1. Introduction
With no permanent human habitation, the Antarctic continent represents one of Earth's most isolated and remote places. An ample reason for this are the severe and unpredictable weather conditions, characterized by temperatures that may plummet below -60 °C in the interior regions of Antarctica during winter, where approximately 99% of the landmass is shrouded in snow or ice [
1]. Recurring freeze-thaw cycles, intense UV radiation, scarce liquid water and substrates, and the lack of vegetation pose the most significant challenges for survival in this environment. However, despite the obstacles that face life in Antarctica, several places within the continent have been implicated as “biodiversity hotspots” and potential reservoirs of undiscovered species [
2]. That is why Antarctica is far from a barren wasteland devoid of microbial life, as was the general notion before the mid-1980s, right at the cusp of a shift towards using molecular techniques for species annotation [
3]. The most hospitable parts of the continent are found at the lower latitudes, especially in the Antarctic Peninsula and Maritime Antarctica.
Studying Antarctic microorganisms holds promise in multiple areas of research because of the unique environment they are found in. These organisms are well adapted to low temperatures, intense UV radiation and low nutrient input. As a result, some of their secondary metabolites and enzymes exhibit considerable potential for industrial applications [
4,
5]. Biotechnology often relies on enzymes capable of performing their catalytic functions at high or low temperatures. Exploring new species inherently bears the prospect of discovering novel antimicrobial compounds; some of the findings in this area have recently been reviewed [
6]. Understanding how microorganisms adapt and how microbial profiles shift in response to rapid environmental changes is crucial for understanding how similar communities react to such perturbations. Some Antarctic studies that track the change in microbial profiles due to the influx of organic carbon sources [
7,
8] or temperature variations [
9] have already been carried out. Such knowledge will allow us to preserve the Antarctic and other similar desert ecosystems better. Additionally, Antarctica's pristine and isolated environments provide an excellent opportunity to study the effects of anthropogenic factors on microbial communities [
10]. Research stations in Antarctica are recognized sources of anthropogenic influence on the continent, attributed to factors such as the deposition of microplastics from equipment [
11] and an increase in antimicrobial-resistant bacteria [
12,
13].
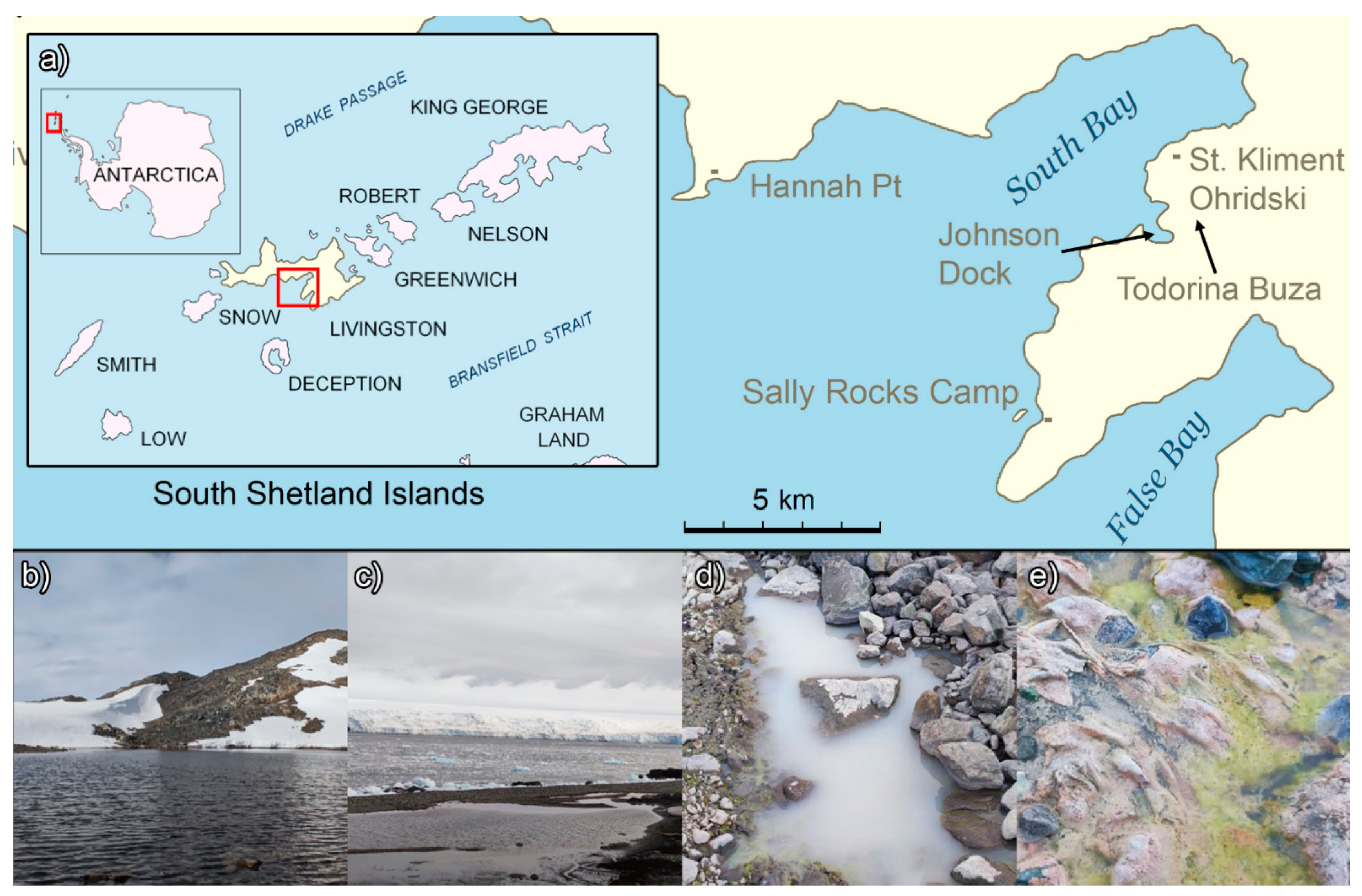
The Bulgarian Antarctic Base “St. Kliment Ohridski” is a research station located in Maritime Antarctica at 62°64'S, 60°36'W, on Livingston Island, which is part of the South Shetland Islands archipelago. It was founded in 1988, but the microbial composition of the diverse ecological niches around the site remains unknown. These environments encompass a lagoon, multiple meltwater ponds and streams, the Todorina Buza tarn, and a recently formed unnamed lake. In 2022, an expedition to the station was carried out to collect samples for direct NGS metagenomic sequencing to assess the microbial composition of various sites around the base and identify noteworthy microorganisms.
2. Materials and Methods
2.1. Sampling Sites
Sampling was conducted in January 2022 near the Bulgarian Antarctic Base “St. Kliment Ohridski” on Livingston Island, Maritime Antarctica (
Figure 1).
The different samples with their descriptions and the corresponding environments are listed in
Table 1.
2.2. DNA Extraction
Total DNA was isolated immediately after the collection to minimize potential shifts in the microbial compositions of the samples. DNA from the solid samples was extracted with ZR Soil Microbe DNA MiniPrep Kit (Zymo Research Corp., USA) according to the manufacturer’s Instructions manual. Two liters of water underwent filtration through a 0.2 µm filter for water samples. Subsequently, the biomass was rinsed off using 700 µl of the Bashing Beads buffer from the ZR Soil Microbe DNA MiniPrep Kit. The eluted biomass was then transferred to the kit's Bashing Beads tubes and subjected to further processing in a manner identical to that employed for solid samples. The eluted DNA samples were quantified via fluorometric measurement (Quantus – Promega Corp., USA) and kept at -20 °C.
2.3. Amplicon-Based Metagenomic Sequencing
Sequencing was carried out by the Novogene company. For bacteria, the V3-V4 16S rRNA regions were amplified using primers 341F (5’-CCTAYGGGRBGCASCAG-3’) and 806R (5’-GGACTACNNGGGTATCTAAT-3’) [
14]. For archaea, the V4-V5 16S rRNA regions were amplified using primers Arch519F (5’- CAGCCGCCGCGGTAA-3’) and Arch915R (5’- GTGCTCCCCCGCCAATTCCT-3’) [
15]. For fungi, the ITS2 region was amplified using primers ITS3-2024F (5’-GCATCGATGAAGAACGCAGC-3’) and ITS4-2409R (5’-TCCTCCGCTTATTGATATGC-3’) [
16]. The corresponding libraries were sequenced on an Illumina platform, generating 2 x 250 bp paired-end reads.
2.4. Data Analysis
Sequence processing, data analysis and visualization were carried out using the QIIME2 pipeline [
17]. Barcode and primer removal, denoising, chimera removal, and pair-end joining were done using the DADA2 plugin [
18] to generate amplicon sequence variants (ASVs). Taxonomy classifiers were trained on the SILVA132 [
19] and UNITE ver9.0 [
20] databases after extracting the sequenced region through
in-silico PCR with the primers used in this study and using QIIME2’s feature-classifier plugin [
21]. Taxonomy was assigned with a 0.8 confidence threshold, and chloroplasts were removed from the dataset [
22]. Alpha diversity indices were calculated using the diversity plugin. For beta diversity analysis, a rooted phylogeny tree was constructed using the phylogeny plugin fasttree method, and then used to calculate the weighted UniFrac distances and a PCoA matrix using the diversity plugins. Plots were made using the matplotlib and seaborn python packages [
23,
24].
3. Results
3.1. Sequencing Quality and ASV Counts
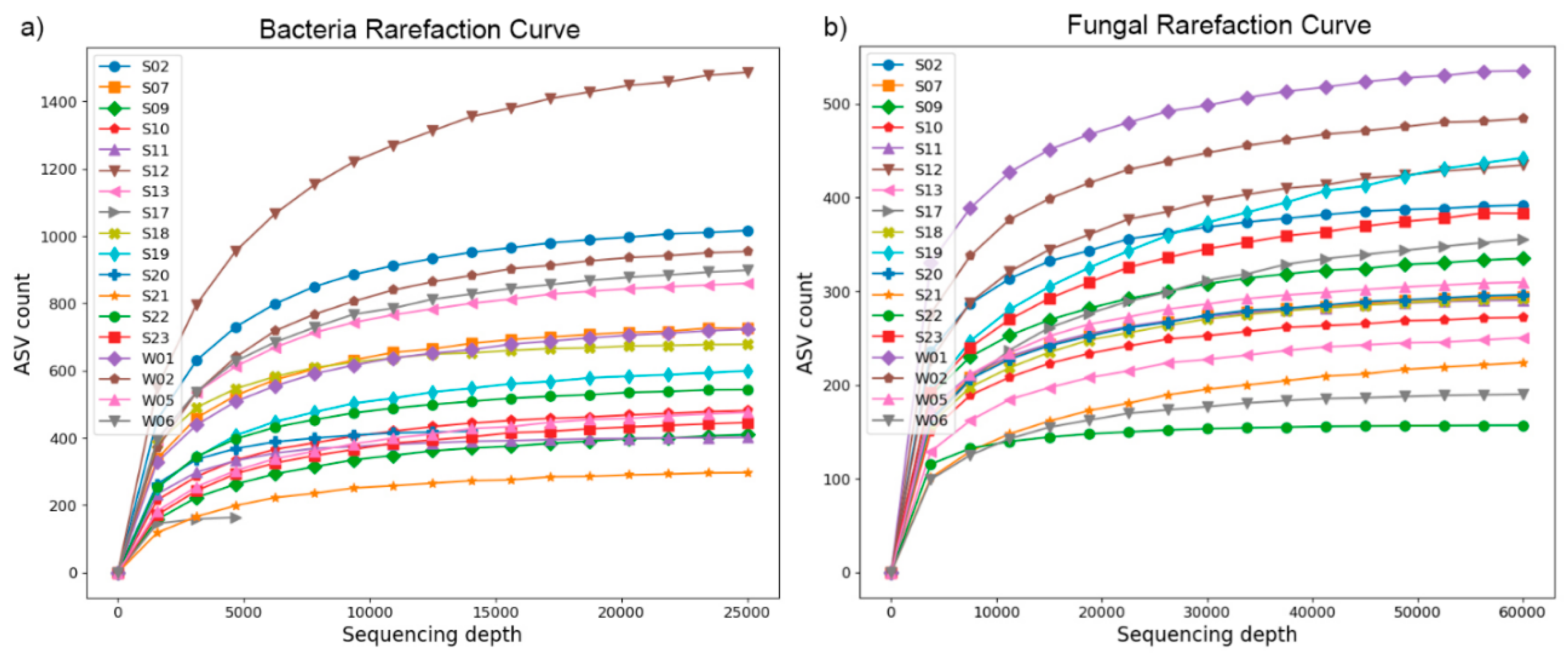
5,583 ASVs were obtained using eubacterial 16S primers, with only 4 ASVs common to all samples. 2696 ASVs were obtained using fungal primers, with 5 of them being common to all samples. Most ASVs obtained using the archaea-specific primers were taxonomically identified as eubacterial. The eubacterial and fungal rarefaction curves are presented in
Figure 2 and show that an adequate sequencing depth was achieved for both domains in all samples. The Q20 and Q30 scores before filtering and denoising were above 97% and 91%, respectively (data not shown).
3.2. Taxonomic Annotation
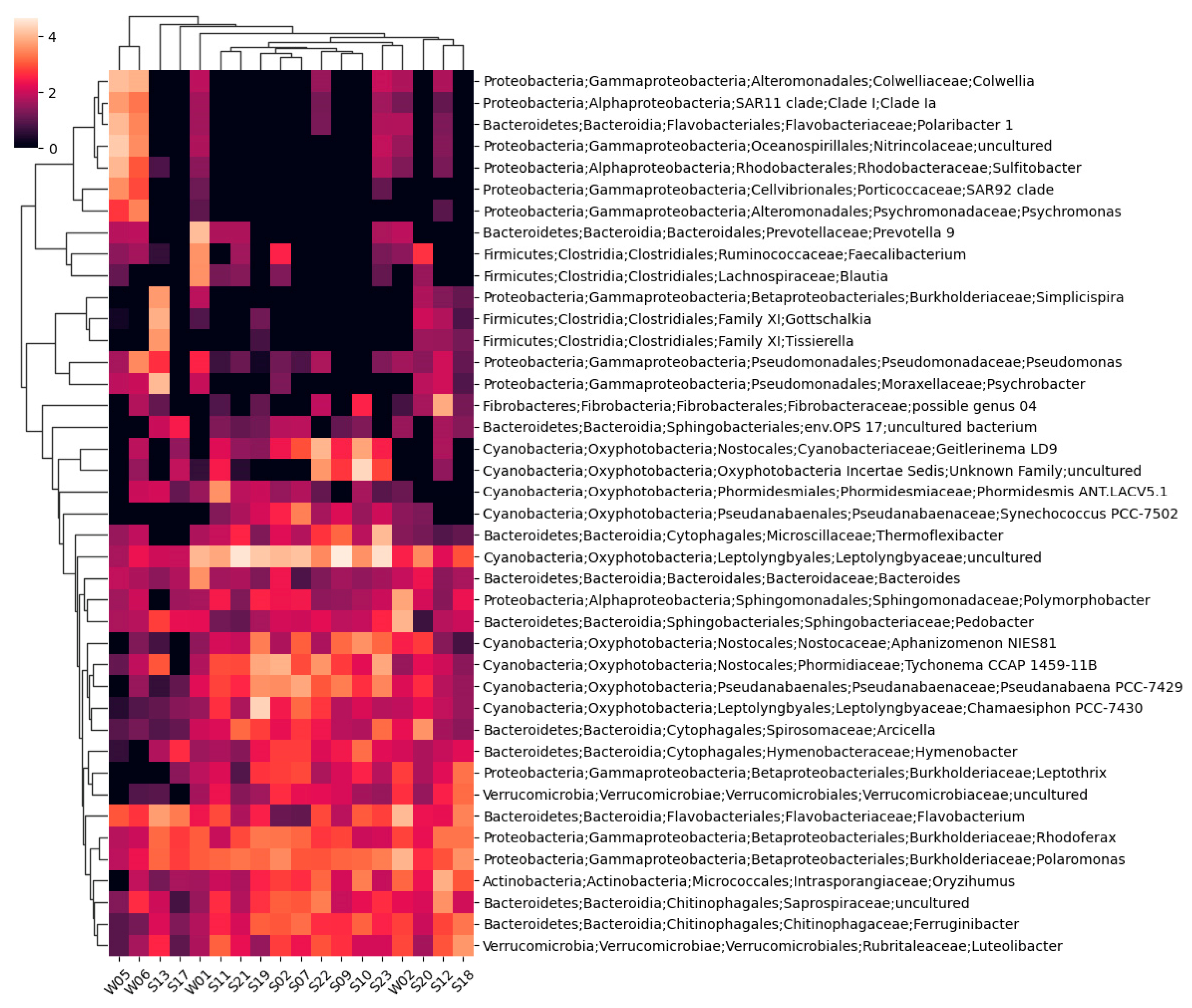
Taxonomic annotations representing at least 4% of eubacterial reads in any sample are shown on the cluster heatmap represented in
Figure 3.
A total of 1894 unique bacterial taxons were identified, 1156 of which were annotated to the species level. Only two taxons could not be assigned to a phylum. The highest number of identified bacterial taxons and annotated species belonged to sample S12, while the lowest were in sample S17.
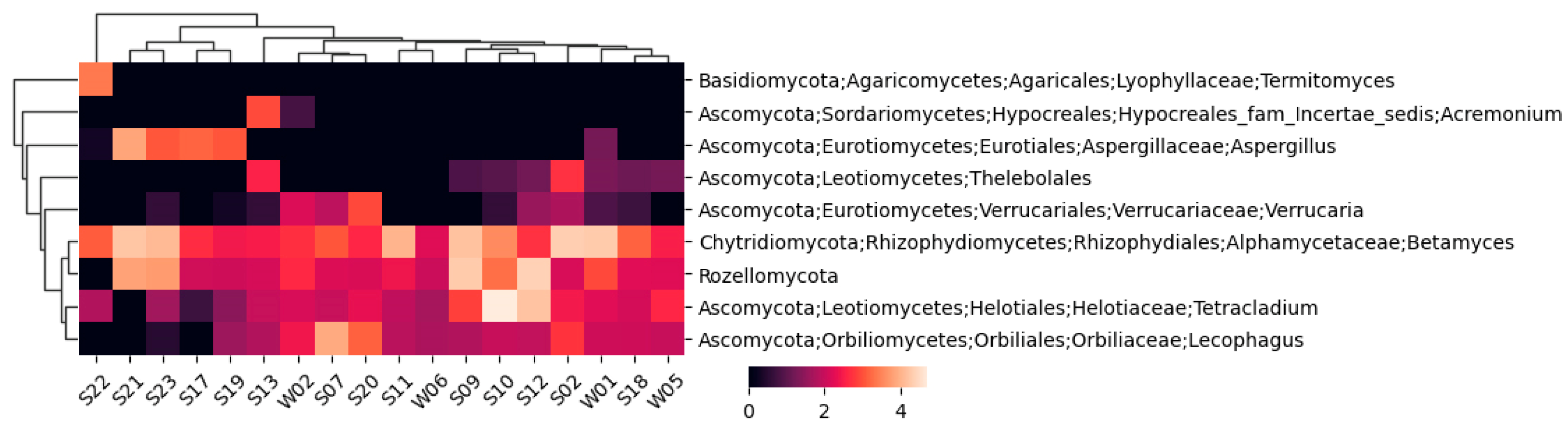
A similar fungal heatmap is given in
Figure 4, but with a 1% cutoff.
For the fungi, 244 unique taxons were identified, 173 of which were annotated to the species level. The highest number of identified fungal taxons and annotated species belonged to sample S19, while the lowest were in sample W06.
The evenness and richness of the bacterial and fungal communities are represented in the rank abundance curves in
Figures 5a,b, respectively.
Sample W06 was isolated from the surface of a rock within the research base and displayed the lowest bacterial evenness and richness of any community. Only three organisms make up more than 50% of sequences in that sample. S12 is a soil sample underneath a vegetation patch and shows the highest richness of any bacterial community. Sample S09 displayed a very high evenness but low richness for the fungal community and is one of the microbial surface mat samples.
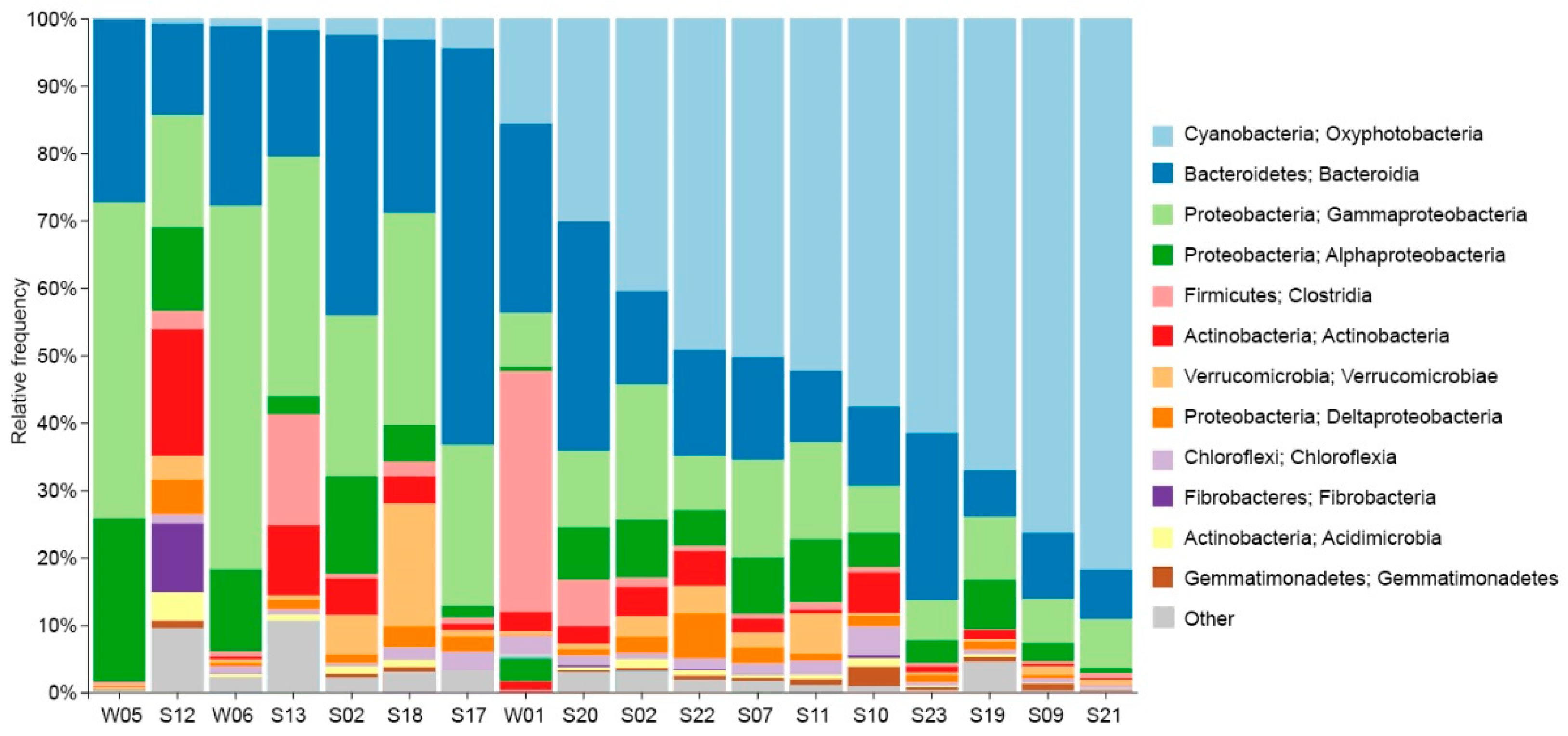
The relative abundances of dominant bacterial classes for each sample, along with their respective phyla, are given in
Figure 6. The most prominent group across most samples was a cyanobacteria belonging to
Leptolyngbyaceae, constituting 79% of reads in one single sample. Moreover, cyanobacteria represented more than 50% of reads of all bacterial surface mats from submerged rocks. Other prevailing groups observed in many samples comprised
Bacteroidetes,
Gammaproteobacteria, and
Alphaproteobacteria.
Bacteroidetes represented 59% of reads in the dryland surface mat sample (S17). The water samples exhibited a high prevalence of
Proteobacteria, up to 71% of the total sequencing reads with assigned taxonomy ranks. The class
Clostridiales represented 36% of reads from the lagoon, a much higher value than in any other sample. Epilithic sample S17 from a rock inside the research base was characterized by a great abundance of
Flavobacterium (37% of sequences),
Hymenobacter, and, to some extent,
Rhodoferax.
50%-98% of fungal reads could not be assigned to a phylum. Sequencing using archaea-specific primers produced a small number of reads, representing <1% of sequences in most samples, while the rest were taxonomically identified as bacteria. The only exceptions were the seawater samples - W05 and W06, where archaea represented 41% and 8% of reads, respectively. Candidatus Nitrosopumilus, Nitrosarchaeum and Marine Group II represented the majority of sequences from the seawater samples. S07 and S12 also had a slightly higher abundance of archaea, with 3% and 4%, respectively, and archaea there were dominated by representatives of Woesearchaeales. The fungal genus Betamyces was prevalent in all samples isolated from the lagoon and the meltwater ponds, with an average of 13% of total reads.
3.3. Alpha Diversity Analysis
Аlpha diversity metrics for bacteria are shown in
Table 2
Sample S12 has the highest bacterial richness and diversity of any sample, although the Simpson index is higher for some other samples. The lowest richness belongs to sample S17, but samples S21 and S23 have the lowest diversity and are both part of microbial surface contaminations.
The corresponding Аlpha diversity metrics for fungi are given in
Table 3
Overall, fungal richness was much lower than bacterial. Freshwater samples W01 and W02 have a higher-than-average richness and diversity, yet phylogenetic diversity is even higher in some microbial surface mat samples. The lowest fungal richness and diversity by any metric were observed in saltwater sample W06.
3.4. Beta Diversity Analysis
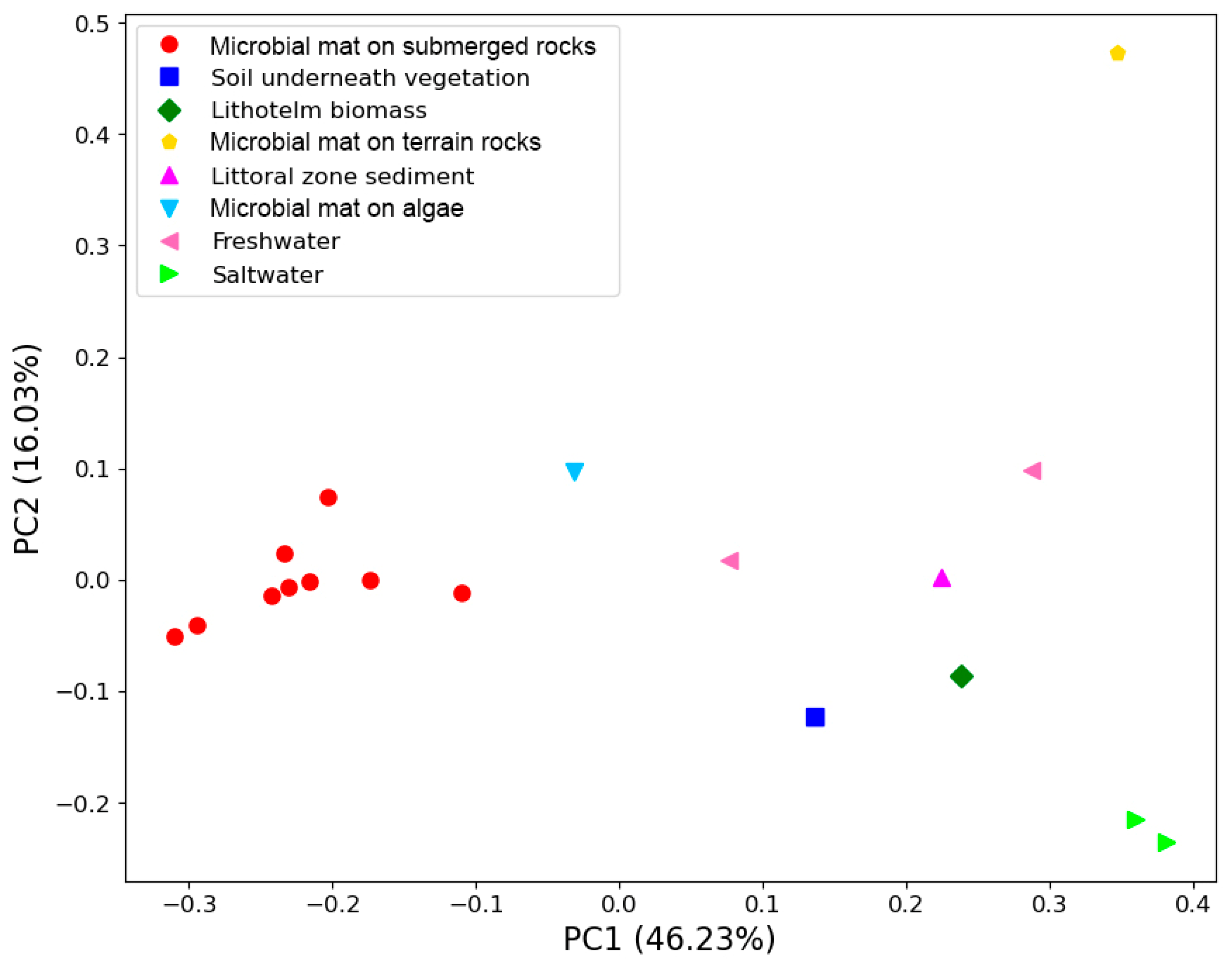
The beta diversity of bacteria is represented using a PCoA plot and weighted UniFrac distances in
Figure 7.
Based on weighted UniFrac distance, all microbial submerged rocks surface mat samples clustered together. The main driver for this clustering was the high abundance of photosynthetic cyanobacteria from the family Oxyphotobacteria. The most abundant ASVs couldn’t be assigned beyond the family Leptolyngbyaceae and ranged from 19 to 78% of reads in these samples. Other genera of cyanobacteria that were common included Tychonema CCAP 1459-11B, Pseudanabaena PCC-7429, Chamaesiphon PCC-7430, Geitlerinema LD9 and Aphanizomenon NIES81. Sample S10 contained one ASV that could not be identified beyond Oxyphotobacteria yet represented 34% of reads. Sample S11 also contained a high abundance of the genus Phormidesmis ANT.LACV5.1.
Sample S12, taken from underneath a patch of vegetation near the nameless lake, boasts the highest alpha diversity of any sample and can’t easily cluster with any other communities based on beta diversity.
4. Discussion
4.1. General Sequencing Statistics Analyses and Observations
Rarefaction curves for bacterial and fungal sequences reach a plateau as sequence depth increases, indicating that samples for bacteria and fungi have attained adequate sampling depth. The observed species richness also strongly correlated with the richness estimators Chao1 and ACE. Overall, the samples exhibited substantial diversity concerning Simpson, Shannon, and phylogenetic diversity indices for bacteria and fungi, with minor exceptions noted in the cases of samples S09 and S21. Utilizing weighted UniFrac distances, beta diversity analysis revealed a robust correlation between bacterial communities and both the sample type and the sampling location, resulting in cohesive clustering based on various taxa. However, no comparable clustering was observed for fungi. The diminished richness and remarkably steep rank abundance gradient observed in sample S17 can be elucidated by the elevated count of chloroplasts, which constituted 94% of the obtained reads. A relatively modest number of bacterial ASVs were retrieved after excluding them from the dataset. Nevertheless, sample S17 reached a plateau on the rarefaction plot, and the observed richness aligned with the richness estimators, confirming its representativeness for the respective community.
Most fungal sequences in all samples could not be assigned to a given phylum. A high number of unidentified fungal species in Antarctic samples is not uncommon and highlights our limited understanding of this domain in both the Arctica and Antarctica [
25]. Only a limited number of sequences accurately characterizing fungi from these regions are currently available within online databases, including UNITE, which was used to train the classifier in this study. A BLAST search was also performed, yielding an equally poor taxonomic identification of ASVs. Still, the unusually high share of unidentified ASVs around the Bulgarian Antarctic base “St. Kliment Ohridski” pointed to a hotspot of novel fungal species that requires further investigation. Investigating such locations can contribute significantly to understanding the fungal diversity in polar regions, offering potential biotechnological applications, including drug discovery. Additionally, the exploration is warranted for the potential presence of human pathogenic strains entombed in permafrost, as previously reported [
26,
27].
A relatively few archaeal reads were retrieved in the present study, with this domain being entirely absent in some samples. A limited number of taxa predominated the majority of archaea in the samples, indicating a low richness in the community. They represent another poorly understood group of organisms in Antarctica, although some have been studied for possible biotechnological application [
28].
4.2. Community Composition of Microbial Surface Mats on Submerged Rocks
The results show a high abundance and richness of cyanobacteria in these samples, all belonging to the family
Oxyphotobacteria, which comprises a large group of oxygenic phototrophs [
29]. That shows that bacterial surface mats on rocks in Antarctic meltwater ponds and meltwater streams near the research base contain a large number of cyanobacterial species that are yet to be identified. Such organisms could hold biotechnological potential to produce pigments, extracellular polysaccharides, antimicrobial compounds and other valuable products [
30,
31,
32].
Phyla other than cyanobacteria expressed very high richness and evenness at the genus level, except for sample S23, where the genus
Thermoflexibacter accounted for 15% of all reads. In Antarctic freshwater bodies, bacteria commonly adopt a distinctive structure within biofilms and microbial mats. Cyanobacteria, particularly in the upper layers, play a dominant role as autotrophic producers for the community. In contrast, a diverse array of other bacteria in the lower layers assimilate the generated products. [
33,
34,
35,
36].
Only about a quarter of fungal reads could be assigned to a phylum.
Betamyces is a genus shown to dominate fungal communities formed on microplastics in a freshwater Arctic lake [
37]. The presence of this genus could be interpreted as an indicator of increased microplastics in the water. Microplastics have also been implicated as a potential vector for the spread of antibiotic-resistant bacteria [
38,
39]. Surprisingly, sample S10 contained a very high abundance of a single genus –
Tetracladium, which accounted for half of all reads and has been shown to have biotechnological potential for producing pectinolytic enzymes [
40]. The phylum
Rozellomycota was also identified, but no further taxonomic annotation was possible, and it was only detected in a significant abundance in sample S09.
Archaeal abundance in the submerged rock surface mat samples was primarily below 0.1% of the reads, except for S07, where they constituted 3% of all reads. In all samples, the genus
Woesearchaeales from phylum
Nanoarchaeota dominated. This genus has been shown to be prevalent in glacial meltwater, and its higher abundance here may be from a similar source [
41].
Woesearchaeales have also been shown to be symbiotic with methanogenic archaea, and some members of the genus have been implicated in methane production [
42,
43].
4.3. Community Composition of a Microbial Surface Mat from an Epilithic Sample within the Research Base
Several
Flavobacterium species have been isolated from polar soils, and multiple ASVs from this study were assigned to this genus [
44]. The predominant presence of
Flavobacterium is the primary factor contributing to the substantial disparity in beta diversity observed in this sample. An excess of algae can alter the composition of the bacterial community towards a distinct profile [
45]. This hypothesis is supported by the elevated prevalence of chloroplast reads in the sample. Epilithic communities can also have a relatively unique set of oligotrophic microorganisms [
46].
Hymenobacter is a genus with
H. roseosalivarius as a type species that was isolated initially from sandstone in the McMurdo Dry Valleys, Antarctica. Still, the genus itself contains species isolated from harsh environments worldwide [
47]. Archaea and identifiable fungi were negligible in this sample.
4.4. Community Composition of the Littoral Zone Sediment from Tarn Todorina Buza
Sediment sample S18 from the shallow littoral zone of Todorina Buza tarn unsurprisingly clustered closest in beta diversity to the water sample from the same tarn. The variation in sediment composition can be attributed to the significantly higher abundance of the genus
Luteolibacter and the notably lower levels of
Pedobacter and
Polymorphobacter, along with, to some extent,
Flavobacterium and
Polaromonas. Overall, the sediment sample boasts a higher alpha diversity than the water column and could serve as a possible reservoir of microorganisms for the water of the tarn.
Flavobacterium is more prevalent in the water column of shallower Antarctic lakes, correlating with its high presence in the sediment, and similar mixing could occur with the other genera [
48]. Members of the genus
Luteolibacter have been isolated from Arctic soils and Antarctic hypersaline brines, which could point to the more significant terrestrial influence at the littoral zone of the tarn [
49,
50]. The only identified fungal genus that represented at least 1% of reads was
Betamyces, while the archaeal fraction of prokaryotes was negligible. It is important to note that fungi don’t constitute a substantial fraction of the biomass in aquatic ecosystems. However, they exhibit high richness, particularly in the littoral zone, which partially accounts for the abundance of novel reads [
51].
4.5. Community Composition of the Soil Sample Underneath a Patch of Vegetation near the Nameless Lake
Most genera accounted for 1-2% of reads each, which reflects the sample's high richness and evenness; only two genera represented more than 10% –
Oryzihumus, and a possible genus from the family
Fibrobacteraceae. This sample exhibited the highest relative abundance of Actinobacteria among all samples in our study. Coupled with the elevated prevalence of Proteobacteria and Bacteroidetes, it mirrors a bacterial composition akin to previous Antarctic rhizoanalyses [
52,
53]. The high diversity could be attributed to the properties of the plant rhizosphere. Studies in more temperate climate zones have shown decreased bacterial diversity in the plant rhizosphere than in surrounding bulk soil due to selective interactions between the plant and the bacterial species [
54]. However, Antarctic plants have also been shown to function as harbors against the unfavorable conditions of cold, oligotrophic deserts by moisture retention, easing temperature stress, preventing desiccation, and providing nutrients to the community [
53].
The rhizosphere sample also contained one of the highest shares of identifiable fungal phyla, accounting for half of all reads. These fungi were shown to belong to
Ascomycota and
Rozellomycota. The
Ascomycota genus
Tetracladium represented most of the phylum, similar to the bacterial surface contamination sample S10 from submerged rocks.
Tetracladium species have been shown to be beneficial root endophytes in terrestrial environments and could thus play an essential role in the rhizosphere of Antarctic plants [
55].
Rozellomycota is a predominantly unfamiliar fungal phylum lacking chitinous cells. They operate as parasites towards molds, algae, crustaceans, and amoebae. Such interactions may confer benefits to the host plant in the rhizosphere. [
56]. The connection between the higher abundance of identifiable fungal phyla and the presence of a plant rhizosphere, which creates a more hospitable microenvironment correlating to conditions closer to those of more temperate regions, has been proposed before [
9]. These conditions could promote the development of more cosmopolitan species, which are more easily identified due to a higher abundance of known sequences available to the classifier.
Archaea in the soil sample exhibit a higher abundance than other sampling sites, constituting 4% of the total reads. Most of them again belonged to the genus
Woesearchaeales within the phylum
Nanoarchaeota. This genus was also prevalent in one of the bacterial surface contamination samples from submerged rocks – S07. Considering that S07 and S12 have very different community profiles, the great variety of environments
Woesearchaeales have been isolated from, and their largely unknown metabolism, it is difficult to assess the source of this particular archaeon or its function in the community [
57].
4.6. Community Composition of a Microbial Mat from the Surface of an Algae
Sample S20, representing the algae surface mat, clusters most closely in composition with the submerged rock surface samples, as anticipated due to their similar nature. This sample also harbors a substantial proportion of Oxyphotobacteria, predominantly represented by an ASV from the family
Leptolyngbyaceae. The primary distinction between both communities lies in the elevated abundance of the genus
Arcicella, constituting 26% of reads in sample S20 while being nearly absent from the rock surface samples. High abundances of this genus in a freshwater lake have been shown to correlate with algal blooms, and their increased presence on the surface of algae is not surprising [
58].
Arcicella species have also been previously found on the rock-water interface of Antarctic freshwater ecosystems [
59], although, in our study, the genus was confined to the surface of algae. Archaea were not detected in this sample.
Once again, only a few fungal reads could be assigned to a phylum level.
Ascomycota accounted for most of them, reaching up to 3% of the total reads. The identified genera from the phylum include
Lecophagus,
Verrucaria and
Tetracladium.
Lecophagus is a genus that includes a variety of predatory fungi, including
L. antarcticus, which infects tardigrades [
60].
4.7. Community Composition of Lithotelm Biomass from Hannah Pt.
Lithotelms are crevices inside rocks that accumulate biomass primarily from the activity of penguins near Hannah Pt. but also contain a lot of trapped seawater. Their unique characteristics contributed to a distinct community profile that does not exhibit specific clustering in weighted UniFrac beta diversity with any other sample. Furthermore, this is one of the most diverse samples in the study, characterized by a substantial number of ASVs that could not be assigned beyond the family level. The most representative genera included
Psychrobacter,
Gottschalkia,
Flavobacterium,
Simplicispira, and
Tissierella, which have all been previously identified within penguin guano using molecular techniques [
61].
Psychrobacter includes halotolerant and psychrotolerant species commonly isolated from Antarctic marine environments [
62,
63]. Recent studies have demonstrated that certain Psychrobacter strains can produce antibiofilm and polyethylene biodegrading enzymes that remain active at low temperatures. Therefore, novel Antarctic species within this genus could harbor significant biotechnological potential [
64,
65]. Members of this genus have also been proposed as probiotic candidates, and they may be part of the active microbiome in penguin intestinal tracks [
66]. Fungi could not be identified to the phylum level, and archaea were absent from the lithotelm sample.
4.8. Saltwater Community Composition
The relatively high abundance of the genus
Cowellia in the sea samples was previously reported in a survey of the sea-surface microlayer (SML) of waters in the same area [
67]. The same study pointed to a high abundance of
Pseudoalteromonas and
Pseudomonas, which could be constrained to the SML. Therefore, we did not see such a high abundance of those genera.
Cowellia is a genus of generalist marine psychrotrophs that have been shown to metabolize a wide range of hydrocarbon compounds and are found in great abundance at oil spills [
68].
Sulfitobacter, an unclassified genus of
Nitrincolaceae,
Polaribacter, SAR11 Clade Ia,
Pseudomonas, and
Psychromonas were some of the other predominant genera in shaping the profile of this community. 98-99% of the fungal reads could not be assigned to a phylum level.
Seawater samples had the greatest share of archaeal reads when using archaea-specific primers. Archaea from South Bay (W06) were dominated almost exclusively by the genera
Nitrosarchaeum and
Candidatus Nitrosopumilus from the phylum
Thaumarchaeota. Still, Marine Group II archaea from Euryarchaeota were also present with about 5% of the reads.
Thaumarchaeota (formerly Marine Group I) have been shown to be more abundant at greater depths than MGII archaea (Lincoln, Wai et al. 2014). Despite this, they represented most of the archaea from the littoral zone sample taken from South Bay, which could be due to mixing from waves at the shoreline. The genus
Nitrosarchaeum does not contain species isolated from marine environments; it only thrives in low-salinity aquatic environments, which could be due to mixing from the freshwater lagoon or the presence of an unknown member of the genus. Both
Nitrosarchaeum and
Candidatus Nitrosopumilus represent mesophilic, neurophilic, aerobic, autotrophic, ammonia-oxidizing archaea [
69,
70]. Members of
Candidatus Nitrosopumilus are among the most abundant archaea in marine environments and play a key role in marine nitrification. MGII were far more prevalent in the Johnson Dock sample (W05), possibly due to being sampled from the pelagic zone. MGII are commonly found in shallow water, potentially playing an important role in carbon cycling [
71].
4.9. Freshwater Community Composition
The bacterial communities of the two freshwater samples did not cluster together based on weighted UniFrac distances. In fact, the freshwater sample W01 from the lagoon clustered separately from all other samples on the third axis of the PCoA plot. Freshwater sample W01 from the lagoon was characterized primarily by the genera
Prevotella,
Faecalibacterium,
Bacteroides,
Bifidobacterium and the family
Lachnospiraceae, which were practically absent from any other sampling location. The
Bacteroides-Prevotella group is a reliable fecal pollution indicator related to the activity of warm-blooded animals [
72]. Furthermore, the genus
Faecalibacterium currently encompasses only a single species,
F. prausnitzii, which, interestingly, is recognized as a predominant bacterium in the human colon [
73]. However, whether the identified ASV in this study represents the existing species within the genus or a novel one remains uncertain. Members of the family
Lachnospiraceae and the genus
Bifidobacterium are some of the most abundant microbes in the intestinal track of humans and animals [
74,
75,
76]. All these genera make up 55% of the total reads, pointing to a significant contamination in the lagoon caused by gut microorganisms. This is most likely a direct result of the known penguin activity in the lagoon. Overall, W01 had the largest relative abundance of Firmicutes in all samples of the present study. The same ASV from the family
Leptolyngbyaceae is once again present in a relatively high abundance. Archaea represented less than 1% of total sequences. 75% of fungal sequences from sample W01 could not be matched to a phylum, and 21% were classified as
Betamyces.
The bacterial composition of freshwater sample W02 from Todorina Buza tarn stood out due to the elevated prevalence of the genera
Pedobacter and
Polymorphobacter, along with a notable presence of
Polaromonas and, to some extent,
Flavobacterium.
Pedobacter and
Polymorphobacter are cosmopolitan genera found in various habitats, and both have been previously identified as part of Antarctic lake communities [
77].
Polaromonas is a genus commonly isolated from polar glaciers and other high-altitude locations [
78]. Notably,
Pedobacter species have demonstrated significant biotechnological potential in producing various compounds, encompassing chitinase, antioxidants, pigments, antimicrobial agents, and more [
79,
80,
81]. 98% of fungal sequences could not be assigned to a phylum, while archaea represented less than 1% of 16S reads.
5. Conclusions
The current study aimed to enhance our understanding of the microorganisms in and around the Bulgarian Polar Base "St. Kliment Ohridski" by employing high-throughput amplicon sequencing to profile community composition and identify microorganisms of potential interest. The results unveiled substantial potential within uncultured Oxyphotobacteria, particularly a member of the family Leptolyngbyaceae, prevalent in nearly all microbial surface mat samples from submerged rocks within the meltwater ponds. The heightened prevalence of unidentified fungi, although a common characteristic in Antarctic metagenomic studies, was particularly pronounced in our results. That underscores the imperative for further research in this domain within polar regions. Interestingly, the genus Tetracladium was one of the identified ASVs and has been shown to have potential for biotechnological applications. Archaea represented a minimal amount of the overall microorganism diversity, with abundance constrained primarily to the sea samples. Overall, our work sheds light on the previously unknown microorganisms near the research base and points to some areas of interest for further studies. We revealed that the lagoon near the base showed clear signs of contamination from various bacteria due to penguin activity, but pathogens were not detected. Finally, the fungal genus Betamyces was identified in the lagoon and adjacent meltwater ponds. Species within this genus have been previously documented as opportunistically growing on microplastics. This observation serves as an indicator of potential pollution with microplastics in the studied environments.
Author Contributions
Conceptualization, V.V.D. and S.G.D.; methodology, V.V.D. and S.G.D.; software, V.V.D.; validation V.V.D., S.P. and S.G.D.; formal analysis, V.V.D. and S.G.D.; investigation, V.V.D. and S.G.D.; resources, S.G.D.; data curation, V.V.D.; writing—original draft preparation, V.V.D.; writing—review and editing, V.V.D., S.P. and S.G.D.; visualization, V.V.D.; supervision, S.G.D.; project administration, S.G.D.; funding acquisition, S.G.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Grant № 70-25-72 from 03.08.2021 г. of the National Center for Polar Studies - Sofia University “St. Kliment Ohridski”.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The raw sequencing reads of this study have been deposited in the Nacional Center for Biotechnology Information of the National Library of Medicine as sequence reads archives under the following accession numbers: PRJNA979344 (for Archaea), PRJNA979782 (for Fungi) and PRJNA979776 (for Bacteria).
Acknowledgments
The authors would like to thank the Bulgarian Polar Base logistics team in Livingston Island for their valuable help and support, which made this research possible. Furthermore, we owe special thanks to Prof. Hristo Pimpirev, chairman of the Bulgarian Antarctic Institute, and Mr. Dragomir Mateev, Science and logistics coordinator of the Bulgarian polar expeditions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Convey:, P. Antarctic ecosystems. 2017. [CrossRef]
- Ji, M.; van Dorst, J.; Bissett, A.; Brown, M.V.; Palmer, A.S.; Snape, I.; Siciliano, S.D.; Ferrari, B.C. Microbial diversity at Mitchell Peninsula, Eastern Antarctica: a potential biodiversity “hotspot”. Polar Biology 2016, 39, 237–249. [Google Scholar] [CrossRef]
- Franzmann, P.D. Examination of Antarctic prokaryotic diversity through molecular comparisons. Biodiversity & Conservation 1996, 5, 1295–1305. [Google Scholar] [CrossRef]
- Bratchkova, A.; Ivanova, V. Bioactive Metabolites Produced by Microorganisms Collected in Antarctica and the Arctic. Biotechnology & Biotechnological Equipment 2011, 25, 1–7. [Google Scholar] [CrossRef]
- Yusof, N.A.; Hashim, N.H.F.; Bharudin, I. Cold Adaptation Strategies and the Potential of Psychrophilic Enzymes from the Antarctic Yeast, Glaciozyma antarctica PI12. Journal of Fungi 2021, 7, 528. [Google Scholar] [CrossRef]
- Núñez-Montero, K.; Barrientos, L. Advances in Antarctic Research for Antimicrobial Discovery: A Comprehensive Narrative Review of Bacteria from Antarctic Environments as Potential Sources of Novel Antibiotic Compounds Against Human Pathogens and Microorganisms of Industrial Importance. Antibiotics 2018, 7, 90. [Google Scholar] [CrossRef] [PubMed]
- Horn, D.J.V.; Okie, J.G.; Buelow, H.N.; Gooseff, M.N.; Barrett, J.E.; Takacs-Vesbach, C.D. Soil Microbial Responses to Increased Moisture and Organic Resources along a Salinity Gradient in a Polar Desert. Applied and Environmental Microbiology 2014, 80, 3034–3043. [Google Scholar] [CrossRef] [PubMed]
- Tiao, G.; Lee, C.K.; McDonald, I.R.; Cowan, D.A.; Cary, S.C. Rapid microbial response to the presence of an ancient relic in the Antarctic Dry Valleys. Nature Communications 2012, 3, 660. [Google Scholar] [CrossRef]
- Yergeau, E.; Newsham, K.K.; Pearce, D.A.; Kowalchuk, G.A. Patterns of bacterial diversity across a range of Antarctic terrestrial habitats. Environmental Microbiology 2007, 9, 2670–2682. [Google Scholar] [CrossRef] [PubMed]
- Cowan, D.A.; Chown, S.L.; Convey, P.; Tuffin, M.; Hughes, K.; Pointing, S.; Vincent, W.F. Non-indigenous microorganisms in the Antarctic: assessing the risks. Trends in Microbiology 2011, 19, 540–548. [Google Scholar] [CrossRef]
- Aves, A.R.; Revell, L.E.; Gaw, S.; Ruffell, H.; Schuddeboom, A.; Wotherspoon, N.E.; LaRue, M.; McDonald, A.J. First evidence of microplastics in Antarctic snow. The Cryosphere 2022, 16, 2127–2145. [Google Scholar] [CrossRef]
- Dimov, S.G.; Strateva, T. Detection of clinically relevant antimicrobial resistance determinants in warm-blooded marine animals in Livingston Island (South Shetland Islands, Antarctica): A field-based molecular genetics study. Marine Pollution Bulletin 2022, 180, 113751. [Google Scholar] [CrossRef]
- Hwengwere, K.; Paramel Nair, H.; Hughes, K.A.; Peck, L.S.; Clark, M.S.; Walker, C.A. Antimicrobial resistance in Antarctica: is it still a pristine environment? Microbiome 2022, 10, 71. [Google Scholar] [CrossRef] [PubMed]
- Hai, R.; Wang, Y.; Wang, X.; Du, Z.; Li, Y. Impacts of Multiwalled Carbon Nanotubes on Nutrient Removal from Wastewater and Bacterial Community Structure in Activated Sludge. PLOS ONE 2014, 9, e107345. [Google Scholar] [CrossRef]
- Coolen, M.J.L.; Hopmans, E.C.; Rijpstra, W.I.C.; Muyzer, G.; Schouten, S.; Volkman, J.K.; Sinninghe Damsté, J.S. Evolution of the methane cycle in Ace Lake (Antarctica) during the Holocene: response of methanogens and methanotrophs to environmental change. Organic Geochemistry 2004, 35, 1151–1167. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications 1990, 18, 315–322. [Google Scholar]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nature Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Research 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Abarenkov, K.Z.; Allan; Piirmann, Timo; Pöhönen, Raivo; Ivanov, Filipp; Nilsson, R. Henrik; Kõljalg, Urmas. UNITE QIIME release for Fungi. UNITE Community: 2022. [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. the Journal of machine Learning research 2011, 12, 2825–2830. [Google Scholar]
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Computing in Science & Engineering 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Waskom, M.L. Seaborn: statistical data visualization. Journal of Open Source Software 2021, 6, 3021. [Google Scholar] [CrossRef]
- Bridge, P.D.; Spooner, B.M. Non-lichenized Antarctic fungi: transient visitors or members of a cryptic ecosystem? Fungal Ecology 2012, 5, 381–394. [Google Scholar] [CrossRef]
- da Silva, T.H.; Câmara, P.E.A.S.; Pinto, O.H.B.; Carvalho-Silva, M.; Oliveira, F.S.; Convey, P.; Rosa, C.A.; Rosa, L.H. Diversity of Fungi Present in Permafrost in the South Shetland Islands, Maritime Antarctic. Microbial Ecology 2022, 83, 58–67. [Google Scholar] [CrossRef]
- Rosa, L.H.; Zani, C.L.; Cantrell, C.L.; Duke, S.O.; Van Dijck, P.; Desideri, A.; Rosa, C.A. Fungi in Antarctica: Diversity, Ecology, Effects of Climate Change, and Bioprospection for Bioactive Compounds. In Fungi of Antarctica: Diversity, Ecology and Biotechnological Applications, Rosa, L.H., Ed. Springer International Publishing: Cham, 2019; pp. 1-17. [CrossRef]
- Cabrera, M.; Blamey, J.M. Biotechnological applications of archaeal enzymes from extreme environments. Biological research 2018, 51. [Google Scholar] [CrossRef] [PubMed]
- Shih, P.M.; Hemp, J.; Ward, L.M.; Matzke, N.J.; Fischer, W.W. Crown group Oxyphotobacteria postdate the rise of oxygen. Geobiology 2017, 15, 19–29. [Google Scholar] [CrossRef]
- Prieto-Barajas, C.M.; Valencia-Cantero, E.; Santoyo, G. Microbial mat ecosystems: Structure types, functional diversity, and biotechnological application. Electronic Journal of Biotechnology 2018, 31, 48–56. [Google Scholar] [CrossRef]
- Silva, T.R.e.; Silva, L.C.F.; de Queiroz, A.C.; Alexandre Moreira, M.S.; de Carvalho Fraga, C.A.; de Menezes, G.C.A.; Rosa, L.H.; Bicas, J.; de Oliveira, V.M.; Duarte, A.W.F. Pigments from Antarctic bacteria and their biotechnological applications. Critical Reviews in Biotechnology 2021, 41, 809–826. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, T.; Hamid, B.; Baba, Z.; Iqbal, S.; Yatoo, A.; Fatima, S.; Nabi, A.; Kanth, R.; Dar, K.; Hussain, N.; et al. Extracellular polymeric substances in psychrophilic cyanobacteria: A potential bioflocculant and carbon sink to mitigate cold stress. Biocatalysis and Agricultural Biotechnology 2022, 42, 102375. [Google Scholar] [CrossRef]
- Davey, M.C.; Clarke, K.J. FINE STRUCTURE OF A TERRESTRIAL CYANOBACTERIAL MAT FROM ANTARCTICA1. Journal of Phycology 1992, 28, 199–202. [Google Scholar] [CrossRef]
- de Los Ríos, A.; Ascaso, C.; Wierzchos, J.; Fernández-Valiente, E.; Quesada, A. Microstructural characterization of cyanobacterial mats from the McMurdo Ice Shelf, Antarctica. Appl Environ Microbiol 2004, 70, 569–580. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Valiente, E.; Camacho, A.; Rochera, C.; Rico, E.; Vincent, W.F.; Quesada, A. Community structure and physiological characterization of microbial mats in Byers Peninsula, Livingston Island (South Shetland Islands, Antarctica). FEMS Microbiology Ecology 2007, 59, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.J.; Schmit, A.; Foster, R.; Littman, S.; Kuypers, M.M.M.; Foreman, C.M. Biofilms on glacial surfaces: hotspots for biological activity. npj Biofilms and Microbiomes 2016, 2, 16008. [Google Scholar] [CrossRef] [PubMed]
- González-Pleiter, M.; Velázquez, D.; Casero, M.C.; Tytgat, B.; Verleyen, E.; Leganés, F.; Rosal, R.; Quesada, A.; Fernández-Piñas, F. Microbial colonizers of microplastics in an Arctic freshwater lake. Science of The Total Environment 2021, 795, 148640. [Google Scholar] [CrossRef] [PubMed]
- Laganà, P.; Caruso, G.; Corsi, I.; Bergami, E.; Venuti, V.; Majolino, D.; La Ferla, R.; Azzaro, M.; Cappello, S. Do plastics serve as a possible vector for the spread of antibiotic resistance? First insights from bacteria associated to a polystyrene piece from King George Island (Antarctica). International Journal of Hygiene and Environmental Health 2019, 222, 89–100. [Google Scholar] [CrossRef]
- Dong, H.; Chen, Y.; Wang, J.; Zhang, Y.; Zhang, P.; Li, X.; Zou, J.; Zhou, A. Interactions of microplastics and antibiotic resistance genes and their effects on the aquaculture environments. Journal of Hazardous Materials 2021, 403, 123961. [Google Scholar] [CrossRef]
- Carrasco, M.; Rozas, J.M.; Alcaíno, J.; Cifuentes, V.; Baeza, M. Pectinase secreted by psychrotolerant fungi: identification, molecular characterization and heterologous expression of a cold-active polygalacturonase from Tetracladium sp. Microbial Cell Factories 2019, 18, 45. [Google Scholar] [CrossRef] [PubMed]
- Purkamo, L.; Ó Dochartaigh, B.; MacDonald, A.; Cousins, C. Following the flow—Microbial ecology in surface- and groundwaters in the glacial forefield of a rapidly retreating glacier in Iceland. Environmental Microbiology 2022, 24, 5840–5858. [Google Scholar] [CrossRef]
- Kuroda, K.; Narihiro, T.; Shinshima, F.; Yoshida, M.; Yamaguchi, H.; Kurashita, H.; Nakahara, N.; Nobu, M.K.; Noguchi, T.Q.P.; Yamauchi, M.; et al. High-rate cotreatment of purified terephthalate and dimethyl terephthalate manufacturing wastewater by a mesophilic upflow anaerobic sludge blanket reactor and the microbial ecology relevant to aromatic compound degradation. Water Research 2022, 219, 118581. [Google Scholar] [CrossRef]
- Ndayisenga, F.; Yu, Z.; Wang, B.; Wu, G.; Zhang, H.; Phulpoto, I.A.; Zhao, J.; Yang, J. Thermophilic-operating environment promotes hydrogen-producing microbial growth in a lignocellulose-fed DF-MEC system for enhanced biohydrogen evolution. Process Safety and Environmental Protection 2022, 167, 213–224. [Google Scholar] [CrossRef]
- Bernardet, J.-F.; Bowman, J.P. Flavobacterium. In Bergey's Manual of Systematics of Archaea and Bacteria. 2015; 1–75. [CrossRef]
- Eigemann, F.; Hilt, S.; Salka, I.; Grossart, H.-P. Bacterial community composition associated with freshwater algae: species specificity vs. dependency on environmental conditions and source community. FEMS Microbiology Ecology 2013, 83, 650–663. [Google Scholar] [CrossRef] [PubMed]
- Amarelle, V.; Carrasco, V.; Fabiano, E. The Hidden Life of Antarctic Rocks. In The Ecological Role of Micro-organisms in the Antarctic Environment, Castro-Sowinski, S., Ed. Springer International Publishing: Cham, 2019; pp. 221-237. [CrossRef]
- Buczolits, S.; Busse, H.-J. Hymenobacter. In Bergey's Manual of Systematics of Archaea and Bacteria, 2015; pp. 1-11. [CrossRef]
- Picazo, A.; Rochera, C.; Villaescusa, J.A.; Miralles-Lorenzo, J.; Velázquez, D.; Quesada, A.; Camacho, A. Bacterioplankton Community Composition Along Environmental Gradients in Lakes From Byers Peninsula (Maritime Antarctica) as Determined by Next-Generation Sequencing. Frontiers in Microbiology 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Pak, S.; Rim, S.; Ren, L.; Jiang, F.; Chang, X.; Liu, P.; Zhang, Y.; Fang, C.; Zheng, C.; et al. Luteolibacter arcticus sp. nov., isolated from high Arctic tundra soil, and emended description of the genus Luteolibacter. International Journal of Systematic and Evolutionary Microbiology 2015, 65, 1922–1928. [Google Scholar] [CrossRef]
- Guglielmin, M.; Azzaro, M.; Buzzini, P.; Battistel, D.; Roman, M.; Ponti, S.; Turchetti, B.; Sannino, C.; Borruso, L.; Papale, M.; et al. A possible unique ecosystem in the endoglacial hypersaline brines in Antarctica. Scientific Reports 2023, 13, 177. [Google Scholar] [CrossRef] [PubMed]
- Grossart, H.-P.; Van den Wyngaert, S.; Kagami, M.; Wurzbacher, C.; Cunliffe, M.; Rojas-Jimenez, K. Fungi in aquatic ecosystems. Nature Reviews Microbiology 2019, 17, 339–354. [Google Scholar] [CrossRef]
- Teixeira, L.C.R.S.; Peixoto, R.S.; Cury, J.C.; Sul, W.J.; Pellizari, V.H.; Tiedje, J.; Rosado, A.S. Bacterial diversity in rhizosphere soil from Antarctic vascular plants of Admiralty Bay, maritime Antarctica. The ISME Journal 2010, 4, 989–1001. [Google Scholar] [CrossRef] [PubMed]
- Prekrasna, I.; Pavlovska, M.; Miryuta, N.; Dzhulai, A.; Dykyi, E.; Convey, P.; Kozeretska, I.; Bedernichek, T.; Parnikoza, I. Antarctic Hairgrass Rhizosphere Microbiomes: Microscale Effects Shape Diversity, Structure, and Function. Microbes and Environments 2022, 37. [Google Scholar] [CrossRef] [PubMed]
- Ling, N.; Wang, T.; Kuzyakov, Y. Rhizosphere bacteriome structure and functions. Nature Communications 2022, 13, 836. [Google Scholar] [CrossRef]
- Lazar, A.; Mushinski, R.M.; Bending, G.D. Landscape scale ecology of Tetracladium spp. fungal root endophytes. Environmental Microbiome 2022, 17, 40. [Google Scholar] [CrossRef]
- Corsaro, D.; Walochnik, J.; Venditti, D.; Hauröder, B.; Michel, R. Solving an old enigma: Morellospora saccamoebae gen. nov., sp. nov. (Rozellomycota), a Sphaerita-like parasite of free-living amoebae. Parasitology Research 2020, 119, 925–934. [Google Scholar] [CrossRef]
- Liu, X.; Li, M.; Castelle, C.J.; Probst, A.J.; Zhou, Z.; Pan, J.; Liu, Y.; Banfield, J.F.; Gu, J.-D. Insights into the ecology, evolution, and metabolism of the widespread Woesearchaeotal lineages. Microbiome 2018, 6, 102. [Google Scholar] [CrossRef] [PubMed]
- Wurzbacher, C.; Attermeyer, K.; Kettner, M.T.; Flintrop, C.; Warthmann, N.; Hilt, S.; Grossart, H.-P.; Monaghan, M.T. DNA metabarcoding of unfractionated water samples relates phyto-, zoo- and bacterioplankton dynamics and reveals a single-taxon bacterial bloom. Environmental Microbiology Reports 2017, 9, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.P.; Swain, A.K.; Thacker, R.W.; Ravindra, R.; Andersen, D.T.; Bej, A.K. Bacterial diversity of the rock-water interface in an East Antarctic freshwater ecosystem, Lake Tawani(P)†. Aquatic Biosystems 2013, 9, 4. [Google Scholar] [CrossRef] [PubMed]
- Loeffelholz, J.; Stahl, L.; Momeni, S.; Turberville, C.; Pienaar, J. Trichoderma infection of limno-terrestrial tardigrades. Journal of Invertebrate Pathology 2021, 186, 107677. [Google Scholar] [CrossRef] [PubMed]
- Grzesiak, J.; Kaczyńska, A.; Gawor, J.; Żuchniewicz, K.; Aleksandrzak-Piekarczyk, T.; Gromadka, R.; Zdanowski, M.K. A smelly business: Microbiology of Adélie penguin guano (Point Thomas rookery, Antarctica). Science of The Total Environment 2020, 714, 136714. [Google Scholar] [CrossRef] [PubMed]
- Kirkinci, S.F.; Edbeib, M.F.; Aksoy, H.M.; Marakli, S.; Kaya, Y. Identification of Dalapon degrading bacterial strain, Psychrobacter sp. TaeBurcu001 isolated from Antarctica. Polar Science 2021, 28, 100656. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Esakkiraj, P.; Bharathi, C.; Ayyanna, R.; Jha, N.; Panigrahi, A.; Karthe, P.; Arul, V. Functional and molecular characterization of a cold-active lipase from Psychrobacter celer PU3 with potential antibiofilm property. International Journal of Biological Macromolecules 2022, 211, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Hou, Y.; Wang, Q.; Wang, Y. Characteristics and polyethylene biodegradation function of a novel cold-adapted bacterial laccase from Antarctic sea ice psychrophile Psychrobacter sp. NJ228. Journal of Hazardous Materials 2022, 439, 129656. [Google Scholar] [CrossRef]
- Wuertz, S.; Beça, F.; Kreuz, E.; Wanka, K.M.; Azeredo, R.; Machado, M.; Costas, B. Two Probiotic Candidates of the Genus Psychrobacter Modulate the Immune Response and Disease Resistance after Experimental Infection in Turbot (Scophthalmus maximus, Linnaeus 1758). Fishes 2023, 8, 144. [Google Scholar] [CrossRef]
- Martinez-Varela, A.; Casas, G.; Piña, B.; Dachs, J.; Vila-Costa, M. Large Enrichment of Anthropogenic Organic Matter Degrading Bacteria in the Sea-Surface Microlayer at Coastal Livingston Island (Antarctica). Frontiers in Microbiology 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Redmond, M.C.; Valentine, D.L. Natural gas and temperature structured a microbial community response to the <i>Deepwater Horizon</i> oil spill. Proceedings of the National Academy of Sciences 2012, 109, 20292–20297. [Google Scholar] [CrossRef]
- Jung, M.-Y.; Islam, M.A.; Gwak, J.-H.; Kim, J.-G.; Rhee, S.-K. Nitrosarchaeum koreense gen. nov., sp. nov., an aerobic and mesophilic, ammonia-oxidizing archaeon member of the phylum Thaumarchaeota isolated from agricultural soil. International Journal of Systematic and Evolutionary Microbiology 2018, 68, 3084–3095. [Google Scholar] [CrossRef]
- Qin, W.; Martens-Habbena, W.; Kobelt, J.N.; Stahl, D.A. Candidatus nitrosopumilus. Bergey’s manual of systematics of archaea and bacteria, 2016; 8818–8827. [Google Scholar]
- Zhang, C.L.; Xie, W.; Martin-Cuadrado, A.-B.; Rodriguez-Valera, F. Marine Group II Archaea, potentially important players in the global ocean carbon cycle. Frontiers in Microbiology 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Okabe, S.; Okayama, N.; Savichtcheva, O.; Ito, T. Quantification of host-specific Bacteroides–Prevotella 16S rRNA genetic markers for assessment of fecal pollution in freshwater. Applied Microbiology and Biotechnology 2007, 74, 890–901. [Google Scholar] [CrossRef] [PubMed]
- Miquel, S.; Martín, R.; Rossi, O.; Bermúdez-Humarán, L.G.; Chatel, J.M.; Sokol, H.; Thomas, M.; Wells, J.M.; Langella, P. Faecalibacterium prausnitzii and human intestinal health. Current Opinion in Microbiology 2013, 16, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Lagier, J.-C.; Million, M.; Hugon, P.; Armougom, F.; Raoult, D. Human Gut Microbiota: Repertoire and Variations. Frontiers in Cellular and Infection Microbiology 2012, 2. [Google Scholar] [CrossRef] [PubMed]
- Bunesova, V.; Vlkova, E.; Rada, V.; Killer, J.; Musilova, S. Bifidobacteria from the gastrointestinal tract of animals: differences and similarities. Beneficial microbes 2014, 5, 377–388. [Google Scholar] [CrossRef]
- Seshadri, R.; Leahy, S.C.; Attwood, G.T.; Teh, K.H.; Lambie, S.C.; Cookson, A.L.; Eloe-Fadrosh, E.A.; Pavlopoulos, G.A.; Hadjithomas, M.; Varghese, N.J.; et al. Cultivation and sequencing of rumen microbiome members from the Hungate1000 Collection. Nature Biotechnology 2018, 36, 359–367. [Google Scholar] [CrossRef]
- Margesin, R.; Shivaji, S. Pedobacter. In Bergey's Manual of Systematics of Archaea and Bacteria. 2015; 1–17. [CrossRef]
- Darcy, J.L.; Lynch, R.C.; King, A.J.; Robeson, M.S.; Schmidt, S.K. Global Distribution of Polaromonas Phylotypes - Evidence for a Highly Successful Dispersal Capacity. PLOS ONE 2011, 6, e23742. [Google Scholar] [CrossRef]
- Wong, C.M.V.L.; Tam, H.K.; Alias, S.A.; González, M.; González-Rocha, G.; Domínguez-Yévenes, M. Pseudomonas and Pedobacter isolates from King George Island inhibited the growth of foodborne pathogens. Polish Polar Research 2011, 3–14. [Google Scholar] [CrossRef]
- Correa-Llantén, D.N.; Amenábar, M.J.; Blamey, J.M. Antioxidant capacity of novel pigments from an Antarctic bacterium. The Journal of Microbiology 2012, 50, 374. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.-S.; Seo, D.-J.; Jung, W.-J. Identification, purification, and expression patterns of chitinase from psychrotolerant Pedobacter sp. PR-M6 and antifungal activity in vitro. Microbial Pathogenesis 2017, 107, 62–68. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).