Submitted:
24 January 2024
Posted:
24 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
3. Results
3.1. Genome Overview
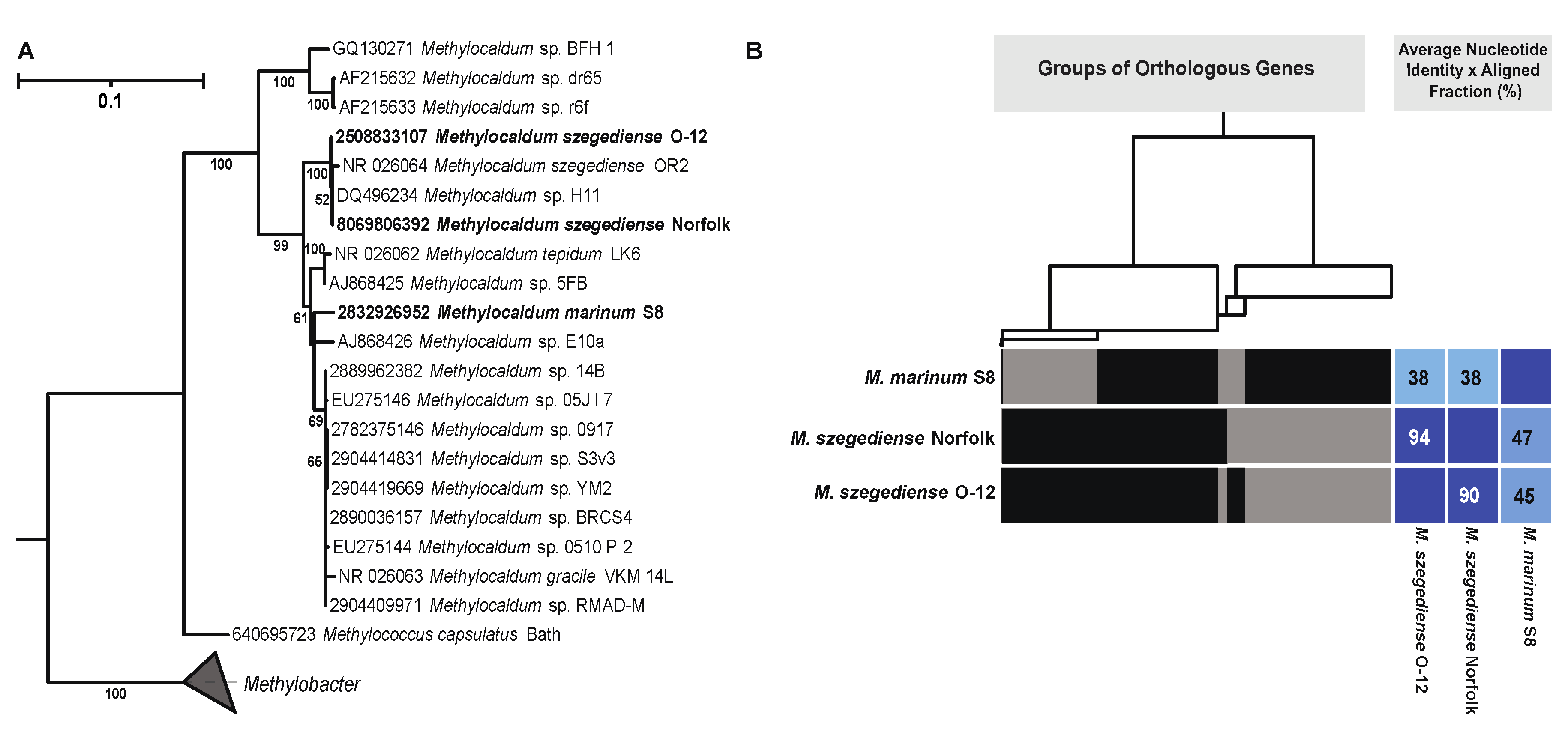
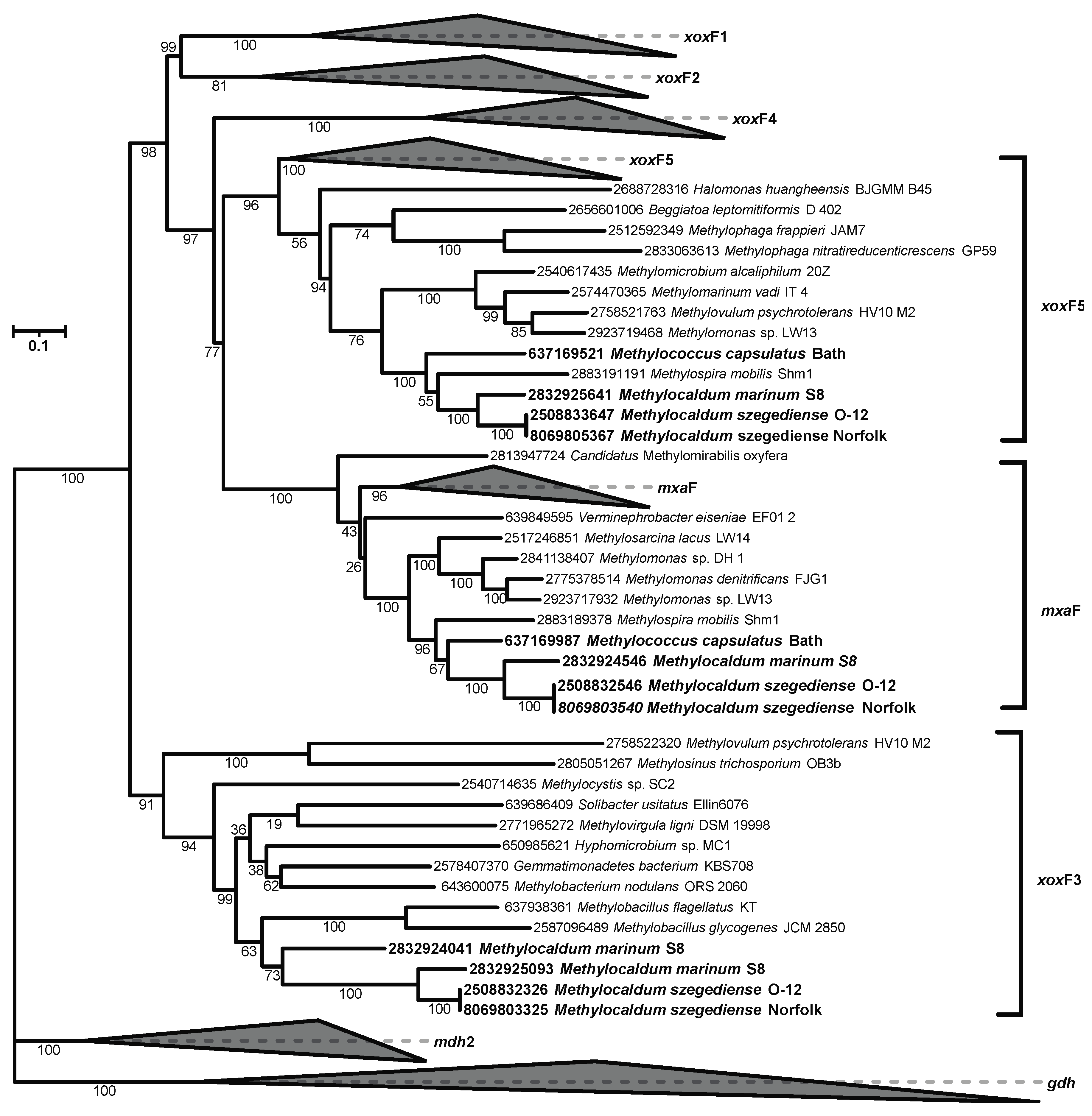
3.2. Methylocaldum Phylogeny
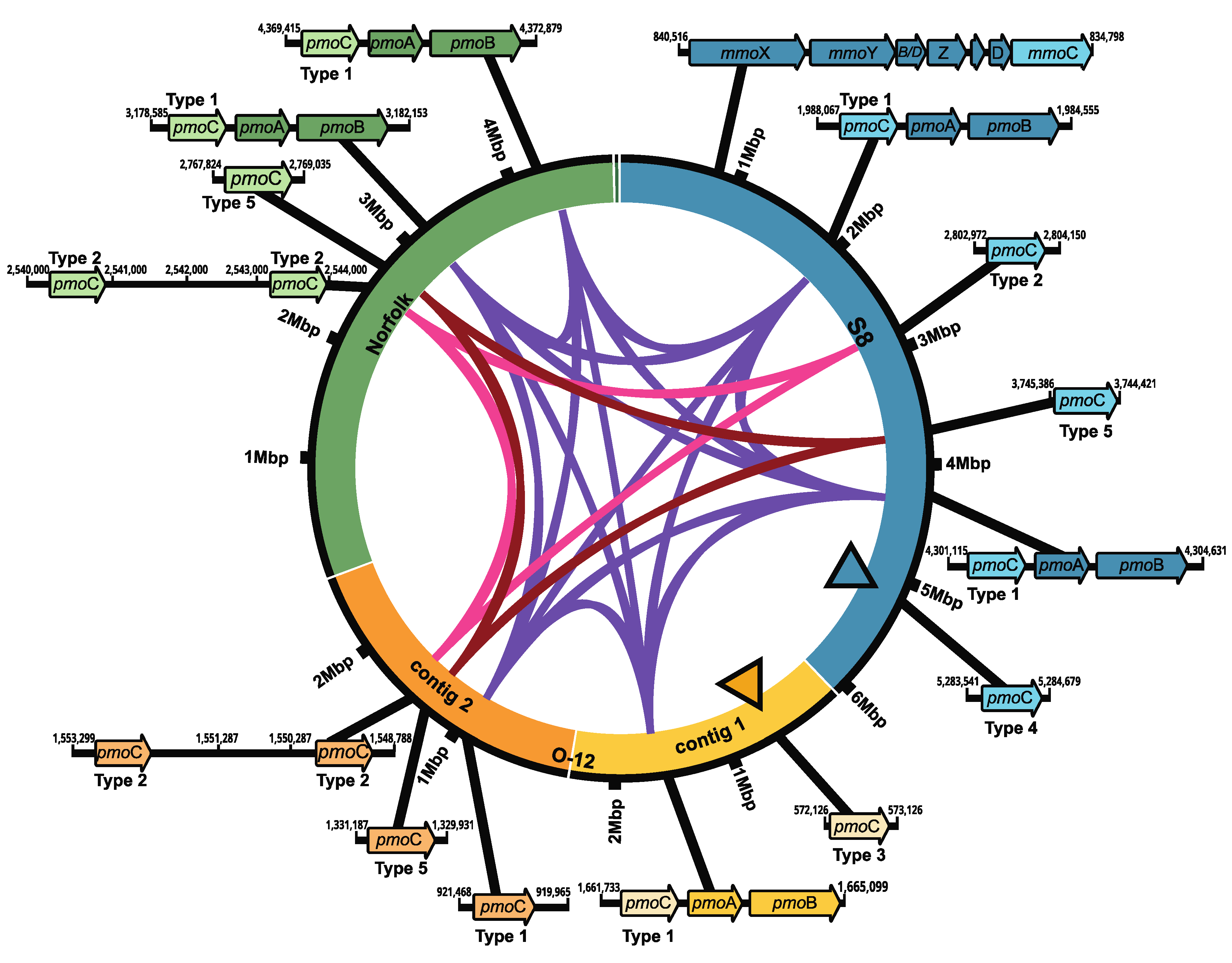
3.3. Synteny of Methane Monooxygenase (MMO) Gene Clusters
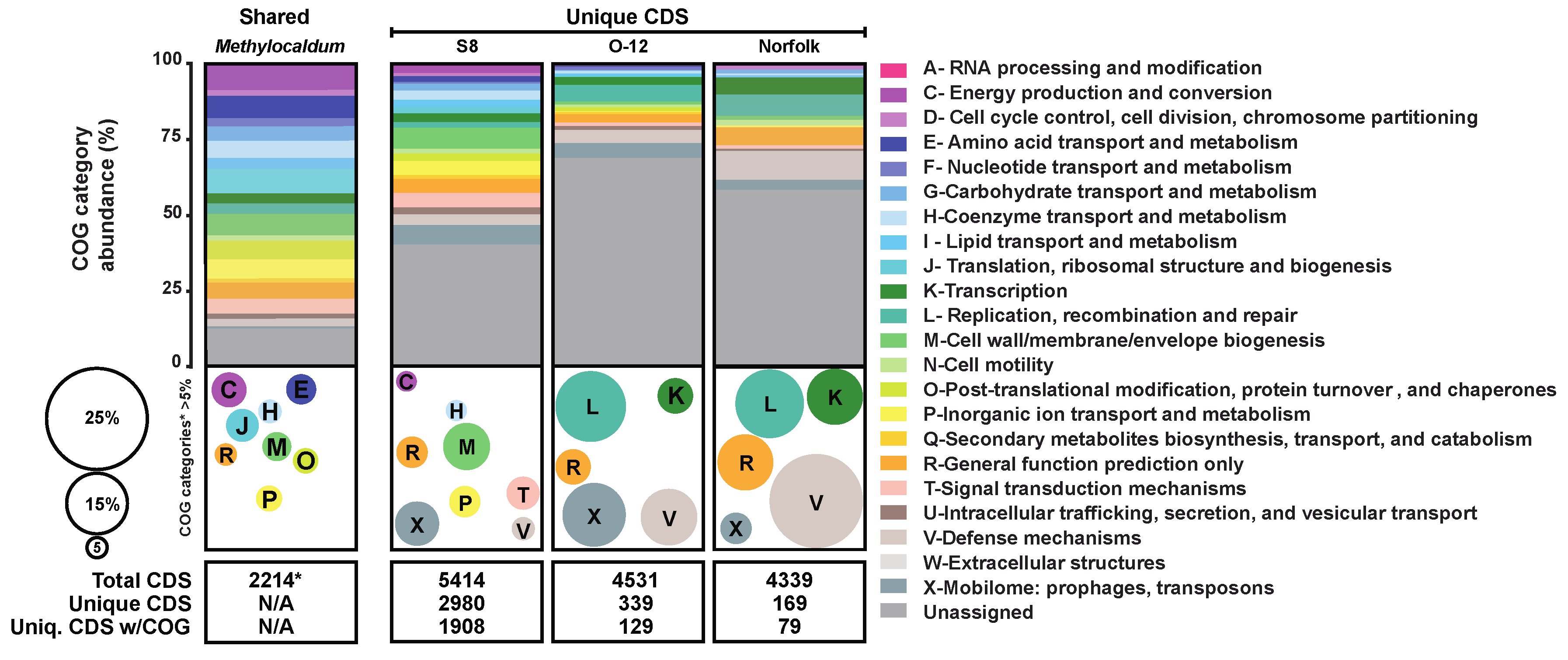
3.4. Comparative Abundance CDS Assigned to COG Categories
3.5. Conjugation
3.6. O-Antigen Biosynthesis
3.7. Catalase
3.8. Cobalamin
3.9. M. marinum S8 Unique Predicted Functions
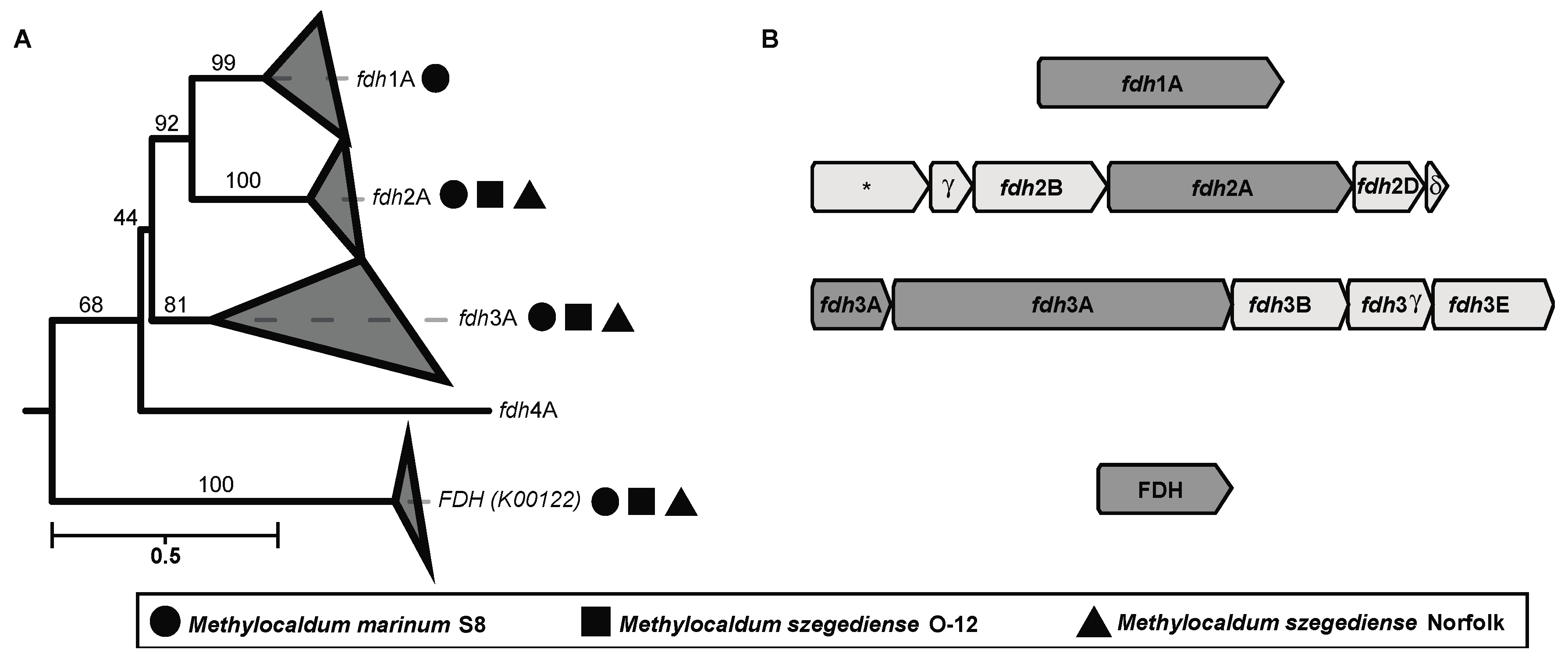
3.10. C1-Oxidation Pathways Are Highly Redundant
3.11. Methylocaldum Potential Single-Carbon Assimilation Pathways
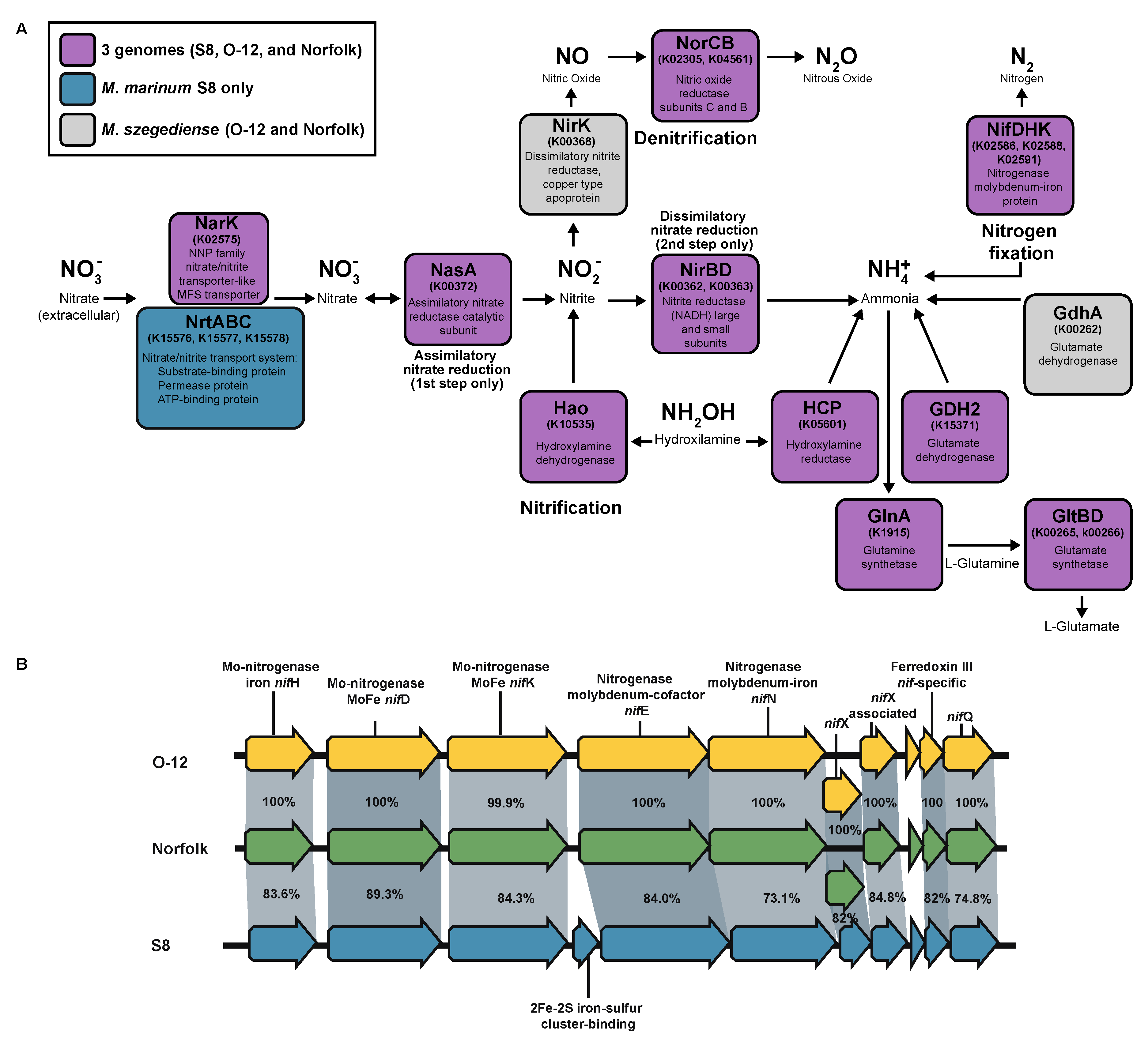
3.12. Nitrogen Metabolism
3.13. Carbon Storage
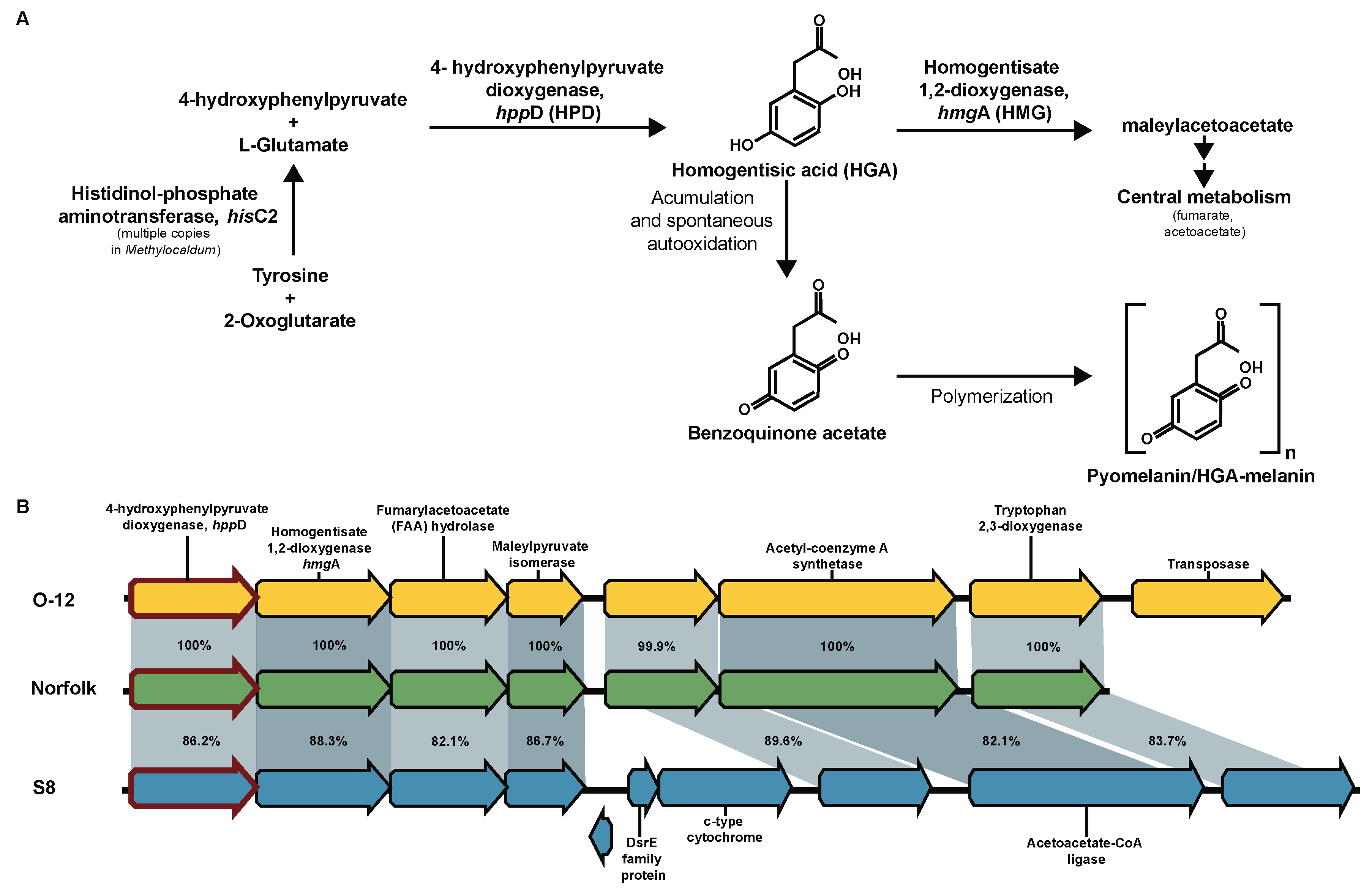
3.14. Pyomelanin/HGA-Melanin Proposed Production
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Dunfield, P.F.; Yuryev, A.; Senin, P.; Smirnova, A.V.; Stott, M.B.; Hou, S.; Ly, B.; Saw, J.H.; Zhou, Z.; Ren, Y. Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature 2007, 450, 879–882. [Google Scholar] [CrossRef]
- Islam, T.; Jensen, S.; Reigstad, L.J.; Larsen, Ø.; Birkeland, N.-K. Methane oxidation at 55 C and pH 2 by a thermoacidophilic bacterium belonging to the Verrucomicrobia phylum. Proc. Natl. Acad. Sci. 2008, 105, 300–304. [Google Scholar] [CrossRef]
- Pol, A.; Heijmans, K.; Harhangi, H.R.; Tedesco, D.; Jetten, M.S.; Op den Camp, H.J. Methanotrophy below pH 1 by a new Verrucomicrobia species. Nature 2007, 450, 874–878. [Google Scholar] [CrossRef]
- Yang, Y.; Shan, J.; Zhang, J.; Zhang, X.; Xie, S.; Liu, Y. Ammonia-and methane-oxidizing microorganisms in high-altitude wetland sediments and adjacent agricultural soils. Appl. Microbiol. Biotechnol. 2014, 98, 10197–10209. [Google Scholar] [CrossRef] [PubMed]
- Bodrossy, L.; Holmes, E.M.; Holmes, A.J.; Kovács, K.L.; Murrell, J.C. Analysis of 16S rRNA and methane monooxygenase gene sequences reveals a novel group of thermotolerant and thermophilic methanotrophs, Methylocaldum gen. nov. Arch. Microbiol. 1997, 168, 493–503. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.Y.; Su, Y.; Zhang, Q.Q.; Bai, Y.; Xia, F.F.; Fang, C.R.; He, R. Vertical profiles of community and activity of methanotrophs in landfill cover soils of different age. J. Appl. Microbiol. 2013, 115, 756–765. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Kong, J.-Y.; Xia, F.-F.; Su, Y.; He, R. Effects of ammonium on the activity and community of methanotrophs in landfill biocover soils. Syst. Appl. Microbiol. 2014, 37, 296–304. [Google Scholar] [CrossRef] [PubMed]
- Sow, S.; Khoo, G.; Chong, L.; Smith, T.; Harrison, P.; Ong, H. Molecular diversity of the methanotrophic bacteria communities associated with disused tin-mining ponds in Kampar, Perak, Malaysia. World J. Microbiol. Biotechnol. 2014, 30, 2645–2653. [Google Scholar] [CrossRef] [PubMed]
- Saidi-Mehrabad, A.; He, Z.; Tamas, I.; Sharp, C.E.; Brady, A.L.; Rochman, F.F.; Bodrossy, L.; Abell, G.C.; Penner, T.; Dong, X. Methanotrophic bacteria in oilsands tailings ponds of northern Alberta. ISME J. 2013, 7, 908–921. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Kim, S.Y.; Kim, P.J.; Madsen, E.L.; Jeon, C.O. Methane emission and dynamics of methanotrophic and methanogenic communities in a flooded rice field ecosystem. FEMS Microbiol. Ecol. 2014, 88, 195–212. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, M.; Kamagata, Y.; Oshima, K.; Hanada, S.; Tamaki, H.; Marumo, K.; Maeda, H.; Nedachi, M.; Hattori, M.; Iwasaki, W. Methylocaldum marinum sp. nov., a thermotolerant, methane-oxidizing bacterium isolated from marine sediments, and emended description of the genus Methylocaldum. Int. J. Syst. Evol. Microbiol. 2014, 64, 3240–3246. [Google Scholar] [CrossRef]
- Eshinimaev, B.T.; Medvedkova, K.; Khmelenina, V.; Suzina, N.; Osipov, G.; Lysenko, A.; Trotsenko, Y.A. New thermophilic methanotrophs of the genus Methylocaldum. Microbiology 2004, 73, 448–456. [Google Scholar] [CrossRef]
- Ward, N.; Larsen, Ø.; Sakwa, J.; Bruseth, L.; Khouri, H.; Durkin, A.S.; Dimitrov, G.; Jiang, L.; Scanlan, D.; Kang, K.H. Genomic insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (Bath). PLoS Biol. 2004, 2, e303. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.M.A.; Chu, K.; Palaniappan, K.; Ratner, A.; Huang, J.; Huntemann, M.; Hajek, P.; Ritter, S.; Varghese, N.; Seshadri, R.; et al. The IMG/M data management and analysis system v.6.0: new tools and advanced capabilities. Nucleic Acids Res. 2021, 49, D751–D763. [Google Scholar] [CrossRef]
- Mukherjee, S.; Stamatis, D.; Bertsch, J.; Ovchinnikova, G.; Sundaramurthi, Jagadish C.; Lee, J.; Kandimalla, M.; Chen, I.M.A.; Kyrpides, N.C.; Reddy, T.B.K. Genomes OnLine Database (GOLD) v.8: overview and updates. Nucleic Acids Res. 2021, 49, D723–D733. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Kuma, K.-i.; Toh, H.; Miyata, T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Kuraku, S.; Zmasek, C.M.; Nishimura, O.; Katoh, K. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 2013, 41, W22–W28. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular evolution and phylogenetics; Oxford University Press, USA: 2000.
- Stecher, G.; Tamura, K.; Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef]
- Eren, A.M.; Esen Ö, C.; Quince, C.; Vineis, J.H.; Morrison, H.G.; Sogin, M.L.; Delmont, T.O. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ 2015, 3, e1319. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; Chen, G.L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Galperin, M.Y.; Wolf, Y.I.; Makarova, K.S.; Vera Alvarez, R.; Landsman, D.; Koonin, E.V. COG database update: focus on microbial diversity, model organisms, and widespread pathogens. Nucleic Acids Res. 2021, 49, D274–D281. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Koonin, E.V.; Lipman, D.J. A genomic perspective on protein families. Science 1997, 278, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Aramaki, T.; Blanc-Mathieu, R.; Endo, H.; Ohkubo, K.; Kanehisa, M.; Goto, S.; Ogata, H. KofamKOALA: KEGG ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 2020, 36, 2251–2252. [Google Scholar] [CrossRef]
- Delmont, T.O.; Eren, A.M. Linking pangenomes and metagenomes: the Prochlorococcus metapangenome. PeerJ 2018, 6, e4320. [Google Scholar] [CrossRef]
- Tamura, K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+ C-content biases. Mol. Biol. Evol. 1992, 9, 678–687. [Google Scholar]
- Whelan, S.; Goldman, N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001, 18, 691–699. [Google Scholar] [CrossRef]
- Chen, L.-X.; Méheust, R.; Crits-Christoph, A.; McMahon, K.D.; Nelson, T.C.; Slater, G.F.; Warren, L.A.; Banfield, J.F. Large freshwater phages with the potential to augment aerobic methane oxidation. Nat. microbiol. 2020, 5, 1504–1515. [Google Scholar] [CrossRef]
- Bragagnolo, N.; Audette, G.F. Solution characterization of the dynamic conjugative entry exclusion protein TraG. Struct. Dyn. 2022, 9. [Google Scholar] [CrossRef]
- Lawley, T.; Klimke, W.; Gubbins, M.; Frost, L. F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 2003, 224, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Bowden, M.G.; Pershad, R.; Kaplan, H.B. The Myxococcus xanthus rfbABC operon encodes an ATP-binding cassette transporter homolog required for O-antigen biosynthesis and multicellular development. J. Bacteriol. 1996, 178, 1631–1639. [Google Scholar] [CrossRef]
- Liu, B.; Furevi, A.; Perepelov, A.V.; Guo, X.; Cao, H.; Wang, Q.; Reeves, P.R.; Knirel, Y.A.; Wang, L.; Widmalm, G. Structure and genetics of Escherichia coli O antigens. FEMS Microbiol. Rev. 2020, 44, 655–683. [Google Scholar] [CrossRef] [PubMed]
- Merino, S.; Gonzalez, V.; Tomás, J.M. The first sugar of the repeat units is essential for the Wzy polymerase activity and elongation of the O-antigen lipopolysaccharide. Future Microbiol. 2016, 11, 903–918. [Google Scholar] [CrossRef] [PubMed]
- Valvano, M.A. Export of O-specific lipopolysaccharide. Front. Biosci. (Landmark Ed.) 2003, 8, 452–471. [Google Scholar] [CrossRef]
- Balsanelli, E.; Serrato, R.V.; De Baura, V.A.; Sassaki, G.; Yates, M.G.; Rigo, L.U.; Pedrosa, F.O.; De Souza, E.M.; Monteiro, R.A. Herbaspirillum seropedicae rfbB and rfbC genes are required for maize colonization. Environ. Microbiol 2010, 12, 2233–2244. [Google Scholar] [CrossRef]
- Jefferies, D.; Shearer, J.; Khalid, S. Role of O-antigen in response to mechanical stress of the E. coli outer membrane: insights from coarse-grained MD simulations. J. Phys. Chem. B. 2019, 123, 3567–3575. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, H.; Huang, L.; Zhang, T.; Zong, B.; Ren, X.; Zhu, Y.; Song, F.; Wang, X.; Chen, H. Effect of O antigen ligase gene mutation on oxidative stress resistance and pathogenicity of NMEC strain RS218. Microb. Pathog. 2019, 136, 103656. [Google Scholar] [CrossRef]
- Manieri, F.Z.; Moreira, C.G. Salmonella Typhimurium O-antigen and VisP play an important role in swarming and osmotic stress response during intracellular conditions. Brazilian Journal of Microbiology 2022, 53, 557–564. [Google Scholar] [CrossRef]
- Lerouge, I.; Vanderleyden, J. O-antigen structural variation: mechanisms and possible roles in animal/plant–microbe interactions. FEMS Microbiol. Rev. 2002, 26, 17–47. [Google Scholar] [CrossRef] [PubMed]
- Eyice, Ö.; Myronova, N.; Pol, A.; Carrión, O.; Todd, J.D.; Smith, T.J.; Gurman, S.J.; Cuthbertson, A.; Mazard, S.; Mennink-Kersten, M.A. Bacterial SBP56 identified as a Cu-dependent methanethiol oxidase widely distributed in the biosphere. ISME J. 2018, 12, 145–160. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, R.A.; Mohammadi, S.S.; Van Erven, T.; Berben, T.; Jetten, M.S.; Pol, A.; den Camp, H.J.O. Methanethiol consumption and hydrogen sulfide production by the thermoacidophilic methanotroph Methylacidiphilum fumariolicum SolV. Front. Microbiol. 2022, 13. [Google Scholar] [CrossRef]
- Börjesson, G. Inhibition of methane oxidation by volatile sulfur compounds (CH3SH and CS2) in landfill cover soils. Waste Manag. Res. 2001, 19, 314–319. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Major, T.A.; Whitman, W.B. Role of the precorrin 6-X reductase gene in cobamide biosynthesis in Methanococcus maripaludis. Archaea 2005, 1, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Rodionov, D.A.; Vitreschak, A.G.; Mironov, A.A.; Gelfand, M.S. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J. Biol. Chem. 2003, 278, 41148–41159. [Google Scholar] [CrossRef]
- Santoro, A.E.; Dupont, C.L.; Richter, R.A.; Craig, M.T.; Carini, P.; McIlvin, M.R.; Yang, Y.; Orsi, W.D.; Moran, D.M.; Saito, M.A. Genomic and proteomic characterization of “Candidatus Nitrosopelagicus brevis”: an ammonia-oxidizing archaeon from the open ocean. Proc. Natl. Acad. Sci. U.S.A. 2015, 112, 1173–1178. [Google Scholar] [CrossRef]
- Heal, K.R.; Qin, W.; Ribalet, F.; Bertagnolli, A.D.; Coyote-Maestas, W.; Hmelo, L.R.; Moffett, J.W.; Devol, A.H.; Armbrust, E.V.; Stahl, D.A. Two distinct pools of B12 analogs reveal community interdependencies in the ocean. Proc. Natl. Acad. Sci. U.S.A. 2017, 114, 364–369. [Google Scholar] [CrossRef]
- Lu, X.; Heal, K.R.; Ingalls, A.E.; Doxey, A.C.; Neufeld, J.D. Metagenomic and chemical characterization of soil cobalamin production. ISME J. 2020, 14, 53–66. [Google Scholar] [CrossRef]
- Shelton, A.N.; Seth, E.C.; Mok, K.C.; Han, A.W.; Jackson, S.N.; Haft, D.R.; Taga, M.E. Uneven distribution of cobamide biosynthesis and dependence in bacteria predicted by comparative genomics. ISME J. 2019, 13, 789–804. [Google Scholar] [CrossRef]
- Chistoserdova, L.; Lidstrom, M.E. Molecular and mutational analysis of a DNA region separating two methylotrophy gene clusters in Methylobacterium extorquens AM1. Microbiology 1997, 143, 1729–1736. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Yu, Z.; Chistoserdova, L. Lanthanide-dependent methanol dehydrogenases of XoxF4 and XoxF5 clades are differentially distributed among methylotrophic bacteria and they reveal different biochemical properties. Front. Microbiol. 2018, 9, 1366. [Google Scholar] [CrossRef] [PubMed]
- Keltjens, J.T.; Pol, A.; Reimann, J.; den Camp, H.J.O. PQQ-dependent methanol dehydrogenases: rare-earth elements make a difference. Appl. Microbiol. Biotechnol. 2014, 98, 6163–6183. [Google Scholar] [CrossRef] [PubMed]
- Chistoserdova, L.; Crowther, G.J.; Vorholt, J.A.; Skovran, E.; Portais, J.-C.; Lidstrom, M.E. Identification of a fourth formate dehydrogenase in Methylobacterium extorquens AM1 and confirmation of the essential role of formate oxidation in methylotrophy. J. Bacteriol. 2007, 189, 9076–9081. [Google Scholar] [CrossRef] [PubMed]
- Chistoserdova, L.; Laukel, M.; Portais, J.-C.; Vorholt, J.A.; Lidstrom, M.E. Multiple formate dehydrogenase enzymes in the facultative methylotroph Methylobacterium extorquens AM1 are dispensable for growth on methanol. J. Bacteriol. 2004, 186, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Laukel, M.; Chistoserdova, L.; Lidstrom, M.E.; Vorholt, J.A. The tungsten-containing formate dehydrogenase from Methylobacterium extorquens AM1: purification and properties. Eur. J. Biochem. 2003, 270, 325–333. [Google Scholar] [CrossRef]
- Khmelenina, V.N.; Murrell, J.C.; Smith, T.J.; Trotsenko, Y.A. Physiology and biochemistry of the aerobic methanotrophs. In Aerobic utilization of hydrocarbons, oils and lipids; Springer: 2018; pp. 1–25.
- Schmitz, R.A.; Peeters, S.H.; Versantvoort, W.; Picone, N.; Pol, A.; Jetten, M.S.; Op den Camp, H.J. Verrucomicrobial methanotrophs: ecophysiology of metabolically versatile acidophiles. FEMS Microbiol. Rev. 2021, 45, fuab007. [Google Scholar] [CrossRef]
- Stein, L.Y.; Roy, R.; Dunfield, P.F. Aerobic methanotrophy and nitrification: processes and connections. eLS 2012. [Google Scholar] [CrossRef]
- Gazioglu, O.; Kareem, B.O.; Afzal, M.; Shafeeq, S.; Kuipers, O.P.; Ulijasz, A.T.; Andrew, P.W.; Yesilkaya, H. Glutamate Dehydrogenase (GdhA) of Streptococcus pneumoniae is required for high temperature adaptation. Infect. Immun. 2021, 89. [Google Scholar] [CrossRef]
- Luangthongkam, P. Biosynthesis of Polyhydroxyalkanoates (PHAs) in methane-utilizing mixed cultures. Ph.D. thesis. St Lucia QLD: The University of Queensland. 2019.
- Takeuchi, M. Methylocaldum. Bergey’s Manual of Systematics of Archaea and Bacteria . 2016; 1–5. [Google Scholar] [CrossRef]
- Medvedkova, K.; Khmelenina, V.; Baskunov, B.; Trotsenko, Y.A. Synthesis of melanin by a moderately thermophilic methanotroph Methylocaldum szegediense depends on cultivation temperature. Microbiology 2008, 77, 112–114. [Google Scholar] [CrossRef]
- Singh, S.; Nimse, S.B.; Mathew, D.E.; Dhimmar, A.; Sahastrabudhe, H.; Gajjar, A.; Ghadge, V.A.; Kumar, P.; Shinde, P.B. Microbial melanin: Recent advances in biosynthesis, extraction, characterization, and applications. Biotechnol. Adv. 2021, 53, 107773. [Google Scholar] [CrossRef]
- Donoso, R.A.; Ruiz, D.; Gárate-Castro, C.; Villegas, P.; González-Pastor, J.E.; de Lorenzo, V.; Gonzalez, B.; Pérez-Pantoja, D. Identification of a self-sufficient cytochrome P450 monooxygenase from Cupriavidus pinatubonensis JMP134 involved in 2-hydroxyphenylacetic acid catabolism, via homogentisate pathway. Microb. Biotechnol. 2021, 14, 1944–1960. [Google Scholar] [CrossRef] [PubMed]
- Galeb, H.A.; Lamantia, A.; Robson, A.; König, K.; Eichhorn, J.; Baldock, S.J.; Ashton, M.D.; Baum, J.V.; Mort, R.L.; Robinson, B.J. The Polymerization of Homogentisic Acid In Vitro as a Model for Pyomelanin Formation. Macromol. Chem. Phys. 2022, 223, 2100489. [Google Scholar] [CrossRef]
- Singh, D.; Kumar, J.; Kumar, A. Isolation of pyomelanin from bacteria and evidences showing its synthesis by 4-hydroxyphenylpyruvate dioxygenase enzyme encoded by hppD gene. Int. J. Biol. Macromol. 2018, 119, 864–873. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




