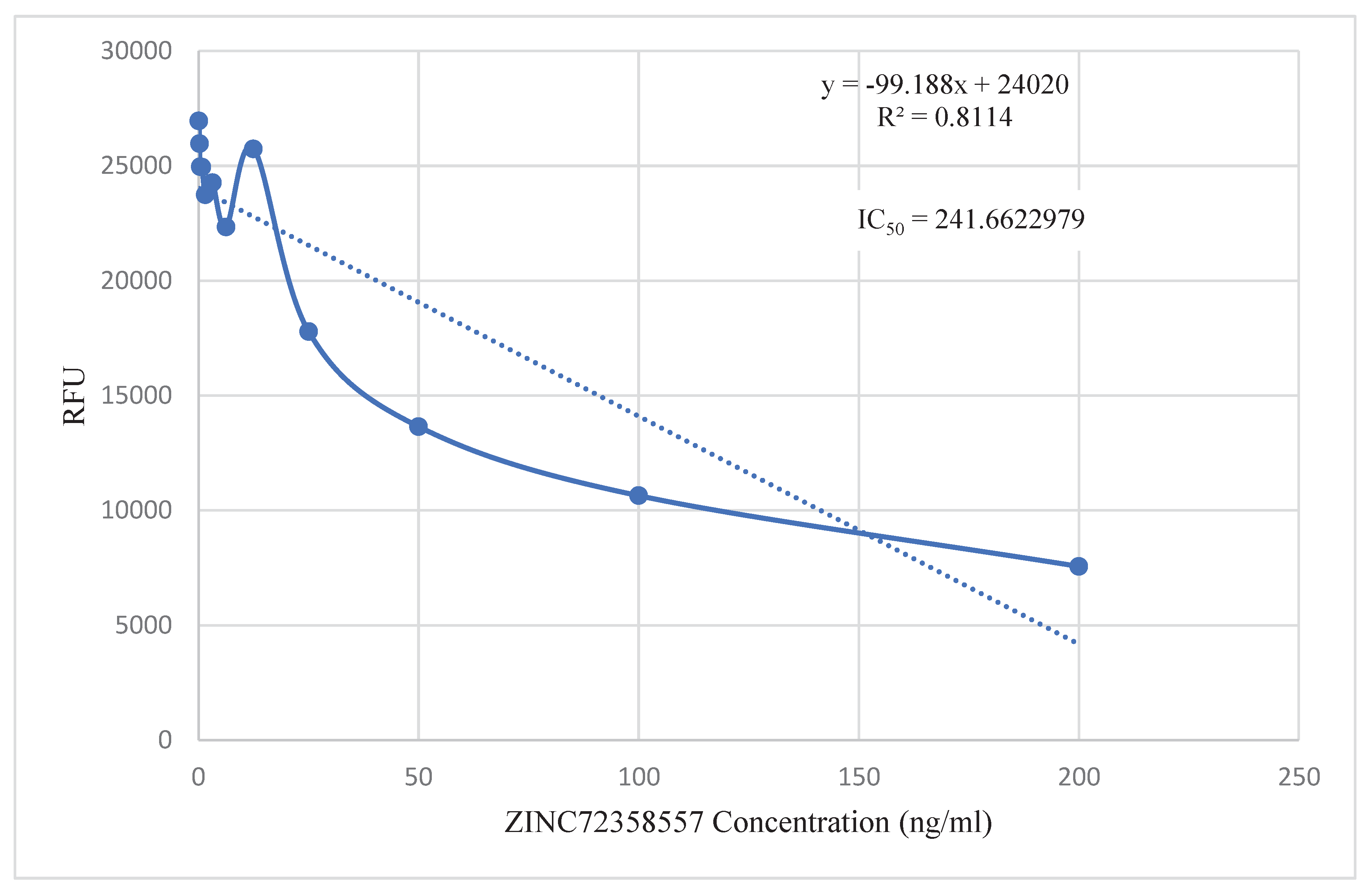
in vitro validation of the lead compounds they discovered in their respective studies. Even though they employed different assays in their studies, they had a common goal that this project shares. The point-to-point calculation found the IC
50 values of the three lead compounds to be within 200 – 400 ng/ml (
Figure 13). These IC
50 values can be compared to that of chloroquine to assess their effectiveness levels. Chloroquine is preferred for this comparison because it is utilized to treat susceptible infections with
P.
falciparum,
P.
ovale,
P.
vivax, and
P.
malariae. It is also characterized by low toxicity, extended duration of action, rapid onset, and high tolerance in humans (Zhou et al., 2020).
According to Agarwal et al. (2017), the highest concentration of chloroquine utilized for analysis of chemosensitivity against the 3D strain of P. falciparum is 50nM. Using the formula (nM) = (ng/mL)/(MW in KD), the IC50 value of chloroquine was converted to 25.793 ng/ml. The IC50 values of the three PfHsp90 inhibitors are extremely high compared to that of chloroquine. Therefore, the inhibitors are required in higher concentrations than chloroquine to treat malaria. Even though this is not ideal, at those particular high concentrations, these three PfHsp90 inhibitors might still be effective against malaria. ZINC72163401, ZINC72358537, and ZINC72358557 can act as antimalarial drugs against Plasmodium malaria. However, further structural optimization studies and clinical testing through in vivo approaches should be performed to ascertain the efficacy of PfHsp90 inhibitors as antimalarial drugs.
Figure 1.
Antimalarial drug resistance.
Figure 1.
Antimalarial drug resistance.
Figure 2.
Hsp90 domain organization and functional cycle. (A) Schematic representation of the V-shaped domain organization of the Hsp90 protein. (B) Schematic representation of the Hsp90 functional cycle begins with the binding of ATP to Hsp90, provoking the association of the protein with a client/unfolded protein. Consequently, a closed conformation is adopted following the NTD dimerization after the lid region of Hsp90 closes over the ATP binding pocket. ATP hydrolysis occurs after repositioning the catalytic loop when the MDs associate. The correctly folded client protein is released upon ATP hydrolysis. This protein folding and ATP hydrolysis cycle reoccur after the Hsp90 homodimer regains its unbound open configuration and is primed. Aha1, HOP, and p23 are other co-chaperones that modulate this functional cycle (Stofberg et al., 2021).
Figure 2.
Hsp90 domain organization and functional cycle. (A) Schematic representation of the V-shaped domain organization of the Hsp90 protein. (B) Schematic representation of the Hsp90 functional cycle begins with the binding of ATP to Hsp90, provoking the association of the protein with a client/unfolded protein. Consequently, a closed conformation is adopted following the NTD dimerization after the lid region of Hsp90 closes over the ATP binding pocket. ATP hydrolysis occurs after repositioning the catalytic loop when the MDs associate. The correctly folded client protein is released upon ATP hydrolysis. This protein folding and ATP hydrolysis cycle reoccur after the Hsp90 homodimer regains its unbound open configuration and is primed. Aha1, HOP, and p23 are other co-chaperones that modulate this functional cycle (Stofberg et al., 2021).
Figure 3.
PfHsp90 domain organization and structure view. (A) Schematic model of the different PfHsp90 domains labeled NTD (N-Terminal Domain), L (Linker Region), MD (Middle Domain), and CTD (C-Terminal Domain). (B) Cartoon representation of PfHsp90 proteins. Red spheres represent ATP bound to NTD. The purple and blue colors in the model depict the two PfHsp90 monomers. (C) Surface representation of PfHsp90’s NTD with 60% transparency and colored according to element type (Dutta et al., 2022).
Figure 3.
PfHsp90 domain organization and structure view. (A) Schematic model of the different PfHsp90 domains labeled NTD (N-Terminal Domain), L (Linker Region), MD (Middle Domain), and CTD (C-Terminal Domain). (B) Cartoon representation of PfHsp90 proteins. Red spheres represent ATP bound to NTD. The purple and blue colors in the model depict the two PfHsp90 monomers. (C) Surface representation of PfHsp90’s NTD with 60% transparency and colored according to element type (Dutta et al., 2022).
Figure 4.
Geldanamycin (GDM) as a PfHsp90 inhibitor. The green stick-like structure represents GDM. The dotted lines of different colors display the interaction points between GDM and PfHsp90. The structure is available in the PDB database (https://www.rcsb.org/) and can be retrieved using the PDB ID 1YET. BIOVIA Discovery Studio 2021 was used to find the suitable binding pocket pose and show the points of interaction between the two molecules.
Figure 4.
Geldanamycin (GDM) as a PfHsp90 inhibitor. The green stick-like structure represents GDM. The dotted lines of different colors display the interaction points between GDM and PfHsp90. The structure is available in the PDB database (https://www.rcsb.org/) and can be retrieved using the PDB ID 1YET. BIOVIA Discovery Studio 2021 was used to find the suitable binding pocket pose and show the points of interaction between the two molecules.
Figure 5.
The 3D structure of prepared PfHsp90 NTD. PfHsp90 NTD retrieved from PDB, ID 3K60. All heteroatoms and water molecules removed and polar hydrogens added. The two chains, A and B, are indicated.
Figure 5.
The 3D structure of prepared PfHsp90 NTD. PfHsp90 NTD retrieved from PDB, ID 3K60. All heteroatoms and water molecules removed and polar hydrogens added. The two chains, A and B, are indicated.
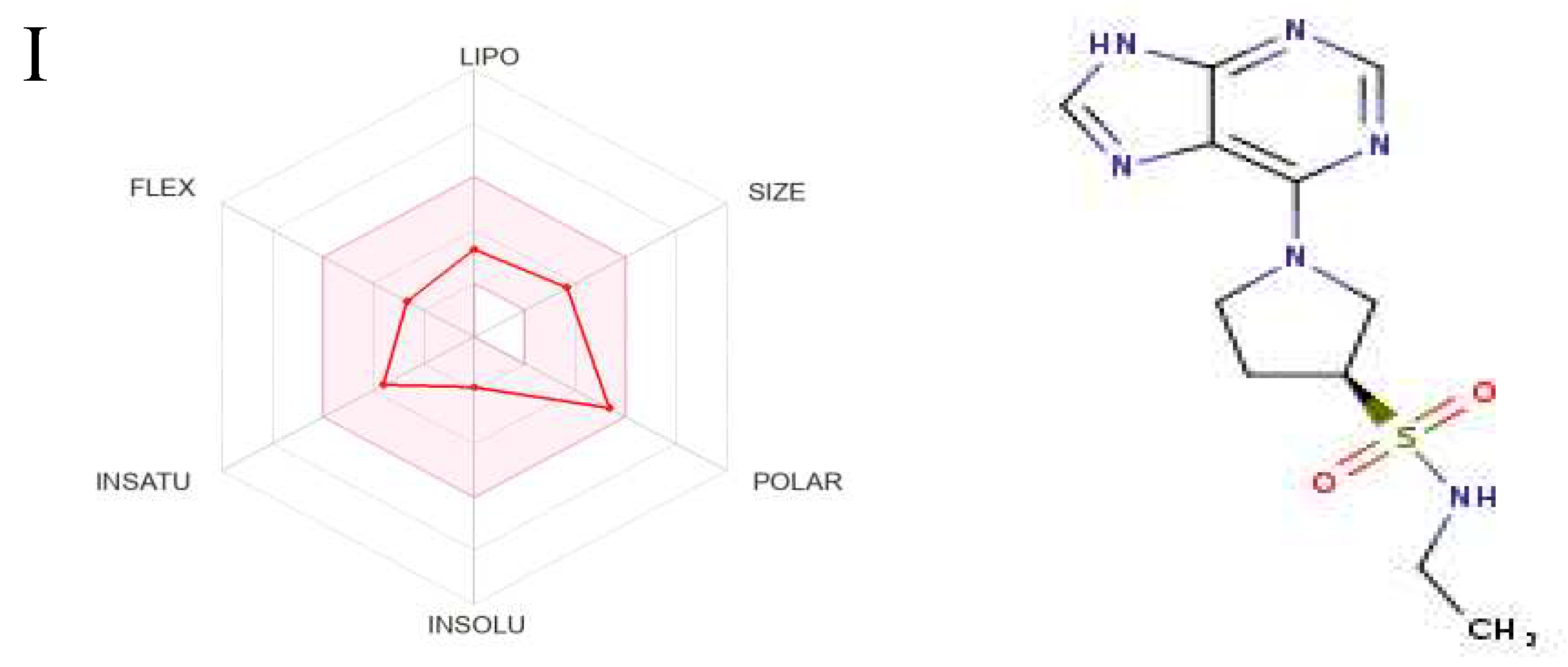
Figure 6.
2D and 3D Structures of GDM. (A) 2D structure. (B) 3D structure.
Figure 6.
2D and 3D Structures of GDM. (A) 2D structure. (B) 3D structure.
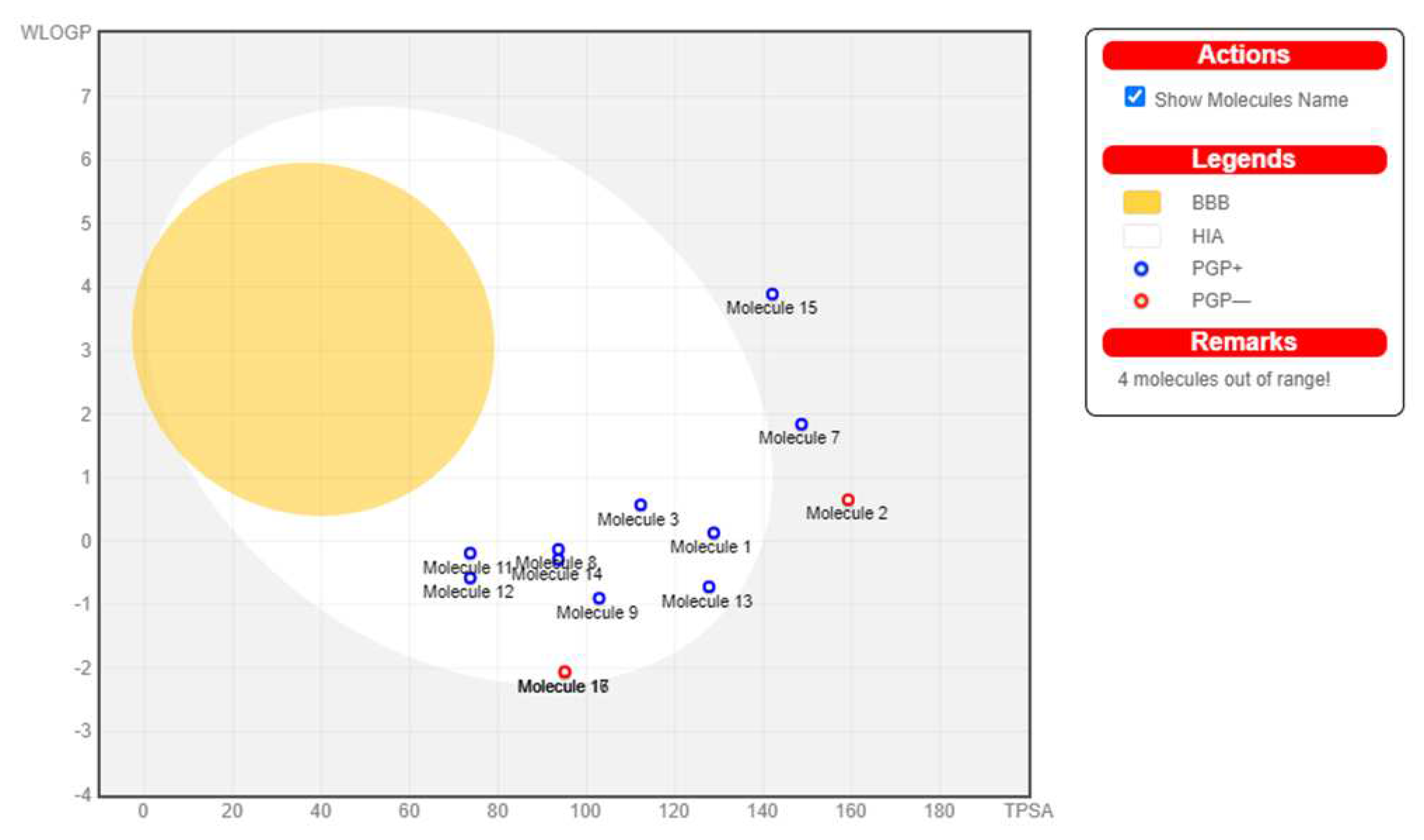
Figure 7.
BOILED-Egg Analysis (GDM). Boiled egg prediction of blood brain barrier permeability and gastrointestinal absorption for the 17 hits. 4 molecules (15, 7, 2, and 16) are out of range, thus excluded. The other molecules are P-glycoprotein (P-gp) substrate, indicated by the blue dot, depicting their ease of excretion from the body.
Figure 7.
BOILED-Egg Analysis (GDM). Boiled egg prediction of blood brain barrier permeability and gastrointestinal absorption for the 17 hits. 4 molecules (15, 7, 2, and 16) are out of range, thus excluded. The other molecules are P-glycoprotein (P-gp) substrate, indicated by the blue dot, depicting their ease of excretion from the body.
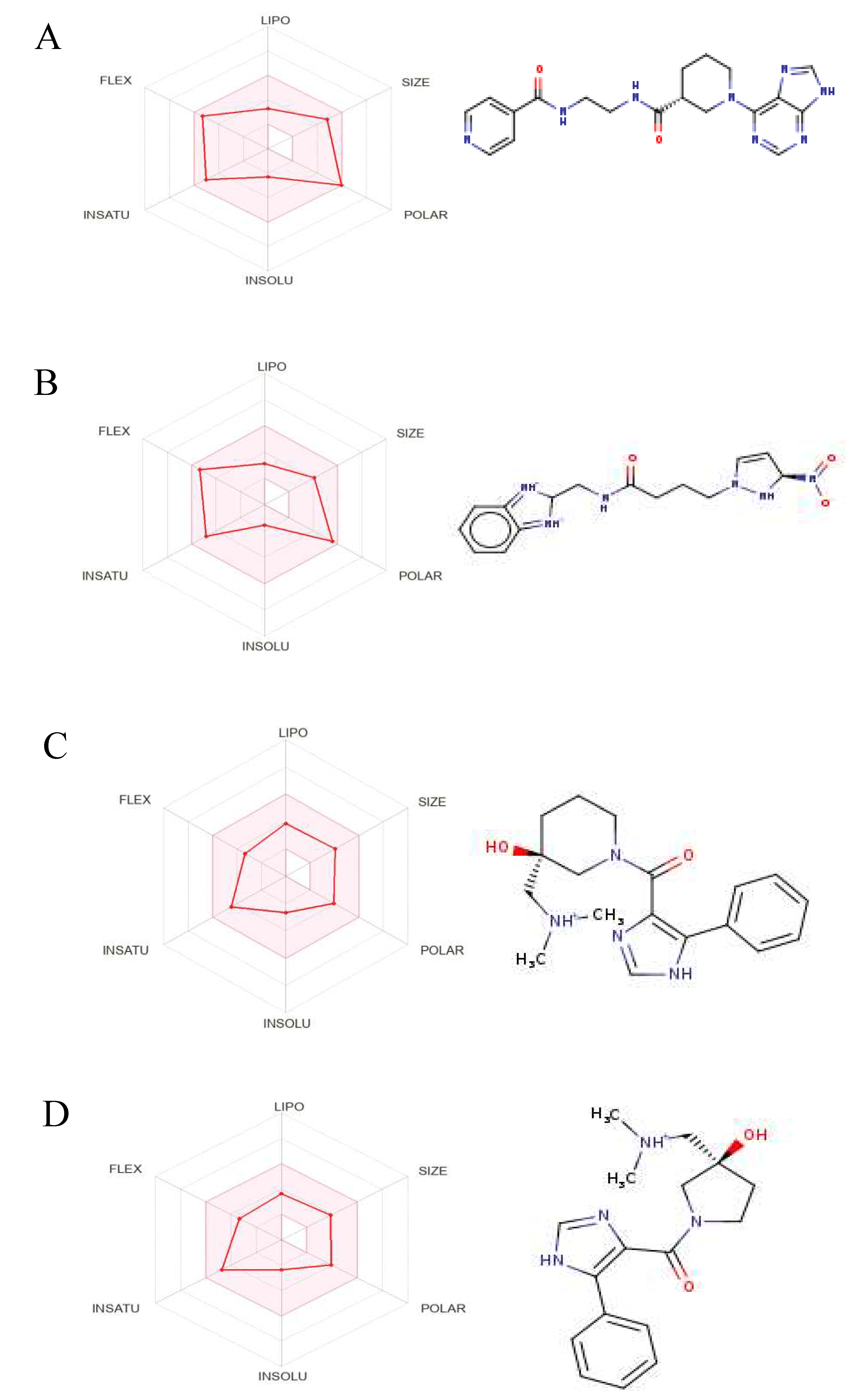
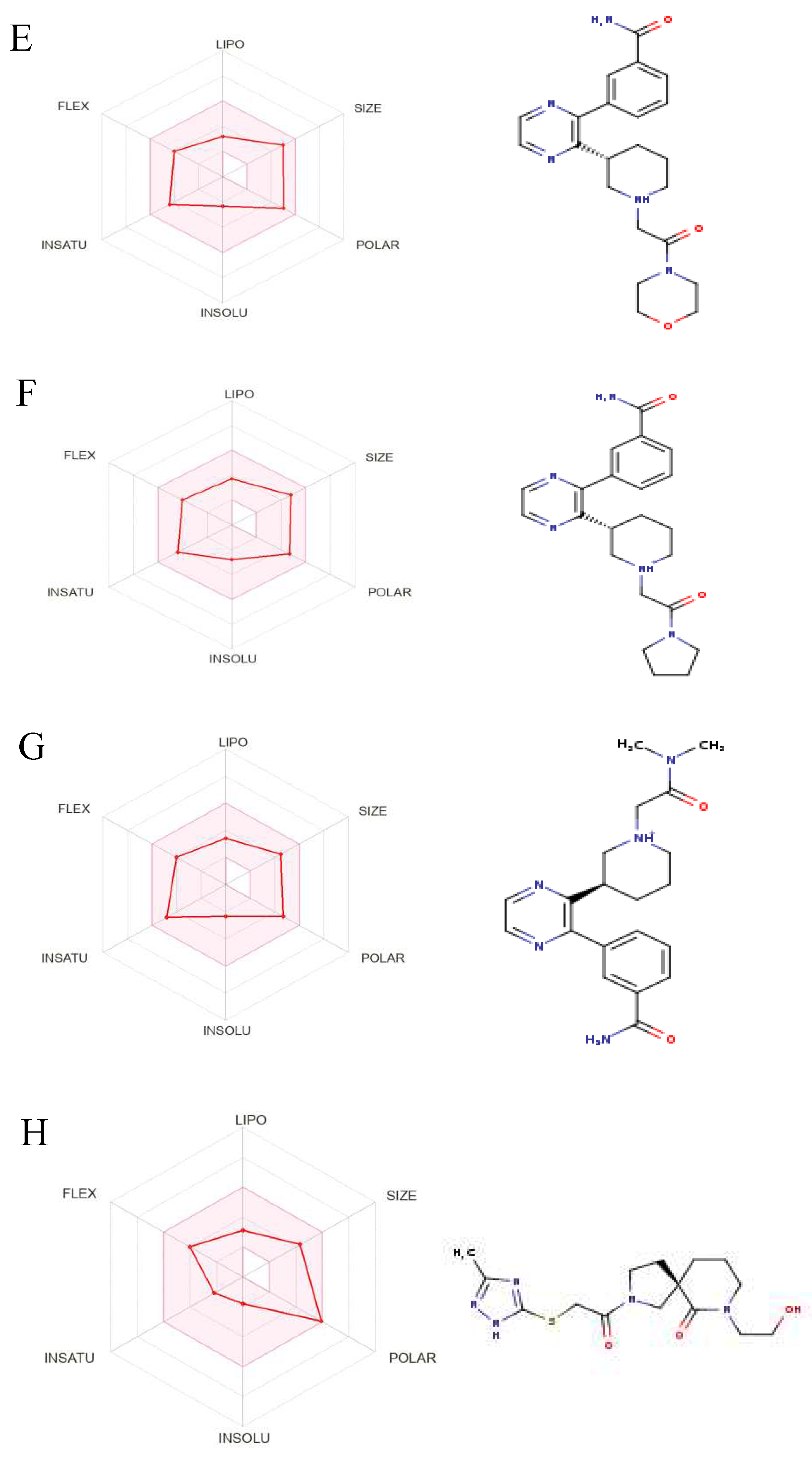
Figure 8.
The Structures and Oral Bioavailability Radars of the 9 Hits (GDM). (A) ZINC09060002, (B) ZINC63526364, (C) ZINC72133064, (D) ZINC72163401, (E) ZINC72358537, (F) ZINC72358557, (G) ZINC72358880, (H) ZINC91416974, (I) ZINC92700801.
Figure 8.
The Structures and Oral Bioavailability Radars of the 9 Hits (GDM). (A) ZINC09060002, (B) ZINC63526364, (C) ZINC72133064, (D) ZINC72163401, (E) ZINC72358537, (F) ZINC72358557, (G) ZINC72358880, (H) ZINC91416974, (I) ZINC92700801.
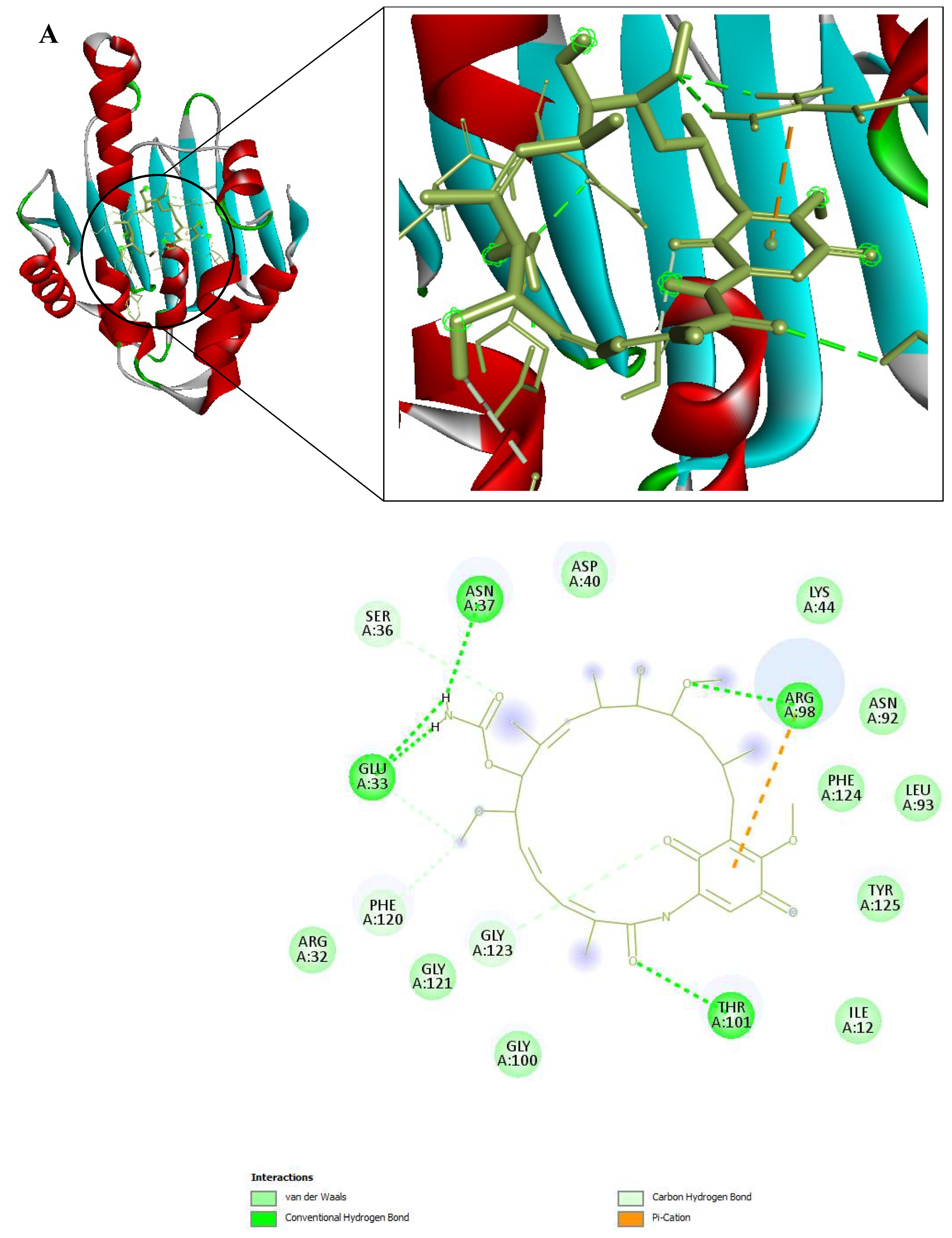
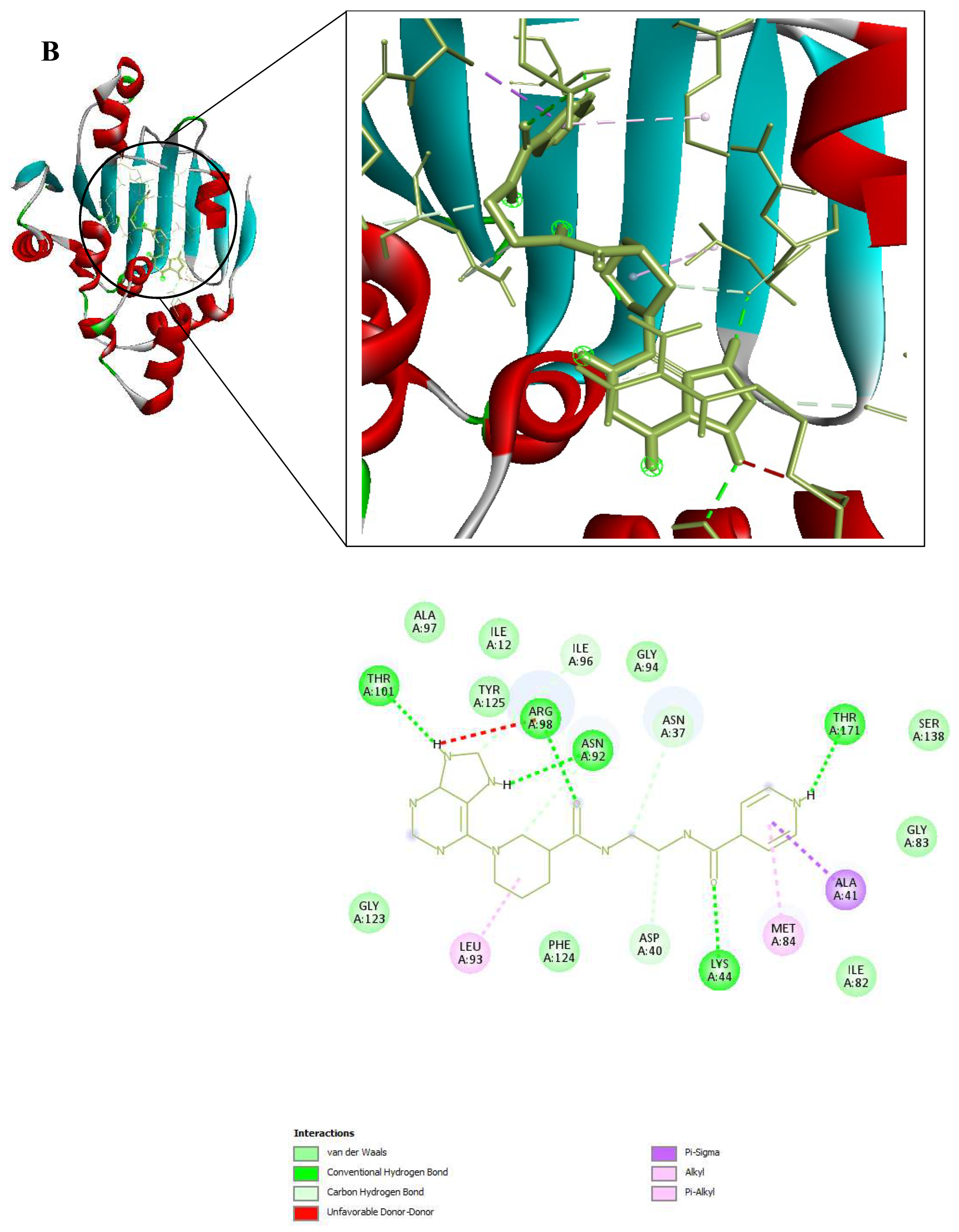
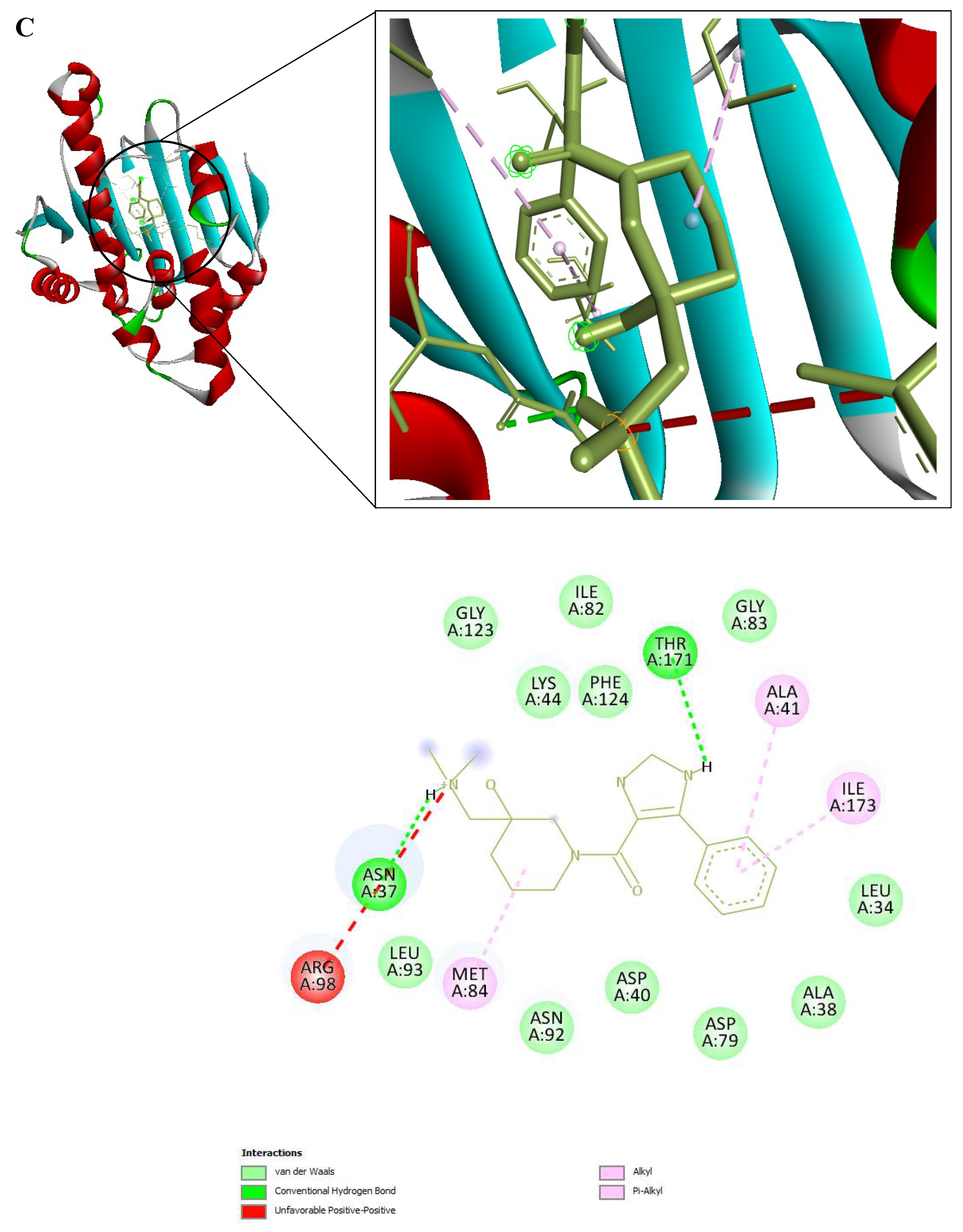
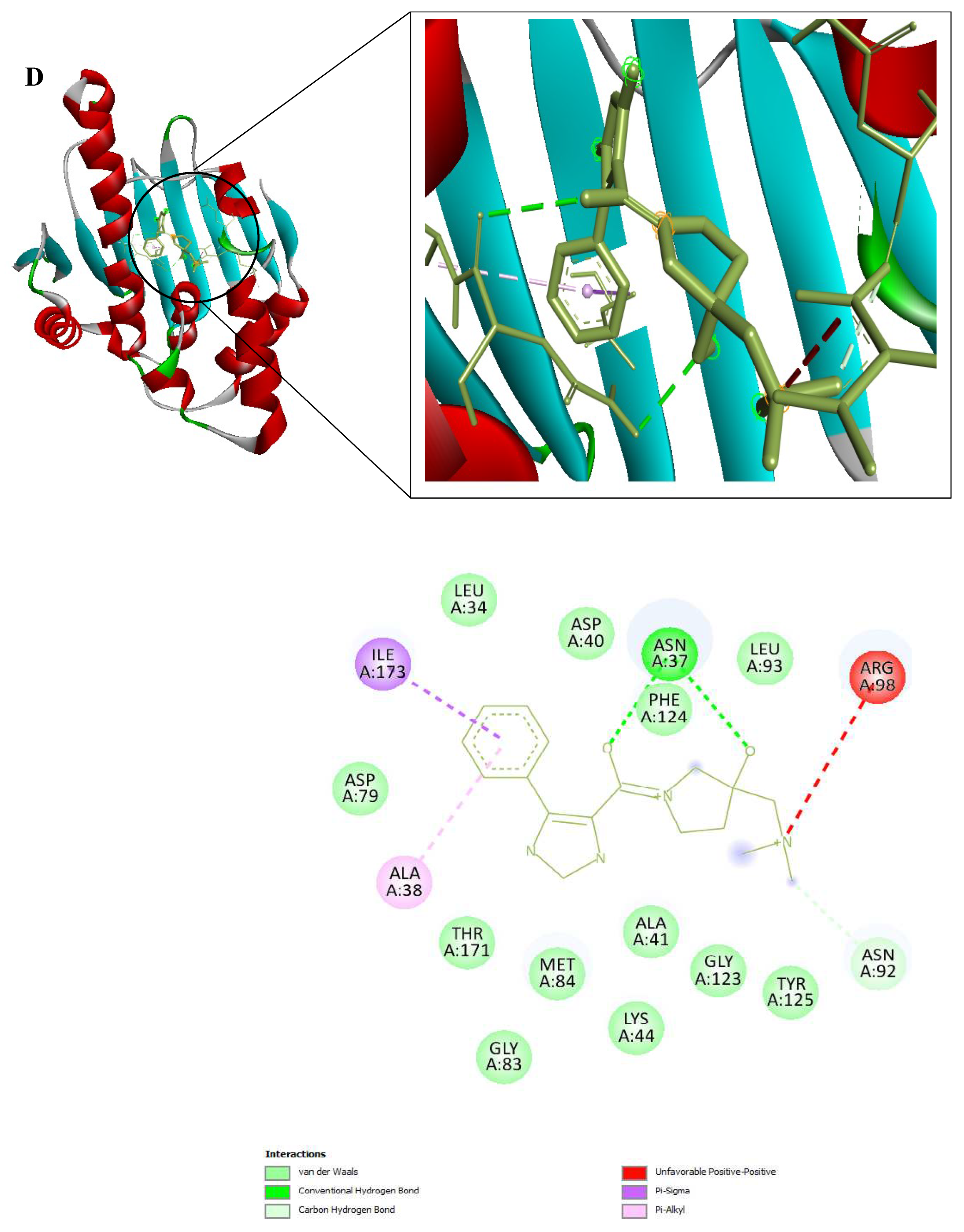
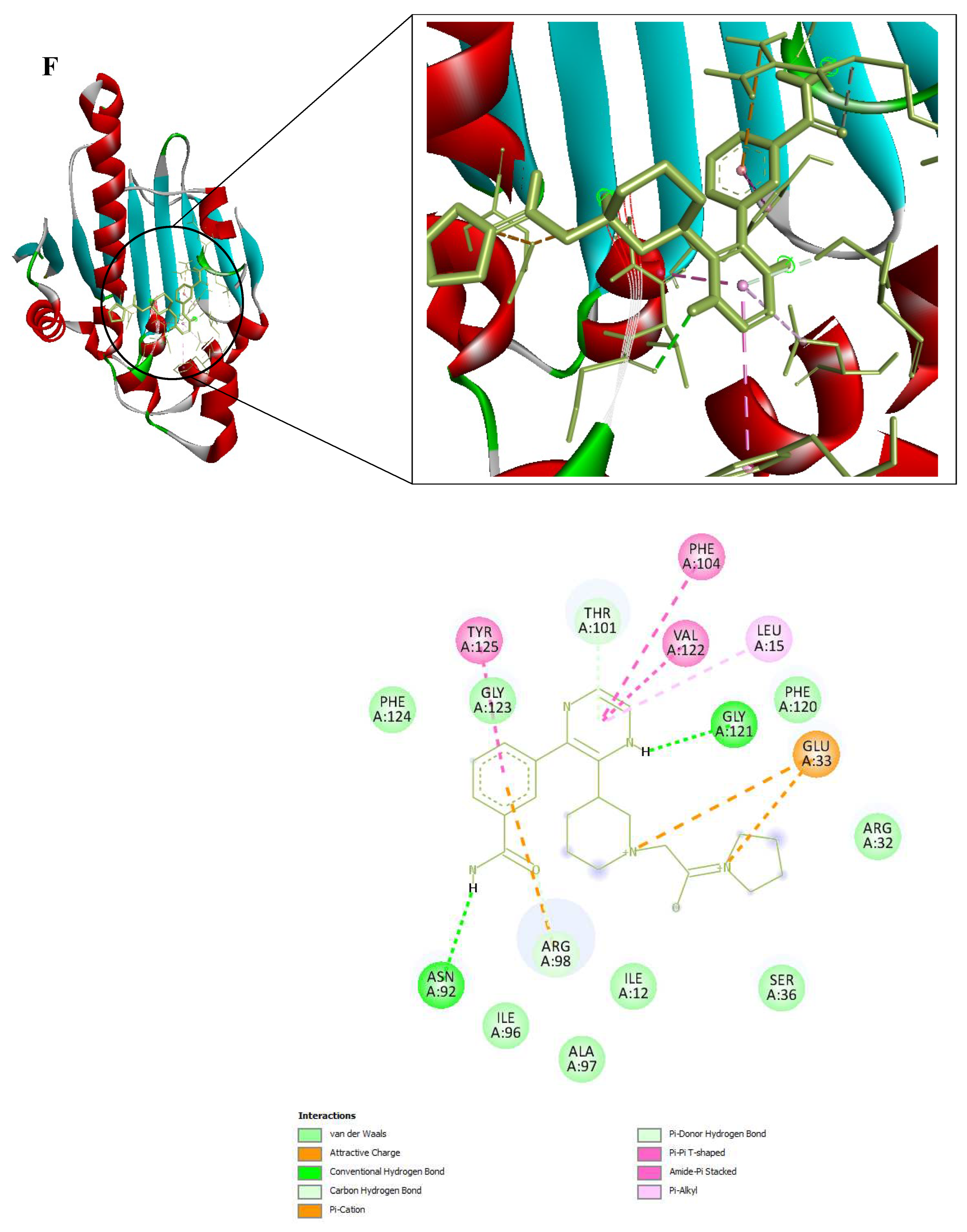
Figure 9.
3D and 2D Interactions of PfHsp90 and the 6 Ligands. (A) PfHsp90 and GDM, with binding affinity of -7.5 kcal/mol. (B) PfHsp90 and ZINC09060002, with binding affinity of -8.2 kcal/mol. (C) PfHsp90 and ZINC72133064, with binding affinity of -7.8 kcal/mol. (D) PfHsp90 and ZINC72163401, with binding affinity of -7.7 kcal/mol. (E) PfHsp90 and ZINC72358537, with binding affinity of -8.1 kcal/mol. (F) PfHsp90 and ZINC72358557, with binding affinity of -7.6 kcal/mol.
Figure 9.
3D and 2D Interactions of PfHsp90 and the 6 Ligands. (A) PfHsp90 and GDM, with binding affinity of -7.5 kcal/mol. (B) PfHsp90 and ZINC09060002, with binding affinity of -8.2 kcal/mol. (C) PfHsp90 and ZINC72133064, with binding affinity of -7.8 kcal/mol. (D) PfHsp90 and ZINC72163401, with binding affinity of -7.7 kcal/mol. (E) PfHsp90 and ZINC72358537, with binding affinity of -8.1 kcal/mol. (F) PfHsp90 and ZINC72358557, with binding affinity of -7.6 kcal/mol.
Figure 10.
RMSD plot of PfHsp90. (PDB ID: 3K60) with GDM as reference ligand and top-five ZINC database compounds as a function of 100ns simulation time. ZINC09060002 (Black), ZINC72133064 (Red), ZINC72163401 (Green), ZINC72358537 (Blue), ZINC72358557 ((Yellow), and GDM (Brown).
Figure 10.
RMSD plot of PfHsp90. (PDB ID: 3K60) with GDM as reference ligand and top-five ZINC database compounds as a function of 100ns simulation time. ZINC09060002 (Black), ZINC72133064 (Red), ZINC72163401 (Green), ZINC72358537 (Blue), ZINC72358557 ((Yellow), and GDM (Brown).
Figure 11.
RMSF plot of GDM as reference ligand and top-five ZINC database compounds. ZINC09060002 (Black), ZINC72133064 (Red), ZINC72163401 (Green), ZINC72358537 (Blue), ZINC72358557 (Yellow), and GDM (Brown).
Figure 11.
RMSF plot of GDM as reference ligand and top-five ZINC database compounds. ZINC09060002 (Black), ZINC72133064 (Red), ZINC72163401 (Green), ZINC72358537 (Blue), ZINC72358557 (Yellow), and GDM (Brown).
Figure 12.
Number of Hydrogen Bonds Plot.
Figure 12.
Number of Hydrogen Bonds Plot.
Figure 13.
Point to Point Plot. IC50 value determination of PfHsp90 inhibitors using point-to-point calculation.
Figure 13.
Point to Point Plot. IC50 value determination of PfHsp90 inhibitors using point-to-point calculation.
Table 1.
Basic Information on GDM.
Table 1.
Basic Information on GDM.
| Molecule |
Name |
PubChem CID |
Molecular Formular (MF) |
Molecular Weight (MW) |
| 1. |
Geldanamycin (GDM) |
5288382 |
C29H40N2O9
|
560.6 |
Table 2.
Basic Information on the Virtual Screening Results using GDM as Ligand.
Table 2.
Basic Information on the Virtual Screening Results using GDM as Ligand.
| No. |
Molecule |
RMSD |
Mass |
RBnds |
| 1 |
ZINC09060002 |
0.363 |
394 |
4 |
| 2 |
ZINC32537723 |
0.380 |
305 |
5 |
| 3 |
ZINC92700801 |
0.291 |
296 |
5 |
| 4 |
ZINC71617232 |
0.257 |
244 |
6 |
| 5 |
ZINC63526364 |
0.342 |
332 |
6 |
| 6 |
ZINC71617229 |
0.270 |
244 |
6 |
| 7 |
ZINC22325332 |
0.352 |
473 |
7 |
| 8 |
ZINC72358557 |
0.332 |
394 |
7 |
| 9 |
ZINC72358537 |
0.331 |
410 |
7 |
| 10 |
ZINC77271253 |
0.240 |
300 |
7 |
| 11 |
ZINC72133064 |
0.279 |
329 |
8 |
| 12 |
ZINC72163401 |
0.410 |
315 |
8 |
| 13 |
ZINC91416974 |
0.373 |
353 |
8 |
| 14 |
ZINC72358880 |
0.330 |
368 |
9 |
| 15 |
ZINC32796276 |
0.300 |
453 |
10 |
| 16 |
ZINC70981147 |
0.245 |
336 |
15 |
| 17 |
ZINC70981147 |
0.249 |
336 |
15 |
Table 3.
Drug-Likeness Test Results of the 17 Molecules.
Table 3.
Drug-Likeness Test Results of the 17 Molecules.
| No. |
Molecule |
Lipinski |
Ghose |
Veber |
Egan |
Muegge |
Drug-Like? |
| 1 |
ZINC09060002 |
Yes |
Yes |
Yes |
Yes |
Yes |
Yes |
| 2 |
ZINC32537723 |
Yes |
Yes |
No |
No |
No |
No |
| 3 |
ZINC92700801 |
Yes |
Yes |
Yes |
Yes |
Yes |
Yes |
| 4 |
ZINC71617232 |
Yes |
No |
Yes |
No |
Yes |
No |
| 5 |
ZINC63526364 |
Yes |
No |
Yes |
Yes |
Yes |
Yes |
| 6 |
ZINC71617229 |
Yes |
No |
Yes |
No |
Yes |
No |
| 7 |
ZINC22325332 |
Yes |
No |
No |
No |
Yes |
No |
| 8 |
ZINC72358557 |
Yes |
Yes |
Yes |
Yes |
Yes |
Yes |
| 9 |
ZINC72358537 |
Yes |
No |
Yes |
Yes |
Yes |
Yes |
| 10 |
ZINC77271253 |
Yes |
No |
Yes |
No |
Yes |
No |
| 11 |
ZINC72133064 |
Yes |
Yes |
Yes |
Yes |
Yes |
Yes |
| 12 |
ZINC72163401 |
Yes |
No |
Yes |
Yes |
Yes |
Yes |
| 13 |
ZINC91416974 |
Yes |
No |
Yes |
Yes |
Yes |
Yes |
| 14 |
ZINC72358880 |
Yes |
Yes |
Yes |
Yes |
Yes |
Yes |
| 15 |
ZINC32796276 |
Yes |
Yes |
No |
No |
Yes |
No |
| 16 |
ZINC70981147 |
Yes |
No |
Yes |
Yes |
Yes |
Yes |
| 17 |
ZINC70981147 |
Yes |
No |
Yes |
Yes |
Yes |
Yes |