Submitted:
21 January 2024
Posted:
22 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and methods
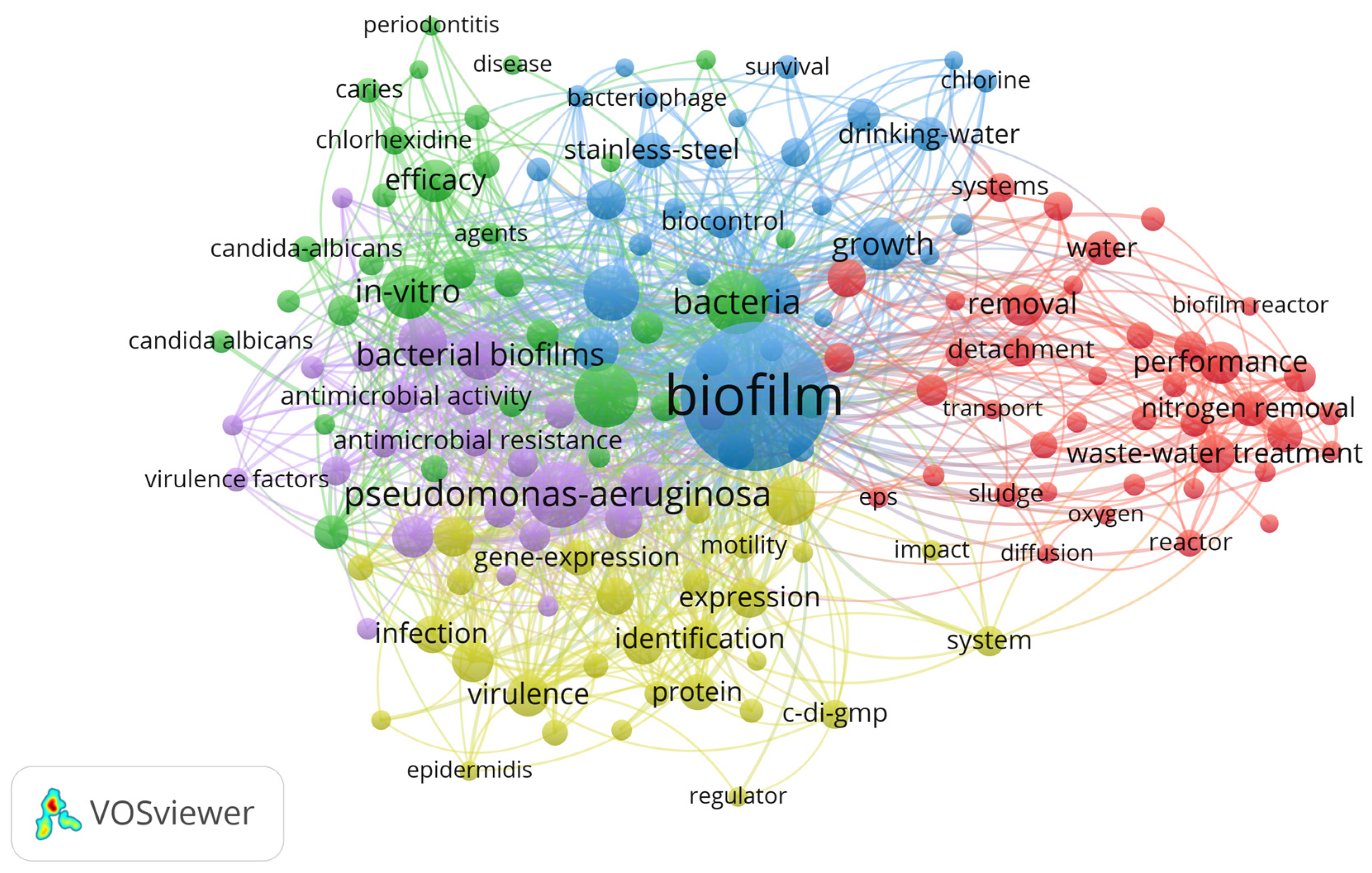
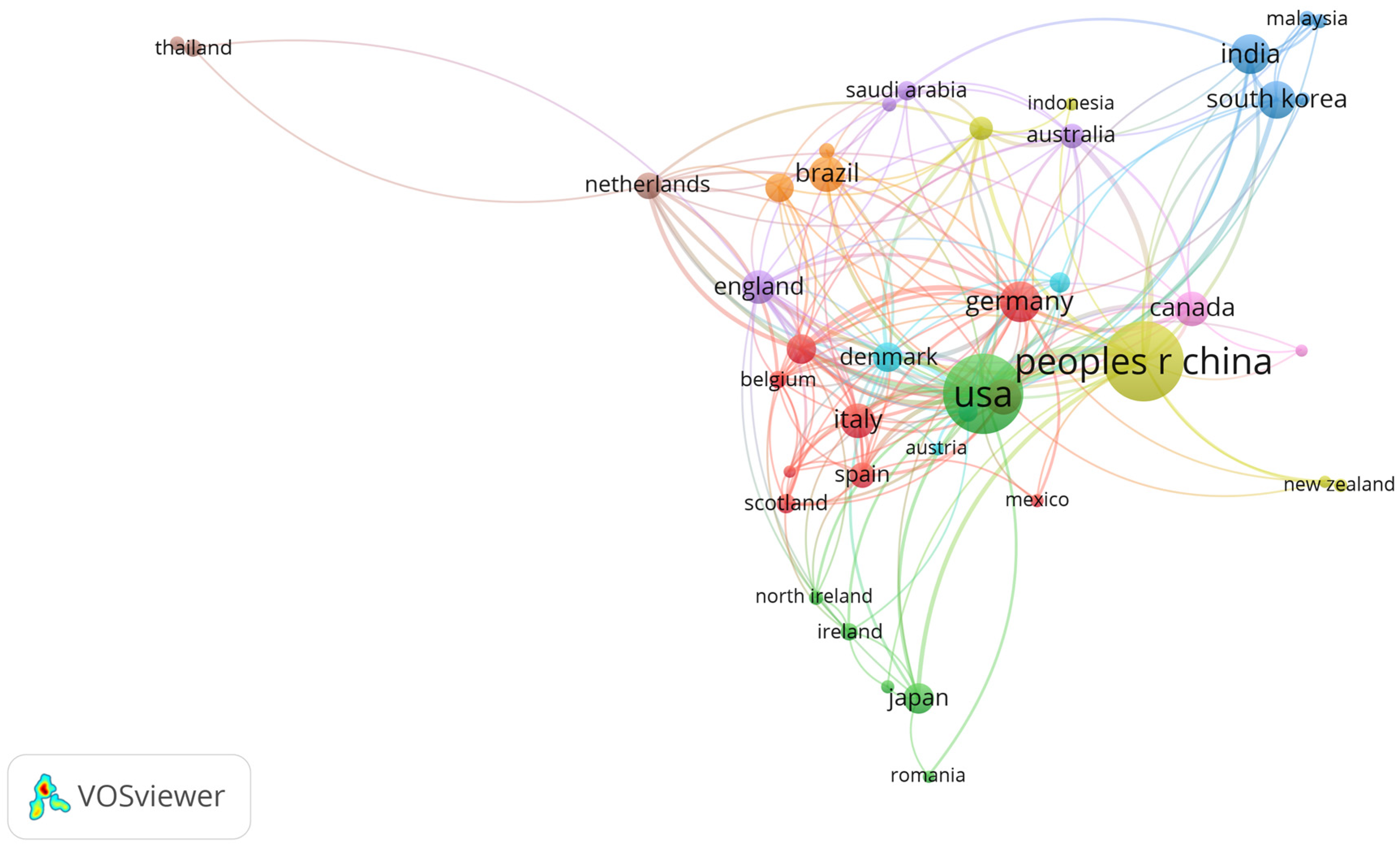
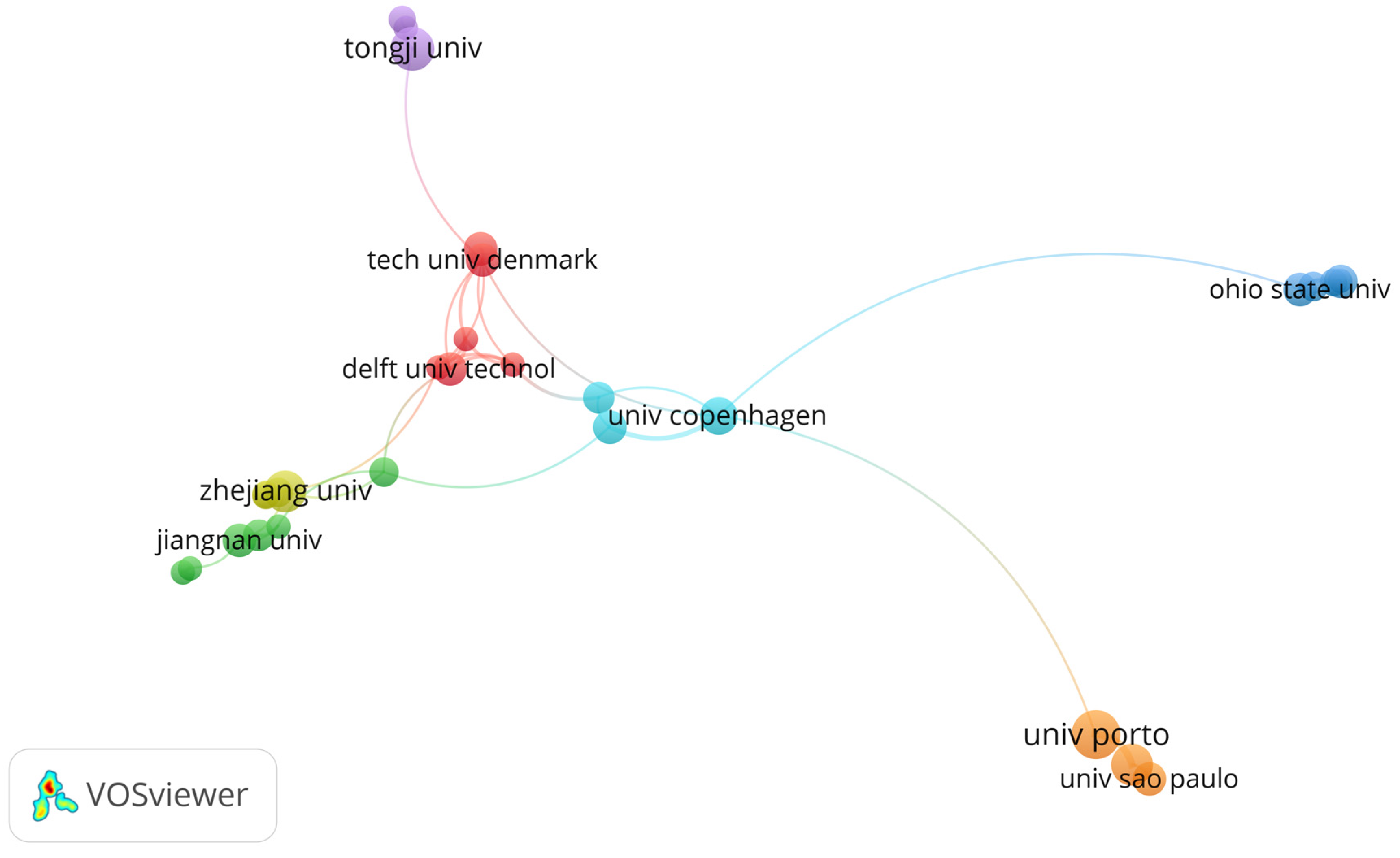
3. Results
4. Discussion
4.1. Biofilm Control Study: Implications and Applications
4.2. Big Data and Machine Learning: Revolutionary Biofilm Control
References
- Salama, Y.; Chennaoui, M.; Sylla, A.; Mountadar, M.; Rihani, M.; Assobhei, O. Characterization, structure, and function of extracellular polymeric substances (EPS) of microbial biofilm in biological wastewater treatment systems: A review. Desalination Water Treat. 2016, 57, 16220–16237. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Neu, T.R.; Wozniak, D.J. The EPS matrix: The “house of biofilm cells”. J. Bacteriol. 2007, 189, 7945–7947. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.-C. EPS—Then and now. Microorganisms 2016, 4, 41. [Google Scholar] [CrossRef] [PubMed]
- Czaczyk, K.; Myszka, K. Biosynthesis of extracellular polymeric substances (EPS) and its role in microbial biofilm formation. Pol. J. Environ. Stud. 2007, 16, 799. [Google Scholar]
- Kreft, J.U.; Wimpenny, J.W. Effect of EPS on biofilm structure and function as revealed by an individual-based model of biofilm growth. Water Sci. Technol. 2001, 43, 135. [Google Scholar] [CrossRef] [PubMed]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Goller, C.C.; Romeo, T. Environmental influences on biofilm development. Bact. Biofilms 2008, 322, 37–66. [Google Scholar]
- Maurya, A.; Raj, A. Recent advances in the application of biofilm in bioremediation of industrial wastewater and organic pollutants. Microorg. Sustain. Environ. Health 2020, 81–118. [Google Scholar] [CrossRef]
- Ding, Y.; Peng, N.; Du, Y.; Ji, L.; Cao, B. Disruption of putrescine biosynthesis in Shewanella oneidensis enhances biofilm cohesiveness and performance in Cr (VI) immobilization. Appl. Environ. Microbiol. 2014, 80, 1498–1506. [Google Scholar] [CrossRef]
- Maurice, N.M.; Bedi, B.; Sadikot, R.T. Pseudomonas aeruginosa biofilms: Host response and clinical implications in lung infections. Am. J. Respir. Cell Mol. Biol. 2018, 58, 428–439. [Google Scholar] [CrossRef]
- Kolpen, M.; Kragh, K.N.; Enciso, J.B.; Faurholt-Jepsen, D.; Lindegaard, B.; Egelund, G.B.; Jensen, A.V.; Ravn, P.; Mathiesen, I.H.M.; Gheorge, A.G. Bacterial biofilms predominate in both acute and chronic human lung infections. Thorax 2022, 77, 1015–1022. [Google Scholar] [CrossRef]
- Stewart, P.S. Biophysics of biofilm infection. Pathog. Dis. 2014, 70, 212–218. [Google Scholar] [CrossRef]
- Trautner, B.W.; Darouiche, R.O. Role of biofilm in catheter-associated urinary tract infection. Am. J. Infect. Control 2004, 32, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Nickel, J.C.; Costerton, J.W.; McLean, R.J.C.; Olson, M. Bacterial biofilms: Influence on the pathogenesis, diagnosis and treatment of urinary tract infections. J. Antimicrob. Chemother. 1994, 33, 31–41. [Google Scholar] [CrossRef]
- Tenke, P.; Kovacs, B.; Jäckel, M.; Nagy, E. The role of biofilm infection in urology. World J. Urol. 2006, 24, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Høiby, N.; Ciofu, O.; Johansen, H.K.; Song, Z.j.; Moser, C.; Jensen, P.Ø.; Molin, S.; Givskov, M.; Tolker-Nielsen, T.; Bjarnsholt, T. The clinical impact of bacterial biofilms. Int. J. Oral Sci. 2011, 3, 55–65. [Google Scholar] [CrossRef]
- Li, X.Z.; Hauer, B.; Rosche, B. Single-species microbial biofilm screening for industrial applications. Appl. Microbiol. Biotechnol. 2007, 76, 1255–1262. [Google Scholar] [CrossRef]
- Garratt, A.; Schmidt, L.; Mackintosh, A.; Fitzpatrick, R. Quality of life measurement: Bibliographic study of patient assessed health outcome measures. Bmj 2002, 324, 1417. [Google Scholar] [CrossRef] [PubMed]
- Simões, M.; Simões, L.C.; Vieira, M.J. A review of current and emergent biofilm control strategies. LWT-Food Sci. Technol. 2010, 43, 573–583. [Google Scholar] [CrossRef]
- Koo, H.; Allan, R.N.; Howlin, R.P.; Stoodley, P.; Hall-Stoodley, L. Targeting microbial biofilms: Current and prospective therapeutic strategies. Nat. Rev. Microbiol. 2017, 15, 740–755. [Google Scholar] [CrossRef]
- Kasza, K.; Gurnani, P.; Hardie, K.R.; Camara, M.; Alexander, C. Challenges and solutions in polymer drug delivery for bacterial biofilm treatment: A tissue-by-tissue account. Adv. Drug Deliv. Rev. 2021, 178, 113973. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Fu, L.; Wei, C.; Fu, Q.; Pan, S. Antibacterial micro/nanomotors: Advancing biofilm research to support medical applications. J. Nanobiotechnol. 2023, 21, 388. [Google Scholar] [CrossRef] [PubMed]
- McQuarrie, J.P.; Boltz, J.P. Moving bed biofilm reactor technology: Process applications, design, and performance. Water Environ. Res. 2011, 83, 560–575. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Ding, Y. Tackling Heavy Metal Pollution: Evaluating Governance Models and Frameworks. Sustainability 2023, 15, 15863. [Google Scholar] [CrossRef]
- Chen, S.; Ding, Y. A bibliography study of Shewanella oneidensis biofilm. FEMS Microbiol. Ecol. 2023, 99, fiad124. [Google Scholar] [CrossRef] [PubMed]
- Pranckutė, R. Web of Science (WoS) and Scopus: The titans of bibliographic information in today’s academic world. Publications 2021, 9, 12. [Google Scholar] [CrossRef]
- Franceschini, F.; Maisano, D.; Mastrogiacomo, L. Empirical analysis and classification of database errors in Scopus and Web of Science. J. Informetr. 2016, 10, 933–953. [Google Scholar] [CrossRef]
- Soosaraei, M.; Khasseh, A.A.; Fakhar, M.; Hezarjaribi, H.Z. A decade bibliometric analysis of global research on leishmaniasis in Web of Science database. Ann. Med. Surg. 2018, 26, 30–37. [Google Scholar] [CrossRef]
- Yu, Y.; Li, Y.; Zhang, Z.; Gu, Z.; Zhong, H.; Zha, Q.; Yang, L.; Zhu, C.; Chen, E. A bibliometric analysis using VOSviewer of publications on COVID-19. Ann. Transl. Med. 2020, 8, 816. [Google Scholar] [CrossRef]
- Van Eck, N.J.; Waltman, L. Citation-based clustering of publications using CitNetExplorer and VOSviewer. Scientometrics 2017, 111, 1053–1070. [Google Scholar] [CrossRef]
- Hjollund, N.H.; Andersen, J.H.; Bech, P. Assessment of fatigue in chronic disease: A bibliographic study of fatigue measurement scales. Health Qual. Life Outcomes 2007, 5, 1–5. [Google Scholar] [CrossRef]
- Steuer, R.E.; Na, P. Multiple criteria decision making combined with finance: A categorized bibliographic study. Eur. J. Oper. Res. 2003, 150, 496–515. [Google Scholar] [CrossRef]
- Zhao, C.-e.; Chen, J.; Ding, Y.; Wang, V.B.; Bao, B.; Kjelleberg, S.; Cao, B.; Loo, S.C.J.; Wang, L.; Huang, W. Chemically functionalized conjugated oligoelectrolyte nanoparticles for enhancement of current generation in microbial fuel cells. ACS Appl. Mater. Interfaces 2015, 7, 14501–14505. [Google Scholar] [CrossRef]
- Zhao, C.e.; Wu, J.; Ding, Y.; Wang, V.B.; Zhang, Y.; Kjelleberg, S.; Loo, J.S.C.; Cao, B.; Zhang, Q. Hybrid conducting biofilm with built-in bacteria for high-performance microbial fuel cells. ChemElectroChem 2015, 2, 654–658. [Google Scholar] [CrossRef]
- Yang, Y.; Ding, Y.; Hu, Y.; Cao, B.; Rice, S.A.; Kjelleberg, S.; Song, H. Enhancing bidirectional electron transfer of Shewanella oneidensis by a synthetic flavin pathway. ACS Synth. Biol. 2015, 4, 815–823. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liu, D.; Ding, Y.; Wang, S. Mechanical performance of strain-hardening cementitious composites (SHCC) with bacterial addition. J. Infrastruct. Preserv. Resil. 2022, 3, 1–11. [Google Scholar] [CrossRef]
- Zhang, Z.; Weng, Y.; Ding, Y.; Qian, S. Use of genetically modified bacteria to repair cracks in concrete. Materials 2019, 12, 3912. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ding, Y.; Qian, S. Influence of bacterial incorporation on mechanical properties of engineered cementitious composites (ECC). Constr. Build. Mater. 2019, 196, 195–203. [Google Scholar] [CrossRef]
- Ding, Y.; Zhou, Y.; Yao, J.; Xiong, Y.; Zhu, Z.; Yu, X.-Y. Molecular evidence of a toxic effect on a biofilm and its matrix. Analyst 2019, 144, 2498–2503. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Zhou, Y.; Yao, J.; Szymanski, C.; Fredrickson, J.; Shi, L.; Cao, B.; Zhu, Z.; Yu, X.-Y. In situ molecular imaging of the biofilm and its matrix. Anal. Chem. 2016, 88, 11244–11252. [Google Scholar] [CrossRef]
- Philipp, L.-A.; Bühler, K.; Ulber, R.; Gescher, J. Beneficial applications of biofilms. Nat. Rev. Microbiol. 2023, 1–15. [Google Scholar] [CrossRef]
- Morikawa, M. Beneficial biofilm formation by industrial bacteria Bacillus subtilis and related species. J. Biosci. Bioeng. 2006, 101, 1–8. [Google Scholar] [CrossRef]
- Rossi, F. Beneficial biofilms for land rehabilitation and fertilization. FEMS Microbiol. Lett. 2020, 367, fnaa184. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Xu, S.; Wang, Y.; Zhang, Y.; Chou, S.-H.; Galperin, M.Y.; He, J. Ways to control harmful biofilms: Prevention, inhibition, and eradication. Crit. Rev. Microbiol. 2021, 47, 57–78. [Google Scholar] [CrossRef] [PubMed]
- Moser, C.; Pedersen, H.T.; Lerche, C.J.; Kolpen, M.; Line, L.; Thomsen, K.; Høiby, N.; Jensen, P.Ø. Biofilms and host response–helpful or harmful. Apmis 2017, 125, 320–338. [Google Scholar] [CrossRef]
- Yan, J.; Bassler, B.L. Surviving as a community: Antibiotic tolerance and persistence in bacterial biofilms. Cell Host Microbe 2019, 26, 15–21. [Google Scholar] [CrossRef]
- Hamdany, A.H.; Ding, Y.; Qian, S. Graphene-Based TiO2 Cement Composites to Enhance the Antibacterial Effect of Self-Disinfecting Surfaces. Catalysts 2023, 13, 1313. [Google Scholar] [CrossRef]
- Hamdany, A.H.; Ding, Y.; Qian, S. Visible light antibacterial potential of graphene-TiO2 cementitious composites for self-sterilization surface. J. Sustain. Cem.-Based Mater. 2023, 12, 972–982. [Google Scholar] [CrossRef]
- Hamdany, A.H.; Ding, Y.; Qian, S. Cementitious Composite Materials for Self-Sterilization Surfaces. ACI Mater. J. 2022, 119, 197–210. [Google Scholar] [CrossRef]
- Hamdany, A.H.; Ding, Y.; Qian, S. Mechanical and antibacterial behavior of photocatalytic lightweight engineered cementitious composites. J. Mater. Civ. Eng. 2021, 33, 04021262. [Google Scholar] [CrossRef]
- Muhammad, M.H.; Idris, A.L.; Fan, X.; Guo, Y.; Yu, Y.; Jin, X.; Qiu, J.; Guan, X.; Huang, T. Beyond risk: Bacterial biofilms and their regulating approaches. Front. Microbiol. 2020, 11, 928. [Google Scholar] [CrossRef]
- Elbourne, A.; Truong, V.K.; Cheeseman, S.; Rajapaksha, P.; Gangadoo, S.; Chapman, J.; Crawford, R.J. The use of nanomaterials for the mitigation of pathogenic biofilm formation. In Methods in Microbiology; Elsevier: Amsterdam, The Netherlands, 2019; Volume 46, pp. 61–92. [Google Scholar]
- Niranjan, V.; Setlur, A.S.; Skariyachan, S.; Chandrashekar, K. Applications of Microbial Consortia consortia and Microbiome microbiome Interactions for Augmenting Sustainable Agrobiology agrobiology. In Sustainable Agrobiology: Design and Development of Microbial Consortia; Springer: Berlin/Heidelberg, Germany, 2023; pp. 275–316. [Google Scholar]
- Kennedy, A.C.; Smith, K.L. Soil microbial diversity and the sustainability of agricultural soils. Plant Soil 1995, 170, 75–86. [Google Scholar] [CrossRef]
- Raju, K.; Chinna Rao, B.; Saikumar, K.; Lakshman Pratap, N. An optimal hybrid solution to local and global facial recognition through machine learning. In A Fusion of Artificial Intelligence and Internet of Things for Emerging Cyber Systems; Springer: Cham, Switzerland, 2022; pp. 203–226. [Google Scholar]
- Coe, J.; Atay, M. Evaluating impact of race in facial recognition across machine learning and deep learning algorithms. Computers 2021, 10, 113. [Google Scholar] [CrossRef]
- Kaur, P.; Krishan, K.; Sharma, S.K.; Kanchan, T. Facial-recognition algorithms: A literature review. Med. Sci. Law 2020, 60, 131–139. [Google Scholar] [CrossRef]
- Bachute, M.R.; Subhedar, J.M. Autonomous driving architectures: Insights of machine learning and deep learning algorithms. Mach. Learn. Appl. 2021, 6, 100164. [Google Scholar] [CrossRef]
- Garcia Cuenca, L.; Sanchez-Soriano, J.; Puertas, E.; Fernandez Andres, J.; Aliane, N. Machine learning techniques for undertaking roundabouts in autonomous driving. Sensors 2019, 19, 2386. [Google Scholar] [CrossRef]
- Grigorescu, S.; Trasnea, B.; Cocias, T.; Macesanu, G. A survey of deep learning techniques for autonomous driving. J. Field Robot. 2020, 37, 362–386. [Google Scholar] [CrossRef]
- Chen, S.; Ding, Y. Machine Learning and Its Applications in Studying the Geographical Distribution of Ants. Diversity 2022, 14, 706. [Google Scholar] [CrossRef]
- Chen, S.; Ding, Y. A Machine Learning Approach to Predicting Academic Performance in Pennsylvania’s Schools. Soc. Sci. 2023, 12, 118. [Google Scholar] [CrossRef]
- Rickert, C.A.; Hayta, E.N.; Selle, D.M.; Kouroudis, I.; Harth, M.; Gagliardi, A.; Lieleg, O. Machine learning approach to analyze the surface properties of biological materials. ACS Biomater. Sci. Eng. 2021, 7, 4614–4625. [Google Scholar] [CrossRef]
- Dimauro, G.; Deperte, F.; Maglietta, R.; Bove, M.; La Gioia, F.; Renò, V.; Simone, L.; Gelardi, M. A novel approach for biofilm detection based on a convolutional neural network. Electronics 2020, 9, 881. [Google Scholar] [CrossRef]
- Ragi, S.; Rahman, M.H.; Duckworth, J.; Kalimuthu, J.; Chundi, P.; Gadhamshetty, V. Artificial intelligence-driven image analysis of bacterial cells and biofilms. IEEE/ACM Trans. Comput. Biol. Bioinform. 2021, 20, 174–184. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




