Submitted:
05 January 2024
Posted:
08 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Isolation and Identification of Protease-Producing Strain
2.1.1. Sampling and Yeast Strains Isolation
2.1.2. Characterization and Identification of Strain PC3
2.2. Protease Production
2.2.1. Substrates Preparation
2.2.2. Fermentation
2.3. Production Optimization
2.4. Analytical Methods
2.4.1. Protease Activity Assay
2.4.2. Protein Assay
2.5. Purification of Protease
2.6. Biochemical Characterization of Clavispora lusitaniae PC3 Protease
2.6.1. Effect of Temperature on Protease Activity
2.6.2. Effects of pH on Protease Activity
2.6.3. Effects of Metal Ions and Chemicals on Enzyme Activity
2.7. Gluten Digestion
2.7.1. Proteolytic Degradation of Gluten
2.7.2. Gluten Determination
3. Results and Discussion
3.1. Isolation and Identification of Yeast Isolate
| Strain Code | Biochemical Characters (ID 32C) | Morphological Characteristics of the Selected Isolate | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Colony Characteristics | Cell Shape | Vegetative Reproduction | Mycelium (RAT) |
Sporulation | ||||||
| PC3 | 5157 3701 17 lac- et ESC+ |
Smooth, glistening, butyrous, White to cream colored, entire margin. | Sub-globose, ovoid to elongate | Budding (unipolar and bipolar) | Pseudo mycelium |
+ | ||||
| Protease production | Strain on YPGA | Microscopic Characteristics | ||||||||
 |
 |
 |
||||||||
| Blast results | ||||||||||
| Id sequences | Query (bp) | Species | Accession number | % max ident. | ||||||
| ITS_2156ZAB059 | ITS 692 | Clavispora lusitaniae strain CBS 6936 T | PP057739 | 676/676(100%) | ||||||
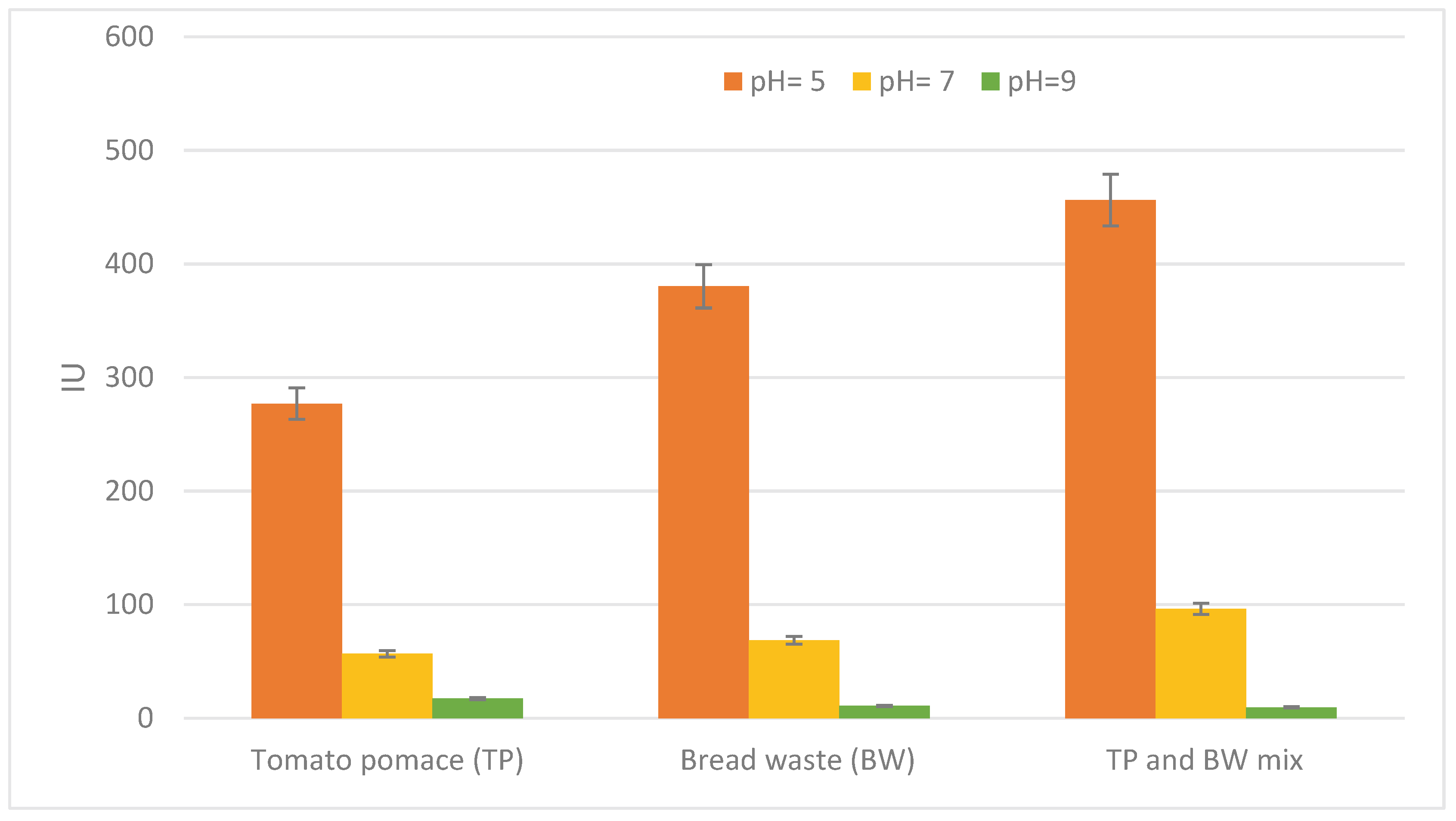
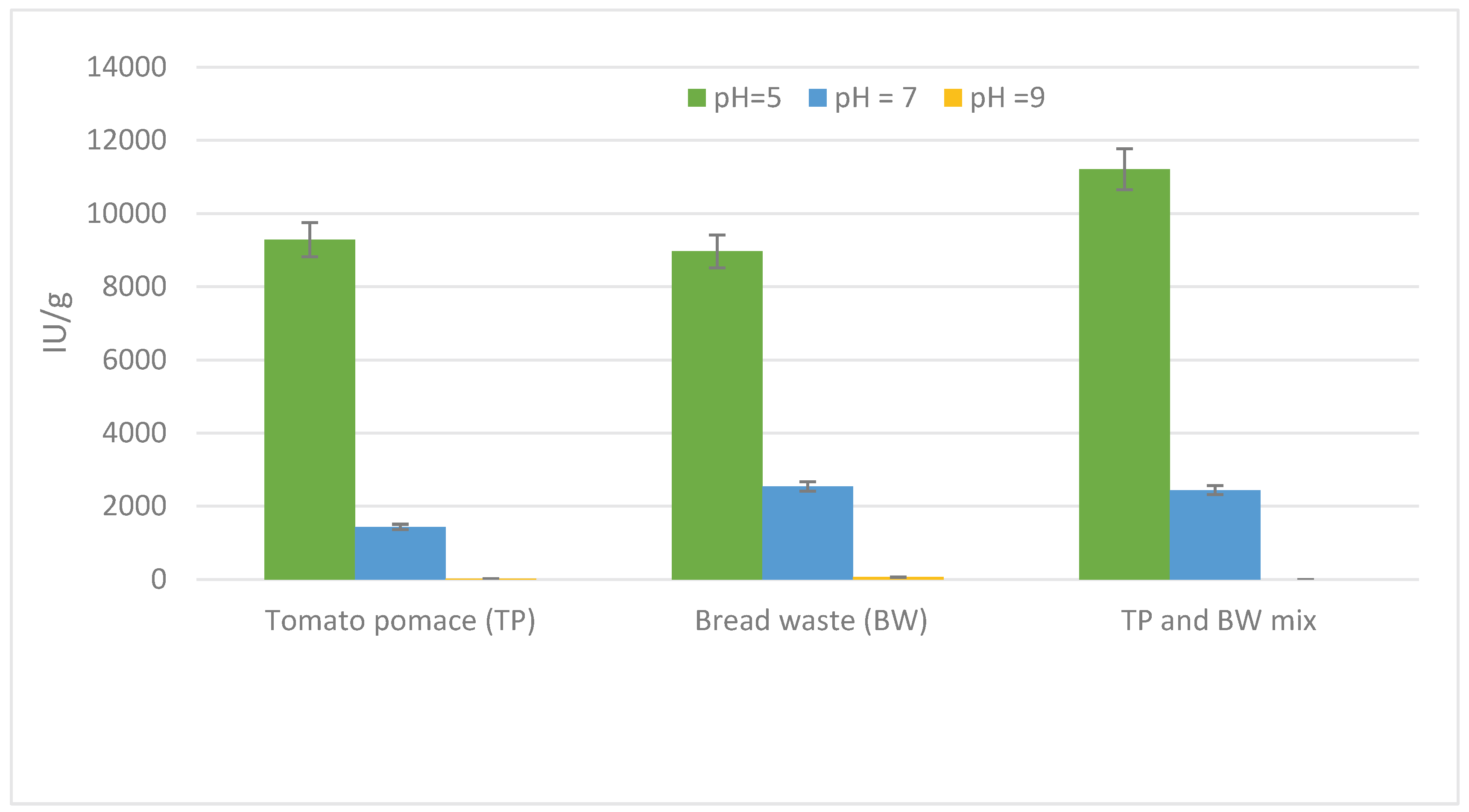
3.2. Fermentation and Substrate Selection
3.3. Optimization of Protease Production from Clavispora lusitaniae PC3
3.3.1. Screening of Significant Factors
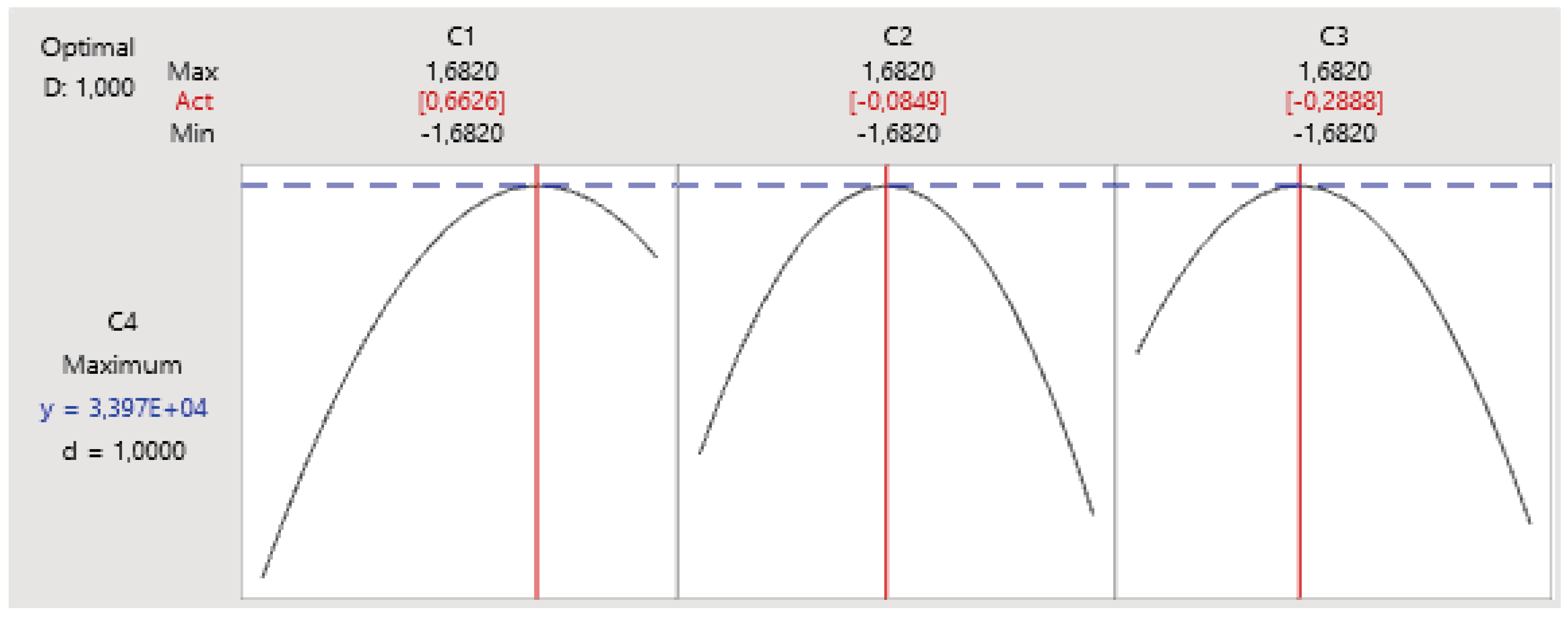
3.3.2. Determination of the Significant Factors' Optimum
| Source | DF | Sum of Squares | Mean Square | F-Value | P-Value |
|---|---|---|---|---|---|
| Model | 9 | 333643381 | 37071487 | 52,40 | 0,000 |
| Linear | 3 | 154916915 | 51638972 | 72,99 | 0,000 |
| B | 1 | 114548527 | 114548527 | 161,91 | 0,000 |
| C | 1 | 8457389 | 8457389 | 11,95 | 0,006 |
| J | 1 | 57185579 | 57185579 | 80,83 | 0,000 |
| Square | 3 | 173532796 | 57844265 | 81,76 | 0,000 |
| B2 | 1 | 46462590 | 46462590 | 65,67 | 0,000 |
| C2 | 1 | 113539208 | 113539208 | 160,48 | 0,000 |
| J2 | 1 | 70951960 | 70951960 | 100,29 | 0,000 |
| 2-factor interaction | 3 | 25845123 | 8615041 | 12,18 | 0,001 |
| B*C | 1 | 5888080 | 5888080 | 8,32 | 0,016 |
| B*J | 1 | 8721639 | 8721639 | 12,33 | 0,006 |
| C*J | 1 | 4624970 | 4624970 | 6,54 | 0,029 |
| Error | 10 | 7074816 | 707482 | ||
| Lack of fit | 6 | 5055665 | 842611 | 1,67 | 0,323 |
| Pure error | 4 | 2019151 | 504788 | ||
| Total | 19 | 340718197 |
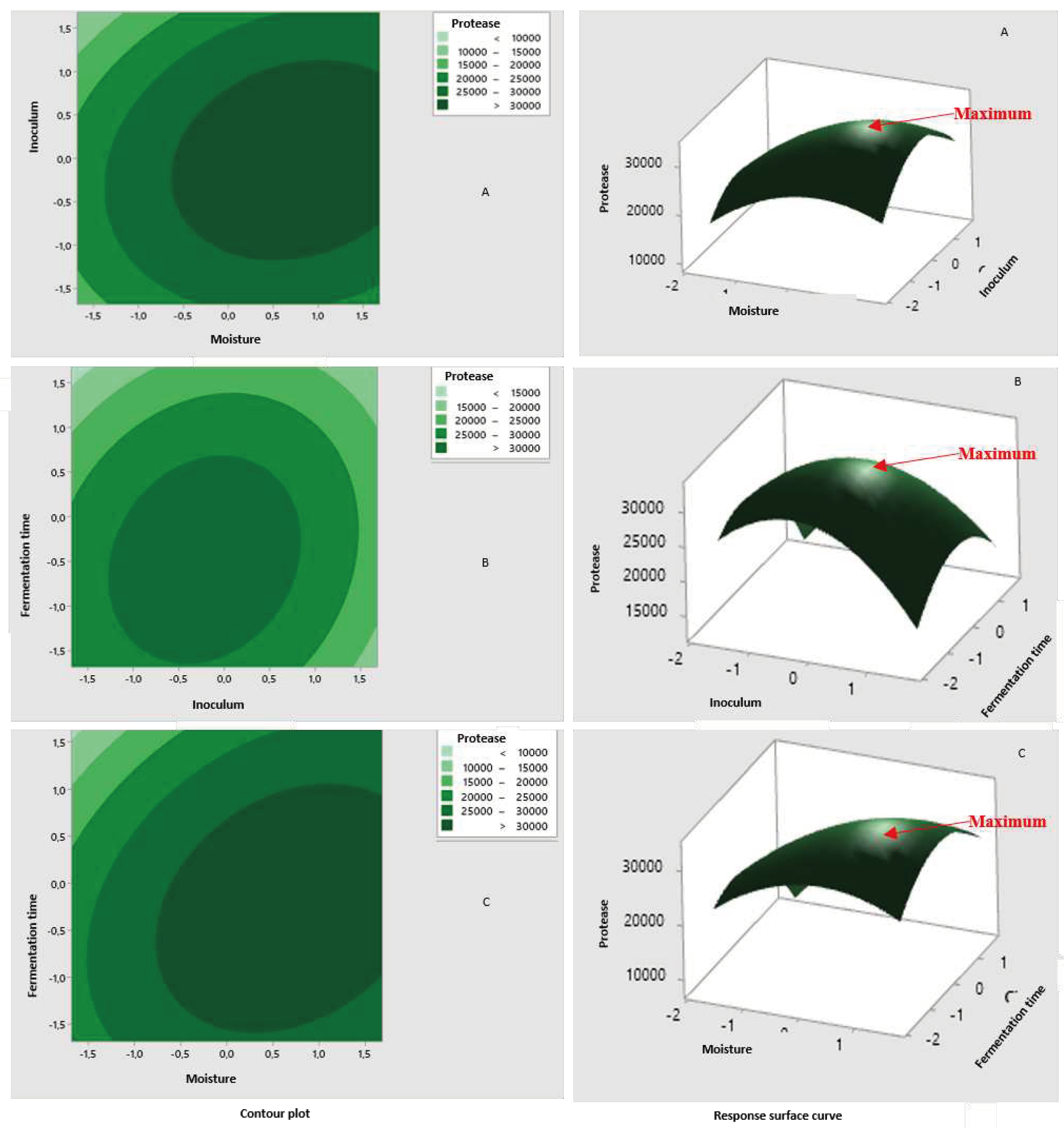
3.3.3. Moisture
3.3.4. Inoculum Size
3.3.5. Fermentation Time
3.4. Purification of Protease
3.5. Protease Characteristics
3.5.1. Effect of pH on Protease Activity
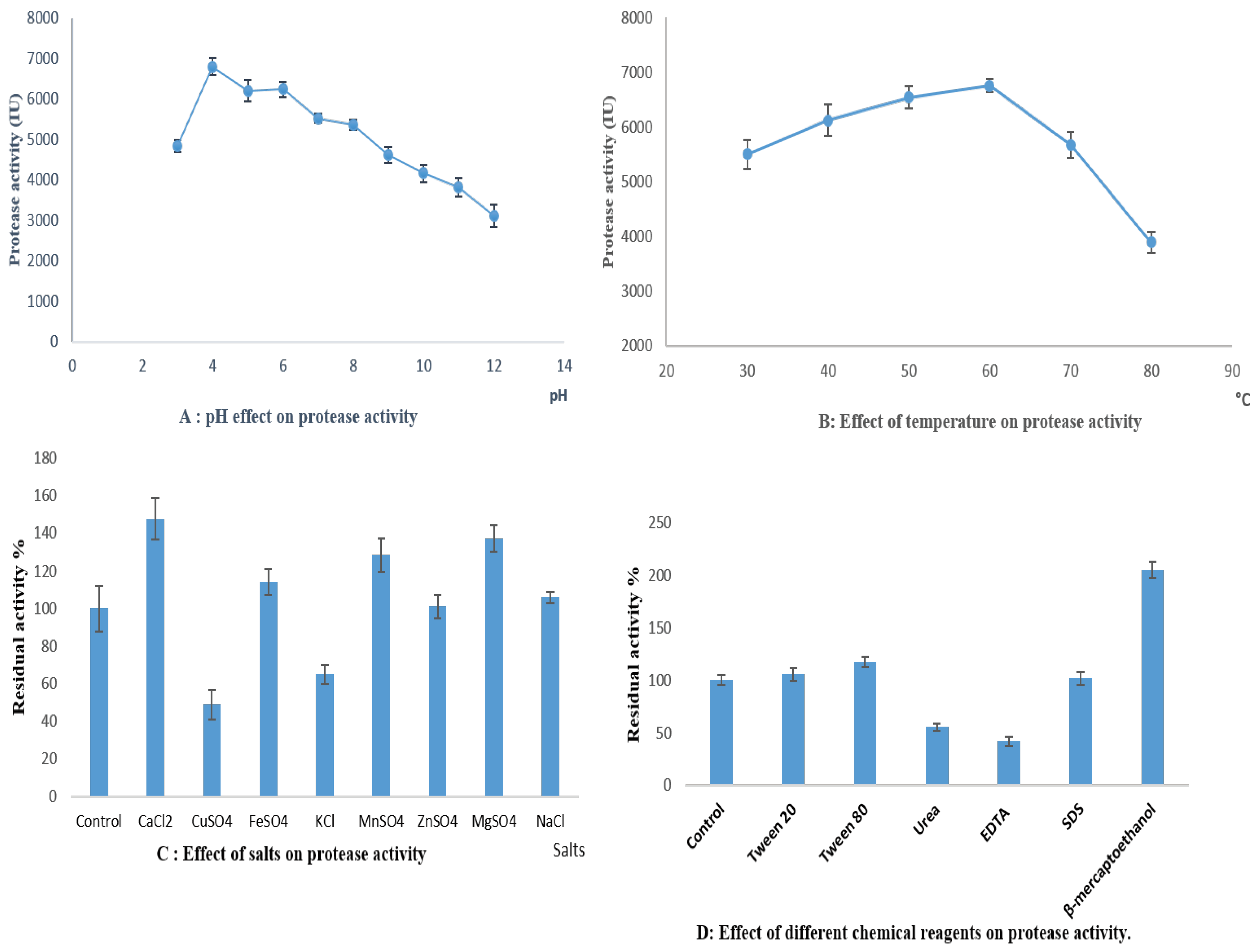
3.5.2. Effect of Temperature on Protease Activity
3.5.3. Effect of Metal Ions and Additives on Protease Activity
3.6. Action of Clavispora lusitaniae Protease on Gluten
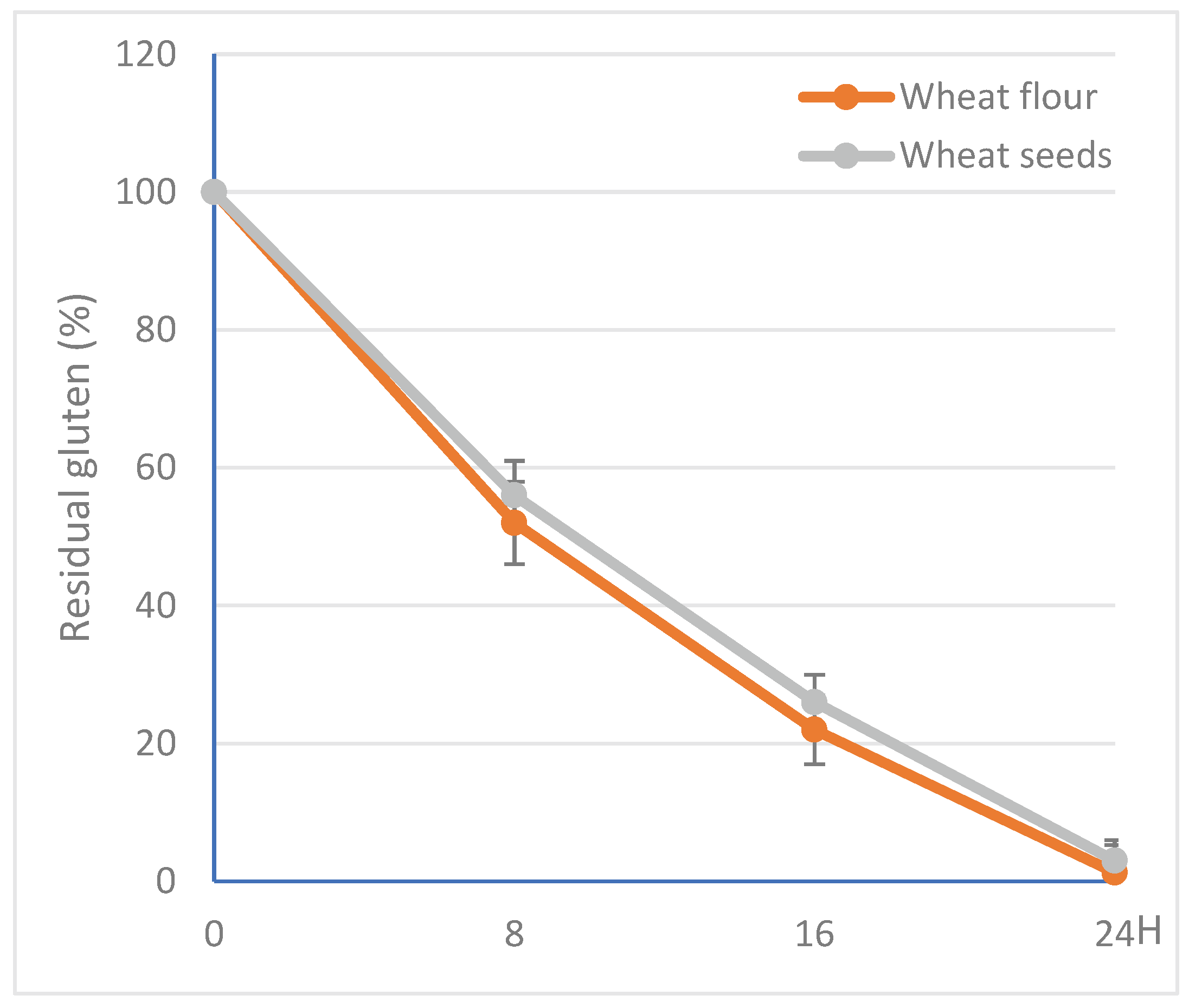
4. Conclusions
Author Contributions
Acknowledgments
Declaration of Competing Interest
References
- Nadeem, F.; Mehmood, T.; Naveed, M.; Shamas, S.; Saman, T.; Anwar, Z. Protease Production from Cheotomium globusum Through Central Composite Design Using Agricultural Wastes and Its Immobilization for Industrial Exploitation. Waste Biomass Valori. 2019, 12, 6529–6539. [Google Scholar] [CrossRef]
- Gimenes, N. C.; Silveira, E.; Tambourgi, E. B. An Overview of Proteases: Production, Downstream Processes and Industrial Applications. Sep. Purif. Rev. 2021, 50, 223–243. [Google Scholar] [CrossRef]
- Alahmad Aljammas, H.; Yazji, S.; Azizieh, A. Optimization of protease production from Rhizomucor miehei Rm4 isolate under solid-state fermentation. J. Genet. Eng. Biotechnol. 2022, 20, 82. [Google Scholar] [CrossRef]
- Bhavikatti, J. S.; Bodducharl, S. M.; Kamagond, R. S.; Desai, S. V.; Shet, A. R. Statistical optimisation of protease production using a freshwater bacterium Chryseobacterium cucumeris SARJS-2 for multiple industrial applications. 3 Biotech. 2020, 10, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Marathe, S.K.; Vashistht, M.A.; Prashanth, A.; Praveen, N.; Chakraborty, S.; Nair, S.S. Isolation, partial purification, biochemical characterization and detergent compatibility of alkaline protease produced by Bacillus subtilis, Alcaligenes faecalis and Pseudomonas aeruginosa obtained from sea water samples. J. Genet. Eng. Biotechnol. 2018, 16, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, N. D.; Barrett, A. J.; Thomas, P. D.; Huang, X.; Bateman, A.; Finn, R. D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the Panther database. Nucleic Acids Res. 2017, 46, D624–D632. [Google Scholar] [CrossRef] [PubMed]
- Song., P.; Zhang, X.; Wang, S.; Xu, W.; Wang, F.; Fu, R.; Wei, F. Microbial proteases and their applications. Front. Microbiol. 2023, 14, 1236368. [Google Scholar] [CrossRef] [PubMed]
- Aruna, V.; Chandrakala, V.; Angajala, G.; Nagarajan, E.R. Proteases: An overview on its recent industrial developments and current scenario in the revolution of biocatalysis. Mater. to day proc. 2023, 92, 565–573. [Google Scholar] [CrossRef]
- Balachandran, C.; Vishali, A.; Arun Nagendran, N.; Baskar, K.; Hashem, A.; Abd_Allah E., F. Optimization of protease production from Bacillus halodurans under solid state fermentation using agrowastes. Saudi J. Biol.Sci 2021, 28, 4263–4269. [Google Scholar] [CrossRef] [PubMed]
- Adetunji, A. I.; Olaitan, M. O.; Erasmus, M.; Olaniran A., O. Microbial proteases: A next generation green catalyst for industrial, environmental and biomedical sustainability. Food Mater. Res. 2023, 3, 1–15. [Google Scholar] [CrossRef]
- Solanki, P.; Putatunda, C.; Kumar, A.; Bhatia, R.; Walia, A. Microbial proteases: ubiquitous enzymes with innumerable uses. 3 Biotech 2021, 11, 428. [Google Scholar] [CrossRef] [PubMed]
- Hashmi, S.; Iqbal, S.; Ahmed, I.; Janjua, H. A. Production, Optimization, and Partial Purification of Alkali- Thermotolerant Proteases from Newly Isolated Bacillus subtilis S1 and Bacillus amyloliquefaciens KSM12. Processes 2022, 10, 6, 1050. [Google Scholar] [CrossRef]
- Wahab, W. A. A.; Ahmed, S. A. Response surface methodology for production, characterization and application of solvent, salt and alkali-tolerant alkaline protease from isolated fungal strain Aspergillus niger WA 2017. Int. J. Biol. Macromol 2018, 115, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Sharma, K. M.; Kumar, R.; Panwar, S.; Kumar, A. Microbial alkaline proteases: Optimization of production parameters and their properties. J. Genet. Eng. Biotechnol. 2017, 15, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Adetunji, A. I.; Olaitan, M. O.; Erasmus, M.; Olaniran, A. O. Microbial proteases: A next generation green catalyst for industrial, environmental and biomedical sustainability. Food Mater. Res. 2023, 3, 12. [Google Scholar] [CrossRef]
- Hesham, A. E. L.; Alrumman, S. A.; Al-Dayel, M. A.; Salah, H. A. Screening and genetic identification of acidic and neutral protease-producing yeasts strains by 26S rRNA gene sequencing. Cytol. Genet. 2017, 51, 221–229. [Google Scholar] [CrossRef]
- Bennamoun, L.; Hiligsmann, S.; Dakhmouche, S.; Ait-Kaki, A.; Labbani, F.-Z. K.; Nouadri, T.; Meraihi, Z.; Turchetti, B.; Buzzini, P.; Thonart, P. Production and Properties of a Thermostable, pH—Stable Exo-Polygalacturonase Using Aureobasidium pullulans Isolated from Saharan Soil of Algeria Grown on Tomato Pomace. Foods 2016, 5, 72. [Google Scholar] [CrossRef]
- Plackett, R. L.; Burman, J. P. The design of optimum multifactorial experiments. Biometrika 1956, 33, 305–325. [Google Scholar] [CrossRef]
- Box, G. E. P.; Wilson, K. B. On the Experimental Attainment of Optimum Conditions. J. R. Stat. Soc. Ser. B (Methodological) 1951, 1, 1–38. [Google Scholar] [CrossRef]
- López-Trujillo, J.; Mellado-Bosque, M.; Ascacio-Valdés, J.A.; Prado-Barragán, L.A.; Hernández-Herrera, J.A.; Aguilera-Carbó, A.F. Temperature and pH optimization for protease production fermented by Yarrowia lipolytica from Agro-Industrial Waste. Fermentation 2023, 9, 819. [Google Scholar] [CrossRef]
- Ogrydziak, D. M. Yeast Extracellular Proteases. Crit. Rev. Biotechnol. 1993, 13, 1–55. [Google Scholar] [CrossRef]
- Nakamura, Y.; Fukuhara, H.; Sano, K. Secreted phytase activities of yeasts. Biosci. Biotechnol. Biochem. 2000, 64, 841–844. [Google Scholar] [CrossRef]
- Dakhmouche Djekrif, S.; Bennamoun, L.; Labbani, F.-Z.K.; Ait Kaki, A.; Nouadri, T.; Pauss, A.; Meraihi, Z.; Gillmann, L. An Alkalothermophilic Amylopullulanase from the Yeast Clavispora lusitaniae ABS7. Purification, Characterization and Potential Application in Laundry Detergent. Catalysts 2021, 11, 12. [Google Scholar] [CrossRef]
- Ranjan, K.; Ahmad, L.; Sahay, S.M. Detergent compatible cold-active alkaline amylases from Clavispora lusitaniae CB13. J. Microbiol. Biotechnol. Food Sci. 2016, 5, 306–310. [Google Scholar] [CrossRef]
- Mohan Kumar, B.V.; Sarabhai, S.; Prabhasankar, P. Targeted degradation of gluten proteins in wheat flour by prolyl endoprotease and its utilization in low immunogenic pasta for gluten sensitivity population. J. Cereal Sci. 2019, 87, 59–67. [Google Scholar] [CrossRef]
- Shaikh, I. A.; Turakani, B.; Malpani, J.; Goudar, S. V.; Mahnashi, M. H.; Hamed Al-Serwi, R.; et al. Extracellular protease production, optimization, and partial purification from Bacillus nakamurai PL4 and its applications. J. King Saud. Univ. Sci. 2023, 35, 102429. [Google Scholar] [CrossRef]
- Svensson, S.E.; Bucuricova, L.; Ferreira, J.A.; Souza Filho, P.F.; Taherzadeh, M.J.; Zamani, A. Valorization of Bread Waste to a Fiber and Protein-Rich Fungal Biomass. Fermentation 2021, 7, 91. [Google Scholar] [CrossRef]
- Benkahoul, M.; Bramki, A.; Belmessikh, A.; Mechakra-Maza, A. Statistical optimization of an acid protease production by a local Aspergillus niger MH109542 using a medium based on decommissioned dates. Asia Pac. J. Mol. Biol. Biotechnol. 2020, 28, 68–76. [Google Scholar] [CrossRef]
- Djekrif-Dakhmouche, S.; Gheribi-Aoulmi, Z.; Meraihi, Z.; Bennamoun, L. Application of a statistical design to the optimization of culture medium for α-amylase production by Aspergillus niger ATCC 16404 grown on orange waste powder. J. Food Eng. 2006, 73, 190–197. [Google Scholar] [CrossRef]
- Shrestha, S.; Chio, C.; Khatiwada, J. R.; Kognou, A. L. M.; Qin, W. Optimization of multiple enzymes production by fermentation using lipid-producing Bacillus sp. Front. Microbiol. 2022, 13, 1049692. [Google Scholar] [CrossRef] [PubMed]
- Shafique, T.; Shafique, J.; Zahid, S.; Kazi, M.; Alnemer, O.; Ahmad, A. Screening, selection and development of Bacillus subtilis apr-IBL04 for hyper production of macromolecule alkaline protease. Saudi J. Biol. Sci. 2021, 28, 1949–1951. [Google Scholar] [CrossRef] [PubMed]
- Lowry, O. H.; Rosebrough, N. J.; Farr, A.; Randall, R.J. Protein measurement with the folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Luoto, S.; Jiang, Z.; Brinck, O.; Sontag-Strohm, T.; Kanerva, P.; Bruins, M.; Edens, L.; Salovaara, H.; Loponen, J. Malt hydrolysates for gluten-free applications: autolytic and proline endopeptidase assisted removal of prolamins from wheat, barley and rye. J. Cereal Sci. 2012, 56, 504–509. [Google Scholar] [CrossRef]
- Sayaslan, A. Wet-milling of wheat flour: industrial processes and small scale test methods. LWT. Food Sci. Technol. 2004, 37, 499–515. [Google Scholar] [CrossRef]
- Lario, L. D.; Pillaca-Pullo, O. S.; Sette, L. D.; Converti, A.; Casati, P.; Spampinato, C.; Pessoa, A. Optimization of protease production and sequence analysis of the purifed enzyme from the cold adapted yeast Rhodotorula mucilaginosa CBMAI 1528. Biotechnol. Rep. 2020, 28, e00546. [Google Scholar] [CrossRef] [PubMed]
- Kim, J. Y. Isolation of the Protease-producing Yeast Pichia anomala CO-1 and Characterization of Its Extracellular Neutral Protease. J. Life Sci. 2019, 29, 1126–1135. [Google Scholar] [CrossRef]
- Ray, M. K.; Uma Devi, K.; Seshu Kumar, G.; Shivaji, S. Extracellular Protease from the Antarctic Yeast Candida humicola. Appl. Environ. Microbiol. 1992, 58, 1918–1923. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Lopez, C. I.; Szabo, R.; Blanchin-Roland, S.; Gaillardin, C. Genetic control of extracellular protease synthesis in the yeast Yarrowia lipolytica. Genetics 2002, 160, 417–427. [Google Scholar] [CrossRef]
- Theron, L.W.; Divol, B. Microbial aspartic proteases: Current and potential applications in industry. Appl. Microbiol. Biotechnol 2014, 98, 8853–8858. [Google Scholar] [CrossRef]
- Schlander, M.; Distler, U.; Tenzer, S.; Thines, E.; Claus, H. Purification and Properties of Yeast Proteases Secreted by Wickerhamomyces anomalus 227 and Metschnikovia pulcherrima 446 during Growth in a White Grape Juice. Fermentation 2016, 3, 2. [Google Scholar] [CrossRef]
- Fernández, M.; Úbeda, J.F.; Briones, A.I. Typing of non-Saccharomyces yeasts with enzymatic activities of interest in wine-making. Int. J. Food. Microbiol. 2000, 59, 29–36. [Google Scholar] [CrossRef]
- Abdelal, A. T. H.; Kennedy, E. H.; Ahearn, G. Purification and characterization of a neutral protease from Saccharomycopsis lipolytica. J. Bacteriol. 1977, 130, 1125–1129. [Google Scholar] [CrossRef] [PubMed]
- Ruchel, R.; Tegeler, R.; Trost, M. A comparison of secretory proteinases from different strains of Candida albicans. Sabouraudia 1982, 20, 233–244. [Google Scholar] [CrossRef] [PubMed]
- James, M.N. Catalytic pathway of aspartic peptidases. In Handbook of Proteolytic Enzymes, 2nd ed.; Beynon, R., Bond, J.S., Eds.; Oxford University Press: Oxford, UK, 2004; pp. 12–18. [Google Scholar] [CrossRef]
- Tunga, R.; Shrivastava, B.; Banerjee, R. Purification and characterization of a protease from solid state cultures of Aspergillus parasiticus. Process Biochem 2003, 38, 1553–1558. [Google Scholar] [CrossRef]
- Wang, R.; Law, R.C.; Webb, C. Protease production and conidiation by Aspergillus oryzae in flour fermentation. Process Biochem. 2005, 40, 217–227. [Google Scholar] [CrossRef]
- Chaud, L. C. S.; Lario, L. D.; Bonugli-Santos, R. C.; Sette, L. D.; Pessoa Junior, A.; Felipe, M.; das Graças, A. Improvement in extracellular protease production by the marine antarctic yeast Rhodotorula mucilaginosa L7. N. Biotechnol. 2016, 33, 807–814. [Google Scholar] [CrossRef] [PubMed]
- Barnett, J. A.; Payne, R. W.; Yarrow, D. Yeasts Characteristics and Identification; Cambridge University Press: London, 1983. [Google Scholar]
- Murao, S.; Kamada, M.; Nakase, T.; Ogura, S.; Oda, K. Studies on the Extracellular Protease of Yeasts Part I. J. agric. chem. Soc. Jpn. 1972, 46, 167–170. [Google Scholar] [CrossRef]
- Tennalli, G. B.; Garawadmath, S.; Sequeira, L.; Murudi, S.; Patil, V.; Divate, M. N.; Hungund, B. S. Media optimization for the production of alkaline protease by Bacillus cereus PW3A using response surface methodology. J. Appl. Biol. Biotechnol. 2022, 10, 17–26. [Google Scholar] [CrossRef]
- Ferracini-Santos, L.; Sato, H. H. Production of alkaline protease from Cellulosimicrobium cellulans. Braz. J. Microbiol. 2009, 40, 54–60. [Google Scholar] [CrossRef]
- Adetunji, A. I.; Olaniran, A. O. Statistical modelling and optimization of protease production by an autochthonous Bacillus aryabhattai Ab15-ES: A response surface methodology approach. Biocatal. Agric. Biotechnol. 2020, 24, 101528. [Google Scholar] [CrossRef]
- Moftah, O. A. S.; Grbavčić, S.; Žuža, M.; Luković, N.; Bezbradica, D.; Knežević-Jugović, Z. Adding Value to the Oil Cake as a Waste from Oil Processing Industry: Production of Lipase and Protease by Candida utilis in Solid State Fermentation. Appl. Biochem. Biotechnol. 2012, 166, 348–364. [Google Scholar] [CrossRef]
- Sindhu, R.; Suprabha, G. N.; Shashidhar, S. Optimization of process parameters for the production of alkaline protease from Penicillium godlewskii SBSS 25 and its application in detergent industry. Afr. J. Microbiol. Res. 2009, 3, 515–522. [Google Scholar]
- Benabda, O.; M’hir, S.; Kasmi, M.; Mnif, W.; Hamdi, M. Optimization of Protease and Amylase Production by Rhizopus oryzae Cultivated on Bread Waste Using Solid-State Fermentation. J. Chem. 2019, 3738181. [Google Scholar] [CrossRef]
- Agrawal, D.; Patidar, P.; Banerjee, T.; Patil, S. Alkaline protease production by a soil isolate of Beauveria feline under SSF condition: parameter optimization and application to soy protein hydrolysis. Process Biochem. 2005, 40, 1131–1136. [Google Scholar] [CrossRef]
- Elmansy, E. A.; Asker, M. S.; El-Kady, E. M.; Hassanein, S. M.; El-Beih, F. M. Production and optimization of α-amylase from thermo-halophilic bacteria isolated from different local marine environments. Bull. Natl. Res. Cent. 2018, 42, 31. [Google Scholar] [CrossRef]
- Takeyama, M. M.; de Carvalho, M. C.; Carvalho, H. S; et al. Pectinases Secretion by Saccharomyces cerevisiae: Optimization in Solid-State Fermentation and Identification by a Shotgun Proteomics Approach. Molecules 2022, 27, 4981. [Google Scholar] [CrossRef] [PubMed]
- Vishwanatha, K. S.; Appu Rao, A. G.; Singh S., A. Acid protease production by solid-state fermentation using Aspergillus oryzae MTCC 5341: optimization of process parameters. J. Ind. Microbiol. Biotechnol. 2010, 37, 129–138. [Google Scholar] [CrossRef]
- Arekemase, M.O.; Alfa, O.P.; Agbabiaka, T.O.; Ajide-Bamigboye, N.T.; Aderoboye, O.Y.; Orogu, J.O.; Ahmed, T. Optimization of amylase produced from bacteria isolated from cassava peel dumpsite using submerged fermentation. Sci. World J. 2020, 15, 64–75. [Google Scholar]
- Rozanov, A. S.; Shekhovtsov, S. V.; Bogacheva, N. V.; Pershina, E. G.; Ryapolova, A.V.; Bytyak, D. S.; Peltek, S. E. Production of subtilisin proteases in bacteria and yeast. Vavilovskii Zh. Genet 2021, 25, 125–134. [Google Scholar] [CrossRef]
- Usman, A.; Mohammed, S.; Mamo, J. Production, optimization, and characterization of an acid protease from a filamentous fungus by solid-state fermentation. Int. J. Microbiol. 2021, 6685963. [Google Scholar] [CrossRef]
- Chimbekujwo, K. I.; Ja’afaru, M. I.; Adeyemo, O. M. Purification, characterization and optimization conditions of protease produced by Aspergillus brasiliensis strain BCW2. Sci. Afr. 2020, 8, e00398. [Google Scholar] [CrossRef]
- Lario, L. D.; Chaud, L.; Almeida M., G.; Converti, A.; Sette L., D.; Pessoa Jr, A. Production, purification, and characterization of an extracellular acid protease from the marine Antarctic yeast Rhodotorula mucilaginosa L7. Fungal Biol. 2015, 119, 1129–1136. [Google Scholar] [CrossRef] [PubMed]
- Mota de Oliveira, J.; Fernandes, P.; Benevides, R. G.; Aparecida de Assis, S. Production, characterization, and immobilization of protease from the yeast Rhodotorula oryzicola. Biotechnol. Appl. Biochem. 2021, 68, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Remold, H.; Fasold, H.; Staib, F. Purification and characterization of a proteolytic enzyme from Candida albicans. Biochim. Biophys. Acta 1968, 167, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Nelson, G.; Young, T. W. Extracellular acid and alkaline proteases from Candida olea. J. Gen. Microbiol. 1987, 133, 1461–1469. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.; Ogrydziak, O.M. Extracellular acid proteases produced by Saccharomycopsis lipolytica. J. Bacteriol. 1983, 154, 23–31. [Google Scholar] [CrossRef]
- Kamada, M.; Oda, K.; Murao, S. The purification of the extracellular acid protease of Rhodotorula glutinis K-24 and its general properties. Agric. Biol. Chem. 1972, 36, 1095e1101. [Google Scholar] [CrossRef]
- Srividya, S. Production and characterization of an acid protease from a local Aspergillus sp. By solid substrate fermentation. Arch. Appl. Sci. Res. 2012, 1, 188–199. [Google Scholar]
- Wei, M.; Chen, P.; Zheng, P.; et al. Purification and characterization of aspartic protease from Aspergillus niger and its efficient hydrolysis applications in soy protein degradation. Microb. Cell Factories 2023, 22, 42. [Google Scholar] [CrossRef]
- Purushothaman, K.; Bhat, S. K.; Annapurna Singh, S.; Kedihithlu Marathe, G.; Appu Rao, G.; Appu, R. Aspartic protease from Aspergillus niger: Molecular characterization and interaction with pepstatin A, Int. J. Biol. Macromol. 2019, 139, 199–212. [Google Scholar] [CrossRef]
- Mohan Kumar, B.V.; Sarabhai, S.; Prabhasankar, P. Targeted degradation of gluten proteins in wheat flour by prolyl endoprotease and its utilization in low immunogenic pasta for gluten sensitivity population. J. Cereal Sci. 2019, 87, 59–67. [Google Scholar] [CrossRef]
- Razzaq, A.; Shamsi, S.; Ali, A.; Ali, Q.; Sajjad, M.; Malik, A.; Ashraf, M. Microbial Proteases Applications. Front. Bioeng. Biotechnol. 2019, 7, 110. [Google Scholar] [CrossRef]
- Mamo, J.; Assefa, F. The role of microbial aspartic protease enzyme in food and beverage industries. J. Food Qual. 2018, 2018, 7957269. [Google Scholar] [CrossRef]
- Mandujano-Gonzalez, V.; Villa-Tanaca, L.; Anducho-Reyes, M. A.; Mercado-Flores, Y. Secreted fungal aspartic proteases: a review. Rev. Iberoam Micol. 2016, 33, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.L.; Zhang, Y. Z.; Gao, P. J.; Luan, X.W. Two different proteases produced by a deep-sea psychrophic bacterial strain, Pseudoalteromonas sp SM9913. Mar. Biol. 2003, 143, 989–993. [Google Scholar] [CrossRef]
- Kim, J.I.; Lee, S.M.; Jung, H.J. Characterization of calcium activated bifunctional peptidase of the psychrotrophic Bacillus cereus. J. Microbiol. 2005, 43, 237–243. [Google Scholar] [PubMed]
- Vazquez, S.; Ruberto, L.; Mac Cormack, W. Properties of extracellular proteases from three psychrotolerant Stenotrophomonas maltophilia isolated from Antarctic soil. Polar Biol. 2005, 28, 319–325. [Google Scholar] [CrossRef]
- Aguilar, dos S. JG; Sato, H. H. Microbial proteases: Production and application in obtaining protein hydrolysates. Food Res. Int. 2018, 103, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, W. C. A.; Martins, M. L. L. Production and properties of an extracellular protease from thermophilic Bacillus sp. Braz. J. Microbiol. 2004, 35, 91–96. [Google Scholar] [CrossRef]
- Gaur, S.; Agrahari, S.; Wadhwa, N. Purification of Protease from Pseudomonas thermaerum GW1 Isolated from Poultry Waste Site. Open Microbiol. J. 2010, 4, 67–74. [Google Scholar] [CrossRef]
- Devi, M. K.; Banu, A. R.; Gnanaprabhal, G. R.; Pradeep, B.V.; Palaniswamy, M. Purification, characterization of alkaline protease enzyme from native isolate Aspergillus niger and its compatibility with commercial detergents. Indian J. Sci. Technol. 2008, 1, 1–6. [Google Scholar] [CrossRef]
- Sharma, N.; Bhatia, S.; Chunduri, V.; Kaur, S.; Sharma, S.; Kapoor, P.; Kumari, A.; Garg, M. Pathogenesis of celiac disease and other gluten related disorders in wheat and strategies for mitigating them. Front. Nutr. 2020, 7, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Walter, T.; Wieser, H.; Koehler, P. Degradation of gluten in wheat bran and bread drink by means of a proline-specific peptidase. J. Nutr. Food Sci. 2014, 4, 2–15. [Google Scholar] [CrossRef]
- Heredia-Sandoval, N.G.; de la Barca, A.C.; Carvajal-Millán, E.; Islas-Rubio, A.R. Amaranth addition to enzymatically modified wheat flour improves dough functionality, bread immunoreactivity and quality. Food Funct 2018, 9, 534–540. [Google Scholar] [CrossRef] [PubMed]
| Factors | Level | Unit | |
|---|---|---|---|
| -1 | +1 | ||
| A:Temperature | 30 | 50 | °C |
| B:Moisture | 50 | 80 | % |
| C:Inoculum | 104 | 108 | Cells/g |
| D:Error | - | - | - |
| E:Glucose | 0 | 0.5 | % |
| F:Corn Steep Liquor | 0 | 0.5 | % |
| G:(NH4)2SO4 | 0 | 0.5 | % |
| H:Error | - | - | - |
| I:NaCl | 0 | 0.1 | g/l |
| J:Time fermentation | 24 | 72 | H |
| K:Error | - | - | - |
| -1.682 | -1 | 0 | +1 | +1.682 | |
|---|---|---|---|---|---|
| B | 53,18 | 60 | 70 | 80 | 86,82 |
| C | 2,08 x 105 | 106 | 107 | 108 | 0,5 x109 |
| J | 39.81 | 48 | 60 | 72 | 80.18 |
| B: Moisture (%), | C: Inoculum (Cells/ml), | J: Fermentation time (H) | |||
| Experiments | A | B | C | (D) | E | F | G | (H) | I | J | (K) | Protease Activity (IU/g) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | +1 | +1 | -1 | +1 | +1 | +1 | -1 | -1 | -1 | +1 | -1 | 18305,3 |
| 2 | -1 | +1 | +1 | -1 | +1 | +1 | +1 | -1 | -1 | -1 | +1 | 6567,55 |
| 3 | +1 | -1 | +1 | +1 | -1 | +1 | +1 | +1 | -1 | -1 | -1 | 13621,2 |
| 4 | -1 | +1 | -1 | +1 | +1 | -1 | +1 | +1 | +1 | -1 | -1 | 9917,55 |
| 5 | -1 | -1 | +1 | -1 | +1 | +1 | -1 | +1 | +1 | +1 | -1 | 15332,15 |
| 6 | -1 | -1 | -1 | +1 | -1 | +1 | +1 | -1 | +1 | +1 | +1 | 25011,8 |
| 7 | +1 | -1 | -1 | -1 | +1 | -1 | +1 | +1 | -1 | +1 | +1 | 30662,8 |
| 8 | +1 | +1 | -1 | -1 | -1 | -1 | +1 | -1 | +1 | -1 | +1 | 12932,05 |
| 9 | +1 | +1 | +1 | -1 | -1 | -1 | +1 | -1 | +1 | +1 | -1 | 21830,3 |
| 10 | -1 | +1 | +1 | +1 | -1 | -1 | -1 | +1 | -1 | +1 | +1 | 15173,05 |
| 11 | +1 | -1 | +1 | +1 | +1 | -1 | -1 | -1 | +1 | -1 | +1 | 8991,2 |
| 12 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | 14138,3 |
| A, B, C, E, F, G, I and J are real variables | ||||||||||||
| D, H and K are dummy variables | ||||||||||||
| Variables | Effect | Coefficient | t-Value | P-Value | Confidence Level % |
|---|---|---|---|---|---|
| Constant | - | 16040 | 26.76 | 0 | - |
| A : Temperature | 3367 | 1684 | 2.81 | 0.067 | 93.3 |
| B : Humidity | -3839 | -1919 | -3.2 | 0.049* | 95.1* |
| C : Inoculum | -4909 | -2454 | -4.09 | 0.026* | 97.4* |
| E : Glucose | -2155 | -1078 | -1.8 | 0.17 | 83 |
| F: Corn steep liquor | -1491 | -745 | -1.24 | 0.302 | 69.8 |
| G : (NH4)2SO4 | 3790 | 1895 | 3.16 | 0.051 | 94.9 |
| I : NaCl | -742 | -371 | -0.62 | 0.058 | 94.2 |
| J : Incubation period | 10025 | 5012 | 8.36 | 0.004* | 99.6* |
| Source | DF | Sum of Squares | Mean Square | F-Value | p-Value |
|---|---|---|---|---|---|
| Model | 8 | 517319398 | 64664925 | 15,00 | 0.024 |
| Error | 3 | 12935581 | 431477314 | ||
| Total | 11 | 530254980 | |||
|
R2 = 0.9756; Adjusted-R2 = 0.9106. | |||||
| Run | B: Moisture | C: Inoculum | J: Fermentation Time | Protease Activity (IU/g) |
|---|---|---|---|---|
| 1 | 1,682 | 0 | 0 | 31856,3 |
| 2 | 0 | 0 | 0 | 33831,6 |
| 3 | 0 | 0 | 1,682 | 21550,6 |
| 4 | 1 | 1 | -1 | 28422,3 |
| 5 | 1 | -1 | 1 | 26156,6 |
| 6 | 0 | 0 | 0 | 31879,4 |
| 7 | 1 | -1 | -1 | 30514,1 |
| 8 | 0 | 0 | 0 | 32626,5 |
| 9 | -1 | -1 | 1 | 19636,1 |
| 10 | -1 | -1 | -1 | 27337,0 |
| 11 | 0 | 0 | 1 | 29101,4 |
| 12 | -1 | 1 | 0 | 22548,0 |
| 13 | 0 | 0 | 0 | 32492,0 |
| 14 | 0 | -1,682 | 0 | 25896,2 |
| 15 | 0 | 1,682 | 0 | 23030,7 |
| 16 | -1,682 | 0 | -1 | 24251,0 |
| 17 | -1 | 1 | 0 | 22731,3 |
| 18 | 0 | 0 | -1 | 31088,8 |
| 19 | 1 | 1 | 1 | 28721,6 |
| 20 | 0 | 0 | -1,682 | 29864,7 |
| Sample | Total Activity (U) | Total Protein (mg) | Specific Activity (U/mg) | Purification Fold | Yield (%) |
|---|---|---|---|---|---|
| Crude extract | 31589.2 | 168 | 188.03 | 1 | 100 |
| Ammonium sulfate (60%) | 29276.1 | 53 | 552.4 | 2.938 | 92.67 |
| Dialysis | 25821.5 | 28 | 922.2 | 4.904 | 81.74 |
| Sephadex G100 | 10406 | 2.8 | 3716.4 | 19.76 | 32.94 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





