Submitted:
13 January 2024
Posted:
15 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
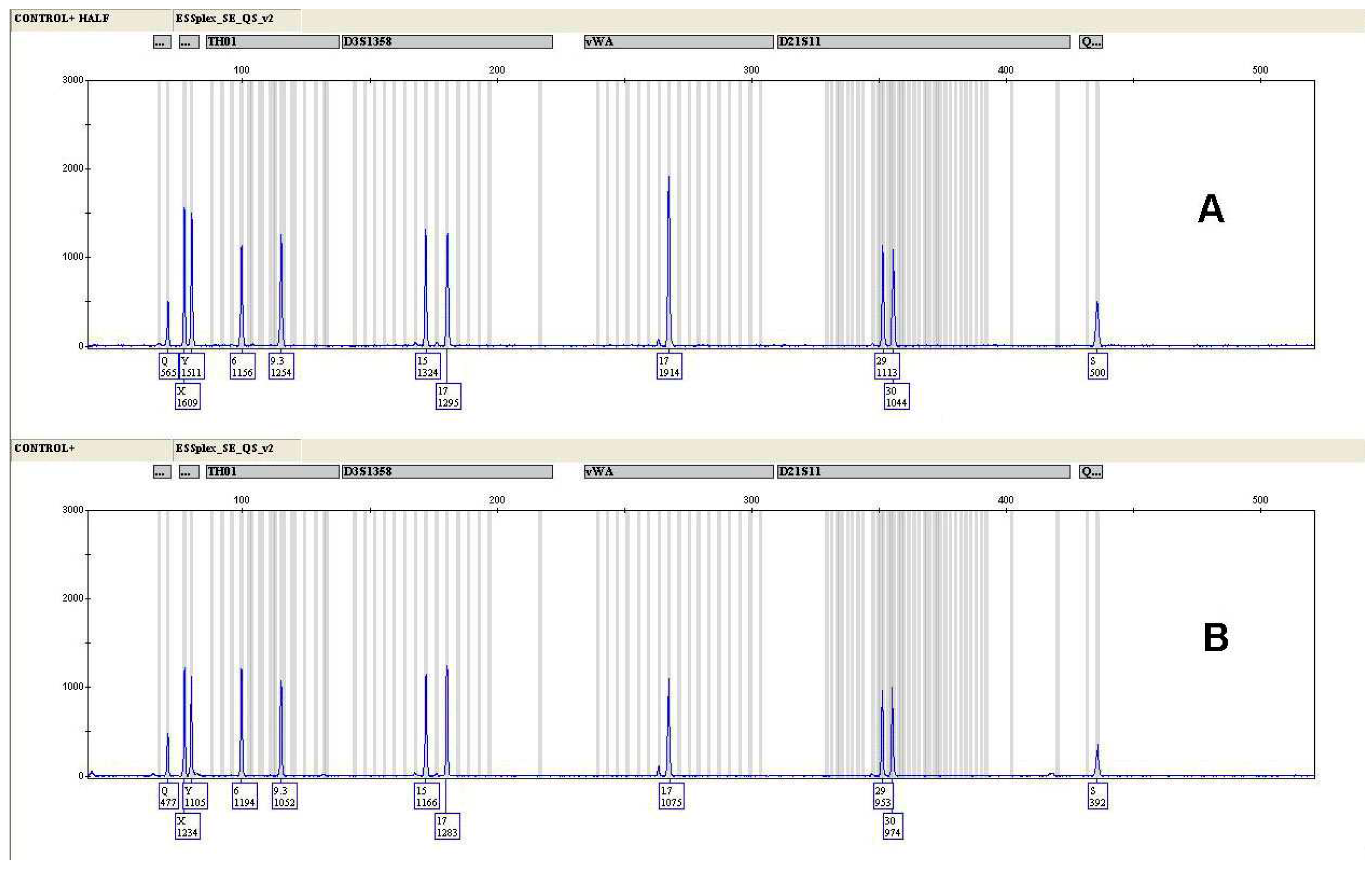
2.1. Male Control DNA 9948 analysis
2.2. Samples analysis
2.2.1. Control samples
2.2.2. Forensic samples
2.2.2.1. Forensic samples pre-processing
2.2.2. Informed consent and ethical aspects
2.3. DNA extraction
2.4. DNA quantification
2.5. DNA amplification and typing
3. Results and Discussion
3.1. Male Control DNA 9948 analysis
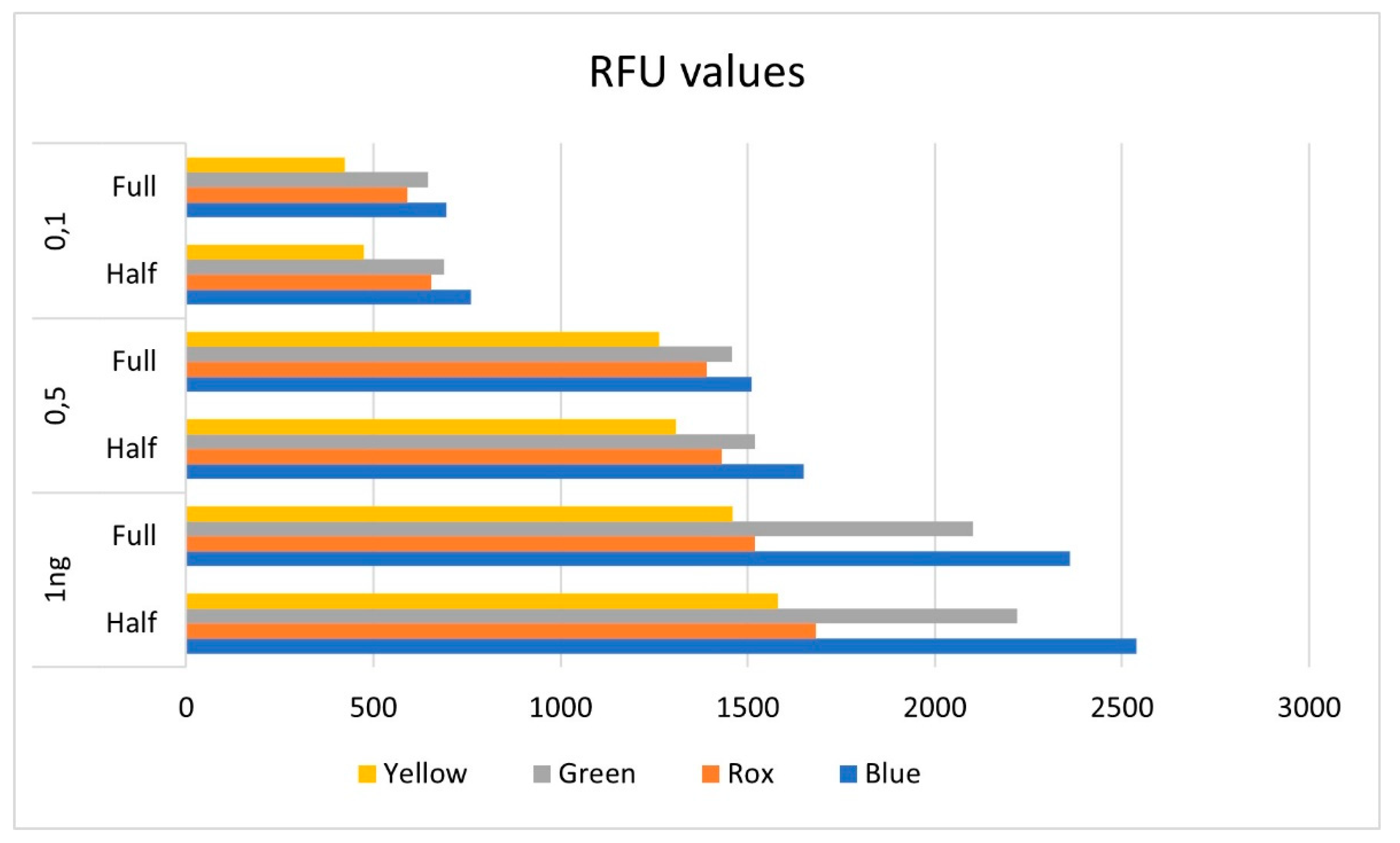
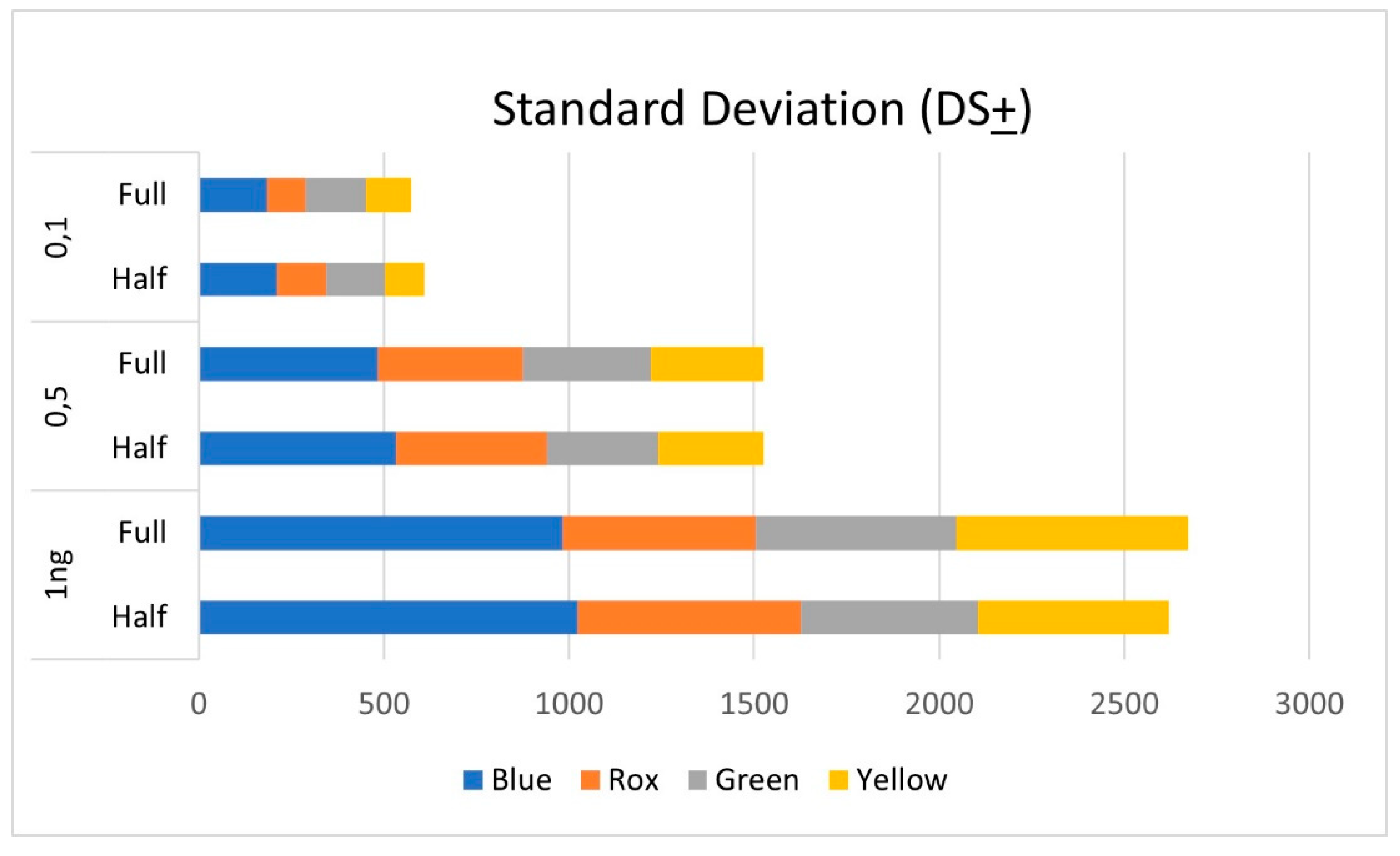
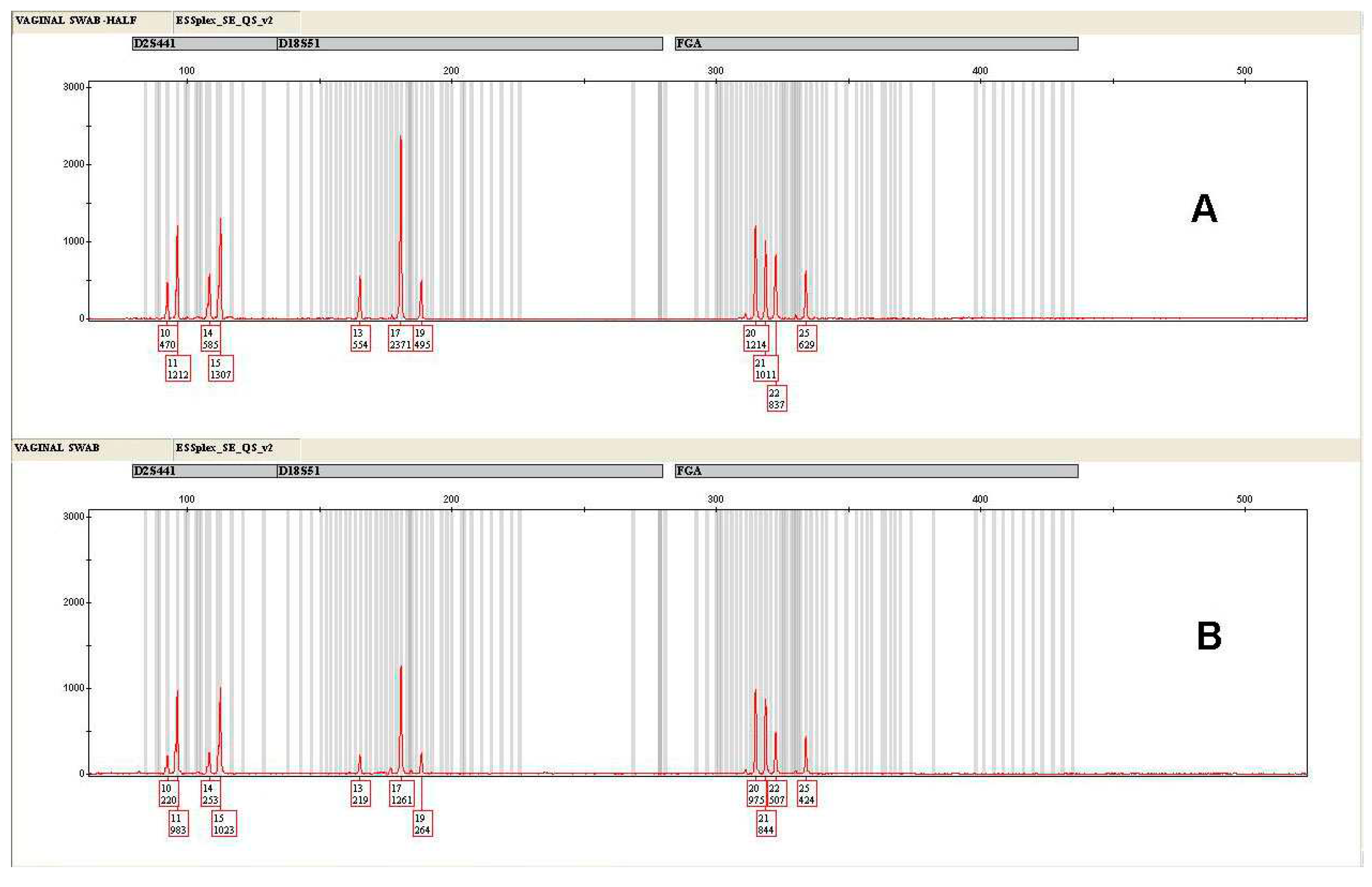
3.2. Control samples
3.3. Forensic caseworks samples
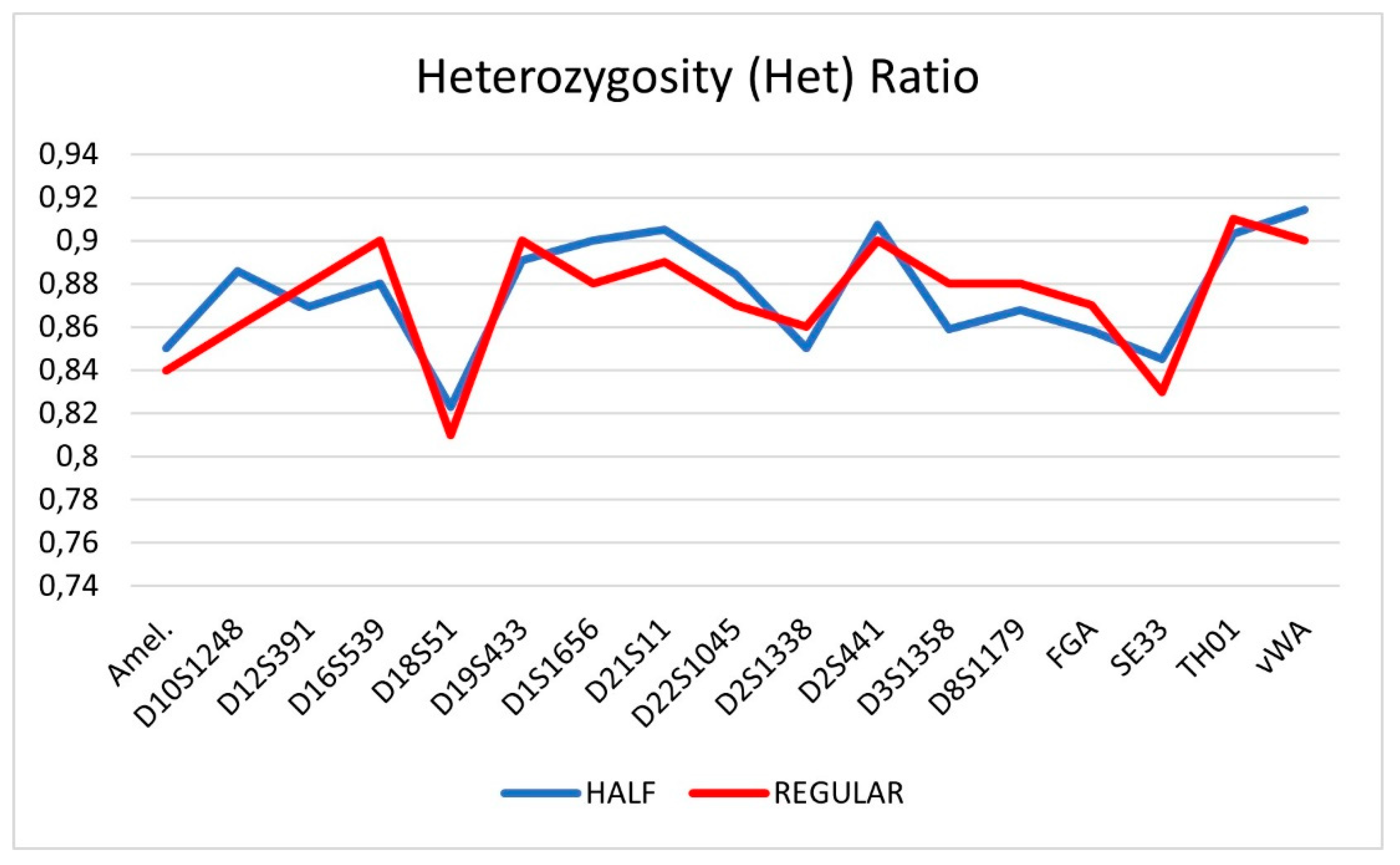
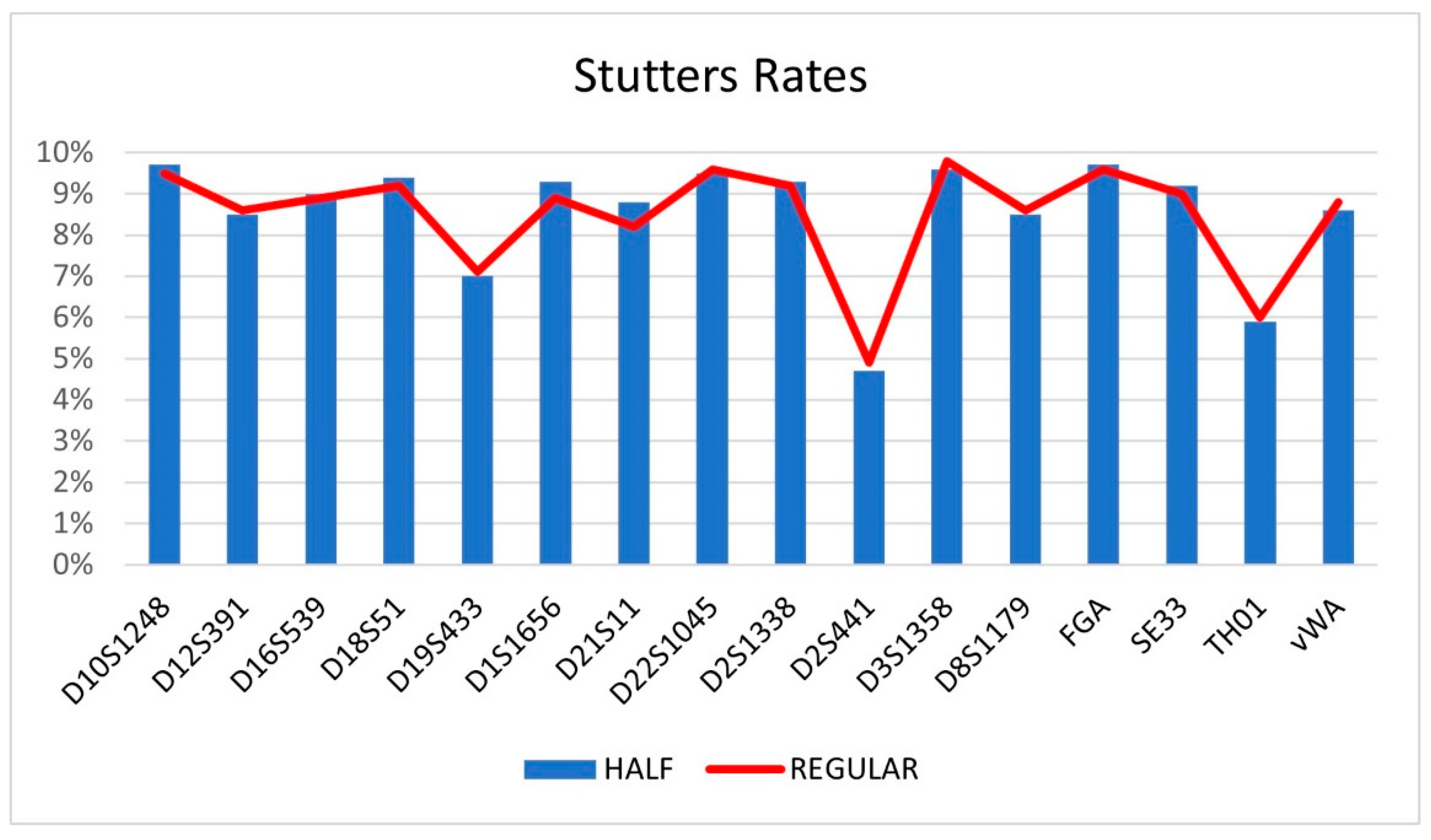
4. Conclusion
References
- Gill, P.; Kimpton, C.P.; Urquhart, A.; Oldroyd, N.; Millican, E.S.; Watson, S.K.; Downes, T.J. Automated short tandem repeats (STR) analysis in forensic casework - a strategy for the future. Electrophoresis 1995, 16, 1543–1552. [Google Scholar] [CrossRef]
- Kimpton, C.; Fisher, D.; Watson, S.; Adams, M.; Urquhart, A.; Lygo, J.; Gill, P. Evaluation of an automated DNA profiling system employing multiplex amplification of four tetrameric STR loci. Int. J. Legal Med. 1994, 106, 302–311. [Google Scholar] [CrossRef]
- Butler, J.M.; Hill, C.R. Biology and genetics of new autosomal STR loci useful for forensic DNA analysis. Forensic Sci Rev 2012, 24, 15. [Google Scholar]
- Hares, D.R. Expanding the CODIS Core Loci in the United States,, Forensic Sci. Int. Genet. 2012, 6, e52–e54. [Google Scholar]
- Schumm, J.W.; Gutierrez-Mateo, C.; Tan, E.; Selden, R. A 27-locus STR assay to meet all United States and European law enforcement agency standards. J Forensic Sci. 2013, 58, 1584–1592. [Google Scholar] [CrossRef]
- Barbaro, A. 2020 STR Typing and Available Multiplex Kits Including Validation Methods. In: Shrivastava P, Dash HR, Lorente JA, Imam, J. , Forensic DNA Typing: Principles, Applications and Advancements. Springer, Singapore.
- Gaines, M.L.; Wojtkiewicz, P.W.; Valentine, J.A.; Brown, C.L. Reduced Volume PCR Amplification Reactions Using the AmpFlSTR Profiler Plus Kit. Journal of Forensic Sciences 2002, 47, 1224–1237. [Google Scholar] [CrossRef] [PubMed]
- Bessekri, M.W.; Aggoune, A.; Lazreg, S.; Bucht, R.; Fuller, V. Comparative study on the effects of reduced PCR reaction volumes and increased cycle number, on the sensitivity and the stochastic threshold of the AmpFlSTR Identifiler® Plus kit. Forensic Science International: Genetics Supplement Series 2013, 4, e306–e307. [Google Scholar] [CrossRef]
- Almheiri, R. Alghafri. Reduced volume for direct PCR amplification of blood reference samples using Identifiler® Direct and Globalfiler™ Express assays. Forensic Science International: Genetics Supplement Series, 2017, 6, e340–e341. [Google Scholar]
- Mahmood, H.K.; Salman, F.N.; Hasan, D.H.; Salih, K.M.; Sadiq, M.A.; Mohammad, B.T.; Mohammed, M.K.; Nahi, S.M.; Baqir, S.S. Validation of Half-Reaction Volumes of the Promega PowerPlex® Forensic Amplification Kits (PowerPlex® 18D Systems, PowerPlex ® 21System, PowerPlex® Fusion System and PowerPlex® Y23 System) in STR Analysis”. Arab Journal of Forensic Sciences & Forensic Medicine 2020, 2, 152–158. [Google Scholar]
- Perry, J.; Munshi, T.; Haizel, T.; Iyavoo, S. Validation of reduced volume VeriFiler™ Express PCR Amplification Kit for buccal swab samples extracted using Prep-n-Go™ Buffer. J Forensic Sci., 2022, 67, 1971–1978. [Google Scholar] [CrossRef] [PubMed]
- Barbaro, A.; Cormaci, P. Validation of PowerPlex® Y23 System (Promega) using reduced reaction volume. Forensic Science International: Genetics Supplement Series 2015, 5, e373–e374. [Google Scholar] [CrossRef]
- Barbaro, A.; Falcone, G. Validation of SureID® 21G Human STR Identification Kit Using Alternative Reaction Protocols. Biometrics And Biostatistics Journal. 2021, 3, 114. [Google Scholar]
- Coble, M.D.; Butler, J.M. Characterization of new miniSTR loci to aid analysis of degraded DNA, J. Forensic Sci. 2009, 50, 43–53. [Google Scholar] [CrossRef]
- Mogensen, H.S.; Frank-Hansen, R.; Morling, N. Test of Investigator ESSPLEX SE QS with quality sensors, For. Sci. Int. Genet. Suppl. Series 2015, 5, e490–e491. [Google Scholar] [CrossRef]
- EZ1 DNA Investigator Handbook – QIAGEN – Rev. 7,2014 Available at: https://www.qiagen.com/us/resources/download.aspx?id=46064856-1b88-4b27-a825d3f616e06c08&lang=en.
- Barbaro, A.; Samar, S.; Falcone, G.; La Marca, A. Highly efficient and automated extraction of DNA from human remains using a modified EZ1 protocol. Forensic Sci Res. 2021, 18, 59–66. [Google Scholar] [CrossRef]
- Investigator Quantiplex Pro RGQ Kit Handbook – QIAGEN – Rev. 1, 2018 Available at: https://www.qiagen.com/us/resources/download.aspx?id=57497d59-7a43-4eaf-8c94086e88742e86&lang=en.
- Vraneš, M.; Scherer, M.; Elliott, K. Development and validation of the Investigator® Quantiplex Pro Kit for qPCR-based examination of the quantity and quality of human DNA in forensic samples. FSI: Genetics Supplement Series, 2017, 6, e518–e519. [Google Scholar] [CrossRef]
- Investigator® ESSplex SE QS Handbook, June 2021 – QIAGEN – Rev.2, 2021. Available at https://www.qiagen.com/us/resources/resourcedetail?id=1fce64fd-9cf1-4750-a4d0-e7aa64113d7b&lang=en. 20 June.
- Butler JM (2012) Advanced Topics in Forensic DNA Typing: Methodology. Elsevier Academic Press, San Diego (USA).
- Developmental validation of the Investigator® ESSplex SE QS Kit, QIAGEN- October 2015. Available at https://www.qiagen.com/th/resources/download.aspx?id=9761730e-def1-4011-bd8a-852b4849dd6d&lang=en. 20 October.
- Coyle, H. Quality Control and Duplication for Concordance in Forensic DNA Samples: Implications for Interpretation in Mixtures. IRJCS:International Research Journal of Computer Science 2015, II, 16–18. [Google Scholar]
- Verdon, T.J.; Mitchell, R.J.; van Oorschot, R.A. The influence of substrate on DNA transfer and extraction efficiency. Forensic Sci Int Genet. 2013, 7, 167–75. [Google Scholar] [CrossRef]
- Lee S.B et al. (2017) DNA Extraction Methods In Forensic Analysis, Encyclopedia of Analytical Chemistry, Wiley & Sons (USA).
- Martins, C.; Lima, G.; Carvalho, M.R.; Cainé, L.; Porto, M.J. DNA quantification by real-time PCR in different forensic samples. Forensic Science International: Genetics Supplement Series 2015, 5, e545–e546. [Google Scholar] [CrossRef]
- Mulero, J.J.; Hennessy, L.K. Next-Generation STR Genotyping Kits for Forensic Applications. Forensic Sci Rev. 2012, 24, 1–13. [Google Scholar] [PubMed]
- Davis, C.P.; King, J.L.; Budowle, B.; Eisenberg, A.J.; Turnbough, M.A. Extraction platform evaluations: A comparison of Automate Express™, EZ1® Advanced XL, and Maxwell® 16 Bench-top DNA extraction systems. Legal Medicine 2012, 14, 36–39. [Google Scholar] [CrossRef] [PubMed]
- Francez, P.A.C.; Penidoa, C.A.F.O.; Costa, G.S.M.B.; de Almeida, R.M.; Pena, E.E.S.; Funabashi, K.; Resque, L.R. Comparison between automated DNA extraction employing the EZ1platform and manual methods using real forensic samples, Rev. Bras. Crimin. 2021, 10, 44–56. [Google Scholar] [CrossRef]

| Locus | HALF | DS (+) | REGULAR | DS (+) |
|---|---|---|---|---|
| Amel. | 0.85 | 0.10 | 0.84 | 0.11 |
| D10S1248 | 0.89 | 0.07 | 0.86 | 0.07 |
| D12S391 | 0.87 | 0.09 | 0.88 | 0.08 |
| D16S539 | 0.88 | 0.10 | 0.9 | 0.11 |
| D18S51 | 0.82 | 0.12 | 0.81 | 0.11 |
| D19S433 | 0.89 | 0.07 | 0.9 | 0.07 |
| D1S1656 | 0.90 | 0.07 | 0.88 | 0.06 |
| D21S11 | 0.91 | 0.11 | 0.89 | 0.12 |
| D22S1045 | 0.88 | 0.08 | 0.87 | 0.07 |
| D2S1338 | 0.85 | 0.12 | 0.86 | 0.11 |
| D2S441 | 0.91 | 0.08 | 0.9 | 0.09 |
| D3S1358 | 0.86 | 0.09 | 0.88 | 0.10 |
| D8S1179 | 0.87 | 0.09 | 0.88 | 0.09 |
| FGA | 0.86 | 0.11 | 0.87 | 0.10 |
| SE33 | 0.85 | 0.09 | 0.83 | 0.08 |
| TH01 | 0.90 | 0.05 | 0.91 | 0.06 |
| vWA | 0.91 | 0.09 | 0.9 | 0.10 |
| Locus | HALF | DS (+) | REGULAR | DS (+) |
|---|---|---|---|---|
| D10S1248 | 9.7 | 0.01 | 9.5 | 0.01 |
| D12S391 | 8.5 | 0.02 | 8.6 | 0.02 |
| D16S539 | 9 | 0.02 | 8.9 | 0.03 |
| D18S51 | 9.4 | 0.03 | 9.2 | 0.02 |
| D19S433 | 7 | 0.01 | 7.1 | 0.01 |
| D1S1656 | 9.3 | 0.02 | 8.9 | 0.02 |
| D21S11 | 8.8 | 0.01 | 8.2 | 0.01 |
| D22S1045 | 9.5 | 0.01 | 9.6 | 0.02 |
| D2S1338 | 9.3 | 0.02 | 9.2 | 0.02 |
| D2S441 | 4.7 | 0.01 | 4.9 | 0.01 |
| D3S1358 | 9.6 | 0.02 | 9.8 | 0.01 |
| D8S1179 | 8.5 | 0.02 | 8.6 | 0.02 |
| FGA | 9.7 | 0.01 | 9.6 | 0.01 |
| SE33 | 9.2 | 0.04 | 9 | 0.03 |
| TH01 | 5.9 | 0.008 | 6 | 0.009 |
| vWA | 8.6 | 0.02 | 8.8 | 0.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





