Submitted:
22 January 2024
Posted:
23 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
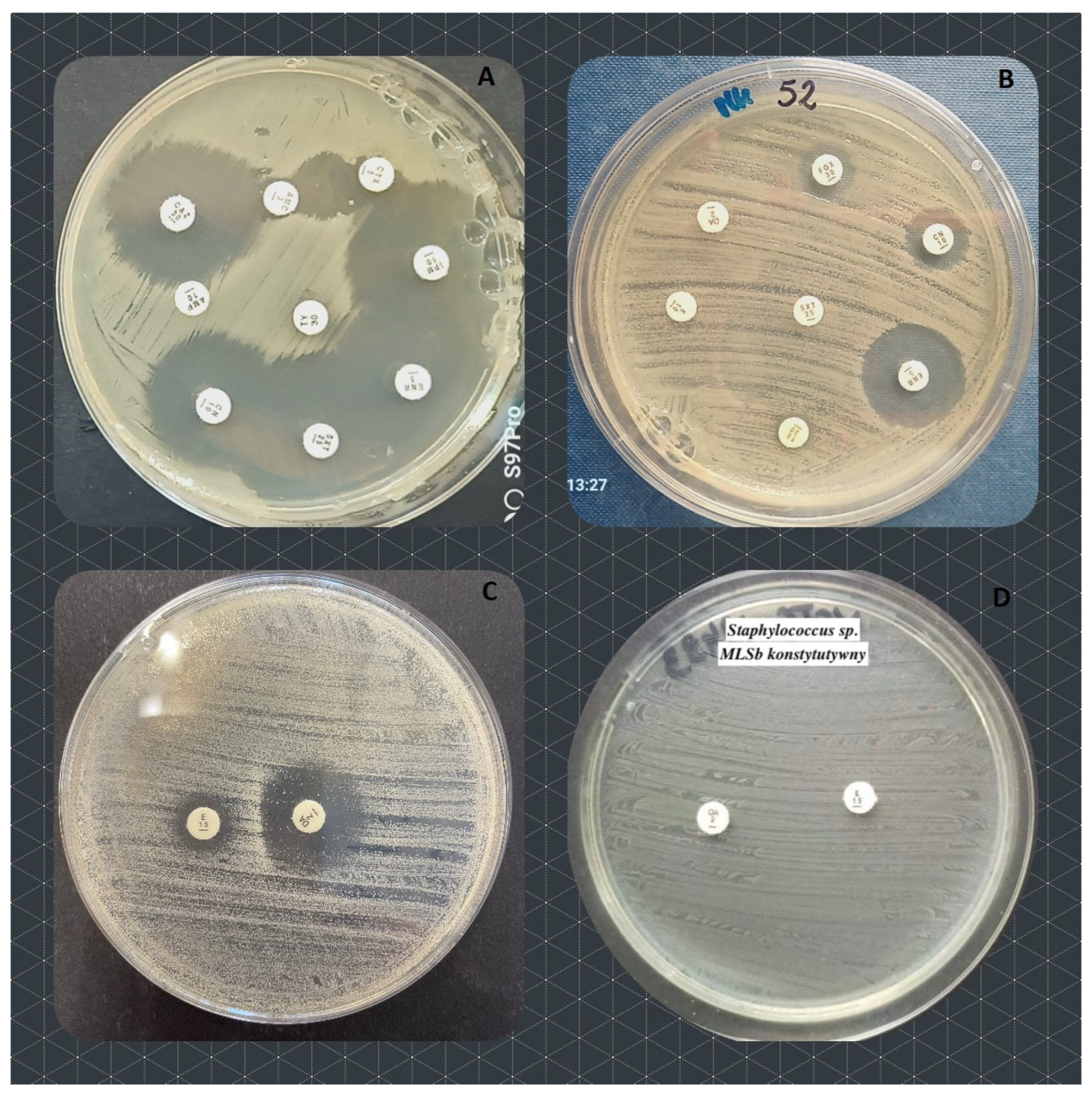
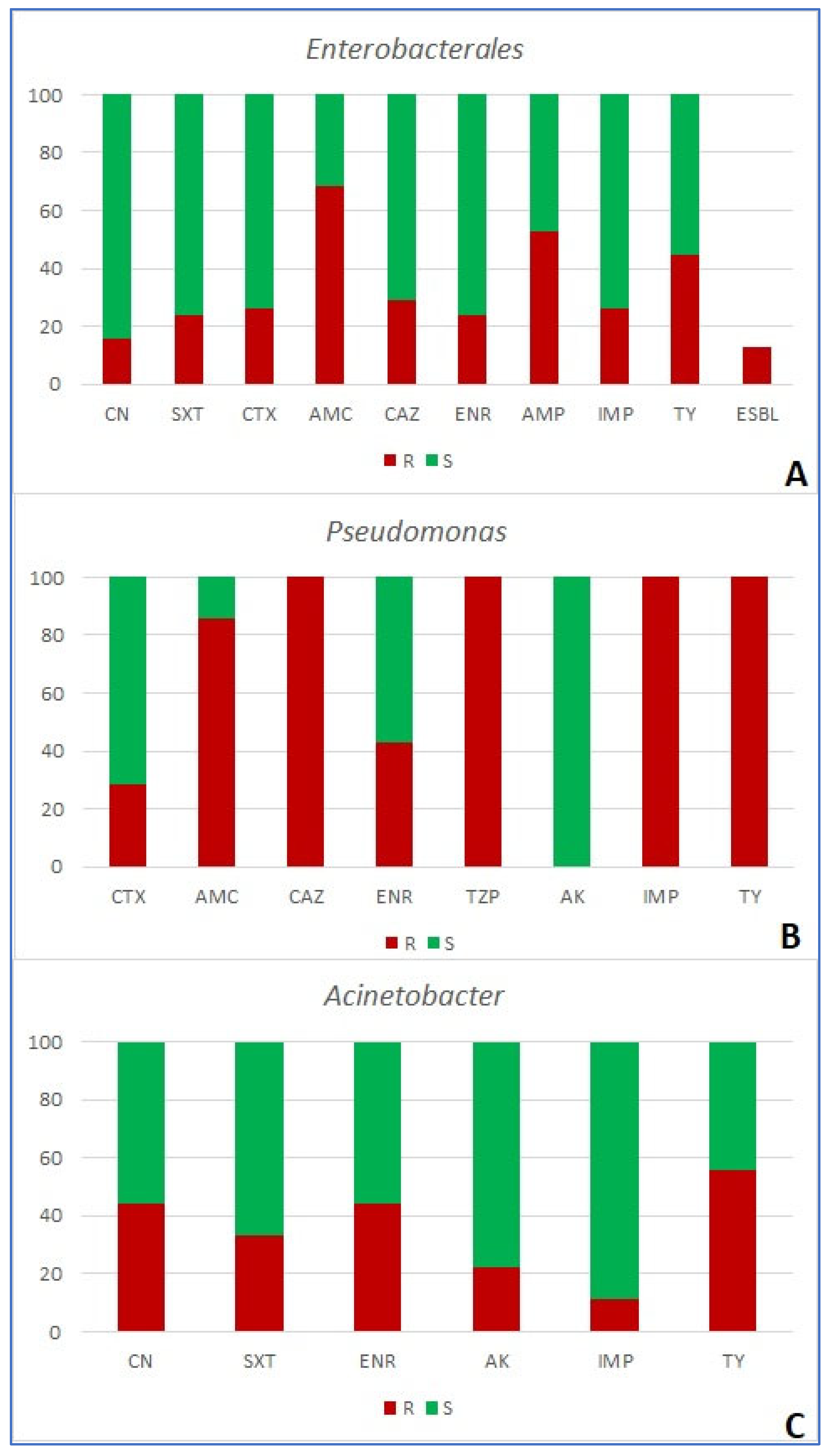
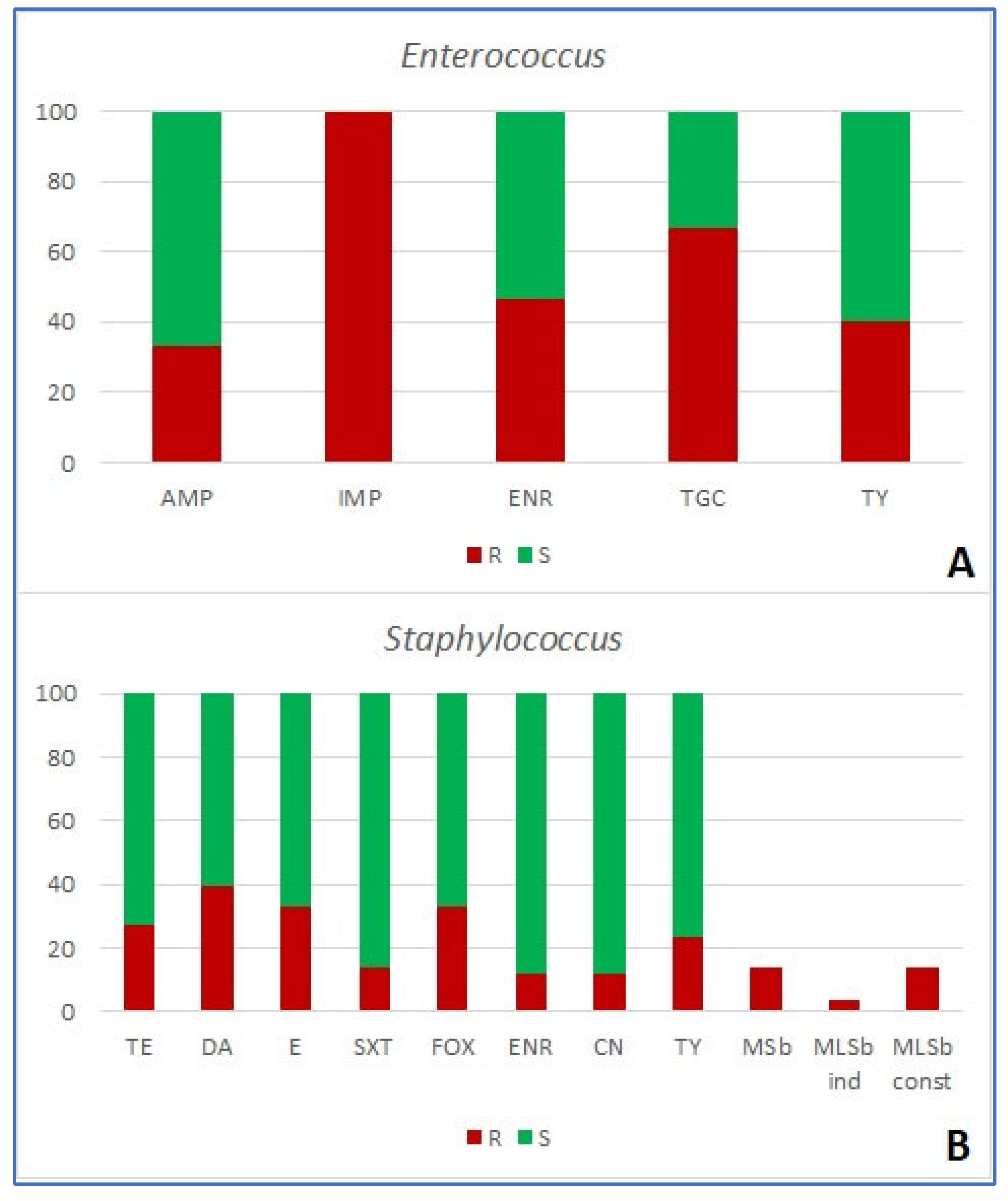
| number of antibiotics bacteria are resistant to (%) | resistance mechanism | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group of bacteria | R0 | R1 | R2 | R3 | R4 | R5 | R6 | R7 | R8 | MDR | ESBL | MRS | MSb | MLSb-i | MLSb-c |
| Enterobacterales (n=38) | 3 (7.9) | 2 (5.3) | 12 (31.6) | 7 (18.4) | 6 (15.8) | 3 (7.9) | 2 (5.3) | 2 (5.3) | 1 (2.6) | 16 (42.1) | 5 (13.2) | - | - | - | - |
| Pseudomonas (n=7) | 0 | 0 | 0 | 1 | 0 | 3 (42.9) | 3 (42.9) | 0 | 0 | 7 (100) | 0 | - | - | - | - |
| Acinetobacter (n=9) | 0 | 5 (55.6) | 1 (11.1) | 1 (11.1) | 1 (11.1) | 1 (11.1) | 0 | - | - | 3 (33.3) | - | - | - | - | - |
| Enterococcus (n=15) | 0 | 3 (20.0) | 3 (20.0) | 2 (13.3) | 7 (46.7) | 0 | - | - | - | 9 (60) | - | - | - | - | - |
| Staphylococcus (n=51) | 14 (27.5) | 15 (29.4) | 8 (15.7) | 3 (5.9) | 5 (9.8) | 0 | 3 (5.9) | 3 (5.9) | 0 | 14 (27.5) | - | 17 (33.3) | 7 (13.7) | 2 (3.9) | 7 (13.7) |
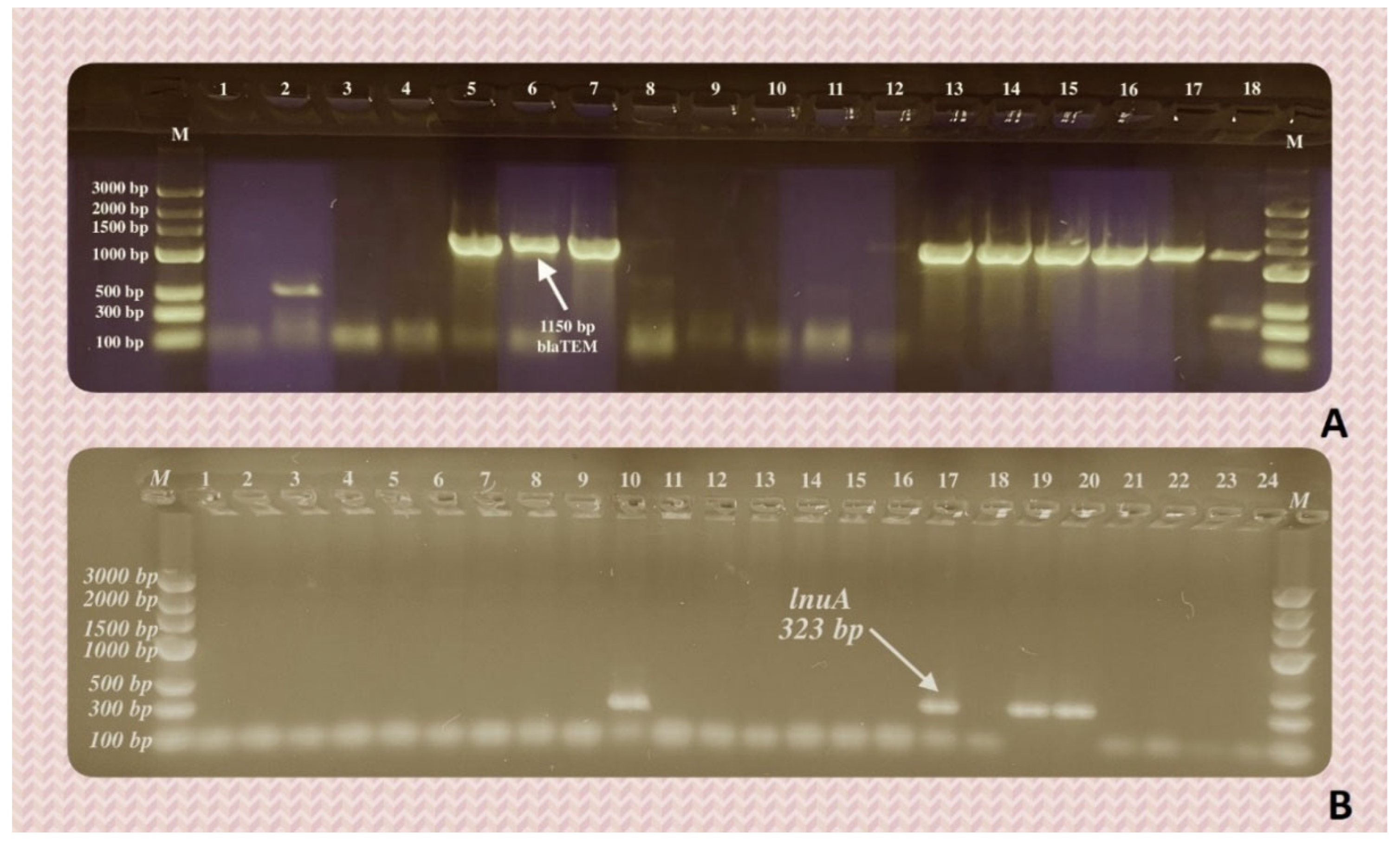
| Animal | n (%) | ||||||
| mecA | msrA | lnuA | strA | tetK | sul3 | ereA | |
| Cat | 0 | 3 (12) | 0 | 7 (28) | 6 (21.4) | 6 (21.4) | 0 |
| Dog | 4 (11.8) | 1 (2.9) | 4 (11.8) | 10 (29.4) | 3 (8.8) | 3 (8.8) | 0 |
| Animal | n (%) | ||||||
| blaTEM | blaSHV | blaCTX-M | blaOXA-1 | sul3 | qnrD | strA | |
| Cat | 3 (25) | 0 | 1 (8.3) | 0 | 3 (25) | 0 | 1 (8.3) |
| Dog | 10 (30.3) | 3 (9.1) | 1 (3) | 1 (3) | 2 (6.1) | 0 | 10 (30.3) |
| Rabbit | 0 | 1 (25) | 0 | 0 | 0 | 0 | 1 (25) |
| Origin | Species | phenotype of resistance (antibiotic class) | resistance genes (type of resistance) |
| feline | Enterococcus faecalis | IMP (β-lactam -carbapeneme) ENR (fluoroquinolone) TGC (tetracycline) TY (macrolide) |
mecA (methicillin) msrA (macrolides) lnuA (lincosamides) tetK (tetracyclines) |
| canine | Enterococcus faecalis | IMP (β-lactam -carbapeneme) ENR(fluoroquinolone) TGC (tetracycline) TY (macrolide) |
msrA (macrolides) strA (aminoglycosides) tetK (tetracyclines) sul3 (sulfonamides) |
| canine | Staphylococcus sciuri | - |
lnuA (lincosamides) strA (aminoglycosides) tetK (tetracyclines) |
| canine | Staphylococus pseudintermedius | TE (tetracycline) DA (lincosamide) E (macrolide) SXT (diaminopyrimidines/sulfonamide) ENR (fluoroquinolone) CN (aminoglycoside) TY (macrolide) |
lnuA (lincosamides) strA (aminoglycosides) tetK (tetracyclines) |
| canine | Escherichia coli | CN (aminoglycoside) AMC (β-lactam/ β-lactamase inhibitor) AMP (β-lactam - aminopenicillin) IMP (β-lactam -carbapeneme) |
blaTEM (ESBL) blaSHV (ESBL) strA (aminoglycosides) |
| canine | Escherichia coli | CTX (β-lactam – 3rd gen. cephalosporin) AMC (β-lactam/ β-lactamase inhibitor) CAZ (β-lactam – 3rd gen. cephalosporin) AMP (β-lactam - aminopenicillin) TY (macrolide) |
blaTEM (ESBL) blaSHV (ESBL) strA (aminoglycosides) |
| canine | Proteus mirabilis | CN (aminoglycoside) SXT (diaminopyrimidines/sulfonamide) CTX (β-lactam – 3rd gen. cephalosporin) AMC (β-lactam/ β-lactamase inhibitor) CAZ (β-lactam – 3rd gen. cephalosporin) ENR (fluoroquinolone) AMP (β-lactam - aminopenicillin) IMP (β-lactam/carbapeneme) |
blaTEM (ESBL) blaOXA-1 (ESBL-carbapenemes) strA (aminoglycosides) |
3. Discussion
4. Materials and Methods
4.1. Collection of Samples
4.2. Isolation and Identification of Bacteria
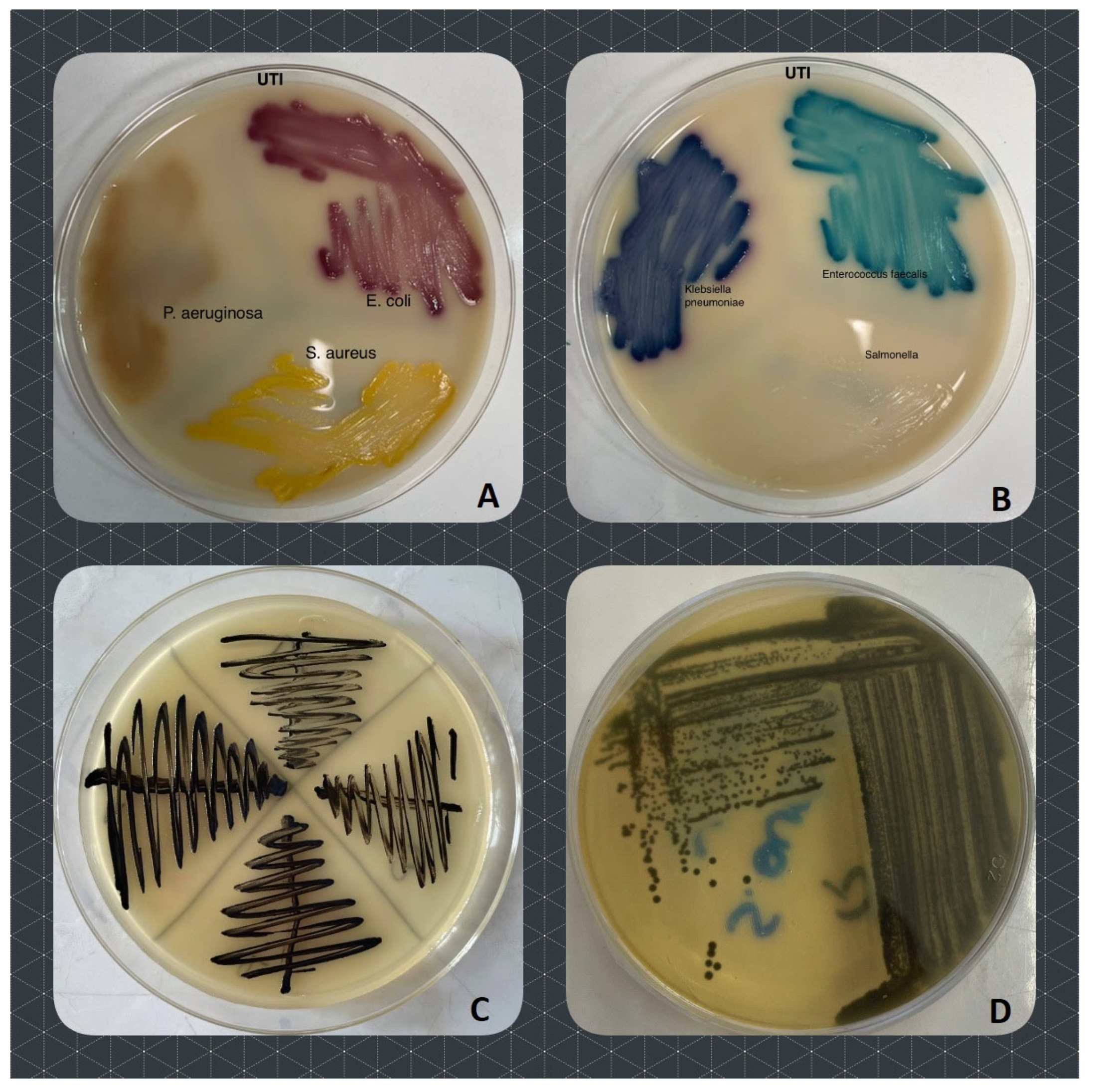
4.3. Antibacterial Susceptibility Tests
| Enterobacterales (E. coli, Klebsiella, Proteus, Enterobacter) | Pseudomonas | Acinetobacter | Enterococcus | Staphylococcus | |
| No of strains in total | 38 | 7 | 9 | 15 | 51 |
| Cats | 7 | 1 | 3 | 5 | 21 |
| Dogs | 28 | 6 | 5 | 10 | 30 |
| Rabbit | 3 | 0 | 1 | 0 | 0 |
| antimicrobial disks abbreviations * | ENR | ENR | ENR | ENR | ENR |
| AMC (ESBL)** | AMC (ESBL)** | AK | AMP | E (MLSb)** | |
| CAZ (ESBL)** | CAZ (ESBL)** | CN | MEM/IMP | DA (MLSb)** | |
| CTX (ESBL)** | CTX (ESBL)** | MEM/IMP | TGC | FOX (MRS)** | |
| AMP | AK | SXT | TY | CN | |
| CN | MEM/IMP | TY | SXT | ||
| SXT | TZP | TE | |||
| MEM/IMP | TY | TY | |||
| TY |
4.4. Assessment of Genes Conferring the Bacterial Resistance to Different Groups of Antimicrobials
| No. | Gene | Primer | Sequence (5’-3’) | Annealing temp. (°C) | Product length (bp) | Reference |
|---|---|---|---|---|---|---|
| 1. | msrA | msrA-F | GGCACAATAAGAGTGTTTAAAGG AAGTTATATCATGAATAGATTGTCCTGTT |
50 | 940 | [19] |
| msrA-R | ||||||
| 2. | ereA | ereA-F | AACACCCTGAACCCAAGGGACG CTTCACATCCGGATTCGCTCGA |
57 | 420 | [20] |
| ereA-R | ||||||
| 3. | lnuA | lnuA-F | GGTGGCTGGGGGGTAGATGTATTAACTGG GCTTCTTTTGAAATACATGGTATTTTTCGATC |
57 | 323 | [19] |
| lnuA-R | ||||||
| 4. | mecA | mecA-F | GTAGAAAATGACTGAACGTCCGATAA CAATTCCACATTGTTTCGGTCTAA |
55 | 310 | [21] |
| mecA-R | ||||||
| 5. | tetK | tetK-F | TCGATAGGAACAGCAGTA CAGCAGATCCTACTCCTT |
55 | 169 | [22] |
| tetK-R | ||||||
| 6. | blaTEM | blaTEM-F | ATTCTTGAAGACGAAAGGGC ACGCTCAGTGGAACGAAAAC |
60 | 1150 | [23] |
| blaTEM-R | ||||||
| 7. | blaSHV | blaSHV-F | CACTCAAGGATGTATTGTG TTAGCGTTGCCAGTGCTCG |
52 | 885 | [23] |
| blaSHV-R | ||||||
| 8. | blaCTX-M | blaCTX-M-F | CGATGTGCAGTACCAGTAA TTAGTGACCAGAATCAGCGG |
55 | 585 | [24] |
| blaCTX-M-R | ||||||
| 9. | blaOXA-1 | blaOXA-1-F | ACACAATACATATCAACTTCGC AGTGTGTTTAGAATGGTGATC |
61 | 813 | [23] |
| blaOXA-1-R | ||||||
| 10. | sul3 | sul3-F | ACCACCGATAGTTTTTCCGA TGCCTTTTTCTTTTAAAGCC |
62 | 199 | [25] |
| sul3-R | ||||||
| 11. | qnrA | qnrA-F | GGGTATGGATATTATTGATAAAG CTAATCCGGCAGCACTATTA |
55 | 580 | [26] |
| qnrA-R | ||||||
| 12. | qnrD | qnrD-F | AGTGAGTGTTTAGCTCAAGGAG CAGTGCCATTCCAGCGATT |
53 | 175 | [27] |
| qnrD-R | ||||||
| 13. | strA | strA-F | TCAATCCCGACTTCTTACCG CACCATGGCAAACAACCATA |
52 | 126 | [28] |
| strA-R |
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Marco-Fuertes, A.; Marin, C.; Lorenzo-Rebenaque, L.; Vega, S.; Montoro-Dasi, L. Antimicrobial Resistance in Companion Animals: A New Challenge for the One Health Approach in the European Union. Veterinary Sciences 2022, 9. [Google Scholar] [CrossRef]
- Pomba, C.; Rantala, M.; Greko, C.; Baptiste, K.E.; Catry, B.; van Duijkeren, E.; Mateus, A.; Moreno, M.A.; Pyörälä, S.; Ružauskas, M.; et al. Public Health Risk of Antimicrobial Resistance Transfer from Companion Animals. Journal of Antimicrobial Chemotherapy 2017, 72, 957–968. [Google Scholar] [CrossRef]
- Kožár, M.; Hamilton, H.; Koščová, J. Types of Wounds and the Prevalence of Bacterial Contamination of Wounds in the Clinical Practice of Small Animals. Folia Veterinaria 2018, 62, 39–47. [Google Scholar] [CrossRef]
- Windahl, U.; Bengtsson, B.; Nyman, A.K.; Holst, B.S. The Distribution of Pathogens and Their Antimicrobial Susceptibility Patterns among Canine Surgical Wound Infections in Sweden in Relation to Different Risk Factors. Acta Veterinaria Scandinavica 2015, 57. [Google Scholar] [CrossRef] [PubMed]
- Scott Weese, J. Antimicrobial Resistance in Companion Animals. Animal health research reviews / Conference of Research Workers in Animal Diseases 2008, 9, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fernández, R.; Durán, I.; Molina-López, R.A.; Darwich, L. Antimicrobial Resistance in Bacteria Isolated From Cats and Dogs From the Iberian Peninsula. Frontiers in Microbiology 2021, 11, 1–12. [Google Scholar] [CrossRef]
- Pires Dos Santos, T.; Damborg, P.; Moodley, A.; Guardabassi, L. Systematic Review on Global Epidemiology of Methicillin-Resistant Staphylococcus Pseudintermedius: Inference of Population Structure from Multilocus Sequence Typing Data. Frontiers in microbiology 2016, 7, 1599. [Google Scholar] [CrossRef]
- Guardabassi, L.; Schwarz, S.; Lloyd, D.H. Pet Animals as Reservoirs of Antimicrobial-Resistant Bacteria: Review. Journal of Antimicrobial Chemotherapy 2004, 54, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Caneschi, A.; Bardhi, A.; Barbarossa, A.; Zaghini, A. The Use of Antibiotics and Antimicrobial Resistance in Veterinary Medicine, a Complex Phenomenon: A Narrative Review. Antibiotics 2023, 12. [Google Scholar] [CrossRef]
- Ganière, J.P.; Médaille, C.; Limet, A.; Ruvoen, N.; André-Fontaine, G. Antimicrobial Activity of Enrofloxacin against Staphylococcus Intermedius Strains Isolated from Canine Pyodermas. Veterinary dermatology 2001, 12, 171–175. [Google Scholar] [CrossRef]
- Actor, J.K. 12 - Clinical Bacteriology. In; Actor, J.K.B.T.-E.I.R.I. and M. (Second E., Ed.; W.B. Saunders: Philadelphia, 2012; pp. 105–120 ISBN 978-0-323-07447-6.
- Carvalho, A.C.; Barbosa, A. V; Arais, L.R.; Ribeiro, P.F.; Carneiro, V.C.; Cerqueira, A.M.F. Resistance Patterns, ESBL Genes, and Genetic Relatedness of Escherichia Coli from Dogs and Owners. Brazilian Journal of Microbiology 2016, 47, 150–158. [Google Scholar] [CrossRef]
- Akhtardanesh, B.; Ghanbarpour, R.; Ganjalikhani, S.; Gazanfari, P. Determination of Antibiotic Resistance Genes in Relation to Phylogenetic Background in Escherichia Coli Isolates from Fecal Samples of Healthy Pet Cats in Kerman City. Veterinary research forum : an international quarterly journal 2016, 7, 301–308. [Google Scholar] [PubMed]
- Lenart-Boron, A.; Augustyniak, K.; Boron, P. Screening of Antimicrobial Resistance and Molecular Detection of Fluoroquinolone Resistance Mechanisms in Chicken Faeces-Derived Escherichia Coli. Veterinarni Medicina 2016, 61, 80–89. [Google Scholar] [CrossRef]
- Gniadkowski, M.; Żabicka, D.; Hryniewicz, W. Rekomendacje Doboru Testów Do Oznaczania Wrażliwości Bakterii Na Antybiotyki i Chemioterapeutyki 2009 Oznaczanie Wrażliwo ś Ci Pałeczek Gram-Ujemnych. 2009, 1–29.
- Drieux, L.; Brossier, F.; Sougakoff, W.; Jarlier, V. Phenotypic Detection of Extended-Spectrum β-Lactamase Production in Enterobacteriaceae: Review and Bench Guide. Clinical Microbiology and Infection 2008, 14, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Fiebelkorn, K.R.; Crawford, S.A.; McElmeel, M.L.; Jorgensen, J.H. Practical Disk Diffusion Method for Detection of Inducible Clindamycin Resistance in Staphylococcus Aureus and Coagulase-Negative Staphylococci. Journal of clinical microbiology 2003, 41, 4740–4744. [Google Scholar] [CrossRef]
- EUCAST European Committee on Antimicrobial Susceptibility Testing Breakpoint Tables for Interpretation of MICs and Zone Diameters European Committee on Antimicrobial Susceptibility Testing Breakpoint Tables for Interpretation of MICs and Zone Diameters. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_5.0_Breakpoint_Table_01.pdf 2023, 0–77.
- Lina, G.; Quaglia, A.; Reverdy, M.E.; Leclercq, R.; Vandenesch, F.; Etienne, J. Distribution of Genes Encoding Resistance to Macrolides, Lincosamides, and Streptogramins among Staphylococci. Antimicrobial agents and chemotherapy 1999, 43, 1062–1066. [Google Scholar] [CrossRef] [PubMed]
- Sutcliffe, J.; Grebe, T.; Tait-Kamradt, A.; Wondrack, L. Detection of Erythromycin-Resistant Determinants by PCR. Antimicrobial agents and chemotherapy 1996, 40, 2562–2566. [Google Scholar] [CrossRef]
- Geha, D.J.; Uhl, J.R.; Gustaferro, C.A.; Persing, D.H. Multiplex PCR for Identification of Methicillin-Resistant Staphylococci in the Clinical Laboratory. Journal of clinical microbiology 1994, 32, 1768–1772. [Google Scholar] [CrossRef]
- Pazda, M.; Rybicka, M.; Stolte, S.; Piotr Bielawski, K.; Stepnowski, P.; Kumirska, J.; Wolecki, D.; Mulkiewicz, E. Identification of Selected Antibiotic Resistance Genes in Two Different Wastewater Treatment Plant Systems in Poland: A Preliminary Study. Molecules 2020, 25. [Google Scholar] [CrossRef]
- Sáenz, Y.; Briñas, L.; Domínguez, E.; Ruiz, J.; Zarazaga, M.; Vila, J.; Torres, C. Mechanisms of Resistance in Multiple-Antibiotic-Resistant Escherichia Coli Strains of Human, Animal, and Food Origins. Antimicrobial Agents and Chemotherapy 2004, 48, 3996–4001. [Google Scholar] [CrossRef]
- Batchelor, M.; Hopkins, K.; Threlfall, E.J.; Clifton-Hadley, F.A.; Stallwood, A.D.; Davies, R.H.; Liebana, E. BlaCTX-M Genes in Clinical Salmonella Isolates Recovered from Humans in England and Wales from 1992 to 2003. Antimicrobial Agents and Chemotherapy 2005, 49, 1319–1322. [Google Scholar] [CrossRef] [PubMed]
- Szczepanowski, R.; Linke, B.; Krahn, I.; Gartemann, K.-H.; Gützkow, T.; Eichler, W.; Pühler, A.; Schlüter, A. Detection of 140 Clinically Relevant Antibiotic-Resistance Genes in the Plasmid Metagenome of Wastewater Treatment Plant Bacteria Showing Reduced Susceptibility to Selected Antibiotics. Microbiology 2009, 155, 2306–2319. [Google Scholar] [CrossRef] [PubMed]
- Cattoir, V.; Poirel, L.; Rotimi, V.; Soussy, C.-J.; Nordmann, P. Multiplex PCR for Detection of Plasmid-Mediated Quinolone Resistance Qnr Genes in ESBL-Producing Enterobacterial Isolates. The Journal of antimicrobial chemotherapy 2007, 60, 394–397. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Li, L.; Ashbolt, N.; Wang, X.; Cui, Y.; Zhu, X.; Xu, Y.; Yang, Y.; Mao, D.; Luo, Y. Arctic Antibiotic Resistance Gene Contamination, a Result of Anthropogenic Activities and Natural Origin. Science of The Total Environment 2018, 621, 1176–1184. [Google Scholar] [CrossRef] [PubMed]
- Walsh, F.; Ingenfeld, A.; Zampicolli, M.; Hilber-Bodmer, M.; Frey, J.E.; Duffy, B. Real-Time PCR Methods for Quantitative Monitoring of Streptomycin and Tetracycline Resistance Genes in Agricultural Ecosystems. Journal of microbiological methods 2011, 86, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Morley, P.S. Surveillance for Nosocomial Infections in Veterinary Hospitals. The Veterinary clinics of North America. Equine practice 2004, 20, 561–576. [Google Scholar] [CrossRef]
| Species | total (n=136) | cats (n=52) | dogs (n=80) | rabbits (n=4) |
|---|---|---|---|---|
| Gram-negative | 65 (47.79) | 23 (44.23) | 38 (47.50) | 4 (100) |
| Acinetobacter baumanni | 1 (0.74) | 1 (1.92) | 0 | 0 |
| Acinetobacter calcoaceticus | 1 (0.74) | 0 | 1 (1.25) | 0 |
| Acinetobacter johnsonii | 1 (0.74) | 1 (1.92) | 0 | 0 |
| Acinetobacter pittii | 2 (1.47) | 0 | 1 (1.25) | 1 (25) |
| Acinetobacter ursingii | 5 (3.68) | 5 (9.62) | 0 | 0 |
| Aeromonas media | 1 (0.74) | 0 | 1 (1.25) | 0 |
| Brevundimonas diminuta | 2 (1.47) | 1 (1.92) | 1 (1.25) | 0 |
| Citrobacter freundi | 3 (2.21) | 2 (3.85) | 1 (1.25) | 0 |
| Enterobacter cloacae | 1 (0.74) | 1 (1.92) | 0 | 0 |
| Enterobacter hormaechei | 2 (1.47) | 0 | 2 (2.50) | 0 |
| Escherichia coli | 11 (8.09) | 1 (1.92) | 8 (10.0) | 2 (50) |
| Hafnia alvei | 1 (0.74) | 0 | 1 (1.25) | 0 |
| Klebsiella pneumoniae | 2 (1.47) | 0 | 1 (1.25) | 1 (25) |
| Klebsiella spp. | 1 (0.74) | 1 (1.92) | 0 | 0 |
| Leclercia adecarboxylata | 1 (0.74) | 0 | 1 (1.25) | 0 |
| Pantoea agglomerans | 2 (1.47) | 0 | 2 (2.50) | 0 |
| Pantoea sp. | 1 (0.74) | 0 | 1 (1.25) | 0 |
| Moraxella osloensis | 1 (0.74) | 1 (1.92) | 0 | 0 |
| Proteus mirabilis | 3 (2.21) | 0 | 3 (3.75) | 0 |
| Proteus vulgaris | 5 (3.68) | 4 (7.69) | 1 (1.25) | 0 |
| Pseudomonas aeruginosa | 3 (2.21) | 0 | 3 (3.75) | 0 |
| Pseudomonas putida | 3 (2.21) | 1 (1.92) | 2 (2.50) | 0 |
| Pseudomonas graminis | 1 (0.74) | 0 | 1 (1.25) | 0 |
| Pseudomonas fulva | 1 (0.74) | 0 | 1 (1.25) | 0 |
| Pseudomonas koreensis | 1 (0.74) | 1 (1.92) | 0 | 0 |
| Psychrobacter sanguinis | 2 (1.47) | 0 | 2 (2.50) | 0 |
| Psychrobacter pulmonis | 1 (0.74) | 0 | 1 (1.25) | 0 |
| Serratia marcescens | 3 (2.21) | 3 (5.77) | 0 | 0 |
| Serratia liquefaciens | 1 (0.74) | 0 | 1 (1.25) | 0 |
| Stenotrophomonas maltophilia | 2 (1.47) | 0 | 2 (2.50) | 0 |
| Gram-positive | 71 (52.21) | 29 (55.77) | 42 (52.50) | 0 |
| Bacillus pumilus | 1 (0.74) | 0 | 1 (1.25) | 0 |
| Curtobacterium flaccumfaciens | 1 (0.74) | 0 | 1 (1.25) | 0 |
| Enterococcus avium | 1 (0.74) | 0 | 1 (1.25) | 0 |
| Enterococcus faecalis | 11 (8.09) | 3 (5.77) | 8 (10) | 0 |
| Enterococcus faecium | 3 (2.21) | 2 (3.85) | 1 (1.25) | 0 |
| Enterococcus hirae | 2 (1.47) | 1 (1.92) | 1 (1.25) | 0 |
| Kocuria rhizophila | 1 (0.74) | 1 (1.92) | 0 | 0 |
| Lactococcus garvieae | 1 (0.74) | 0 | 1 (1.25) | 0 |
| Lysinibacillus fusiformis | 1 (0.74) | 0 | 1 (1.25) | 0 |
| Macrococcus canis | 1 (0.74) | 0 | 1 (1.25) | 0 |
| Micrococcus luteus | 1 (0.74) | 0 | 1 (1.25) | 0 |
| Microbacterium maritypicum | 1 (0.74) | 0 | 1 (1.25) | 0 |
| Microbacterium oxydans | 1 (0.74) | 0 | 1 (1.25) | 0 |
| Microbacterium paraoxydans | 1 (0.74) | 1 (1.92) | 0 | 0 |
| Microbacterium phyllosphaerae | 1 (0.74) | 0 | 1 (1.25) | 0 |
| Peribacillus simplex | 1 (0.74) | 1 (1.92) | 0 | 0 |
| Staphylococcus aureus | 5 (3.68) | 4 (7.69) | 1 (1.25) | 0 |
| Staphylococcus capitis | 2 (1.47) | 2 (3.85) | 0 | 0 |
| Staphylococcus cohnii | 1 (0.74) | 1 (1.92) | 0 | 0 |
| Staphylococcus devriesei | 1 (0.74) | 0 | 1 (1.25) | 0 |
| Staphylococcus condimenti | 2 (1.47) | 2 (3.85) | 0 | 0 |
| Staphylococcus epidermidis | 2 (1.47) | 1 (1.92) | 1 (1.25) | 0 |
| Staphylococcus felis | 2 (1.47) | 2 (3.85) | 0 | 0 |
| Staphylococcus haemolyticus | 1 (0.74) | 0 | 1 (1.25) | 0 |
| Staphylococcus pasteuri | 1 (0.74) | 0 | 1 (1.25) | 0 |
| Staphylococcus lentus | 1 (0.74) | 1 (1.92) | 0 | 0 |
| Staphylococcus pseudintermedius | 13 (9.56) | 3 (5.77) | 10 (12.5) | 0 |
| Staphylococcus schleiferi | 2 (1.47) | 0 | 2 (2.50) | 0 |
| Staphylococcus saprophyticus | 1 (0.74) | 0 | 1 (1.25) | 0 |
| Staphylococcus warneri | 1 (0.74) | 1 (1.92) | 0 | 0 |
| Staphylococcus sciuri | 1 (0.74) | 0 | 1 (1.25) | 0 |
| Streptococcus canis | 5 (3.68) | 2 (3.85) | 3 (3.75) | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





