Submitted:
09 January 2024
Posted:
11 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Relevant Sections
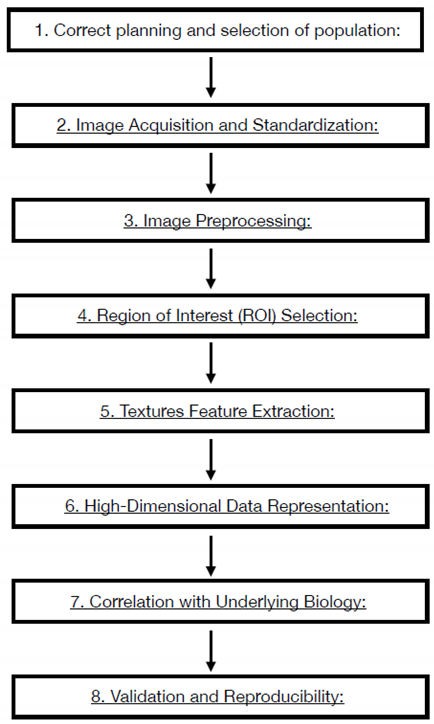
2.1. Radiomics Workflow
- Correct planning and selection of population: The Radiomics study must answer to a precise clinical question still open and debated, the study must be built with enough data to demonstrate the clinical hypothesis. Attrite data errors are common, needing a precise model validation, measuring the predict performance of the model. The training sample is necessary to measure the capability of the software to work properly and , should be around one-third of the sample size.
- Image Acquisition and Standardization: Radiomics workflow begins with the acquisition of medical images through various modalities such as CT, MRI, or PET. Standardization of imaging protocols is crucial to ensure consistency across studies and institutions. This involves establishing uniform parameters for image acquisition, resolution, contrast type and parameters of administration. Anonymization process is a delicate point and necessary to undergo law prescription and patient privacy respect. The necessity of homogenous data is critical. Artificial intelligence algorithms that work on the modality acquisition software help to produce and homogenize data obtained from different device using optimization process derived from database and tailoring the acquisition parameters to patient to obtain homogeneous quality images.
- Image Preprocessing: Raw medical images often undergo preprocessing steps to enhance the quality of data for analysis. Preprocessing may involve noise reduction, normalization, and correction for artifacts to ensure the reliability of extracted features. MRI filters for homogenization are proposed in many papers and should be used to obtain the possibility to extract texture parameters homogenously.
- Region of Interest (ROI) Selection: The process involves defining the Region of Interest (ROI), which represents the area of the image containing the lesion or tissue of interest. Accurate ROI delineation is critical, and it can be performed manually or through automated/semi-automated (region growing or thresholding) segmentation techniques. In the field of automated methods deep learning Artificial Intelligence software are developing very promising results. All the methods described have weak points: manual segmentation Is time consuming, manual and semi-automated segmentation present an observer-bias; automated softwares are not still robust enough to be objective. The use of automated software could introduce variability in obtaining Radiomics features as many of them are good enough to isolate entire organs but not enough for single tumor lesions segmentation. 3D segmentation will perform a more precise measure but introduce time spanding procedur if manual or semiautomated and great technical difficulty to automated software. Manual and semiautomatic segmentation with readers correlation seems to be the best way to obtain correct data.
- Textures Feature Extraction: Radiomics features are quantitative measurements extracted from ROIs, encompassing a range of parameters such as intensity, morphology, texture. Intensity features include statistical measures like mean, median, and standard deviation, while texture features capture patterns and variations within the image. The process to extrapolate these parameters are different, using the Image Biomarker Standardization Initiative (IBSI) guidelines is mandatory to obtain reproducible measurements. An example of texture parameters different orders is:
- First order statistics compose by 19 features.
- Shaped based (3d) compose by 16 features.
- Shaped based (2d) compose by 10 features.
- Gray Level Co-occurrence Matrix compose by 24 features.
- Gray Level Run Length Matrix compose by 16 features.
- Gray Level Size Zone Matrix compose by 16 features.
- Neighbouring Gray Tone Difference Matrix compose by 5 features.
- Gray Level Dependence Matrix compose by 14 features.
- 6.
- High-Dimensional Data Representation: The extracted features result in a high-dimensional dataset, where each data point represents a specific quantitative aspect of the image. This high-dimensional data is the basis for subsequent analyses and need to be accurate selected to obtain correct results. The number of feature correlate with the possibility of overfitting of the model. The selection and reduction of features dimension is an important step to produce generalizable and reproducible results. In literature, there is not generalized definitive rules to assess the correct reduction and selection of parameters. In this step the role of artificial software analysis seems to obtain the principal role to manage the correct pathway. First step is the exclusion of non-reproducible value, in particular, data coming from manual or semiautomated selection of ROI. Test-retest robustness of data extracted from features should be assessed. In this step the use of phantom could help defining correct evaluation.
- 7.
- Correlation with Underlying Biology: Radiomic features are not arbitrary; they correlate with underlying biological and pathological characteristics. The challenge is to identify and understanding these correlations, linking the quantitative features to the physiological, genetic, and molecular attributes of the imaged tissue. Artificial intelligence software elaboration allows an automated integration of data from images and biological features. These algorithms aid in predicting outcomes, classifying diseases, and uncovering relationships that might be challenging for human observers to discern.
- 8.
- Validation and Reproducibility: Rigorous validation is crucial to ensure the reliability and reproducibility of radiomic features. Validation consists in assessing the performance of radiomic models using independent datasets and considering factors such as intra- and inter-observer variability. Understanding and applying these principles allow radiologists to navigate the essence of radiomics, harnessing the quantitative power of medical imaging for enhanced diagnostic and prognostic capabilities. As radiomics continues to evolve, adherence to these principles ensures the credibility and utility of findings in this new technology. Some standardization processes are suggesting with the Image biomarker standardization initiative (IBSI). The IBSI provide standardized image biomarker nomenclature and definitions, a standardized general image processing workflow, tools for verifying radiomics software implementations and reporting guidelines for radiomic studies [9,10].
2.2. Radiomics Standardization and Verification Process
2.3. Ethical issue and Data Privacy
2.4. Explicability of Radiomics Software
2.5. Biological Correlation and Radiogenomics
2.6. Radiomics Mean Application, Towards Patient Custom Care
2.7. Organs Software Applications
- -
- Neuroimaging Applications: Radiomics is widely used in stroke imaging disease. One of the main application is ischemic stroke. Ischemic stroke is defined as a neurological deficit lasting more than 24h with a correspondent radiological imaging. Radiomics softwares help finding differences between ischemic stroke images and transient ischemic attack. Knowing that ischemic stroke is 85% of the stroke subtype with a high morbility of disability, it is highly important to have a rapid confirmation of pathology and Brain angioCT is the exam of choice with an important role of time in therapy success. Rapid Systemic thrombolytic treatment (no more than 4.5 h from symptoms) with plasminogen activator and local interventional radiology treatment with thrombectomy is the way to reduce morbidity and mortality. In the very acute phase of ischemic stroke attack CT imaging interpretation could be challenging. Some texture feature are reported to better correlate to ischemic suffering tissue. The identification of a penumbra cerebral tissue that could be reperfused without damage improve the decision making process widening the window treatment. The penumbra core area could be evaluated from an artificial intelligence algorithm calculating the apparent diffusion coefficient and cerebral blood flow maps obtained with a contrast enhanced CT perfusion exam in patient with symptoms from no more than 9 h. The radiomics nomogram obtained strongly predicts clinical outcomes at 7 days and at 3 months. These evaluation selected patient that could be treated with thombolysis even if symptoms were recognized inside a 9 hours windows, reducing potential complication of hemorrhage damage of reperfused tissue. Same interesting evaluation is demonstrated by some papers using radiomics on MR imaging. Some other papers investigate the ability of MR imaging radiomics evaluation to predict the hemorrhagic complication at distance, for example 72h after trombolytic treatment, in these papers the results were compared with only radiological visual evaluation determining a high advantage given by Radiomics evaluation. Selecting correct patients for endovascular recanalization expecially when thrombus is in the distal portion lower the number of procedure fail and reduce risks in frail patients. Radiomics evaluation have an high impact in stroke patient management and have completed replaced the morphological evaluation of thrombus with CT characteristics such as length, volume, or permeability. Other application of Brain Radiomics is to characterize brain metastasis and to differentiate them from primitive tumor; Radiomics well correlate in differentiating tumor subtypes, assessing treatment response, and predicting patient outcomes in brain oncology. Promising results are in evaluating neurodegenerative diseases, often diffuse pathology with difficult possibility of characterization [15,16,17,19,20,21,22,28].
- -
- Cardiovascular Imaging: Radiomics has many promised applications in cardiovascular imaging. From image quality and dose optimization to cardiac function evaluation, segmentation and automatic reconstruction. That point will soon improve radiological workflow reducing time consuming operation and allowing radiologist time to concentrate on interpreting images and not in reconstruction operations.
- -
- Pulmonary Imaging: In pulmonary imaging, radiomics is valuable for characterizing lung nodules, distinguishing between benign and malignant lesions, and assessing the progression of respiratory diseases. Radiomic analysis of lung images contributes to early detection and personalized management strategies [27,28,29,30,31].
- -
- Gastrointestinal and Oncologic Imaging: Radiomics is widely applied in oncologic imaging for the characterization of tumors, lymph nodes, and metastases. In gastrointestinal imaging, it aids in the identification of biomarkers associated with disease progression and treatment response. Some softwares have a promised impact on evaluation of prostate cancer multiparametric MRI values, which need a preliminary long, spending time, images elaboration before Radiologist report with p-rads criteria [32,33,34,35,36,37,38,39,40,41].
3. Discussion and Future Perspectives
References
- Lambin P, Rios-Velazquez E, Leijenaar R et al (2012) Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 48(4):441–446. [CrossRef]
- Tomaszewski MR, Gillies RJ (2021) The biological meaning of radiomic features. Radiology 298(3):505–516. [CrossRef]
- Gillies RJ, Kinahan PE (2016) Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278(2):563–577. [CrossRef]
- Lambin P, Leijenaar RTH, Deist TM et al (2017) Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 14(12):749–762. [CrossRef]
- O’Connor JP, Aboagye EO, Adams JE et al (2017) Imaging biomarker roadmap for cancer studies. Nat Rev Clin Oncol 14:169–186. [CrossRef]
- Van Timmeren JE, Cester D, Tanadini-Lang S, Alkadhi H, Baessler B (2020) Radiomics in medical imaging-"how-to" guide and critical reflection. Insights Imaging 11(1):91. [CrossRef]
- Song J, Yin Y, Wang H, Chang Z, Liu Z, Cui L (2020) A review of origi- nal articles published in the emerging field of radiomics. Eur J Radiol 127:108991. [CrossRef]
- Fusco R, Granata V, Grazzini G, Pradella S, Borgheresi A, Bruno A, Palumbo P, Bruno F, Grassi R, Giovagnoni A, Grassi R, Miele V, Barile A. Radiomics in medical imaging: pitfalls and challenges in clinical management. Jpn J Radiol. 2022 Sep;40(9):919-929. Epub 2022 Mar 28. PMID: 35344132. [CrossRef]
- Zwanenburg A, Leger S, Vallieres M, Lock S. Image biomarker standardisation initiative. arXiv preprint arXiv:1612.07003.
- Lambin P. Radiomics Digital Phantom, CancerData(2016). [CrossRef]
- Neri, E., Aghakhanyan, G., Zerunian, M. et al. Explainable AI in radiology: a white paper of the Italian Society of Medical and Interventional Radiology. Radiol med 128, 755–764 (2023). [CrossRef]
- Radiomics of Brain MRI: Utility in Prediction of Metastatic Tumor Type. Helge C. Kniep, Frederic Madesta, Tanja Schneider, Uta Hanning, Michael H. Schönfeld, Gerhard Schön, Jens Fiehler, Tobias Gauer, René Werner, and Susanne Gellissen. Radiology 2019 290:2, 479-487. [CrossRef]
- Granata V, Fusco R, De Muzio F, Brunese MC, Setola SV, Ottaiano A, Cardone C, Avallone A, Patrone R, Pradella S, Miele V, Tatangelo F, Cutolo C, Maggialetti N, Caruso D, Izzo F, Petrillo A. Radiomics and machine learning analysis by computed tomography and magnetic resonance imaging in colorectal liver metastases prognostic assessment. Radiol Med. 2023 Sep 11. Epub ahead of print. PMID: 37697033. [CrossRef]
- Granata V, Fusco R, De Muzio F, Cutolo C, Mattace Raso M, Gabelloni M, Avallone A, Ottaiano A, Tatangelo F, Brunese MC, Miele V, Izzo F, Petrillo A. Radiomics and Machine Learning Analysis Based on Magnetic Resonance Imaging in the Assessment of Colorectal Liver Metastases Growth Pattern. Diagnostics (Basel). 2022 Apr 29;12(5):1115. PMID: 35626271; PMCID: PMC9140199. [CrossRef]
- Vivek Verma, Charles B. Simone, Sunil Krishnan, Steven H. Lin, Jinzhong Yang, Stephen M. Hahn, The Rise of Radiomics and Implications for Oncologic Management, JNCI: Journal of the National Cancer Institute, Volume 109, Issue 7, July 2017, djx055. [CrossRef]
- Fritz, Benjamin, et al. "Radiomics and deep learning for disease detection in musculoskeletal radiology: an overview of novel MRI-and CT-based approaches." Investigative radiology 58.1 (2023): 3-13.
- Crivelli, Paola, et al. "A new challenge for radiologists: radiomics in breast cancer." BioMed research international 2018 (2018). [CrossRef]
- Rogers, William, et al. "Radiomics: from qualitative to quantitative imaging." The British journal of radiology 93.1108 (2020): 20190948. [CrossRef]
- Wagner, Matthias W., et al. "Radiomics, machine learning, and artificial intelligence—what the neuroradiologist needs to know." Neuroradiology (2021): 1-11.
- Pontillo, G., et al. "A combined radiomics and machine learning approach to overcome the clinicoradiologic paradox in multiple sclerosis." American Journal of Neuroradiology 42.11 (2021): 1927-1933. [CrossRef]
- Chen, Qian, et al. "Radiomics in stroke neuroimaging: techniques, applications, and challenges." Aging and disease 12.1 (2021): 143.Kniep, Helge C., et al. "Radiomics of brain MRI: utility in prediction of metastatic tumor type." Radiology 290.2 (2019): 479-487.
- Ponsiglione, Andrea, et al. "Cardiac CT and MRI radiomics: systematic review of the literature and radiomics quality score assessment." European Radiology (2022): 1-10. [CrossRef]
- Spadarella, Gaia, et al. "Radiomics in cardiovascular disease imaging: from pixels to the heart of the problem." Current Cardiovascular Imaging Reports 15.2 (2022): 11-21. [CrossRef]
- Raisi-Estabragh, Zahra, et al. "Cardiac magnetic resonance radiomics: basic principles and clinical perspectives." European Heart Journal-Cardiovascular Imaging 21.4 (2020): 349-356. [CrossRef]
- Cetin, Irem, et al. "Radiomics signatures of cardiovascular risk factors in cardiac MRI: results from the UK Biobank." Frontiers in Cardiovascular Medicine 7 (2020): 591368. [CrossRef]
- Hassani, Cameron, et al. "Radiomics in pulmonary lesion imaging." American Journal of Roentgenology 212.3 (2019): 497-504.
- Wilson, Ryan, and Anand Devaraj. "Radiomics of pulmonary nodules and lung cancer." Translational lung cancer research 6.1 (2017): 86. [CrossRef]
- Lafata, Kyle J., et al. "An exploratory radiomics approach to quantifying pulmonary function in CT images." Scientific reports 9.1 (2019): 11509. [CrossRef]
- Ather, S., T. Kadir, and F. Gleeson. "Artificial intelligence and radiomics in pulmonary nodule management: current status and future applications." Clinical radiology 75.1 (2020): 13-19. [CrossRef]
- Ferrari, R., Mancini-Terracciano, C., Voena, C., Rengo, M., Zerunian, M., Ciardiello, A., Grasso, S., Mare, V., Paramatti, R., Russomando, A., Santacesaria, R., Satta, A., Solfaroli Camillocci, E., Faccini, R., & Laghi, A. (2019). MR-based artificial intelligence model to assess response to therapy in locally advanced rectal cancer. European Journal of Radiology, 118. [CrossRef]
- Cozzi D, Bicci E, Cavigli E, Danti G, Bettarini S, Tortoli P, Mazzoni LN, Busoni S, Pradella S, Miele V. Radiomics in pulmonary neuroendocrine tumours (NETs). Radiol Med. 2022 Jun;127(6):609-615. Epub 2022 May 10. PMID: 35538389. [CrossRef]
- Caruso D et al. "Radiomics in oncology, part 1: Technical principles and gastrointestinal application in CT and MRI." Cancers 13.11 (2021): 2522. [CrossRef]
- De Santis D et al. "Radiomics analysis in gastrointestinal imaging: a narrative review." Digestive Medicine Research (2023): 1-13.
- Wesdorp, Nina J., et al. "Advanced analytics and artificial intelligence in gastrointestinal cancer: a systematic review of radiomics predicting response to treatment." European journal of nuclear medicine and molecular imaging 48 (2021): 1785-1794. [CrossRef]
- Wang, Yue, et al. "Potential value of CT radiomics in the distinction of intestinal-type gastric adenocarcinomas." European Radiology 30 (2020): 2934-2944. [CrossRef]
- Granata V, Fusco R, De Muzio F, Cutolo C, Setola SV, Dell’Aversana F, Grassi F, Belli A, Silvestro L, Ottaiano A, Nasti G, Avallone A, Flammia F, Miele V, Tatangelo F, Izzo F, Petrillo A. Radiomics and machine learning analysis based on magnetic resonance imaging in the assessment of liver mucinous colorectal metastases. Radiol Med. 2022 Jul;127(7):763-772. Epub 2022 Jun 2. PMID: 35653011. [CrossRef]
- Chiti G, Grazzini G, Flammia F, Matteuzzi B, Tortoli P, Bettarini S, Pasqualini E, Granata V, Busoni S, Messserini L, Pradella S, Massi D, Miele V. Gastroenteropancreatic neuroendocrine neoplasms (GEP-NENs): a radiomic model to predict tumor grade. Radiol Med. 2022 Sep;127(9):928-938. Epub 2022 Aug 2. PMID: 35917099. [CrossRef]
- Palatresi D, Fedeli F, Danti G, Pasqualini E, Castiglione F, Messerini L, Massi D, Bettarini S, Tortoli P, Busoni S, Pradella S, Miele V. Correlation of CT radiomic features for GISTs with pathological classification and molecular subtypes: preliminary and monocentric experience. Radiol Med. 2022 Feb;127(2):117-128. Epub 2022 Jan 12. PMID: 35022956. [CrossRef]
- Galluzzo A, Boccioli S, Danti G, De Muzio F, Gabelloni M, Fusco R, Borgheresi A, Granata V, Giovagnoni A, Gandolfo N, Miele V. Radiomics in gastrointestinal stromal tumours: an up-to-date review. Jpn J Radiol. 2023 May 12. Epub ahead of print. PMID: 37171755. [CrossRef]
- Santini D, Danti G, Bicci E, Galluzzo A, Bettarini S, Busoni S, Innocenti T, Galli A, Miele V. Radiomic Features Are Predictive of Response in Rectal Cancer Undergoing Therapy. Diagnostics (Basel). 2023 Aug 2;13(15):2573. PMID: 37568936; PMCID: PMC10417449. [CrossRef]
- Brunese MC, Fantozzi MR, Fusco R, De Muzio F, Gabelloni M, Danti G, Borgheresi A, Palumbo P, Bruno F, Gandolfo N, Giovagnoni A, Miele V, Barile A, Granata V. Update on the Applications of Radiomics in Diagnosis, Staging, and Recurrence of Intrahepatic Cholangiocarcinoma. Diagnostics (Basel). 2023 Apr 20;13(8):1488. PMID: 37189589; PMCID: PMC10137417. [CrossRef]
- Pesapane, F., Rotili, A., Penco, S., Nicosia, L., & Cassano, E. (2022). Digital twins in radiology. Journal of Clinical Medicine, 11(21), 6553.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





