Submitted:
09 January 2024
Posted:
10 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
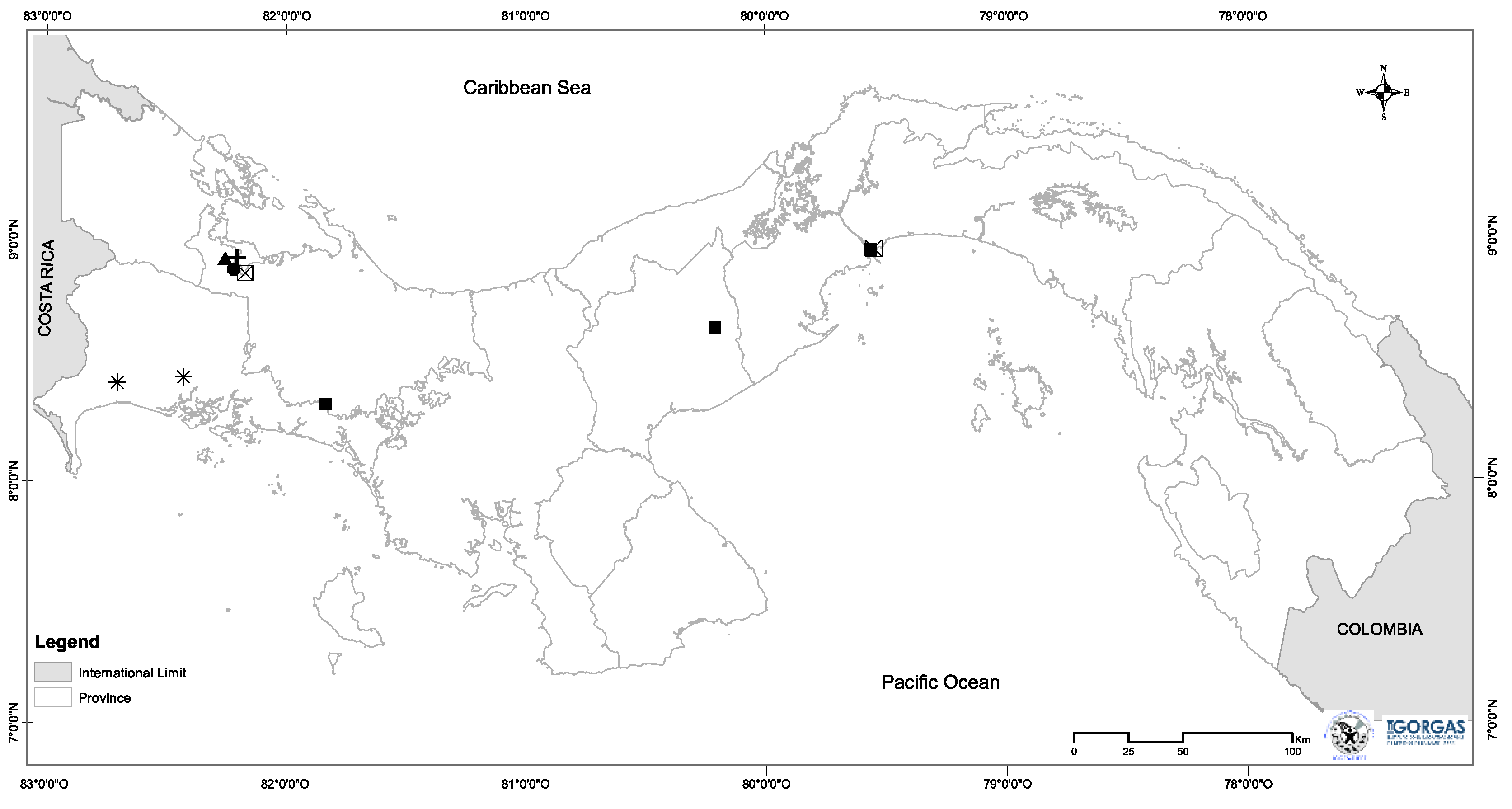
2.1. The Materials Rodent surveys and sample collection
2.2. DNA extraction
2.3. DNA amplification
2.4. DNA Library preparation
2.5. Data analysis
2.6. Bacterial diversity, and community composition
3. Results
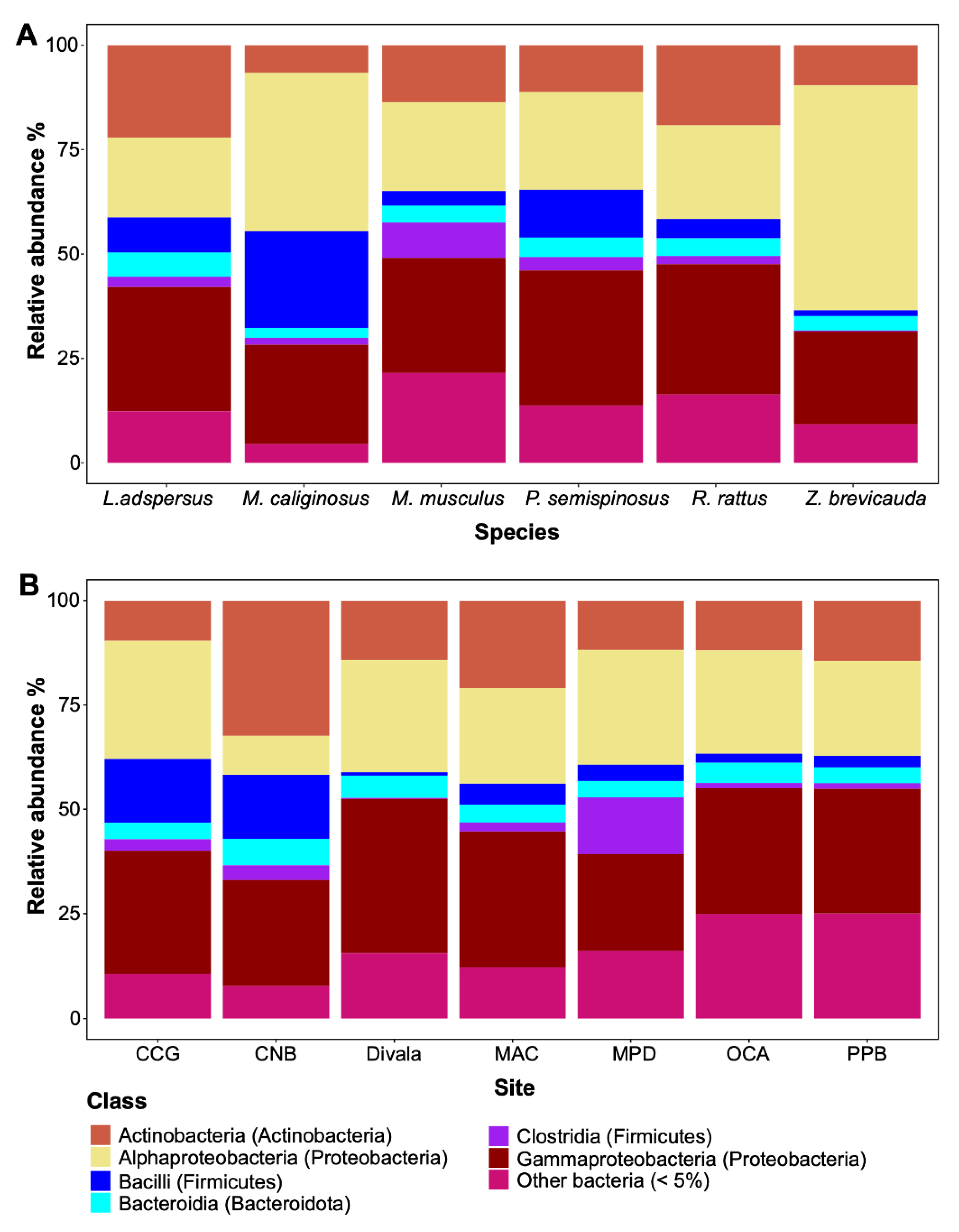
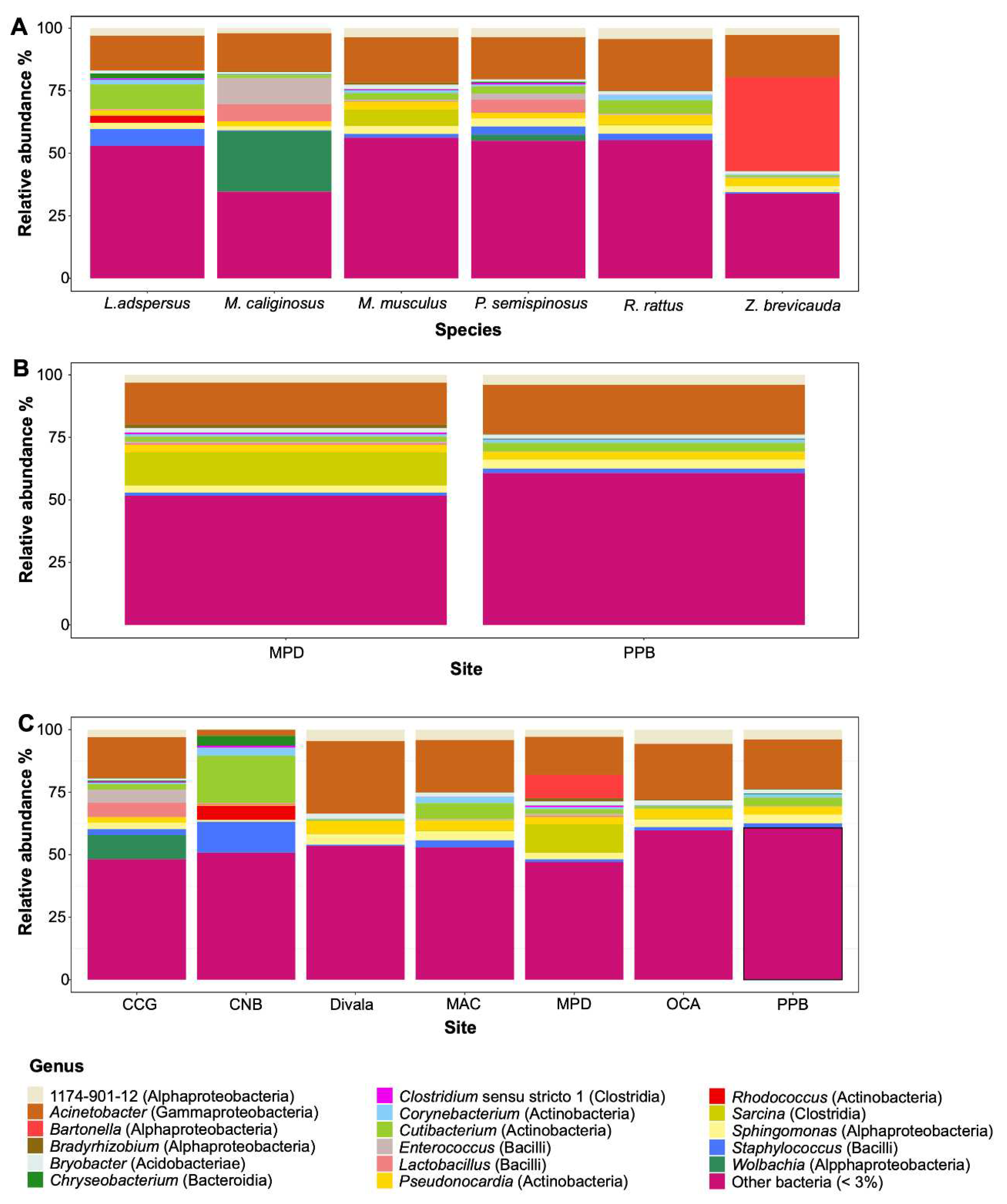
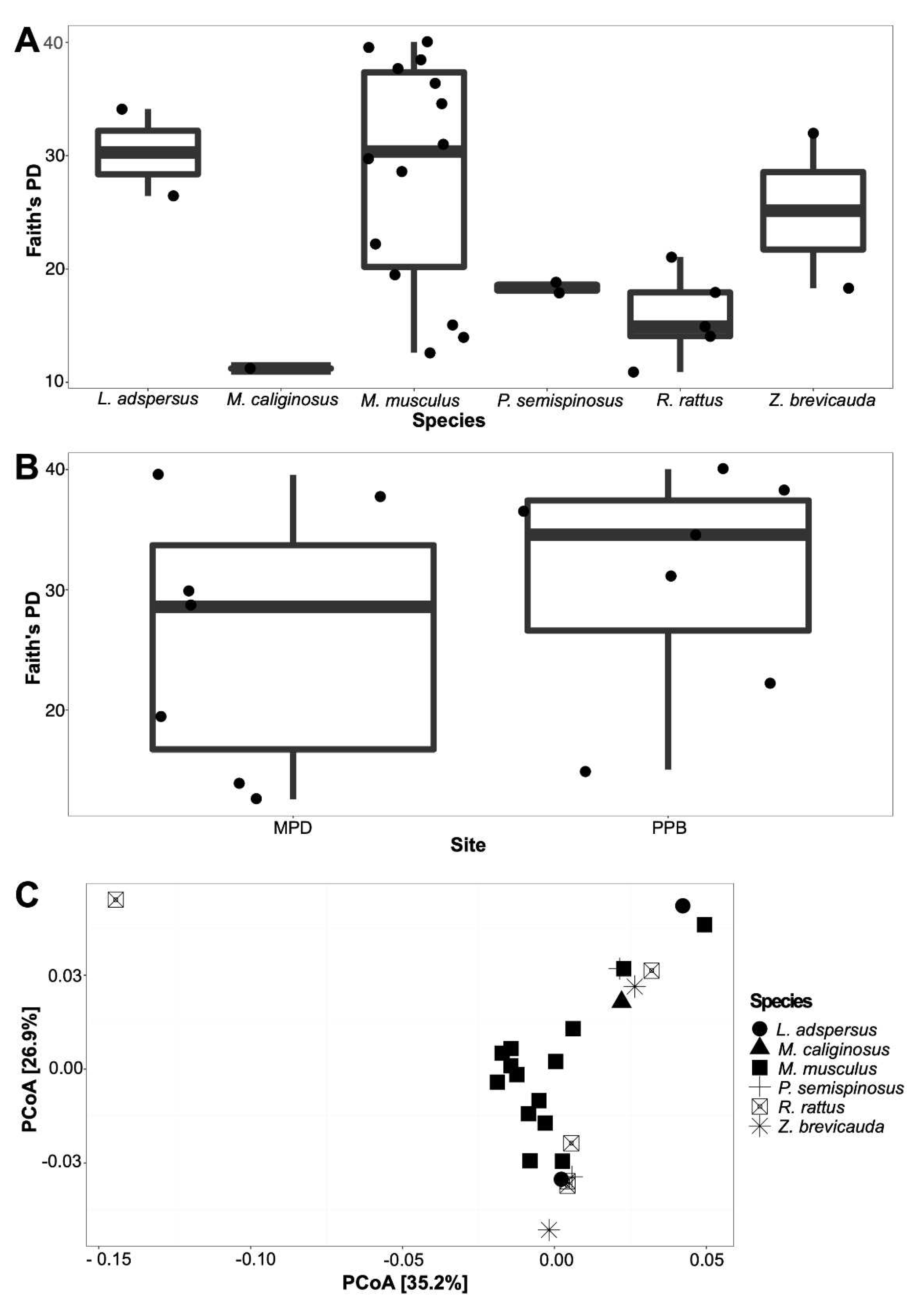
3.1. The spleen microbiome by rodent species and locality
4. Discussion
4.1. Bacterial composition in the spleen of six species of rodents, role in rodents and implications in zoonotic diseases
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Ethics
References
- Morens, D.M.; Folkers, G.K.; Anthony, S. Fauci The challenge of emerging and re-emerging infectious diseases. Nature 2004, 430, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Rabozzi, G.; Bonizzi, L.; Crespi, E.; Somaruga, C.; Sokooti, M.; Tabibi, R.; Vellere, F.; Brambilla, G.; Colosio, C. Emerging zoonoses: The “one health approach. ” Saf. Health Work 2012, 3, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Vouga, M.; Greub, G. Emerging bacterial pathogens: the past and beyond. Clin Microbiol Infect 2015, 1, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Barac, A.; Poljak, M.; Ong, D.S.Y. Editorial: Innovative Approaches in Diagnosis of Emerging/Re-emerging Infectious Diseases. Front. Microbiol. 2020, 11, 10–12. [Google Scholar] [CrossRef] [PubMed]
- Magouras, I.; Brookes, V.J.; Jori, F.; Martin, A.; Pfeiffer, D.U.; Dürr, S. Emerging Zoonotic Diseases: Should We Rethink the Animal–Human Interface? Front. Vet. Sci. 2020, 7, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.T.; Sobur, M.A.; Islam, M.S.; Ievy, S.; Hossain, M.J.; Zowalaty, M.E.E.; Rahman, A.M.M.T.; Ashour, H.M. Zoonotic diseases: Etiology, impact, and control. Microorganisms 2020, 8, 1–34. [Google Scholar] [CrossRef] [PubMed]
- Woolhouse, M.E.J.; Gowtage-Sequeria, S. Host range and emerging and reemerging pathogens. Emerg. Infect. Dis. 2005, 11, 1842–1847. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.; Kutz, S. Introduction to the Special Issue on ‘Emerging Zoonoses and Wildlife. ’ Int. J. Parasitol. Parasites Wildl. 2019, 9, 322. [Google Scholar] [CrossRef]
- Wang, W.-H.; Thitithanyanont, A.; Urbina, A.N.; Wang, S.-F. Emerging and re-emerging virus. Pathogens 2021, 10. [Google Scholar] [CrossRef]
- Cantas, L.; Suer, K. Review: The important bacterial zoonoses in “One Health” concept. Front. Public Heal. 2014, 2, 1–8. [Google Scholar] [CrossRef]
- Kruse, H.; Kirkemo, A.M.; Handeland, K. Wildlife as source of zoonotic infections. Emerg. Infect. Dis. 2004, 10, 2067–2072. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.K.; Hitchens, P.L.; Pandit, P.S.; Rushmore, J.; Evans, T.S.; Young, C.C.W.; Doyle, M.M. Global shifts in mammalian population trends reveal key predictors of virus spillover risk. Proc. R. Soc. B Biol. Sci. 2020. [Google Scholar] [CrossRef] [PubMed]
- Meerburg, B.G.; Singleton, G.R.; Kijlstra, A. Rodent-borne diseases and their risks for public health Rodent-borne diseases and their risks for public health; 2009; Vol. 35; ISBN 1040841090.
- Dahmana, H.; Granjon, L.; Diagne, C.; Davoust, B.; Fenollar, F.; Mediannikov, O. Rodents as hosts of pathogens and related zoonotic disease risk. Pathogens 2020, 9, 2–21. [Google Scholar] [CrossRef] [PubMed]
- Mawanda, P.; Rwego, I.; Kisakye, J.J.; Sheil, D. Rodents as potential hosts and reservoirs of parasites along the edge of a central african forest: Bwindi impenetrable national park, South Western Uganda. Afr. Health Sci. 2020, 20, 1168–1178. [Google Scholar] [CrossRef] [PubMed]
- Luis, A.D.; Hayman, D.T.S.; O’Shea, T.J.; Cryan, P.M.; Gilbert, A.T.; Pulliam, J.R.C.; Mills, J.N.; Timonin, M.E.; Willis, C.K.R.; Cunningham, A.A.; et al. A comparison of bats and rodents as reservoirs of zoonotic viruses: Are bats special? Proc. R. Soc. B Biol. Sci. 2013, 280. [Google Scholar] [CrossRef] [PubMed]
- Han, B.A.; Schmidt, J.P.; Bowden, S.E.; Drake, J.M. Rodent reservoirs of future zoonotic diseases. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 7039–7044. [Google Scholar] [CrossRef]
- Assefa, A.; Chelmala, S. Correction to: Comparison of rodent community between natural and modified habitats in Kafta-Sheraro National Park and its adjoining villages, Ethiopia: implication for conservation. J. Basic Appl. Zool. 2019, 80. [Google Scholar] [CrossRef]
- Williams, E.P.; Spruill-harrell, B.M.; Taylor, M.K.; Lee, J.; Nywening, A. V; Yang, Z.; Nichols, J.H.; Camp, J. V; Owen, R.D.; Jonsson, C.B. Emergence : Lessons Learned from Bat- and Rodent-Borne. Viruses 2021, 13. [Google Scholar] [CrossRef]
- Hassell, J.M.; Begon, M.; Ward, M.J.; Fèvre, E.M. Urbanization and Disease Emergence: Dynamics at the Wildlife–Livestock–Human Interface. Trends Ecol. Evol. 2017, 32, 55–67. [Google Scholar] [CrossRef]
- Wasserberg, G.; Abramsky, Z.; Kotler, B.P.; Ostfeld, R.S.; Yarom, I.; Warburg, A. Anthropogenic disturbances enhance occurrence of cutaneous leishmaniasis in Israel deserts: Patterns and mechanisms. Ecol. Appl. 2003, 13, 868–881. [Google Scholar] [CrossRef]
- Friggens, M.M.; Beier, P. Anthropogenic disturbance and the risk of flea-borne disease transmission. Oecologia 2010, 164, 809–820. [Google Scholar] [CrossRef] [PubMed]
- Young, H.S.; McCauley, D.J.; Dirzo, R.; Nunn, C.L.; Campana, M.G.; Agwanda, B.; Otarola-Castillo, E.R.; Castillo, E.R.; Pringle, R.M.; Veblen, K.E.; et al. Interacting effects of land use and climate on rodent-borne pathogens in central Kenya. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372. [Google Scholar] [CrossRef]
- Young, H.S.; Dirzo, R.; Helgen, K.M.; McCauley, D.J.; Billeter, S.A.; Kosoy, M.Y.; Osikowicz, L.M.; Salkeld, D.J.; Young, T.P.; Dittmar, K. Declines in large wildlife increase landscape-level prevalence of rodent-borne disease in Africa. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 7036–7041. [Google Scholar] [CrossRef]
- Haddad, N.M.; Brudvig, L.A.; Clobert, J.; Davies, K.F.; Gonzalez, A.; Holt, R.D.; Lovejoy, T.E.; Sexton, J.O.; Austin, M.P.; Collins, C.D.; et al. Habitat fragmentation and its lasting impact on Earth’s ecosystems. Sci. Adv. 2015, 1, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Andermann, T.; Faurby, S.; Turvey, S.T.; Antonelli, A.; Silvestro, D. The past and future human impact on mammalian diversity. Sci. Adv. 2020, 6, 1–18. [Google Scholar] [CrossRef]
- Armién, A.G.; Armién, B.; Koster, F.; Pascale, J.M.; Avila, M.; Gonzalez, P.; De La Cruz, M.; Zaldivar, Y.; Mendoza, Y.; Gracia, F.; et al. Hantavirus infection and habitat associations among rodent populations in agroecosystems of Panama: Implications for human disease risk. Am. J. Trop. Med. Hyg. 2009, 81, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Jahan, N.A.; Lindsey, L.L.; Larsen, P.A. The Role of Peridomestic Rodents as Reservoirs for Zoonotic Foodborne Pathogens. Vector-Borne Zoonotic Dis. 2021, 21, 133–148. [Google Scholar] [CrossRef]
- Gonzalez, P.; Salazar, J.R.; Salinas, T.P.; Avila, M.; Colella, J.P.; Dunnum, J.L.; Glass, G.E.; Gonzalez, G.; Juarez, E.; Lindblade, K.; et al. Two Decades of Wildlife Pathogen Surveillance: Case Study of Choclo orthohantavirus and Its Wild Reservoir Oligoryzomys costaricensis. Viruses 2023, 15. [Google Scholar] [CrossRef]
- Armién, B.; Muñoz, C.; Cedeño, H.; Salazar, J.R.; Salinas, T.P.; González, P.; Trujillo, J.; Sánchez, D.; Mariñas, J.; Hernández, A.; et al. Hantavirus in Panama: Twenty Years of Epidemiological Surveillance Experience. Viruses 2023, 15, 1–14. [Google Scholar] [CrossRef]
- Mills, J.N.; Yates, T.L.; Childs, J.E.; Parmenter, R.R.; Ksiazek, T.G.; Rollin, P.E.; Peters, C.J. Guidelines for Working with Rodents Potentially Infected with Hantavirus. J. Mammal. 1995, 76, 716. [Google Scholar] [CrossRef]
- Hanshew, A.S.; Mason, C.J.; Raffa, K.F.; Currie, C.R. Minimization of chloroplast contamination in 16S rRNA gene pyrosequencing of insect herbivore bacterial communities. J. Microbiol. Methods 2013, 95, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Kembel, S.W.; O’Connor, T.K.; Arnold, H.K.; Hubbell, S.P.; Wright, S.J.; Green, J.L. Relationships between phyllosphere bacterial communities and plant functional traits in a neotropical forest. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 13715–13720. [Google Scholar] [CrossRef] [PubMed]
- Bekele, A.; Leirs, H. Population ecology of rodents of maize fields and grassland in central Ethiopia. Belgian J. Zool. 1997, 127 Suppl, 39–48. [Google Scholar]
- Meheretu, Y.; Sluydts, V.; Welegerima, K.; Bauer, H.; Teferi, M.; Yirga, G.; Mulungu, L.; Haile, M.; Nyssen, J.; Deckers, J.; et al. Rodent abundance, stone bund density and its effects on crop damage in the Tigray highlands, Ethiopia. Crop Prot. 2014, 55, 61–67. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 1–17. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. QIIME 2: Reproducible, interactive, scalable, and extensible microbiome data science. PeerJ Prepr. 2018, 6, e27295v1. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.A.; Holmes, S.P. DADA2: High resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 4–5. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; Sankaran, K.; Fukuyama, J.A.; McMurdie, P.J.; Holmes, S.P. Bioconductor Workflow for Microbiome Data Analysis: from raw reads to community analyses. F1000Research 2016, 5, 1492. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “all-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014, 42, 643–648. [Google Scholar] [CrossRef] [PubMed]
- R Development Core R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2008.
- Clarke, K.R. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 1993, 18, 117–143. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; Mcglinn, D.; Minchin, P.R.; O’hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R package Version 2.0-10. 2017, 2. [Google Scholar]
- Wickham, H. ggplot2 - Elegant Graphics for Data Analysis | Hadley Wickham | Springer . 2017. [Google Scholar] [CrossRef]
- Liyai, R.; Kimita, G.; Masakhwe, C.; Abuom, D.; Mutai, B.; Onyango, D.M.; Waitumbi, J. The spleen bacteriome of wild rodents and shrews from Marigat, Baringo County, Kenya. PeerJ 2021, 9, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Balish, E.; Warner, T. Enterococcus faecalis induces inflammatory bowel disease in interleukin-10 knockout mice. Am. J. Pathol. 2002, 160, 2253–2257. [Google Scholar] [CrossRef]
- Marrow, J.; Whittington, J.K.; Mitchell, M.; Hoyer, L.L.; Maddox, C. Prevalence and antibiotic-resistance characteristics of Enterococcus spp. isolated from free-living and captive raptors in central illinois. J. Wildl. Dis. 2009, 45, 302–313. [Google Scholar] [CrossRef]
- Gutiérrez, R.; Krasnov, B.; Morick, D.; Gottlieb, Y.; Khokhlova, I.S.; Harrus, S. Bartonella infection in rodents and their flea ectoparasites: An overview. Vector-Borne Zoonotic Dis. 2015, 15, 27–39. [Google Scholar] [CrossRef]
- Benga, L.; Feßler, A.T.; Benten, W.P.M.; Engelhardt, E.; Köhrer, K.; Schwarz, S.; Sager, M. Acinetobacter species in laboratory mice: species survey and antimicrobial resistance. Lab. Anim. 2019, 53, 470–477. [Google Scholar] [CrossRef]
- Maragakis, L.L.; Perl, T.M. Acinetobacter baumannii: Epidemiology, antimicrobial resistance, and treatment options. Clin. Infect. Dis. 2008, 46, 1254–1263. [Google Scholar] [CrossRef]
- Pérez-Martínez, L.; Venzal, J.M.; González-Acuña, D.; Portillo, A.; Blanco, J.R.; Oteo, J.A. Bartonella rochalimae and other Bartonella spp. in fleas, Chile. Emerg Infect Dis. 2009, 1150–2. [Google Scholar] [CrossRef]
- Theonest, N.O.; Carter, R.W.; Amani, N.; Doherty, S.L.; Hugho, E.; Keyyu, J.D.; Mable, B.K.; Shirima, G.M.; Tarimo, R.; Thomas, K.M.; et al. Molecular detection and genetic characterization of Bartonella species from rodents and their associated ectoparasites from northern Tanzania. PLoS One 2019, 14, 1–17. [Google Scholar] [CrossRef]
- Landaeta-Aqueveque, C.; Moreno Salas, L.; Henríquez, A.L.; Silva-de la Fuente, M.C.; González-Acuña, D. Parasites of Native and Invasive Rodents in Chile: Ecological and Human Health Needs. Front. Vet. Sci. 2021, 8, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Saengsawang, P.; Morand, S.; Desquesnes, M.; Yangtara, S.; Inpankaew, T. Molecular detection of Bartonella species in rodents residing in urban and suburban areas of central Thailand. Microorganisms 2021, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Daly, J.S.; Worthington, M.G.; Brenner, D.J.; Moss, C.W.; Hollis, D.G.; Weyant, R.S.; Steigerwalt, A.G.; Weaver, R.E.; Daneshvar, M.I.; O’Connor, S.P. Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J. Clin. Microbiol. 1993, 31, 872–881. [Google Scholar] [CrossRef] [PubMed]
- Fenollar, F.; Sire, S.; Raoult, D. Bartonella vinsonii subsp. arupensis as an agent of blood culture-negative endocarditis in a human. J. Clin. Microbiol. 2005, 43, 945–947. [Google Scholar] [CrossRef]
- Welch, D.F.; Carroll, K.C.; Hofmeister, E.K.; Persing, D.H.; Robison, D.A.; Steigerwalt, A.G.; Brenner, D.J. Isolation of a new subspecies, Bartonella vinsonii subsp. arupensis, from a cattle rancher: Identity with isolates found in conjunction with Borrelia burgdorferi and Babesia microti among naturally infected mice. J. Clin. Microbiol. 1999, 37, 2598–2601. [Google Scholar] [CrossRef] [PubMed]
- Probert, W.; Louie, J.K.; Tucker, J.R.; Longoria, R.; Hogue, R.; Moler, S.; Graves, M.; Palmer, H.J.; Cassady, J.; Fritz, C.L. Meningitis due to a “Bartonella washoensis”-like human pathogen. J. Clin. Microbiol. 2009, 47, 2332–2335. [Google Scholar] [CrossRef]
- Corvec, S. Clinical and biological features of Cutibacterium (Formerly Propionibacterium) avidum, an underrecognized microorganism. Clin. Microbiol. Rev. 2018, 31, 1–42. [Google Scholar] [CrossRef]
- Elston, M.J.; Dupaix, J.P.; Opanova, M.I.; Atkinson, R.E. Cutibacterium acnes (formerly Proprionibacterium acnes) and Shoulder Surgery. Hawai’i J. Heal. Soc. Welf. 2019, 78, 3–5. [Google Scholar]
- Ellerbroek, L.; Mac, K.N.; Peters, J.; Hultquist, L. Hazard Potential from Antibiotic-resistant Commensals like Enterococci. J. Vet. Med. 2004. [Google Scholar] [CrossRef]
- Mallon, D.J.P.; Corkill, J.E.; Hazel, S.M.; Sian Wilson, J.; French, N.P.; Bennett, M.; Hart, C.A. Excretion of vancomycin-resistant enterococci by wild mammals. Emerg. Infect. Dis. 2002, 8, 636–638. [Google Scholar] [CrossRef] [PubMed]
- Marcelino, L.P.; Valentini Junior, D.F.; Machado, S.M.D.S.; Schaefer, P.G.; Rivero, R.C.; Osvaldt, A.B. Sarcina ventriculi a rare pathogen. Autops. Case Reports 2021, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Owens, L.A.; Colitti, B.; Hirji, I.; Pizarro, A.; Jaffe, J.E.; Moittié, S.; Bishop-Lilly, K.A.; Estrella, L.A.; Voegtly, L.J.; Kuhn, J.H.; et al. A Sarcina bacterium linked to lethal disease in sanctuary chimpanzees in Sierra Leone. Nat. Commun. 2021, 12. [Google Scholar] [CrossRef]
- Kim, H.K.; Missiakas, D.; Schneewind, O. Mouse models for infectious diseases caused by Staphylococcus aureus. Physiol. Behav. 2018, 176, 139–148. [Google Scholar] [CrossRef]
- Poinsot, D.; Charlat, S.; Merçot, H. On the mechanism of Wolbachia-induced cytoplasmic incompatibility: Confronting the models with the facts. BioEssays 2003, 25, 259–265. [Google Scholar] [CrossRef]
- Hedges, L.M.; Brownlie, J.C.; O’Neill, S.L.; Johnson, K.N. Wolbachia and Virus Protection in Insects. Science (80-. ). 2008, 322, 702–702. [Google Scholar] [CrossRef]
- Castillo, A.M.; Saltonstall, K.; Arias, C.F.; Chavarria, K.A.; Ramírez-Camejo, L.A.; Mejía, L.C.; De León, L.F. The microbiome of neotropical water striders and its potential role in codiversification. Insects 2020, 11, 1–15. [Google Scholar] [CrossRef]
- Castillo, A.M.; Chavarria, K.A.; Saltonstall, K.; Arias, C.F.; Mejía, L.C. Salinity effects on the microbiome of a Neotropical water strider. Hydrobiologia 2021. [Google Scholar] [CrossRef]
- Moran, N.A.; McCutcheon, J.P.; Nakabachi, A. Genomics and Evolution of Heritable Bacterial Symbionts. Annu. Rev. Genet. 2008, 42, 165–190. [Google Scholar] [CrossRef]
- Tang, X.; Xie, G.; Shao, K.; Bayartu, S.; Chen, Y.; Gao, G. Influence of salinity on the bacterial community composition in Lake Bosten, a large oligosaline lake in arid Northwestern China. Appl. Environ. Microbiol. 2012, 78, 4748–4751. [Google Scholar] [CrossRef]
- Schuler, H.; Egan, S.P.; Hood, G.R.; Busbee, R.W.; Driscoe, A.L.; Ott, J.R. Diversity and distribution of Wolbachia in relation to geography, host plant affiliation and life cycle of a heterogonic gall wasp. BMC Evol. Biol. 2018, 18, 1–15. [Google Scholar] [CrossRef] [PubMed]
| Species | Sites | n |
|---|---|---|
| L. adspersus | Comarca Ngäbe-Buglé (CNB) | 1 |
| L. adspersus | Oajaca Chiguirí Arriba (OCA) | 1 |
| M. caliginosus | Cañazas Chiriquí Grande (CCG) | 1 |
| M. musculus | Mercado Público de David (MPD) | 7 |
| M. musculus | Panama Port Balboa (PPB) | 7 |
| P. semispinosus | Cañazas Chiriquí Grande (CCG) | 2 |
| R. rattus | Oajaca Chiguirí Arriba (OCA) | 1 |
| R. rattus | Mercado de Abasto Curundú (MAC) | 4 |
| Z. brevicauda | Divalá | 1 |
| Z. brevicauda | Mercado Público de David (MPD) | 1 |
| Bacteria | Species of Rodents | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genus |
L. adspersus |
M. caliginosus |
M. musculus |
P. semispinosus |
R. rattus |
Z. brevicauda |
||||
| CNB | OCA | CCG | MPD | PPB | CCG | OCA | MAC | Divalá | MPD | |
| n=1 | n=1 | n=1 | n=7 | n=7 | n=2 | n=1 | n=4 | n=1 | n=1 | |
| 1174-901-12 (Alphaproteobacteria) | 0.00 | 5.97 | 1.97 | 3.16 | 3.97 | 3.54 | 5.31 | 4.11 | 4.59 | 0.66 |
| Acinetobacter (Gammaproteobacteria) | 2.46 | 25.19 | 15.24 | 16.70 | 19.62 | 16.67 | 19.36 | 20.90 | 29.01 | 5.16 |
| Bartonella (Alphaproteobacteria) | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 74.87 |
| Bradyrhizobium (Alphaproteobacteria) | 0.00 | 0.36 | 0.38 | 1.45 | 0.31 | 0.25 | 0.27 | 0.22 | 0.00 | 0.22 |
| Bryobacter (Acidobacteria) | 0.00 | 2.33 | 0.71 | 1.73 | 1.60 | 0.91 | 1.55 | 1.44 | 2.03 | 0.30 |
| Chryseobacterium (Bacteroidia) | 3.83 | 0.11 | 0.31 | 0.12 | 0.29 | 0.54 | 0.00 | 0.03 | 0.03 | 0.11 |
| Clostridium sensu stricto 1 (Clostridia) | 0.90 | 0.14 | 0.00 | 0.65 | 0.09 | 0.66 | 0.00 | 0.14 | 0.00 | 0.00 |
| Corynebacterium (Actinobacteria) | 3.28 | 0.38 | 0.24 | 0.88 | 1.29 | 0.67 | 0.07 | 2.52 | 0.19 | 0.56 |
| Cutibacterium (Actinobacteria) | 18.85 | 0.92 | 1.01 | 2.05 | 3.50 | 2.87 | 1.02 | 6.41 | 0.64 | 0.93 |
| Enterococcus (Bacilli) | 0.16 | 0.00 | 10.56 | 0.64 | 0.17 | 2.50 | 0.39 | 0.46 | 0.06 | 0.63 |
| Lactobacillus (Bacilli) | 0.62 | 0.00 | 6.79 | 0.58 | 0.01 | 5.27 | 0.00 | 0.07 | 0.00 | 0.00 |
| Pseudonocardia (Actinobacteria) | 0.33 | 4.06 | 2.08 | 3.17 | 3.04 | 2.22 | 3.44 | 3.97 | 5.10 | 1.10 |
| Rhodococcus (Actinobacteria) | 5.65 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.02 | 0.00 | 0.00 |
| Sarcina (Clostridia) | 0.00 | 0.24 | 0.00 | 13.10 | 0.11 | 0.06 | 0.71 | 0.26 | 0.19 | 0.07 |
| Sphingomonas (Alphaproteobacteria) | 0.71 | 4.11 | 1.51 | 2.85 | 3.47 | 3.12 | 1.90 | 3.66 | 4.18 | 0.58 |
| Staphylococcus (Bacilli) | 12.36 | 1.07 | 0.31 | 1.19 | 1.82 | 3.26 | 1.58 | 2.86 | 0.49 | 0.59 |
| Wolbachia (Alphaproteobacteria) | 0.00 | 0.00 | 24.26 | 0.00 | 0.00 | 2.47 | 0.00 | 0.00 | 0.00 | 0.00 |
| Other bacteria (< 3 %) | 50.85 | 55.12 | 34.63 | 51.73 | 60.71 | 54.99 | 64.40 | 52.93 | 53.49 | 14.22 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





