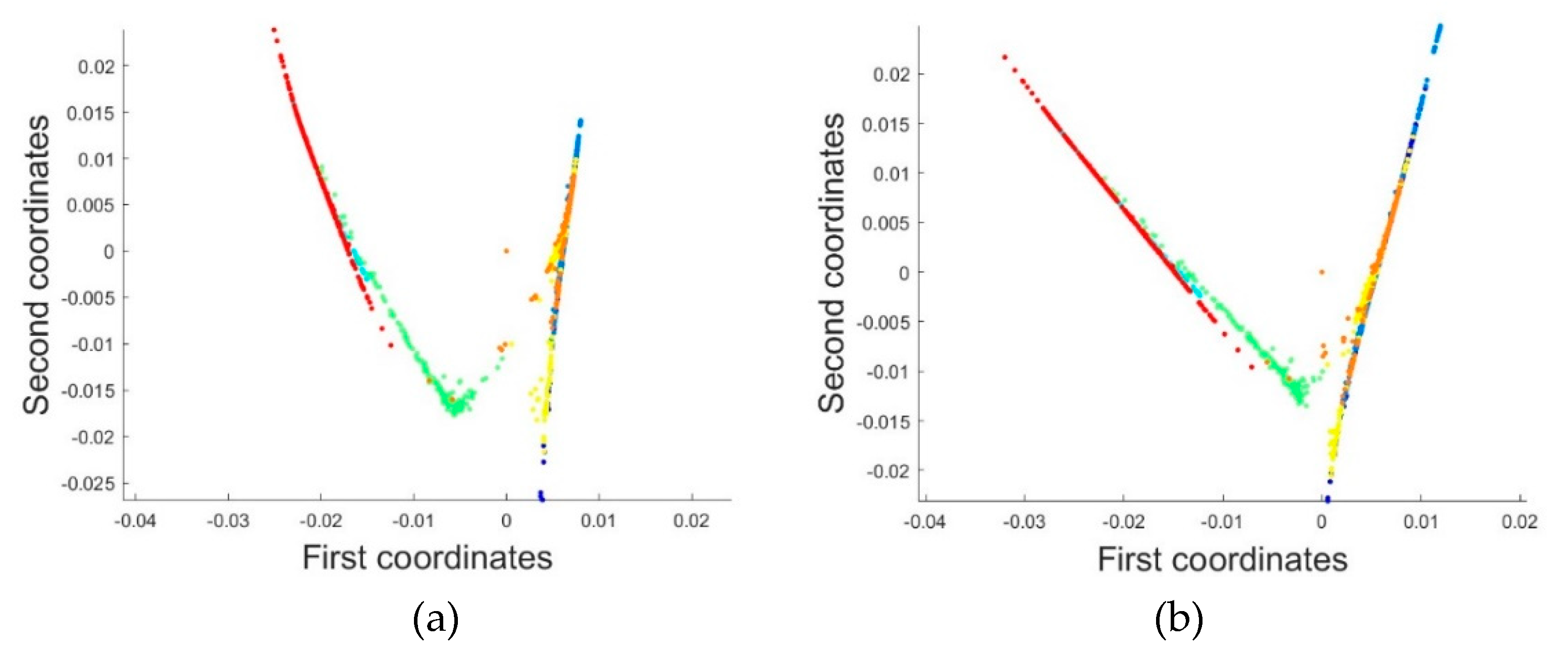
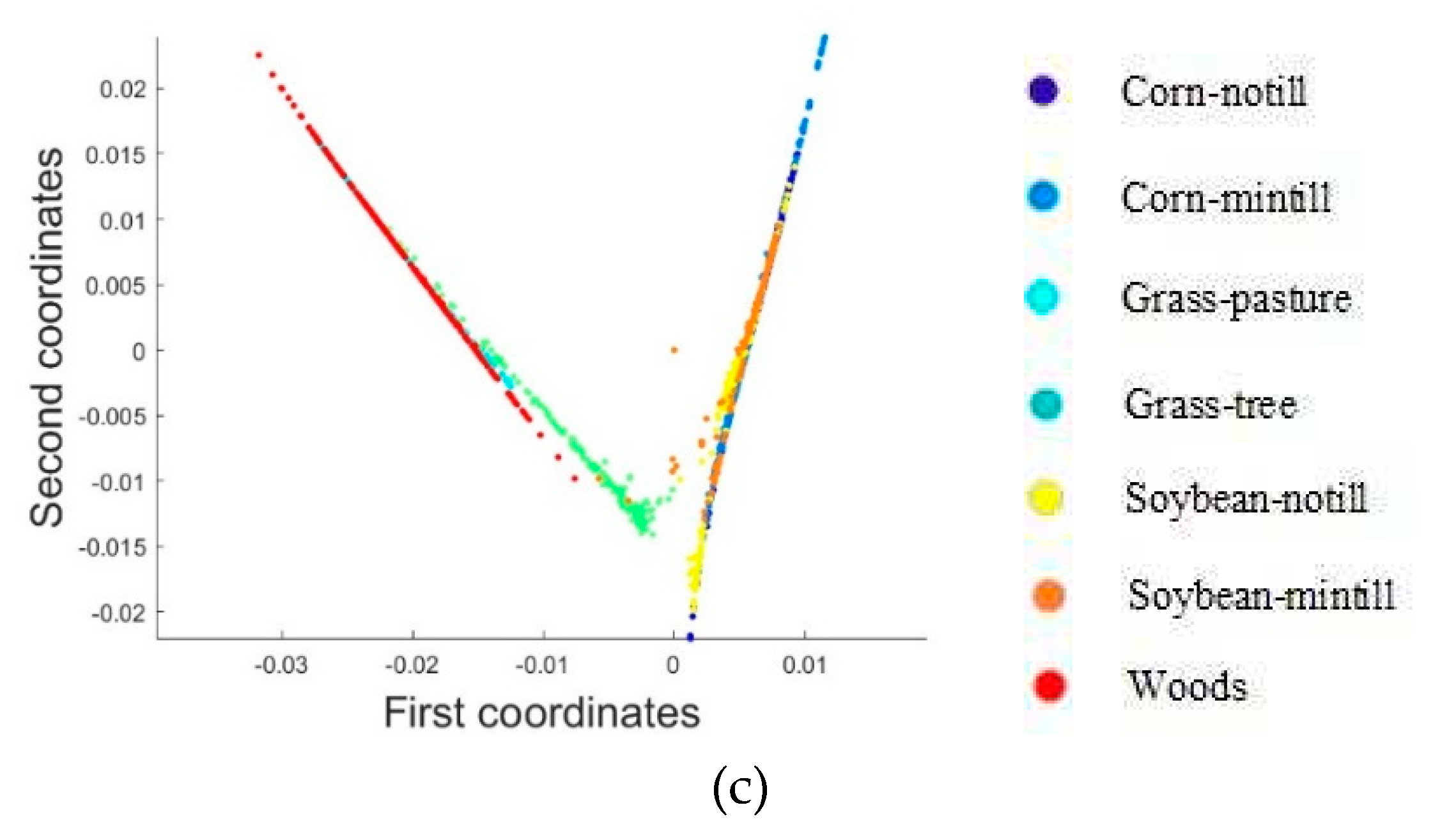
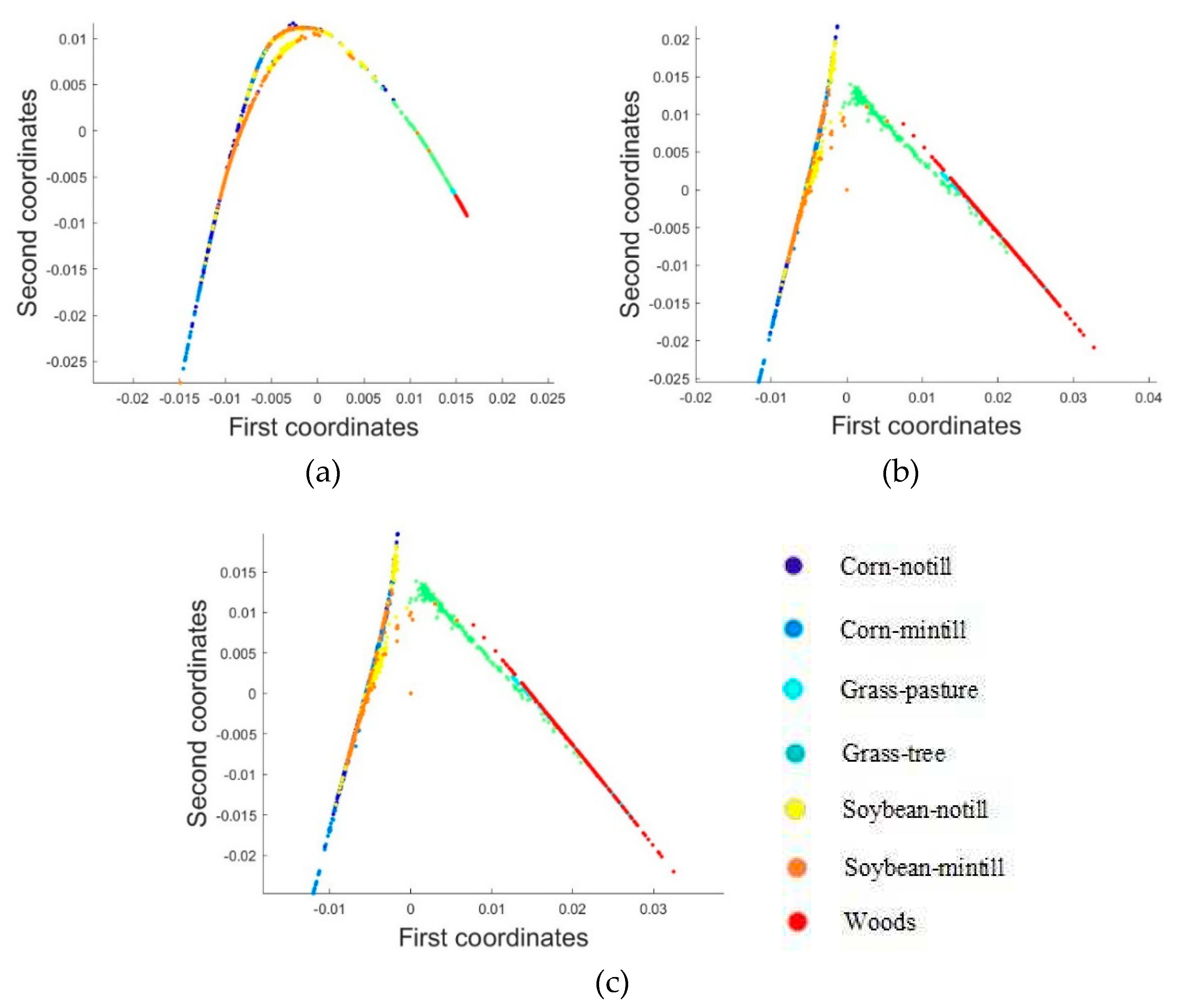
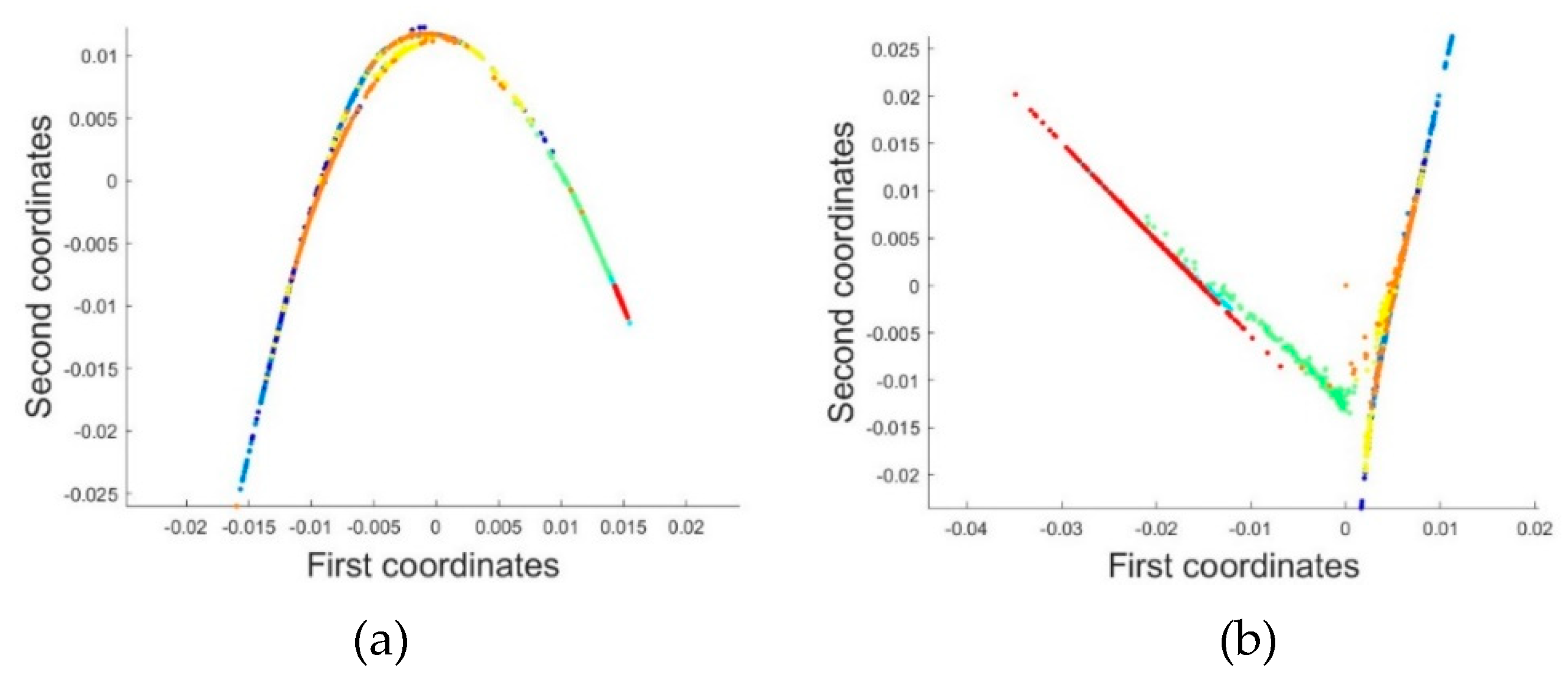
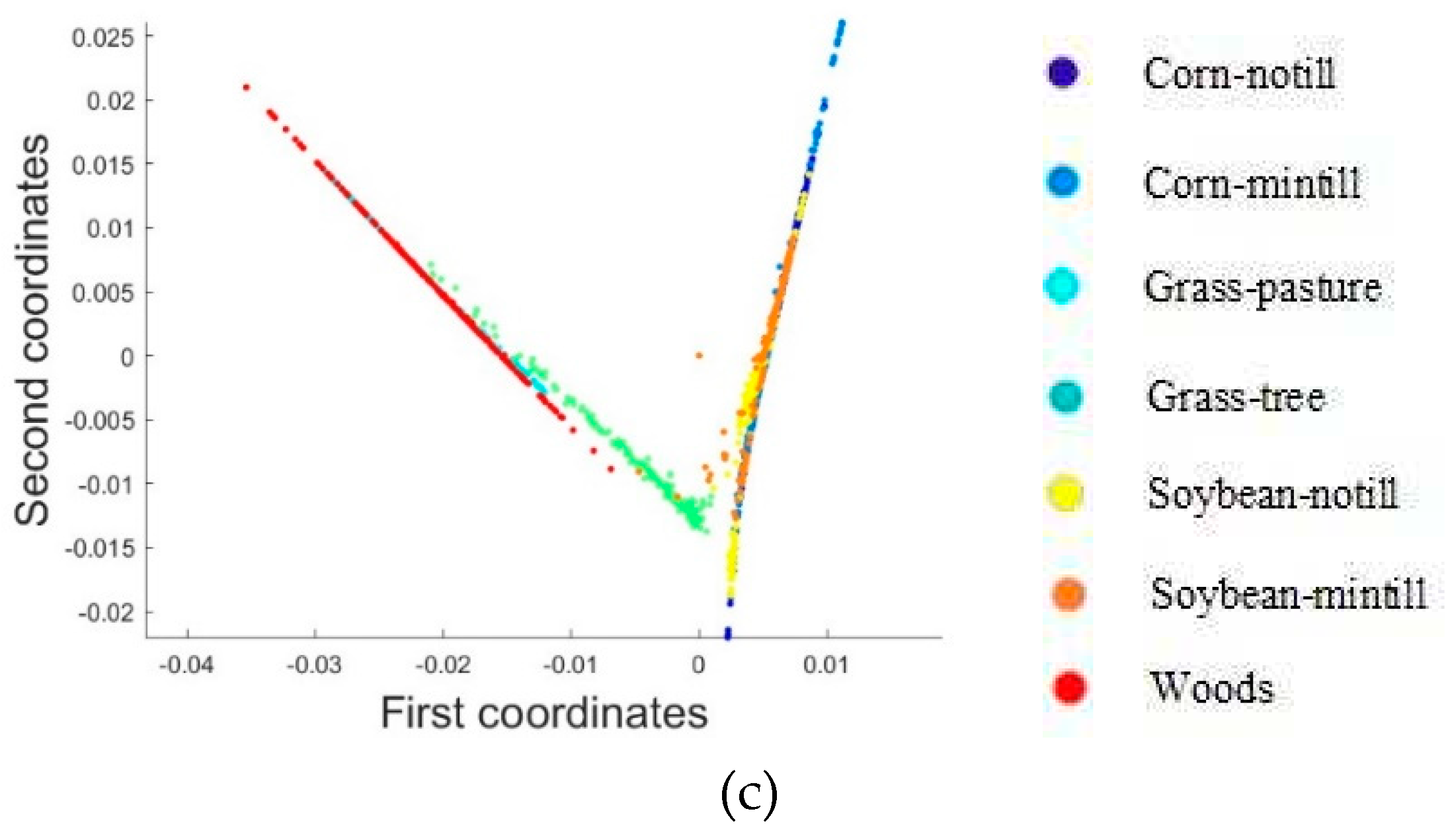
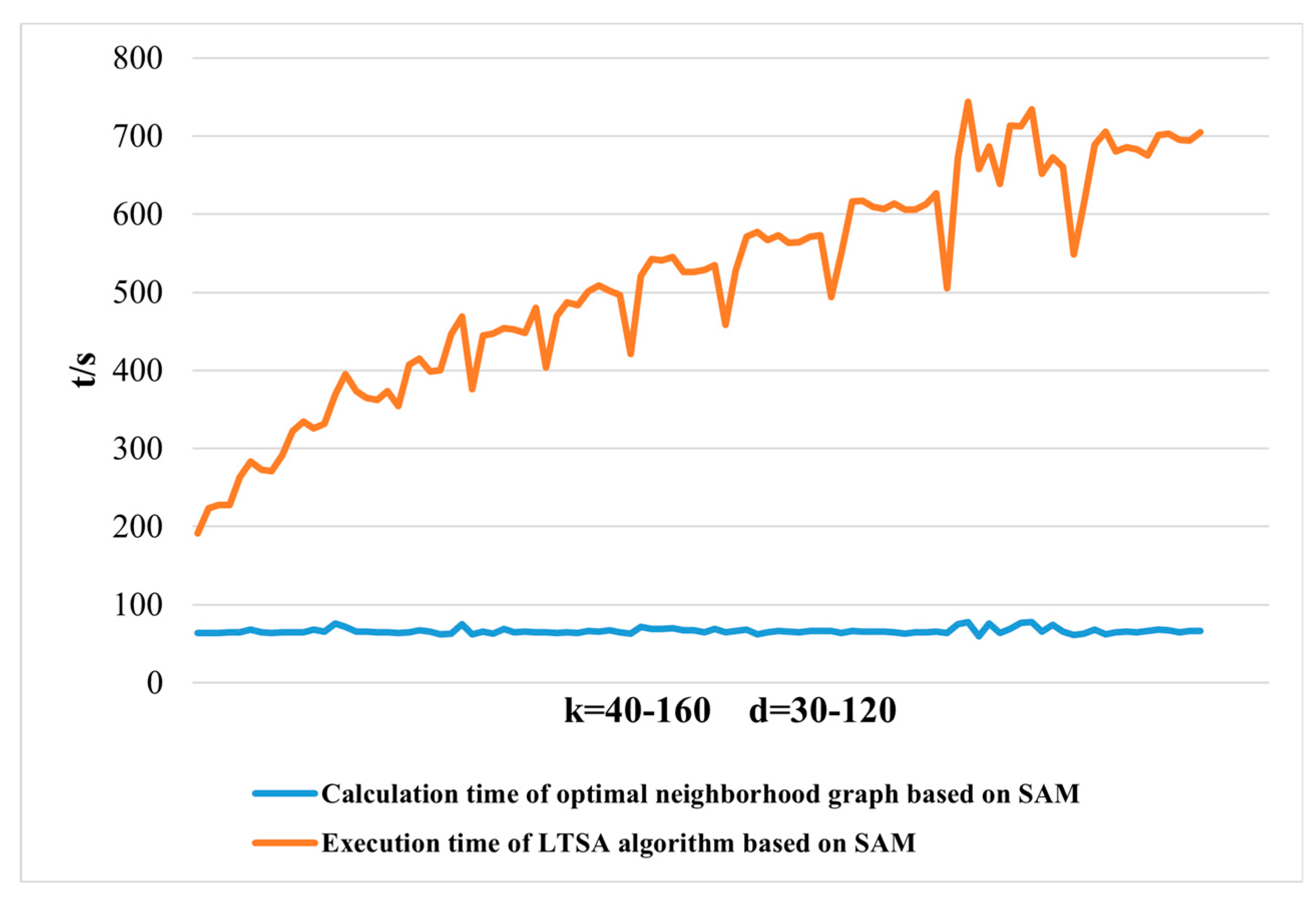
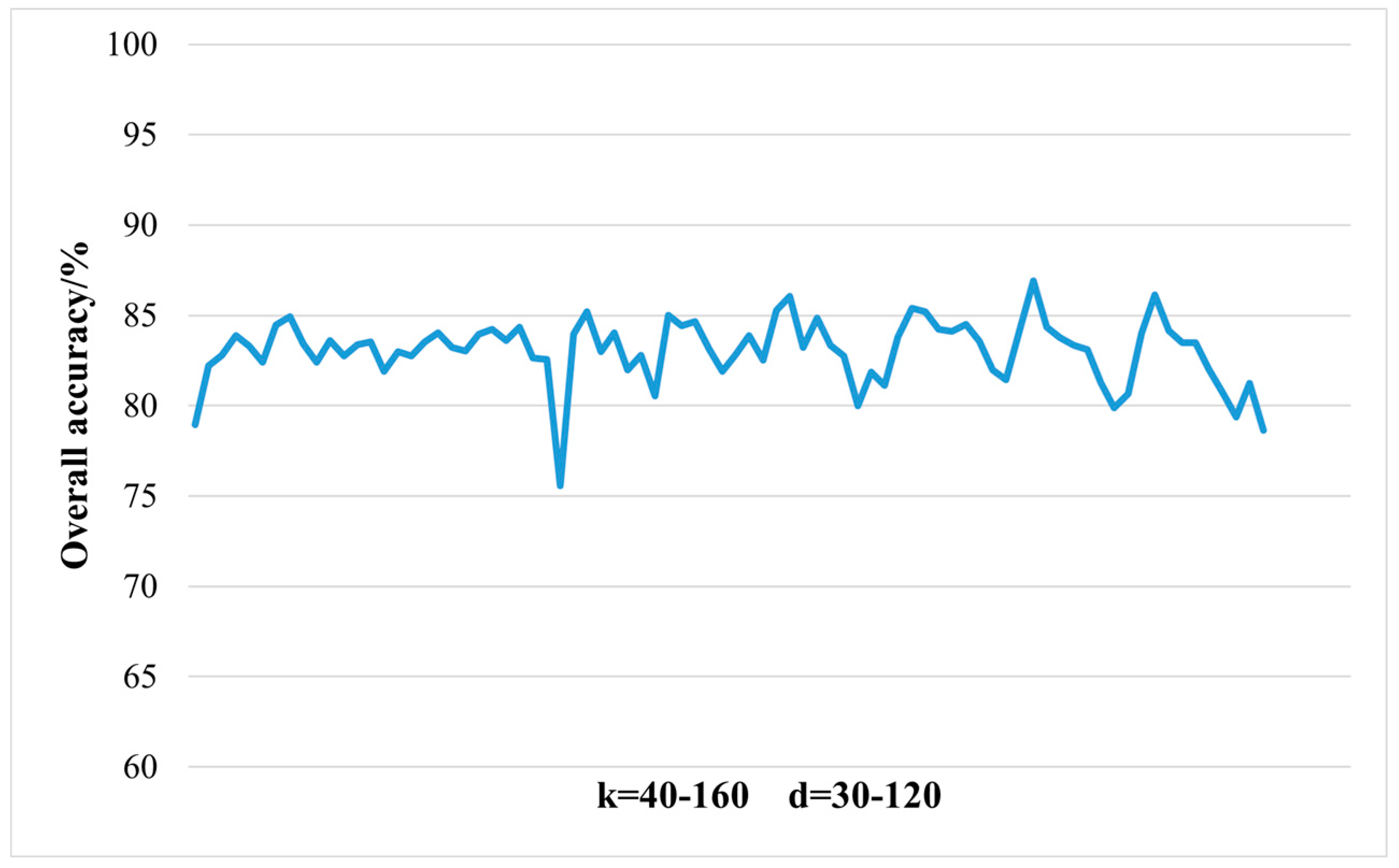
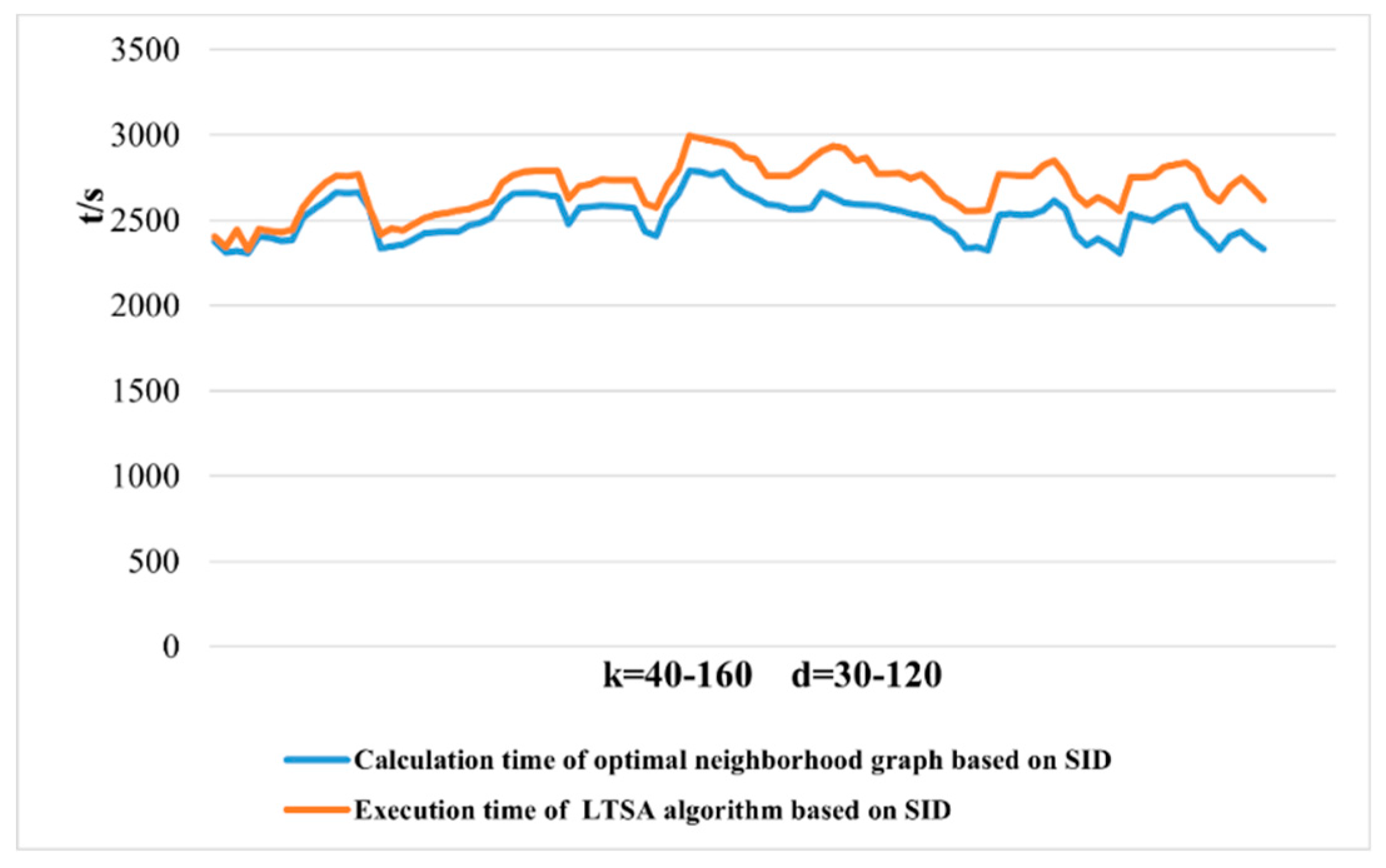
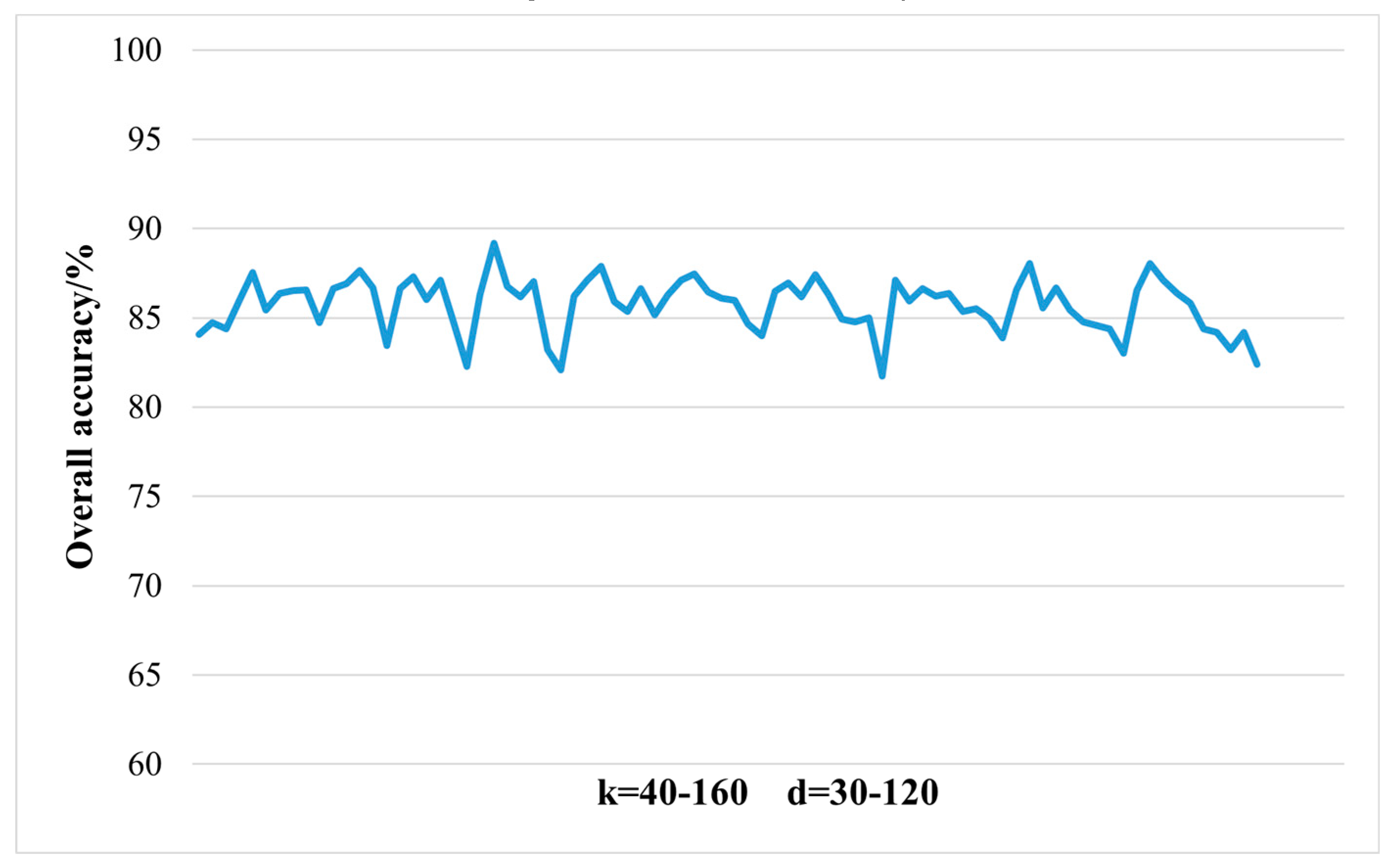
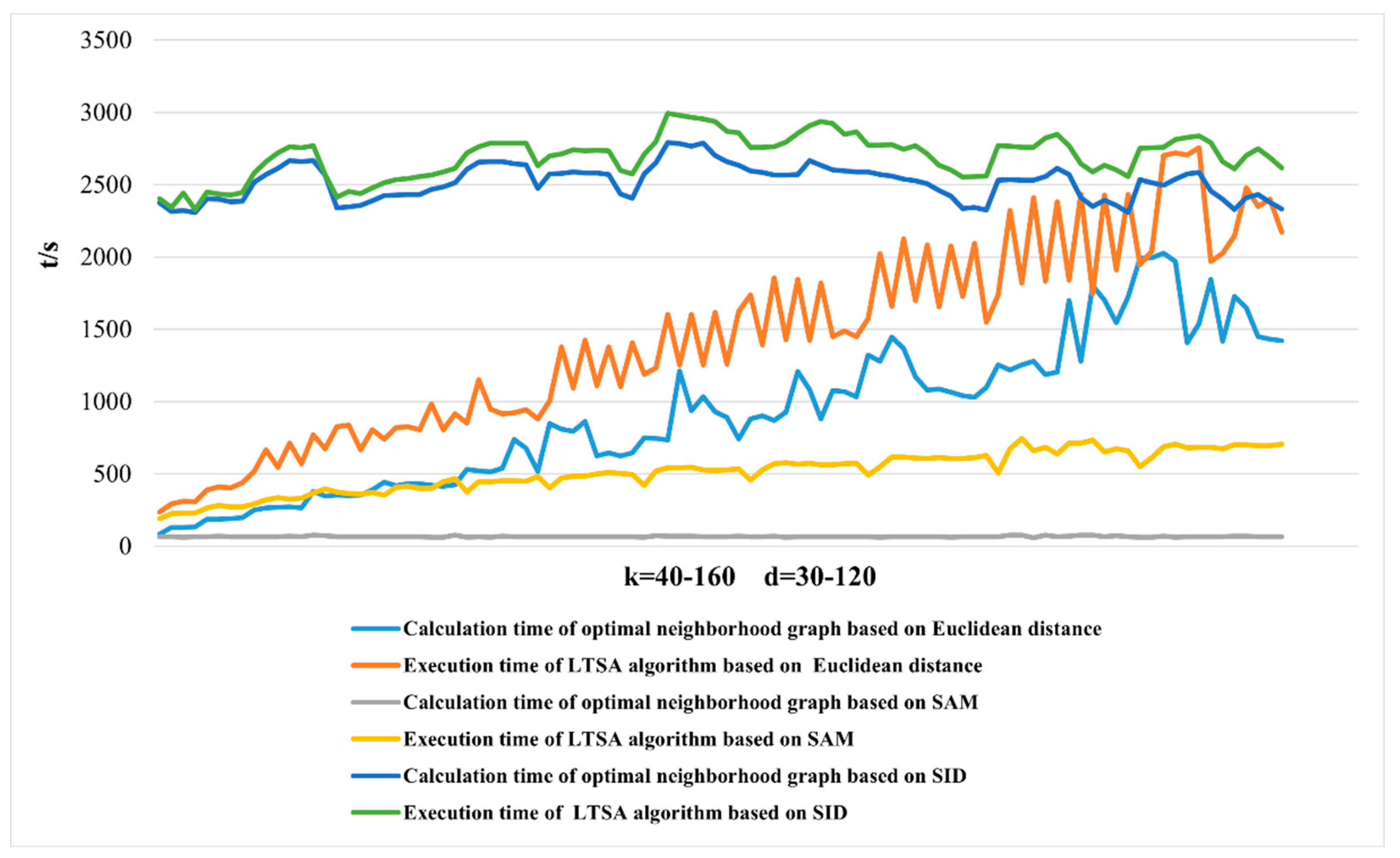
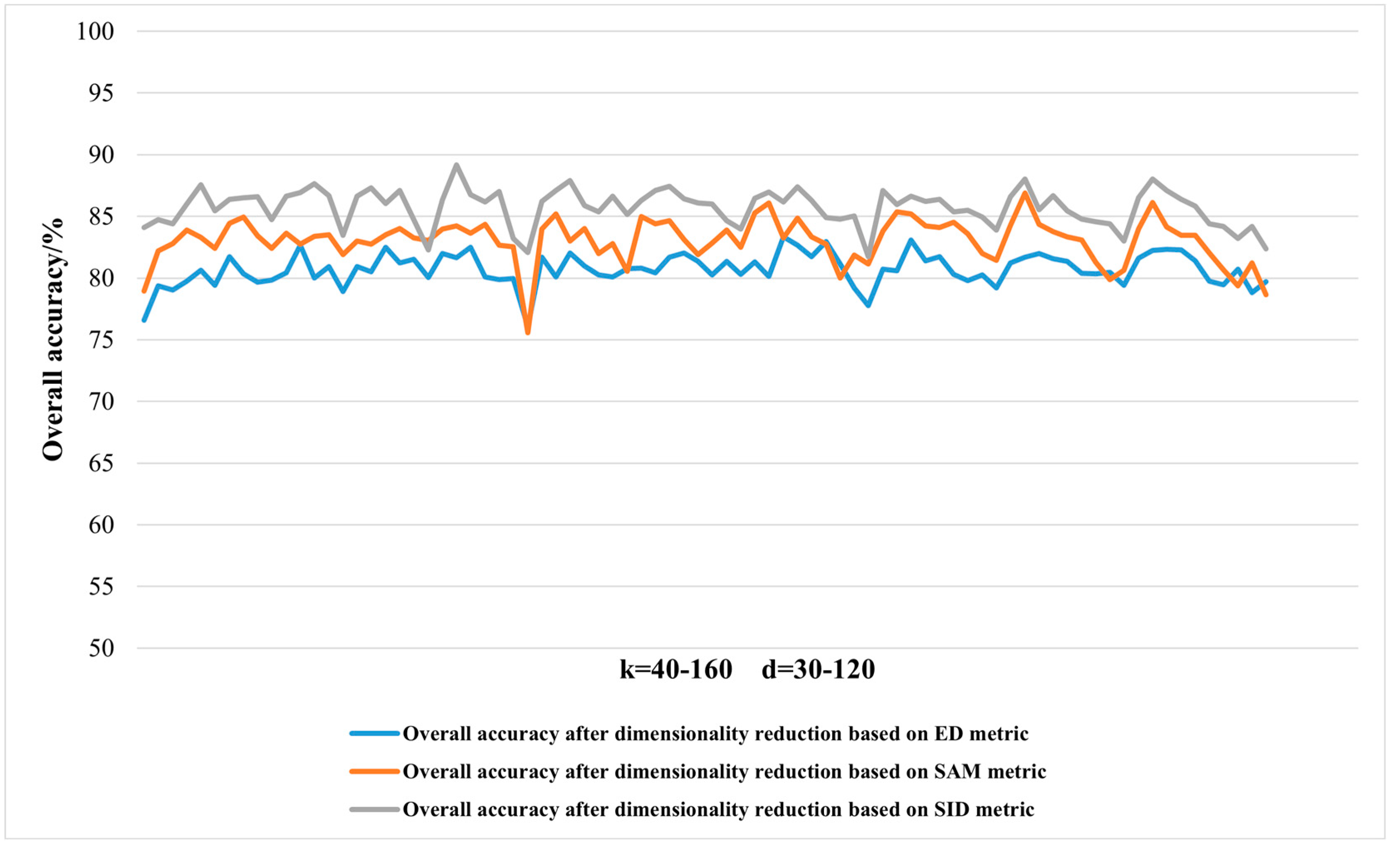
2.3.1. Dimensionality Reduction Method for Hyperspectral Remote Sensing Data Based on LTSA Algorithm
The fundamental concept of the Local Tangent Space Alignment (LTSA) algorithm is to use the tangent space of a sample point's neighborhood to represent its local geometric structure, and then align these local tangent spaces to construct global coordinates of the manifold [
37]. For a given sample point, LTSA uses its neighboring region to build a local tangent space to characterize the local geometric structure. The local tangent space provides a low-dimensional linear estimate of the nonlinear manifold's local geometric structure, preserving the local coordinates of sample points in the neighborhood. Subsequently, the local tangent coordinates are rearranged in the low-dimensional space through different local affine transformations to achieve a better global coordinate system. This method is particularly suitable for hyperspectral data as it can more effectively capture the spectral characteristics and local structures of ground objects. The process of dimensionality reduction of hyperspectral remote sensing images data using the LTSA algorithm primarily includes the following steps:
Step 1: Nearest Neighbor Search: For each sample point, identify its nearest neighbors in the high-dimensional space to construct a nearest neighbor graph.
Step 2: Local Tangent Space: Calculate the tangent space between the nearest neighbors for each sample point. The tangent space represents the local linear relationships, describing how the nearest neighbors are arranged relative to each other.
Step 3: Global Tangent Space: Using the nearest neighbor graph, construct the global tangent space, taking into account the local linear relationships between all nearest neighbors.
Step 4: Eigenvector Calculation: Perform eigenvalue decomposition on the global tangent space to obtain eigenvectors.
Step 5: Dimensionality Reduction Mapping: Arrange the eigenvectors in ascending order of their corresponding eigenvalues, select the first few eigenvectors to form a mapping matrix, and use this matrix to map the high-dimensional data to a low-dimensional space.
Step 6: Output Results: The dimensionally reduced data can be obtained through the mapping matrix, where each sample's coordinates in the low-dimensional space represent its position in the manifold structure.
The following is a detailed implementation process of using the LTSA algorithm to reduce the dimensionality of hyperspectral remote sensing images:
Suppose -dimensional manifold is embedded in an -dimensional space , where the -dimensional space is a high-dimensional space containing noise. Given a sample set distributed in this noisy -dimensional space:
where
represents the intrinsic representation of
, and
is a mapping function, with
representing the noise.
Given a hyperspectral remote sensing imagery dataset , where is the number of bands and is the number of pixel points. The first step of the algorithm is to find the optimal neighborhood for each pixel point. Let be a matrix composed of the nearest k pixel points to pixel point , including itself, measured using ED. The algorithm calculates the best d-dimensional approximate affine space for samples in to approximate the points in , minimizing the reconstruction error:
where
is a standard orthogonal matrix of local affine transformation with
columns, and
is the local manifold coordinates of
. After minimization, the local manifold coordinates of each pixel in the neighborhood
are obtained as
, where
represents the left singular vectors corresponding to the first
largest singular values of the centralized neighborhood matrix
.
is the central pixel point of the neighborhood matrix
, and
is a unit column vector of length
.
By constructing the local tangent spaces, the local manifold coordinates of all neighborhoods are obtained, and these overlapping local manifold coordinate systems are arranged to form the global coordinate system . LTSA (Local Tangent Space Alignment) posits that is obtained based on the local representative structure , that is, the global coordinates are used to reflect as much as possible the local spatial geometric structure expressed by the local coordinates . Therefore, it is necessary to satisfy , where is the local affine transformation to be found, playing the role of arranging the global coordinates. The reconstruction residuals for each pixel point represent the local reconstruction error, and the global manifold coordinates are calculated by minimizing the local reconstruction residuals. By minimizing the error matrix , the optimal solution is obtained as . To preserve as much local geometric structure as possible in the low-dimensional space, LTSA aims for the post-dimensionality reduction sample representation and the local affine transformation to minimize the reconstruction residual:
where
is the Moore-Penrose generalized inverse matrix of
. Let
be a 0-1 selection matrix satisfy
, and then find
such that the total reconstruction error of all samples is minimized:
. Where
, and
. To ensure that
has a unique solution, LTSA restricts
, and
is an
-dimensional identity matrix.
From the above process, it can be seen that given , LTSA first finds nearest neighbors for each sample (including itself) using ED metric, forming a neighborhood region containing itself for each sample . Then, within this region , PCA is performed without dimensionality reduction, and then is transformed into by PCA, where becomes . Subsequently, it is assumed that there is a linear relationship between the dimensionality reduction result and , aiming to minimize the error between the two. The affine relationship is represented by , and the residual is also represented by , thus turning it into a nonlinear method. The local aspect refers to what is known as the local tangent space, and the subsequent nonlinear dimensionality reduction is what is referred to as alignment, ultimately forming a problem in the following form:
The square of the Frobenius norm is transformed into the sum of the squares of the vector's L2 norms. Where is redefined as the -th row of , differing from the previous section, thus:
The classical Lagrangian multiplier method is applied to derive and set the derivative to zero. The resulting solution is then substituted back to obtain .
Therefore, it can be seen that minimizing the original reconstruction error is equivalent to minimizing . Consequently, the eigenvectors corresponding to the 2nd to -th smallest eigenvalues of form the global aligned manifold coordinate system .
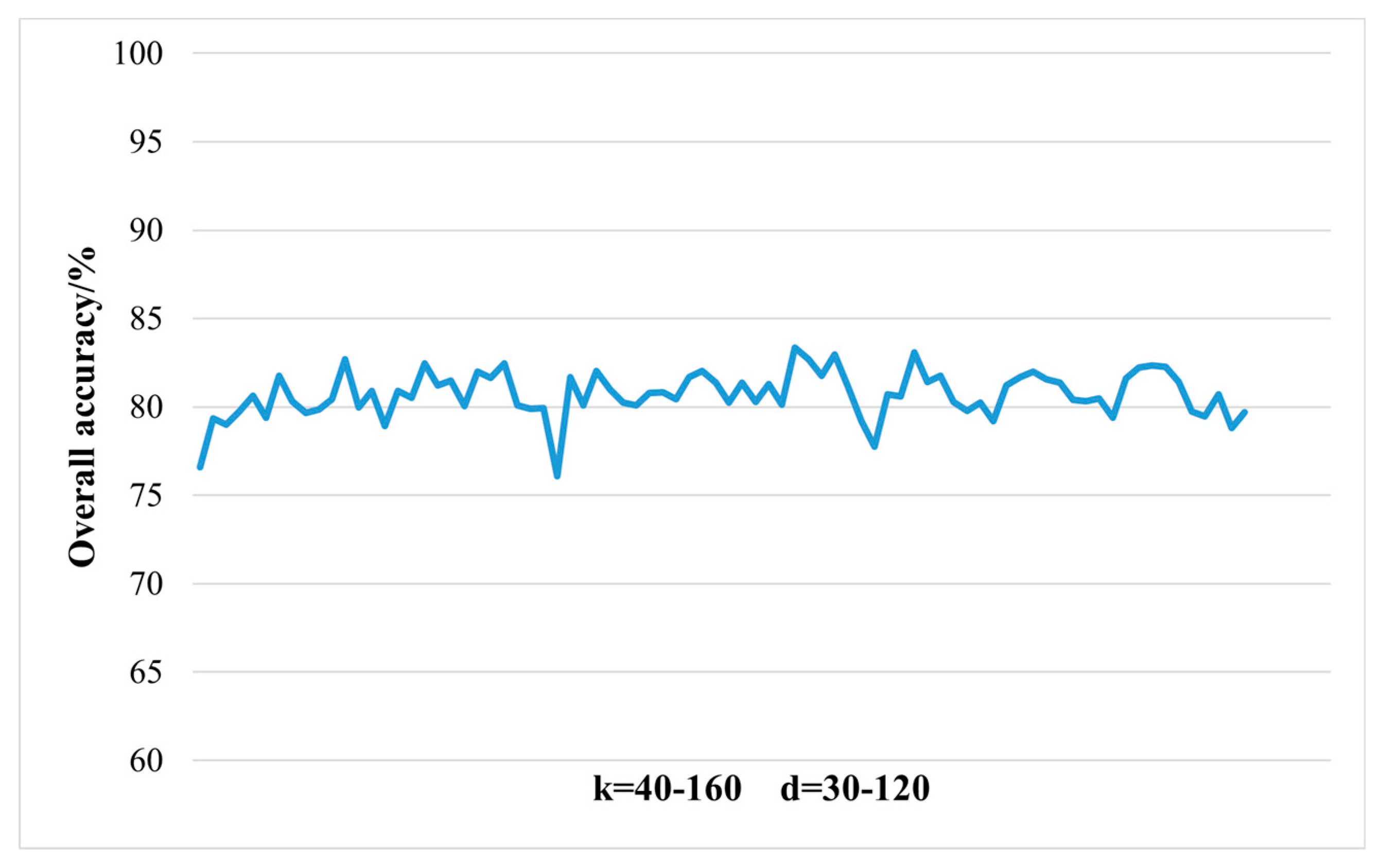
During the feature extraction process, the LTSA algorithm minimizes data redundancy on the basis of retaining effective information, reduces data dimensions to avoid the "curse of dimensionality," and decreases the occurrence of "same object different spectrum" and "different object same spectrum" phenomena. The transformed low-dimensional features represent a larger amount of information with fewer data points.
2.3.2. Nearest Neighbor Graph Calculation Method Based on Different Distance Metrics
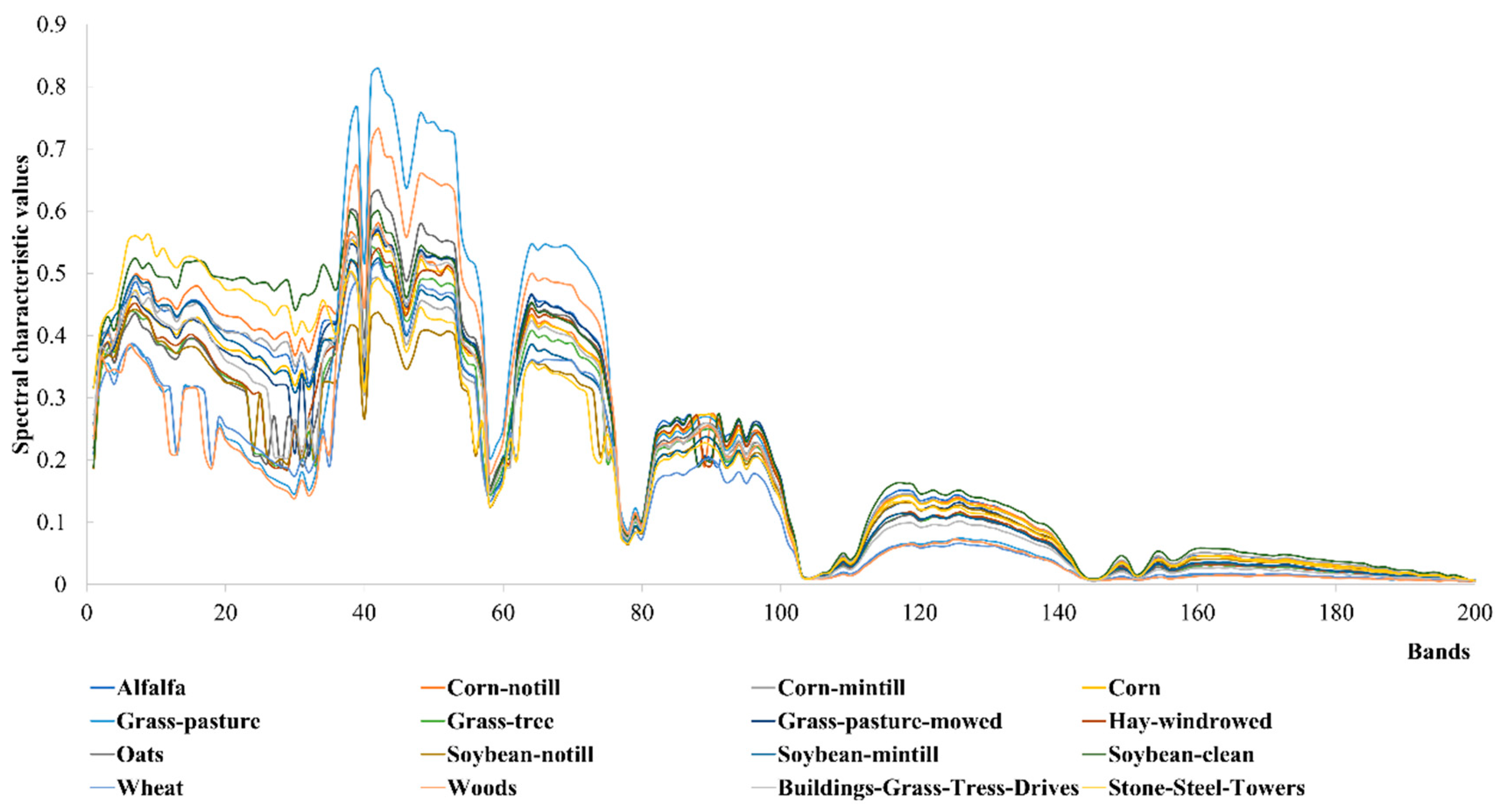
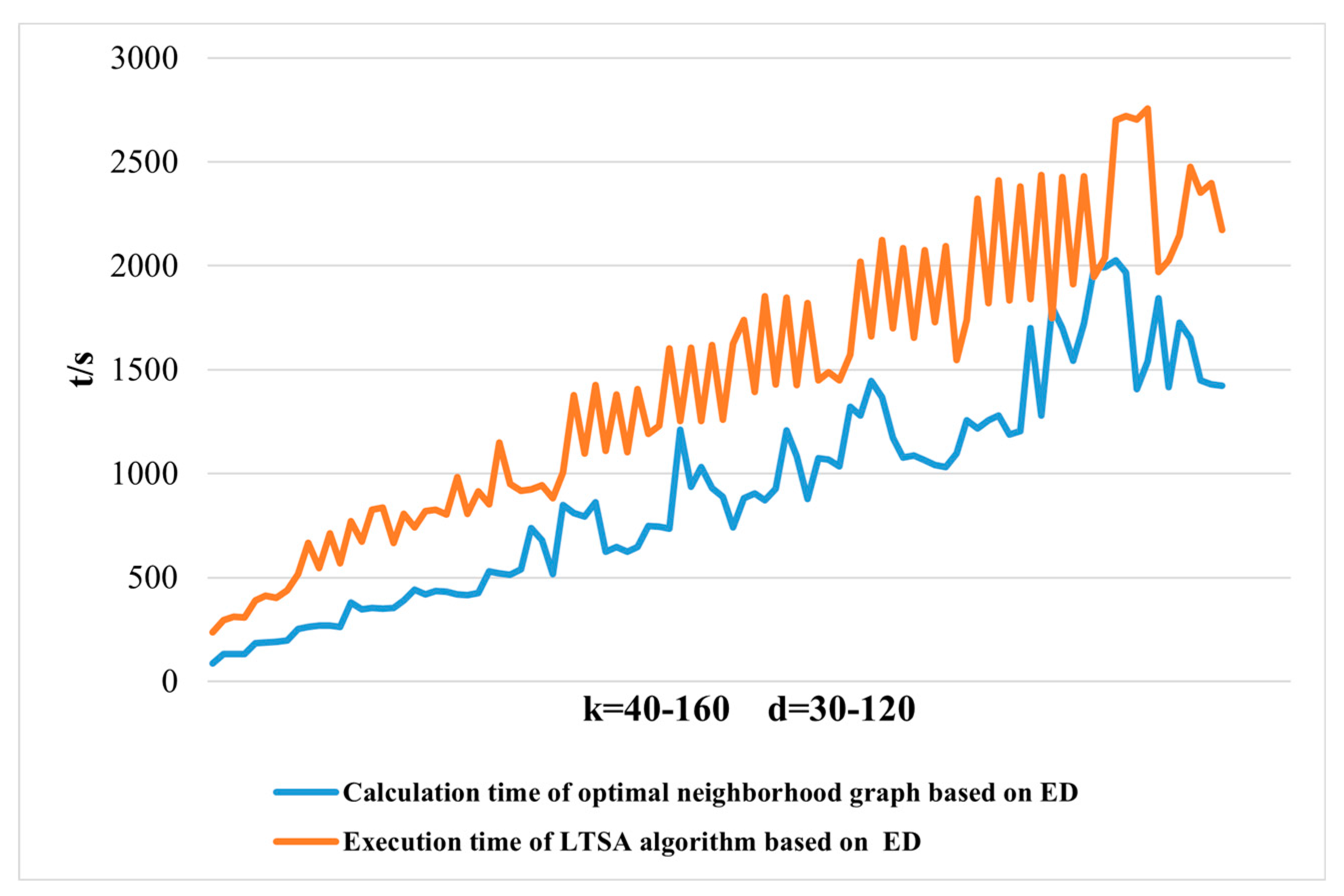
When applying the LTSA algorithm for dimensionality reduction of hyperspectral remote sensing images data, the first step involves determining the optimal neighborhood for each pixel. Most manifold learning algorithms compute the nearest neighbor graph based on ED metric, which fails to accurately capture the potential local non-linear structural features of hyperspectral remote sensing images. Consequently, scholars have introduced SAM and SID into the Isomap non-linear manifold learning algorithm as alternatives to ED for calculating the optimal neighborhood [
40]. However, since the Isomap algorithm focuses on the global structure of sample points and does not consider the local geometric relationships of data points, it does not reflect the local true topology of non-linear high-dimensional data. For hyperspectral remote sensing data, where different pixels represent different ground objects spectral features, the data distribution should form cloud-like clustering structures. Data from the same ground objects class cluster together, while data from different classes form multiple cloud-like clusters of varying sizes, distinguishable in the original high-dimensional space [
41]. Considering the inherent non-linear characteristics and spatial distribution features of hyperspectral remote sensing images, researchers have introduced the concept of spectral angle into the LTSA non-linear manifold learning algorithm [
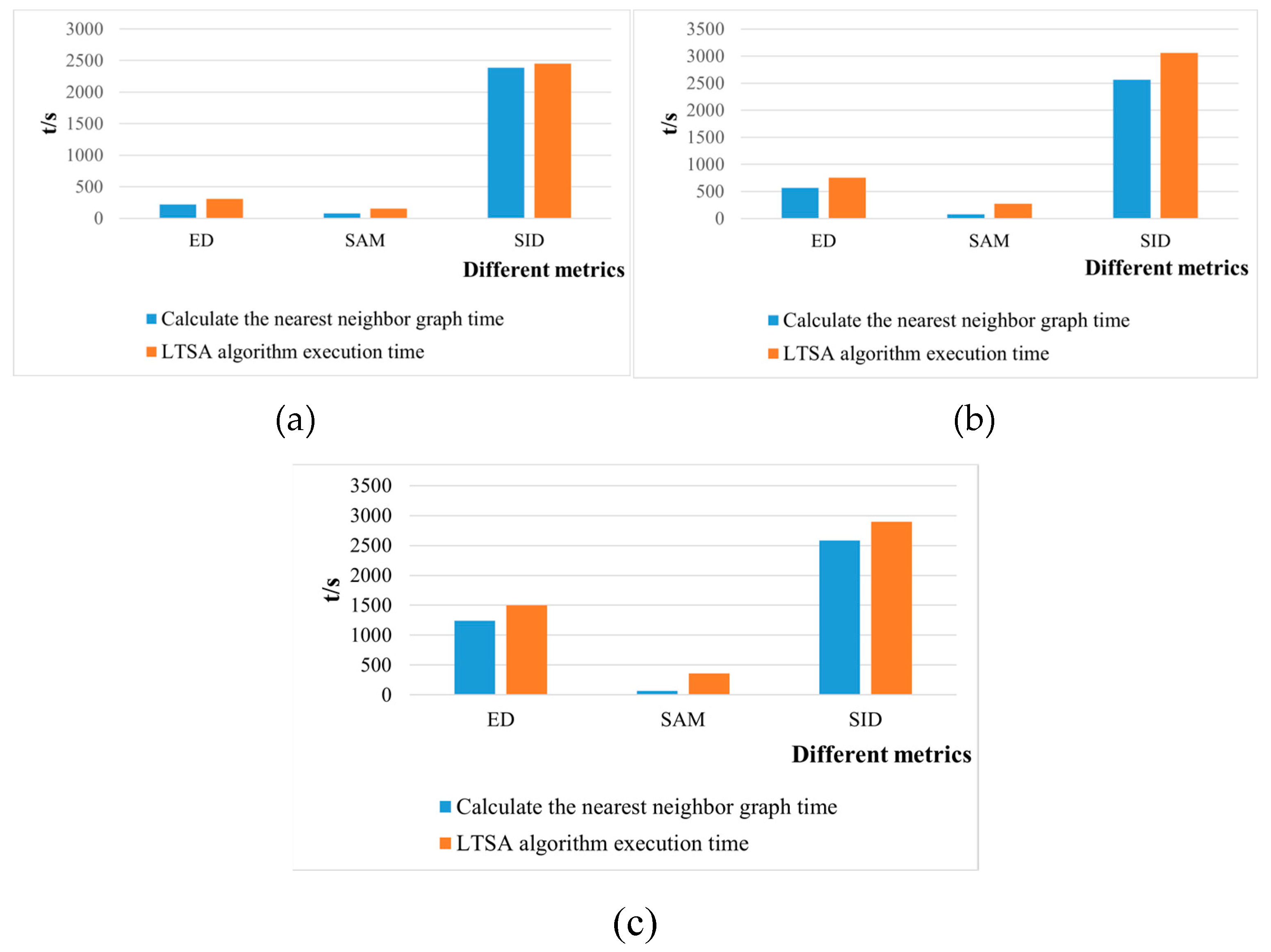
42]. This paper attempts to incorporate SID into the LTSA algorithm. It compares and analyzes the effects of calculating the nearest neighbor graph using ED, SAM, and SID metrics, and studies the dimensionality reduction capability of the LTSA manifold learning algorithm under these three different metrics. The specific implementation steps of the nearest neighbor graph are as follows:
Step 1: Distance Calculation. Use different distance metrics to calculate the distance between each data point and all other data points;
Step 2: Sorting. Sort the distances for each data point and find the k nearest neighbors;
Step 3: Storing Results. Store the distance matrix and the index matrix of the nearest neighbors.
In the LTSA algorithm, the construction of the nearest neighbor graph utilizes the ED metric. It determines the k nearest neighbors by calculating the ED between the sample point and its neighboring sample points, thereby constructing the nearest neighbor graph. Given a dataset , the k nearest neighbors of a data point are identified by calculating its ED to other sample points in the dataset, where k is the number of nearest neighbors to be found.
Suppose there are two spectral vectors and , each with the same number of bands N. The “distance” d between the two spectral curves can be calculated using the ED definition:
Therefore, the ED metric can be used to calculate the optimal neighborhood of a sample point, and thus construct the nearest neighbor graph for each sample point. The implementation steps of the nearest neighbor graph algorithm using the ED metric are as follows:
Step 1: Define a function [D, ni] = find_nn(X, k), which returns the ED matrix D and the nearest neighbor index matrix ni when given a dataset X and the defined number of nearest neighbors k.
Step 2: Obtain the size of the dataset n=size(X,1); and create matrices D and ni to store the computed ED values and their corresponding nearest neighbor indices, respectively.
Step 3: Use a single-layer nested loop to calculate the ED values between each sample point and all other sample points. First, use the function bsxfun() to compute the matrix DD of squared ED between two pixels and all other pixels, then sort the obtained distance matrix sort(DD).
Step 4: Find the indices of the k nearest neighbors and store the corresponding ED matrix values in ascending order in the distance matrix D and the index matrix ni .
Where D is the computed distance matrix, storing the distance information of each data point to its nearest neighbors. The size of matrix D is typically (n, k), where n is the number of data points, and k is the number of nearest neighbors. ni is the computed nearest neighbor index matrix, storing the indices of the nearest neighbors for each data point. The size of ni is also generally (n, k), with k being the number of nearest neighbors. These two matrices will be involved in subsequent LTSA algorithm computations for dimensionality reduction of hyperspectral remote sensing images.
- 2.
Nearest neighbor graph calculation based on spectral angle mapping (SAM) metric
SAM is an algorithm based on the overall similarity of spectral curves, which evaluates their similarity by calculating the generalized angle between the test spectrum and the target spectrum. When the angle between two spectra is less than a given threshold, they are considered similar; when the angle is greater than the threshold, they are considered dissimilar. The basic principle of spectral angle matching is to treat spectra as vectors and project them into an N-dimensional space, where N is the total number of selected bands. In this N-dimensional space, the spectrum of each pixel in the spectral image is treated as a high-dimensional vector with both direction and magnitude. The angle between spectra, known as the spectral angle, is used to quantify the angle between two spectral vectors, measuring their similarity or difference. The similarity between spectra is measured by calculating the angle between two vectors; the smaller the spectral angle, the more similar the two spectra are, with a spectral angle close to zero indicating identical spectra.
Suppose there are two spectral vectors and , each with the same number of bands N. The spectral angle between the two spectral curves can be calculated using the definition of cosine similarity:
where
is the dot product of A and B,
and
are the norms of A and B, respectively.
represents the spectral angle between the two spectra.
and
are the magnitudes of the two spectral vectors, respectively.
Consequently, the spectral angle measure between two spectral feature curves can be used to calculate the optimal neighborhood of a sample point, leading to the construction of the nearest neighbor graph for each sample point. The implementation steps of the nearest neighbor graph algorithm using the spectral angle measure are as follows:
Step 1: Define a function [D, ni] = calculate_SAM_for_pixels_test(X, k), which returns the SAM value matrix D and the nearest neighbor index matrix ni when given a dataset X and the defined number of nearest neighbors k.
Step 2: Obtain the size of the dataset n=size(X,1); and create matrices D and ni to store the computed SAM values and their corresponding nearest neighbor indices, respectively.
Step 3: Use a double-layer nested loop to calculate the SAM values for each sample point with all other sample points. First, obtain the spectral values spectrum_x and spectrum_y of two pixel points, then compute the dot product of the spectral feature values of the two pixel points, followed by calculating the norms and of each pixel point, finally calculate the SAM value and sort it.
Step 4: Find the indices of the k nearest neighbors and store the corresponding SAM values in ascending order in the SAM matrix D and the index matrix ni.
- 3.
Nearest neighbor graph calculation based on Spectral Information Divergence (SID) metric
Unlike SAM, SID is a stochastic method that considers spectral probability distributions. It is a spectral classification method based on information theory to measure the differences between two spectra. Treating spectral vectors as random variables, this method starts from the shape of spectral curves and uses probability and statistical theory to analyze and calculate the information entropy contained in each data point. It compares the magnitude of information entropy to judge the similarity between two different curves. SID can reflect the differences or degrees of change between different bands in hyperspectral data; the smaller the value of SID, the more similar the two sets of spectra are. Larger SID indicates significant spectral variations between different bands, while smaller divergence signifies consistent spectra. SID can describe the diversity of spectral features within a pixel or area and can consider the differences in reflected energy values, thereby more comprehensively assessing spectral similarity.
SID treats each pixel as a random variable and uses its spectral histogram to define a probability distribution. It then measures the spectral similarity between two pixels through the differences in probability behavior between spectra. This advantage is unachievable by any deterministic metric. ED, commonly used in classic pattern classification to measure spatial distance between two data samples, and SAM in remote sensing imagery are deterministic, as each data sample itself is a deterministic data vector, unlike the random variables considered by SID. Therefore, SID can be viewed as a stochastic or probabilistic method.
Suppose
and
represent the target spectrum and the test spectrum, respectively, with N being the number of bands, the SID for a pixel or region's spectral data can be calculated as follows [
43]:
Similarly,
where
and
represent the probabilities of the two spectra, respectively, and
,
are the self-information values of the two spectra derived from information theory.
Consequently, the SID between two spectral feature curves can be used to calculate the optimal neighborhood of a sample point, leading to the construction of the nearest neighbor graph for each sample point. The implementation steps of the nearest neighbor graph algorithm using SID measure are as follows:
Step 1: Define a function [D, ni] = calculate_SID_for_pixels_test(X, k), which returns the SID value matrix D and the nearest neighbor index matrix ni when given a dataset X and the defined number of nearest neighbors k.
Step 2: Obtain the size of the dataset n=size(X,1); and create matrices D and ni to store the computed SID values and their corresponding nearest neighbor indices, respectively.
Step 3: Use a double-layer nested loop to calculate the SID values for each sample point with all other sample points. First, obtain the spectral feature values spectrum_x and spectrum_y of two pixel points, then calculate the probability vectors and of the spectral feature values of the two pixel points, followed by computing and , and finally calculate the SID value and sort it.
Step 4: Find the indices of the k nearest neighbors and store the corresponding SID values in ascending order in the SID matrix D and the index matrix ni.