Submitted:
28 December 2023
Posted:
04 January 2024
You are already at the latest version
Abstract
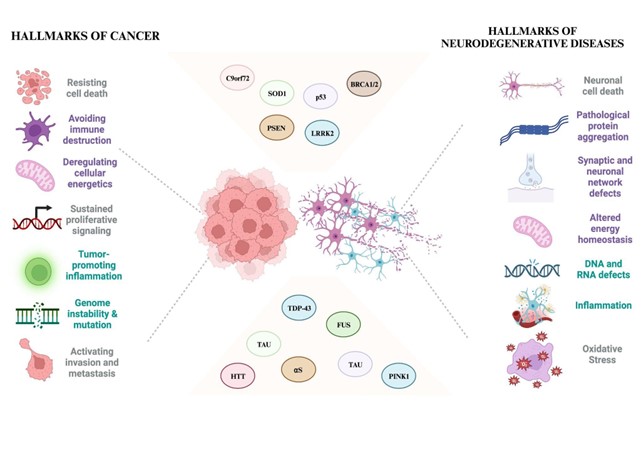
Keywords:
1. Introduction
1.1. Converging Pathways: Unveiling Shared Risk Factors in Cancer and Neurodegeneration
1.2. Parallel Pathogenesis, Divergent Destinies: Double Hit
2. Exploring Genetic Intersections: Parkinson's Associated Genes and Potential Links to Cancer Pathways
3. Exploring Genetic Intersections: Alzheimer's Associated Genes and Potential Links to Cancer Pathways
4. Exploring Genetic Intersections: amyotrophic lateral sclerosis (ALS)'s-Associated Genes and Potential Links to Cancer Pathways
5. Exploring Genetic Intersections: Huntington's Disease’s Associated Genes and Potential Links to Cancer Pathways
| GENE | FUNCTION | ROLE IN NEURODEGENERATION | ROLE IN CANCER |
|---|---|---|---|
|
HTT |
Signaling, transporting materials,binding proteins and other structures, and protecting against apoptosis. |
Primary role is linked to neuronal damage in HD. | Alterations in the HTT gene could impact cell survival, DNA repair mechanisms, or cellular processes relevant to tumorigenesis |
|
BRCA1 BRCA2 |
The BRCA1 protein has multiple functions in different cellular processes, including DNA repair, transcriptional activation, cell cycle regulation and chromatin remodeling. BRCA2 plays a role in transcriptional and cell cycle regulation, DNA repair, mitophagy and replication fork stabilization. |
Direct links between BRCA1/2 mutations and the disease are not well-established; exploring their functions in DNA repair mechanisms may shed light on potential intersections with HD pathology. |
Mutations in BRCA1 and BRCA2 significantly elevate the risk of developing breast, ovarian, and other cancers. |
| TDP-43 | Cell cycle regulation | Abnormal aggregates of TDP-43 have been detected in the brain tissues of individuals with the disease. However, the exact role of TDP-43 in the development or progression of HD is still being researched | Alterations in TDP-53 expression or function have been observed in various cancer types, affecting cellular processes like RNA metabolism, splicing, and stability. Dysregulation of TDP-53 has been linked to tumor growth, invasion, and metastasis in some cancers. |
6. Conclusion and Future Prospective
Abbreviations
- ❖ Aβ: amyloid-β
- ❖ AD: Alzheimer's disease
- ❖ αS: α-synuclein
- ❖ PD: Parkinson's disease
- ❖ PTEN: phosphatase and tensin homolog
- ❖ PINK1 or PARK6: PTEN-induced kinase 1
- ❖ LRRK2 or PARK8: leucine-rich repeat kinase 2
- ❖ MAPT: microtubule-associated protein tau
- ❖ APP: amyloid precursor protein
- ❖ PSEN1/2: presenilin 1/2
- ❖ CDK5: cyclin-dependent kinase 5
- ❖ HD: Huntington's disease
- ❖ siRNA: short-interfering RNA
- ❖ L-DOPA: levodopa
- ❖ PolyQ: polyglutamine
- ❖ Huntingtin (HTT)
- ❖ BRCA1: breast cancer type 1
- ❖ BRCA2: breast cancer type 2
- ❖ APOE: Apolipoprotein E
- ❖ TP53: tumor protein p53
- ❖ Presenilin 1: PSEN
- ❖ Presenilin 2: PSEN2
- ❖ Amyloid precursor protein: APP
- ❖ Beta-secretase 1: BACE1
- ❖ PTEN-induced putative kinase 1: PINK1
- ❖ Caspase-3: CASP3
- ❖ Amyotrophic lateral sclerosis: ALS
- ❖ Fused in Sarcoma: FUS
- ❖ Superoxide dismutase 1: SOD1
Author Contributions
Acknowledgments
References
- Driver, J.A. Understanding the link between cancer and neurodegeneration. Journal of Geriatric Oncology 2012, 3, 58–67. [Google Scholar] [CrossRef]
- Plun-Favreau, H.; Lewis, P.A.; Hardy, J.; Martins, L.M.; Wood, N.W. Cancer and neurodegeneration: between the devil and the deep blue sea. PLoS Genet. 2010, 6, e1001257. [Google Scholar] [CrossRef] [PubMed]
- Fisher, J.C. Multiple-mutation theory of carcinogenesis. Nature 1958, 181, 651–652. [Google Scholar] [CrossRef] [PubMed]
- Crusz, S.M.; Balkwill, F.R. Inflammation and cancer: advances and new agents. Nat. Rev. Clin. Oncol. 2015, 12, 584–596. [Google Scholar] [CrossRef]
- Gao, H.-M.; Hong, J.-S. Why neurodegenerative diseases are progressive: uncontrolled inflammation drives disease progression. Trends Immunol. 2008, 29, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Walker, L.C.; LeVine, H. Corruption and spread of pathogenic proteins in neurodegenerative diseases. J. Biol. Chem. 2012, 287, 33109–33115. [Google Scholar] [CrossRef] [PubMed]
- Houck, A.L.; Seddighi, S.; Driver, J.A. At the crossroads between neurodegeneration and cancer: A review of overlapping biology and its implications. Curr. Aging Sci. 2018, 11, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.; Park, M. Molecular crosstalk between cancer and neurodegenerative diseases. Cell. Mol. Life Sci. 2020, 77, 2659–2680. [Google Scholar] [CrossRef]
- Ganguly, G.; Chakrabarti, S.; Chatterjee, U.; Saso, L. Proteinopathy, oxidative stress and mitochondrial dysfunction: cross talk in Alzheimer’s disease and Parkinson’s disease. Drug Des. Devel. Ther. 2017, 11, 797–810. [Google Scholar] [CrossRef]
- Sankowski, R.; Mader, S.; Valdés-Ferrer, S.I. Systemic inflammation and the brain: novel roles of genetic, molecular, and environmental cues as drivers of neurodegeneration. Front. Cell. Neurosci. 2015, 9, 28. [Google Scholar] [CrossRef]
- Joseph, C.; Mangani, A.S.; Gupta, V.; Chitranshi, N.; Shen, T.; Dheer, Y.; Kb, D.; Mirzaei, M.; You, Y.; Graham, S.L.; Gupta, V. Cell cycle deficits in neurodegenerative disorders: uncovering molecular mechanisms to drive innovative therapeutic development. Aging Dis. 2020, 11, 946–966. [Google Scholar] [CrossRef]
- Rojas, N.G.; Cesarini, M.; Etcheverry, J.L.; Prat, G.A.D.; Arciuch, V.A.; Gatto, E.M. Neurodegenerative diseases and cancer: sharing common mechanisms in complex interactions. J. Integr. Neurosci. 2020, 19, 187–199. [Google Scholar] [CrossRef]
- Klus, P.; Cirillo, D.; Botta Orfila, T.; Gaetano Tartaglia, G. Neurodegeneration and cancer: where the disorder prevails. Sci. Rep. 2015, 5, 15390. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhou, T.; Ziegler, A.C.; Dimitrion, P.; Zuo, L. Oxidative stress in neurodegenerative diseases: from molecular mechanisms to clinical applications. Oxid. Med. Cell. Longev. 2017, 2017, 2525967. [Google Scholar] [CrossRef]
- Madabhushi, R.; Pan, L.; Tsai, L.-H. DNA damage and its links to neurodegeneration. Neuron 2014, 83, 266–282. [Google Scholar] [CrossRef] [PubMed]
- Mogavero, M.P.; Silvani, A.; DelRosso, L.M.; Salemi, M.; Ferri, R. Focus on the Complex Interconnection between Cancer, Narcolepsy and Other Neurodegenerative Diseases: A Possible Case of Orexin-Dependent Inverse Comorbidity. Cancers (Basel) 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Engel, P.A. Is age-related failure of metabolic reprogramming a principal mediator in idiopathic Parkinson’s disease? Implications for treatment and inverse cancer risk. Med. Hypotheses 2016, 93, 154–160. [Google Scholar] [CrossRef]
- Coarelli, G.; Diallo, A.; Thion, M.S.; Rinaldi, D.; Calvas, F.; Boukbiza, O.L.; Tataru, A.; Charles, P.; Tranchant, C.; Marelli, C.; Ewenczyk, C.; Tchikviladzé, M.; Monin, M.-L.; Carlander, B.; Anheim, M.; Brice, A.; Mochel, F.; Tezenas du Montcel, S.; Humbert, S.; Durr, A. Low cancer prevalence in polyglutamine expansion diseases. Neurology 2017, 88, 1114–1119. [Google Scholar] [CrossRef]
- Ibáñez, K.; Boullosa, C.; Tabarés-Seisdedos, R.; Baudot, A.; Valencia, A. Molecular evidence for the inverse comorbidity between central nervous system disorders and cancers detected by transcriptomic meta-analyses. PLoS Genet. 2014, 10, e1004173. [Google Scholar] [CrossRef]
- Lane, C.A.; Hardy, J.; Schott, J.M. Alzheimer’s disease. Eur. J. Neurol. 2018, 25, 59–70. [Google Scholar] [CrossRef]
- McColgan, P.; Tabrizi, S.J. Huntington’s disease: a clinical review. Eur. J. Neurol. 2018, 25, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Nutt, J.G.; Wooten, G.F. Clinical practice. Diagnosis and initial management of Parkinson’s disease. N. Engl. J. Med. 2005, 353, 1021–1027. [Google Scholar] [CrossRef]
- Bano, D.; Zanetti, F.; Mende, Y.; Nicotera, P. Neurodegenerative processes in Huntington’s disease. Cell Death Dis. 2011, 2, e228. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, S.; Safia, *!!! REPLACE !!!*; Haque, E.; Mir, S.S. Neurodegenerative diseases: multifactorial conformational diseases and their therapeutic interventions. J. Neurodegener. Dis. 2013, 2013, 563481. [Google Scholar] [CrossRef] [PubMed]
- Prasher, D.; Greenway, S.C.; Singh, R.B. The impact of epigenetics on cardiovascular disease. Biochem. Cell Biol. 2020, 98, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, A.; Preeti, K.; Tryphena, K.P.; Srivastava, S.; Singh, S.B.; Khatri, D.K. Proteostasis in Parkinson’s disease: Recent development and possible implication in diagnosis and therapeutics. Ageing Res. Rev. 2023, 84, 101816. [Google Scholar] [CrossRef] [PubMed]
- Hernaiz, A.; Toivonen, J.M.; Bolea, R.; Martín-Burriel, I. Epigenetic Changes in Prion and Prion-like Neurodegenerative Diseases: Recent Advances, Potential as Biomarkers, and Future Perspectives. Int. J. Mol. Sci. 2022, 23. [Google Scholar] [CrossRef]
- Chesnokova, E.; Beletskiy, A.; Kolosov, P. The role of transposable elements of the human genome in neuronal function and pathology. Int. J. Mol. Sci. 2022, 23. [Google Scholar] [CrossRef]
- Griess, B.; Tom, E.; Domann, F.; Teoh-Fitzgerald, M. Extracellular superoxide dismutase and its role in cancer. Free Radic. Biol. Med. 2017, 112, 464–479. [Google Scholar] [CrossRef]
- Maldonado, E.; Morales-Pison, S.; Urbina, F.; Solari, A. Aging hallmarks and the role of oxidative stress. Antioxidants (Basel) 2023, 12. [Google Scholar] [CrossRef]
- Missiroli, S.; Genovese, I.; Perrone, M.; Vezzani, B.; Vitto, V.A.M.; Giorgi, C. The role of mitochondria in inflammation: from cancer to neurodegenerative disorders. J. Clin. Med. 2020, 9. [Google Scholar] [CrossRef]
- Corrado, M.; Scorrano, L.; Campello, S. Mitochondrial dynamics in cancer and neurodegenerative and neuroinflammatory diseases. Int. J. Cell Biol. 2012, 2012, 729290. [Google Scholar] [CrossRef] [PubMed]
- Jagtap, Y.A.; Kumar, P.; Kinger, S.; Dubey, A.R.; Choudhary, A.; Gutti, R.K.; Singh, S.; Jha, H.C.; Poluri, K.M.; Mishra, A. Disturb mitochondrial associated proteostasis: Neurodegeneration and imperfect ageing. Front. Cell Dev. Biol. 2023, 11, 1146564. [Google Scholar] [CrossRef] [PubMed]
- Wahabi, K.; Perwez, A.; Rizvi, M.A. Parkin in Parkinson’s Disease and Cancer: a Double-Edged Sword. Mol. Neurobiol. 2018, 55, 6788–6800. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Sun, W.; Wang, Y.-F.; Li, J.; Li, D.-W. Association of p53 with Neurodegeneration in Parkinson’s Disease. Parkinsons Dis 2022, 2022, 6600944. [Google Scholar] [CrossRef] [PubMed]
- Drapalo, K.; Jozwiak, J. Parkin, PINK1 and DJ1 as possible modulators of mTOR pathway in ganglioglioma. Int. J. Neurosci. 2018, 128, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Barodia, S.K.; Creed, R.B.; Goldberg, M.S. Parkin and PINK1 functions in oxidative stress and neurodegeneration. Brain Res. Bull. 2017, 133, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Checler, F.; Alves da Costa, C. p53 in neurodegenerative diseases and brain cancers. Pharmacol. Ther. 2014, 142, 99–113. [Google Scholar] [CrossRef] [PubMed]
- Ramazi, S.; Daddzadi, M.; Sahafnejad, Z.; Allahverdi, A. Epigenetic regulation in lung cancer. MedComm 2023, 4, e401. [Google Scholar] [CrossRef]
- Cives, M.; Mannavola, F.; Lospalluti, L.; Sergi, M.C.; Cazzato, G.; Filoni, E.; Cavallo, F.; Giudice, G.; Stucci, L.S.; Porta, C.; Tucci, M. Non-Melanoma Skin Cancers: Biological and Clinical Features. Int. J. Mol. Sci. 2020, 21. [Google Scholar] [CrossRef]
- Salemi, M.; Mogavero, M.P.; Lanza, G.; Mongioì, L.M.; Calogero, A.E.; Ferri, R. Examples of Inverse Comorbidity between Cancer and Neurodegenerative Diseases: A Possible Role for Noncoding RNA. Cells 2022, 11. [Google Scholar] [CrossRef] [PubMed]
- Park, M.H.; Jo, M.; Kim, Y.R.; Lee, C.-K.; Hong, J.T. Roles of peroxiredoxins in cancer, neurodegenerative diseases and inflammatory diseases. Pharmacol. Ther. 2016, 163, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, V.; Santoro, A.; Monti, D.; Crupi, R.; Di Paola, R.; Latteri, S.; Cuzzocrea, S.; Zappia, M.; Giordano, J.; Calabrese, E.J.; Franceschi, C. Aging and Parkinson’s Disease: Inflammaging, neuroinflammation and biological remodeling as key factors in pathogenesis. Free Radic. Biol. Med. 2018, 115, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Vendramini-Costa, D.B.; Carvalho, J.E. Molecular link mechanisms between inflammation and cancer. Curr. Pharm. Des. 2012, 18, 3831–3852. [Google Scholar] [CrossRef] [PubMed]
- Morris, L.G.T.; Veeriah, S.; Chan, T.A. Genetic determinants at the interface of cancer and neurodegenerative disease. Oncogene 2010, 29, 3453–3464. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Ma, Y.; Luo, Y.; Song, Y.; Xiong, G.; Ma, Y.; Sun, X.; Kan, C. Metabolic diseases and healthy aging: identifying environmental and behavioral risk factors and promoting public health. Front. Public Health 2023, 11, 1253506. [Google Scholar] [CrossRef] [PubMed]
- Migliore, L.; Coppedè, F. Genetic and environmental factors in cancer and neurodegenerative diseases. Mutation Research/Reviews in Mutation Research 2002, 512, 135–153. [Google Scholar] [CrossRef] [PubMed]
- Picca, A.; Calvani, R.; Coelho-Junior, H.J.; Landi, F.; Bernabei, R.; Marzetti, E. Mitochondrial dysfunction, oxidative stress, and neuroinflammation: intertwined roads to neurodegeneration. Antioxidants (Basel) 2020, 9. [Google Scholar] [CrossRef]
- Thanan, R.; Oikawa, S.; Hiraku, Y.; Ohnishi, S.; Ma, N.; Pinlaor, S.; Yongvanit, P.; Kawanishi, S.; Murata, M. Oxidative stress and its significant roles in neurodegenerative diseases and cancer. Int. J. Mol. Sci. 2014, 16, 193–217. [Google Scholar] [CrossRef]
- Inflammatory Mechanisms and Oxidative Stress as Key Factors Respo...: Ingenta Connect. Available online: https://www.ingentaconnect.com/content/ben/cnsnddt/2016/00000015/00000003/art00008 (accessed on 16 December 2023).
- Jeppesen, D.K.; Bohr, V.A.; Stevnsner, T. DNA repair deficiency in neurodegeneration. Prog. Neurobiol. 2011, 94, 166–200. [Google Scholar] [CrossRef]
- Martin, L.J. DNA damage and repair: relevance to mechanisms of neurodegeneration. J. Neuropathol. Exp. Neurol. 2008, 67, 377–387. [Google Scholar] [CrossRef]
- Maynard, S.; Fang, E.F.; Scheibye-Knudsen, M.; Croteau, D.L.; Bohr, V.A. DNA damage, DNA repair, aging, and neurodegeneration. Cold Spring Harb. Perspect. Med. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Surguchov, A.; Surguchev, A. Synucleins: new data on misfolding, aggregation and role in diseases. Biomedicines 2022, 10. [Google Scholar] [CrossRef] [PubMed]
- Picca, A.; Guerra, F.; Calvani, R.; Romano, R.; Coelho-Júnior, H.J.; Bucci, C.; Marzetti, E. Mitochondrial dysfunction, protein misfolding and neuroinflammation in parkinson’s disease: roads to biomarker discovery. Biomolecules 2021, 11. [Google Scholar] [CrossRef]
- Bai, Y.; Zhang, S.; Dong, H.; Liu, Y.; Liu, C.; Zhang, X. Advanced techniques for detecting protein misfolding and aggregation in cellular environments. Chem. Rev. 2023, 123, 12254–12311. [Google Scholar] [CrossRef] [PubMed]
- Reddy, G.P.; Reddy, L.V.; Kim, S. Cancer biology and pathology. In Cancer: prevention, early detection, treatment and recovery; Stein, G.S., Luebbers, K.P., Eds.; Wiley, 2019; pp. 13–52. ISBN 9781118962886. [Google Scholar]
- Jankovic, J.; Tan, E.K. Parkinson’s disease: etiopathogenesis and treatment. J. Neurol. Neurosurg. Psychiatr. 2020, 91, 795–808. [Google Scholar] [CrossRef] [PubMed]
- Montanari, M.; Imbriani, P.; Bonsi, P.; Martella, G.; Peppe, A. Beyond the Microbiota: Understanding the Role of the Enteric Nervous System in Parkinson’s Disease from Mice to Human. Biomedicines 2023, 11. [Google Scholar] [CrossRef] [PubMed]
- Rugbjerg, K.; Friis, S.; Lassen, C.F.; Ritz, B.; Olsen, J.H. Malignant melanoma, breast cancer and other cancers in patients with Parkinson’s disease. Int. J. Cancer 2012, 131, 1904–1911. [Google Scholar] [CrossRef]
- Constantinescu, R.; Romer, M.; Kieburtz, K. ; DATATOP Investigators of the Parkinson Study Group Malignant melanoma in early Parkinson’s disease: the DATATOP trial. Mov. Disord. 2007, 22, 720–722. [Google Scholar] [CrossRef] [PubMed]
- Fiala, K.H.; Whetteckey, J.; Manyam, B.V. Malignant melanoma and levodopa in Parkinson’s disease: causality or coincidence? Parkinsonism Relat. Disord. 2003, 9, 321–327. [Google Scholar] [CrossRef]
- Sun, L.-M.; Liang, J.-A.; Chang, S.-N.; Sung, F.-C.; Muo, C.-H.; Kao, C.-H. Analysis of Parkinson’s disease and subsequent cancer risk in Taiwan: a nationwide population-based cohort study. Neuroepidemiology 2011, 37, 114–119. [Google Scholar] [CrossRef]
- Sandyk, R. Accelerated growth of malignant melanoma by levodopa in parkinson’s disease and role of the pineal gland. International Journal of Neuroscience 1992, 63, 137–140. [Google Scholar] [CrossRef]
- Thakur, G.; Kumar, V.; Lee, K.W.; Won, C. Structural insights and development of LRRK2 inhibitors for parkinson’s disease in the last decade. Genes (Basel) 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Vetchinova, A.S.; Kapkaeva, M.R.; Ivanov, M.V.; Kutukova, K.A.; Mudzhiri, N.M.; Frumkina, L.E.; Brydun, A.V.; Sukhorukov, V.S.; Illarioshkin, S.N. Mitochondrial Dysfunction in Dopaminergic Neurons Derived from Patients with LRRK2- and SNCA-Associated Genetic Forms of Parkinson’s Disease. Curr. Issues Mol. Biol. 2023, 45, 8395–8411. [Google Scholar] [CrossRef] [PubMed]
- Wallings, R.; Manzoni, C.; Bandopadhyay, R. Cellular processes associated with LRRK2 function and dysfunction. FEBS J. 2015, 282, 2806–2826. [Google Scholar] [CrossRef] [PubMed]
- Hur, E.-M.; Lee, B.D. LRRK2 at the crossroad of aging and parkinson’s disease. Genes (Basel) 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.P.; Mata, I.F.; Farrer, M.J. LRRK2: a common pathway for parkinsonism, pathogenesis and prevention? Trends Mol. Med. 2006, 12, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Ortega, R.A.; Wang, C.; Raymond, D.; Bryant, N.; Scherzer, C.R.; Thaler, A.; Alcalay, R.N.; West, A.B.; Mirelman, A.; Kuras, Y.; Marder, K.S.; Giladi, N.; Ozelius, L.J.; Bressman, S.B.; Saunders-Pullman, R. Association of dual LRRK2 G2019S and GBA variations with parkinson disease progression. JAMA Netw. Open 2021, 4, e215845. [Google Scholar] [CrossRef] [PubMed]
- Pan, T.; Li, X.; Jankovic, J. The association between Parkinson’s disease and melanoma. Int. J. Cancer 2011, 128, 2251–2260. [Google Scholar] [CrossRef]
- Chittoor-Vinod, V.G.; Nichols, R.J.; Schüle, B. Genetic and environmental factors influence the pleomorphy of LRRK2 parkinsonism. Int. J. Mol. Sci. 2021, 22. [Google Scholar] [CrossRef]
- Ejma, M.; Madetko, N.; Brzecka, A.; Guranski, K.; Alster, P.; Misiuk-Hojło, M.; Somasundaram, S.G.; Kirkland, C.E.; Aliev, G. The Links between Parkinson’s Disease and Cancer. Biomedicines 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Kelm-Nelson, C.A.; Brauer, A.F.L.; Barth, K.J.; Lake, J.M.; Sinnen, M.L.K.; Stehula, F.J.; Muslu, C.; Marongiu, R.; Kaplitt, M.G.; Ciucci, M.R. Characterization of early-onset motor deficits in the Pink1-/- mouse model of Parkinson disease. Brain Res. 2018, 1680, 1–12. [Google Scholar] [CrossRef] [PubMed]
- van der Merwe, C.; Jalali Sefid Dashti, Z.; Christoffels, A.; Loos, B.; Bardien, S. Evidence for a common biological pathway linking three Parkinson’s disease-causing genes: parkin, PINK1 and DJ-1. Eur. J. Neurosci. 2015, 41, 1113–1125. [Google Scholar] [CrossRef]
- Tassone, A.; Meringolo, M.; Ponterio, G.; Bonsi, P.; Schirinzi, T.; Martella, G. Mitochondrial bioenergy in neurodegenerative disease: huntington and parkinson. Int. J. Mol. Sci. 2023, 24. [Google Scholar] [CrossRef]
- Narendra, D.P.; Jin, S.M.; Tanaka, A.; Suen, D.-F.; Gautier, C.A.; Shen, J.; Cookson, M.R.; Youle, R.J. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010, 8, e1000298. [Google Scholar] [CrossRef] [PubMed]
- Gispert, S.; Ricciardi, F.; Kurz, A.; Azizov, M.; Hoepken, H.-H.; Becker, D.; Voos, W.; Leuner, K.; Müller, W.E.; Kudin, A.P.; Kunz, W.S.; Zimmermann, A.; Roeper, J.; Wenzel, D.; Jendrach, M.; García-Arencíbia, M.; Fernández-Ruiz, J.; Huber, L.; Rohrer, H.; Barrera, M.; Auburger, G. Parkinson phenotype in aged PINK1-deficient mice is accompanied by progressive mitochondrial dysfunction in absence of neurodegeneration. PLoS ONE 2009, 4, e5777. [Google Scholar] [CrossRef] [PubMed]
- Brunelli, F.; Valente, E.M.; Arena, G. Mechanisms of neurodegeneration in Parkinson’s disease: keep neurons in the PINK1. Mech. Ageing Dev. 2020, 189, 111277. [Google Scholar] [CrossRef]
- Thomas, K.J.; McCoy, M.K.; Blackinton, J.; Beilina, A.; van der Brug, M.; Sandebring, A.; Miller, D.; Maric, D.; Cedazo-Minguez, A.; Cookson, M.R. DJ-1 acts in parallel to the PINK1/parkin pathway to control mitochondrial function and autophagy. Hum. Mol. Genet. 2011, 20, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Sliter, D.A.; Martinez, J.; Hao, L.; Chen, X.; Sun, N.; Fischer, T.D.; Burman, J.L.; Li, Y.; Zhang, Z.; Narendra, D.P.; Cai, H.; Borsche, M.; Klein, C.; Youle, R.J. Parkin and PINK1 mitigate STING-induced inflammation. Nature 2018, 561, 258–262. [Google Scholar] [CrossRef]
- Imbriani, P.; Tassone, A.; Meringolo, M.; Ponterio, G.; Madeo, G.; Pisani, A.; Bonsi, P.; Martella, G. Loss of Non-Apoptotic Role of Caspase-3 in the PINK1 Mouse Model of Parkinson’s Disease. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef]
- Wang, M.; Luan, S.; Fan, X.; Wang, J.; Huang, J.; Gao, X.; Han, D. The emerging multifaceted role of PINK1 in cancer biology. Cancer Sci. 2022, 113, 4037–4047. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-L.; Wang, Z.-X.; Ying, C.-Z.; Zhang, B.-R.; Pu, J.-L. Decoding the Role of Familial Parkinson’s Disease-Related Genes in DNA Damage and Repair. Aging Dis. 2022, 13, 1405–1412. [Google Scholar] [CrossRef]
- Koros, C.; Simitsi, A.-M.; Bougea, A.; Papagiannakis, N.; Antonelou, R.; Pachi, I.; Angelopoulou, E.; Prentakis, A.; Zachou, A.; Chrysovitsanou, C.; Beratis, I.; Fragkiadaki, S.; Kontaxopoulou, D.; Eftymiopoulou, E.; Stanitsa, E.; Potagas, C.; Papageorgiou, S.G.; Karavasilis, E.; Velonakis, G.; Prassopoulos, V.; Stefanis, L. Double Trouble: Association of Malignant Melanoma with Sporadic and Genetic Forms of Parkinson’s Disease and Asymptomatic Carriers of Related Genes: A Brief Report. Medicina (Kaunas) 2023, 59. [Google Scholar] [CrossRef] [PubMed]
- O’Flanagan, C.H.; O’Neill, C. PINK1 signalling in cancer biology. Biochim. Biophys. Acta 2014, 1846, 590–598. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, A.; Kataoka, K.; Hong, M.; Sakaguchi, M.; Huh, N. BRPK, a novel protein kinase showing increased expression in mouse cancer cell lines with higher metastatic potential. Cancer Lett. 2003, 201, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Arena, G.; Valente, E.M. PINK1 in the limelight: multiple functions of an eclectic protein in human health and disease. J. Pathol. 2017, 241, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Inzelberg, R.; Jankovic, J. Are Parkinson disease patients protected from some but not all cancers? Neurology 2007, 69, 1542–1550. [Google Scholar] [CrossRef]
- MacKeigan, J.P.; Murphy, L.O.; Blenis, J. Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nat. Cell Biol. 2005, 7, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.A.; Hewish, M.; Sims, D.; Lord, C.J.; Ashworth, A. Parallel high-throughput RNA interference screens identify PINK1 as a potential therapeutic target for the treatment of DNA mismatch repair-deficient cancers. Cancer Res. 2011, 71, 1836–1848. [Google Scholar] [CrossRef]
- Ibáñez, P.; Bonnet, A.M.; Débarges, B.; Lohmann, E.; Tison, F.; Pollak, P.; Agid, Y.; Dürr, A.; Brice, A. Causal relation between alpha-synuclein gene duplication and familial Parkinson’s disease. Lancet 2004, 364, 1169–1171. [Google Scholar] [CrossRef]
- Zhou, L.-X.; Zheng, H.; Tian, Y.; Luo, K.-F.; Ma, S.-J.; Wu, Z.-W.; Tang, P.; Jiang, J.; Wang, M.-H. SNCA inhibits epithelial-mesenchymal transition and correlates to favorable prognosis of breast cancer. Carcinogenesis 2022, 43, 1071–1082. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Xu, Z.; Hu, X.; Qian, L.; Li, Z.; Zhou, Y.; Dai, S.; Zeng, S.; Gong, Z. SNCA Is a Functionally Low-Expressed Gene in Lung Adenocarcinoma. Genes (Basel) 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Zanotti, L.C.; Malizia, F.; Cesatti Laluce, N.; Avila, A.; Mamberto, M.; Anselmino, L.E.; Menacho-Márquez, M. Synuclein proteins in cancer development and progression. Biomolecules 2023, 13. [Google Scholar] [CrossRef] [PubMed]
- Bernardini, J.P.; Lazarou, M.; Dewson, G. Parkin and mitophagy in cancer. Oncogene 2017, 36, 1315–1327. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Suzuki, T.; Chiba, T.; Shimura, H.; Hattori, N.; Mizuno, Y. Parkin is linked to the ubiquitin pathway. J. Mol. Med. 2001, 79, 482–494. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Suzuki, T.; Hattori, N.; Mizuno, Y. Ubiquitin, proteasome and parkin. Biochim. Biophys. Acta 2004, 1695, 235–247. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-L.; Feng, S.-T.; Wang, Z.-Z.; Yuan, Y.-H.; Chen, N.-H.; Zhang, Y. Parkin, an E3 ubiquitin ligase, plays an essential role in mitochondrial quality control in parkinson’s disease. Cell. Mol. Neurobiol. 2021, 41, 1395–1411. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Denison, S.; Lai, J.-P.; Philips, L.A.; Montoya, D.; Kock, N.; Schüle, B.; Klein, C.; Shridhar, V.; Roberts, L.R.; Smith, D.I. Parkin gene alterations in hepatocellular carcinoma. Genes Chromosomes Cancer 2004, 40, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Denison, S.R.; Wang, F.; Becker, N.A.; Schüle, B.; Kock, N.; Phillips, L.A.; Klein, C.; Smith, D.I. Alterations in the common fragile site gene Parkin in ovarian and other cancers. Oncogene 2003, 22, 8370–8378. [Google Scholar] [CrossRef]
- Naren, P.; Cholkar, A.; Kamble, S.; Khan, S.S.; Srivastava, S.; Madan, J.; Mehra, N.; Tiwari, V.; Singh, S.B.; Khatri, D.K. Pathological and therapeutic advances in parkinson’s disease: mitochondria in the interplay. J Alzheimers Dis 2023, 94, S399–S428. [Google Scholar] [CrossRef]
- Olanow, C.W.; McNaught, K.S.P. Ubiquitin-proteasome system and Parkinson’s disease. Mov. Disord. 2006, 21, 1806–1823. [Google Scholar] [CrossRef] [PubMed]
- Jin, W. Novel Insights into PARK7 (DJ-1), a Potential Anti-Cancer Therapeutic Target, and Implications for Cancer Progression. J. Clin. Med. 2020, 9. [Google Scholar] [CrossRef]
- Perl, D.P. Neuropathology of Alzheimer’s disease. Mt Sinai J Med 2010, 77, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Nudelman, K.N.H.; McDonald, B.C.; Lahiri, D.K.; Saykin, A.J. Biological hallmarks of cancer in alzheimer’s disease. Mol. Neurobiol. 2019, 56, 7173–7187. [Google Scholar] [CrossRef] [PubMed]
- Nudelman, K.N.H.; Risacher, S.L.; West, J.D.; McDonald, B.C.; Gao, S.; Saykin, A.J. ; Alzheimer’s Disease Neuroimaging Initiative Association of cancer history with Alzheimer’s disease onset and structural brain changes. Front. Physiol. 2014, 5, 423. [Google Scholar] [CrossRef]
- Fernandez, H.R.; Varma, A.; Flowers, S.A.; Rebeck, G.W. Cancer chemotherapy related cognitive impairment and the impact of the alzheimer’s disease risk factor APOE. Cancers (Basel) 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Mahley, R.W. Central nervous system lipoproteins: apoe and regulation of cholesterol metabolism. Arterioscler. Thromb. Vasc. Biol. 2016, 36, 1305–1315. [Google Scholar] [CrossRef] [PubMed]
- Darwish, N.M.; Al-Hail, M.K.; Mohamed, Y.; Al Saady, R.; Mohsen, S.; Zar, A.; Al-Mansoori, L.; Pedersen, S. The role of apolipoproteins in the commonest cancers: A review. Cancers (Basel) 2023, 15. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Chen, J.; Ma, Y.; Chen, H. Apolipoproteins: New players in cancers. Front. Pharmacol. 2022, 13, 1051280. [Google Scholar] [CrossRef]
- Miao, G.; Zhuo, D.; Han, X.; Yao, W.; Liu, C.; Liu, H.; Cao, H.; Sun, Y.; Chen, Z.; Feng, T. From degenerative disease to malignant tumors: Insight to the function of ApoE. Biomed. Pharmacother. 2023, 158, 114127. [Google Scholar] [CrossRef]
- Grant, W.B. A multicountry ecological study of risk-modifying factors for prostate cancer: apolipoprotein E epsilon4 as a risk factor and cereals as a risk reduction factor. Anticancer Res. 2010, 30, 189–199. [Google Scholar] [PubMed]
- Paniri, A.; Hosseini, M.M.; Akhavan-Niaki, H. Alzheimer’s Disease-Related Epigenetic Changes: Novel Therapeutic Targets. Mol. Neurobiol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Clark, J.S.; Kayed, R.; Abate, G.; Uberti, D.; Kinnon, P.; Piccirella, S. Post-translational Modifications of the p53 Protein and the Impact in Alzheimer’s Disease: A Review of the Literature. Front. Aging Neurosci. 2022, 14, 835288. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.D.; Ehrlich, B.E. Cellular mechanisms and treatments for chemobrain: insight from aging and neurodegenerative diseases. EMBO Mol. Med. 2020, 12, e12075. [Google Scholar] [CrossRef] [PubMed]
- Lardelli, M. An alternative view of familial alzheimer’s disease genetics. 2023. [Google Scholar] [CrossRef]
- Yang, Y.; Bagyinszky, E.; An, S.S.A. Presenilin-1 (PSEN1) Mutations: Clinical Phenotypes beyond Alzheimer’s Disease. Int. J. Mol. Sci. 2023, 24. [Google Scholar] [CrossRef]
- Vetrivel, K.S.; Zhang, Y.; Xu, H.; Thinakaran, G. Pathological and physiological functions of presenilins. Mol. Neurodegener. 2006, 1, 4. [Google Scholar] [CrossRef]
- Hunter, S.; Brayne, C. Understanding the roles of mutations in the amyloid precursor protein in Alzheimer disease. Mol. Psychiatry 2018, 23, 81–93. [Google Scholar] [CrossRef]
- Palihati, N.; Tang, Y.; Yin, Y.; Yu, D.; Liu, G.; Quan, Z.; Ni, J.; Yan, Y.; Qing, H. Clusterin is a potential therapeutic target in alzheimer’s disease. Mol. Neurobiol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Cheng, L.; Dai, H.; Zhang, R.; Wang, M.; Shi, T.; Sun, M.; Cheng, X.; Wei, Q. Variants in Notch signalling pathway genes, PSEN1 and MAML2, predict overall survival in Chinese patients with epithelial ovarian cancer. J. Cell. Mol. Med. 2018, 22, 4975–4984. [Google Scholar] [CrossRef]
- Ham, S.; Kim, T.K.; Ryu, J.; Kim, Y.S.; Tang, Y.-P.; Im, H.-I. Comprehensive MicroRNAome Analysis of the Relationship Between Alzheimer Disease and Cancer in PSEN Double-Knockout Mice. Int. Neurourol. J. 2018, 22, 237–245. [Google Scholar] [CrossRef]
- Holohan, K.N.; Lahiri, D.K.; Schneider, B.P.; Foroud, T.; Saykin, A.J. Functional microRNAs in Alzheimer’s disease and cancer: differential regulation of common mechanisms and pathways. Front. Genet. 2012, 3, 323. [Google Scholar] [CrossRef] [PubMed]
- Laird, F.M.; Cai, H.; Savonenko, A.V.; Farah, M.H.; He, K.; Melnikova, T.; Wen, H.; Chiang, H.-C.; Xu, G.; Koliatsos, V.E.; Borchelt, D.R.; Price, D.L.; Lee, H.-K.; Wong, P.C. BACE1, a major determinant of selective vulnerability of the brain to amyloid-beta amyloidogenesis, is essential for cognitive, emotional, and synaptic functions. J. Neurosci. 2005, 25, 11693–11709. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, H.; Li, R.; Sterling, K.; Song, W. Amyloid β-based therapy for Alzheimer’s disease: challenges, successes and future. Signal Transduct. Target. Ther. 2023, 8, 248. [Google Scholar] [CrossRef] [PubMed]
- Liebsch, F.; Kulic, L.; Teunissen, C.; Shobo, A.; Ulku, I.; Engelschalt, V.; Hancock, M.A.; van der Flier, W.M.; Kunach, P.; Rosa-Neto, P.; Scheltens, P.; Poirier, J.; Saftig, P.; Bateman, R.J.; Breitner, J.; Hock, C.; Multhaup, G. Aβ34 is a BACE1-derived degradation intermediate associated with amyloid clearance and Alzheimer’s disease progression. Nat. Commun. 2019, 10, 2240. [Google Scholar] [CrossRef]
- Zhai, K.; Huang, Z.; Huang, Q.; Tao, W.; Fang, X.; Zhang, A.; Li, X.; Stark, G.R.; Hamilton, T.A.; Bao, S. Pharmacological inhibition of BACE1 suppresses glioblastoma growth by stimulating macrophage phagocytosis of tumor cells. Nat. Cancer 2021, 2, 1136–1151. [Google Scholar] [CrossRef] [PubMed]
- Farris, F.; Matafora, V.; Bachi, A. The emerging role of β-secretases in cancer. J. Exp. Clin. Cancer Res. 2021, 40, 147. [Google Scholar] [CrossRef] [PubMed]
- Dash, B.P.; Naumann, M.; Sterneckert, J.; Hermann, A. Genome Wide Analysis Points towards Subtype-Specific Diseases in Different Genetic Forms of Amyotrophic Lateral Sclerosis. Int. J. Mol. Sci. 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, R.L.; Schijven, D.; van Rheenen, W.; van Eijk, K.R.; O’Brien, M.; Kahn, R.S.; Ophoff, R.A.; Goris, A.; Bradley, D.G.; Al-Chalabi, A.; van den Berg, L.H.; Luykx, J.J.; Hardiman, O.; Veldink, J.H. Project MinE GWAS Consortium; Schizophrenia Working Group of the Psychiatric Genomics Consortium Genetic correlation between amyotrophic lateral sclerosis and schizophrenia. Nat. Commun. 2017, 8, 14774. [Google Scholar] [CrossRef]
- Pang, W.; Hu, F. Cellular and physiological functions of C9ORF72 and implications for ALS/FTD. J. Neurochem. 2021, 157, 334–350. [Google Scholar] [CrossRef]
- Nassif, M.; Woehlbier, U.; Manque, P.A. The enigmatic role of C9ORF72 in autophagy. Front. Neurosci. 2017, 11, 442. [Google Scholar] [CrossRef]
- Brown, C.A.; Lally, C.; Kupelian, V.; Flanders, W.D. Estimated prevalence and incidence of amyotrophic lateral sclerosis and SOD1 and c9orf72 genetic variants. Neuroepidemiology 2021, 55, 342–353. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Westergard, T.; Pasinelli, P.; Trotti, D. Pathogenic determinants and mechanisms of ALS/FTD linked to hexanucleotide repeat expansions in the C9orf72 gene. Neurosci. Lett. 2017, 636, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, I.R.A.; Frick, P.; Neumann, M. The neuropathology associated with repeat expansions in the C9ORF72 gene. Acta Neuropathol. 2014, 127, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Lafita-Navarro, M.C.; Conacci-Sorrell, M. Nucleolar stress: From development to cancer. Semin. Cell Dev. Biol. 2023, 136, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Hautbergue, G.M. RNA nuclear export: from neurological disorders to cancer. Adv. Exp. Med. Biol. 2017, 1007, 89–109. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, I.; Azuma, Y.; Yamaguchi, M. Cancer-related genes and ALS. Front Biosci (Landmark Ed) 2019, 24, 1241–1258. [Google Scholar] [CrossRef] [PubMed]
- Ingre, C.; Roos, P.M.; Piehl, F.; Kamel, F.; Fang, F. Risk factors for amyotrophic lateral sclerosis. Clin. Epidemiol. 2015, 7, 181–193. [Google Scholar] [CrossRef] [PubMed]
- Settembre, C.; Perera, R.M. Lysosomes as coordinators of cellular catabolism, metabolic signalling and organ physiology. Nat. Rev. Mol. Cell Biol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Hegde, M.L. New Mechanisms of DNA Repair Defects in Fused in Sarcoma-Associated Neurodegeneration: Stage Set for DNA Repair-Based Therapeutics? J. Exp. Neurosci. 2019, 13, 1179069519856358. [Google Scholar] [CrossRef]
- Dormann, D.; Haass, C. Fused in sarcoma (FUS): an oncogene goes awry in neurodegeneration. Mol. Cell. Neurosci. 2013, 56, 475–486. [Google Scholar] [CrossRef]
- Sukhanova, M.V.; Singatulina, A.S.; Pastré, D.; Lavrik, O.I. Fused in Sarcoma (FUS) in DNA Repair: Tango with Poly(ADP-ribose) Polymerase 1 and Compartmentalisation of Damaged DNA. Int. J. Mol. Sci. 2020, 21. [Google Scholar] [CrossRef]
- Sun, Z.; Diaz, Z.; Fang, X.; Hart, M.P.; Chesi, A.; Shorter, J.; Gitler, A.D. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol. 2011, 9, e1000614. [Google Scholar] [CrossRef]
- Deng, H.; Gao, K.; Jankovic, J. The role of FUS gene variants in neurodegenerative diseases. Nat. Rev. Neurol. 2014, 10, 337–348. [Google Scholar] [CrossRef]
- Shang, Y.; Huang, E.J. Mechanisms of FUS mutations in familial amyotrophic lateral sclerosis. Brain Res. 2016, 1647, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, Y.H.; Wang, H. Genetic Association between Amyotrophic Lateral Sclerosis and Cancer. Genes (Basel) 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Haile, S.; Lal, A.; Myung, J.-K.; Sadar, M.D. FUS/TLS is a co-activator of androgen receptor in prostate cancer cells. PLoS ONE 2011, 6, e24197. [Google Scholar] [CrossRef]
- Ghanbarpanah, E.; Kohanpour, M.A.; Hosseini-Beheshti, F.; Yari, L.; Keshvari, M. Structure and function of FUS gene in prostate cancer. Bratisl. Lek. Listy 2018, 119, 660–663. [Google Scholar] [CrossRef]
- Bukowska, B.; Michalowicz, J.; Pieniazek, D.; Sicinska, P.; Duda, W. Superoxide Dismutases and Their Inhibitors-the Role in Some Diseases. cei 2006, 2, 379–397. [Google Scholar] [CrossRef]
- Rosa, A.C.; Corsi, D.; Cavi, N.; Bruni, N.; Dosio, F. Superoxide dismutase administration: A review of proposed human uses. Molecules 2021, 26. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.J.; McKeown, S.R.; Rashid, S. Mutant SOD1 mediated pathogenesis of Amyotrophic Lateral Sclerosis. Gene 2016, 577, 109–118. [Google Scholar] [CrossRef]
- Kirby, J.; Halligan, E.; Baptista, M.J.; Allen, S.; Heath, P.R.; Holden, H.; Barber, S.C.; Loynes, C.A.; Wood-Allum, C.A.; Lunec, J.; Shaw, P.J. Mutant SOD1 alters the motor neuronal transcriptome: implications for familial ALS. Brain 2005, 128, 1686–1706. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Su, X.; Burley, S.K.; Zheng, X.F.S. Nuclear SOD1 in growth control, oxidative stress response, amyotrophic lateral sclerosis, and cancer. Antioxidants (Basel) 2022, 11. [Google Scholar] [CrossRef]
- Gomez, M.; Germain, D. Cross talk between SOD1 and the mitochondrial UPR in cancer and neurodegeneration. Mol. Cell. Neurosci. 2019, 98, 12–18. [Google Scholar] [CrossRef]
- Roos, R.A.C. Huntington’s disease: a clinical review. Orphanet J. Rare Dis. 2010, 5, 40. [Google Scholar] [CrossRef]
- DiFiglia, M.; Sapp, E.; Chase, K.O.; Davies, S.W.; Bates, G.P.; Vonsattel, J.P.; Aronin, N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 1997, 277, 1990–1993. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Trojanowski, J.Q.; Lee, V.M.-Y. Protein transmission in neurodegenerative disease. Nat. Rev. Neurol. 2020, 16, 199–212. [Google Scholar] [CrossRef]
- Waldvogel, H.J.; Kim, E.H.; Tippett, L.J.; Vonsattel, J.-P.G.; Faull, R.L.M. The neuropathology of huntington’s disease. Curr. Top. Behav. Neurosci. 2015, 22, 33–80. [Google Scholar] [CrossRef] [PubMed]
- Eje, O. 29 Journal of Chemical Reviews Article info: Journal of Chemical Reviews.
- Turner, M.R.; Goldacre, R.; Goldacre, M.J. Reduced cancer incidence in Huntington’s disease: record linkage study clue to an evolutionary trade-off? Clinical genetics 2013, 83, 588–590. [Google Scholar] [CrossRef]
- Thion, M.S.; Tézenas du Montcel, S.; Golmard, J.-L.; Vacher, S.; Barjhoux, L.; Sornin, V.; Cazeneuve, C.; Bièche, I.; Sinilnikova, O.; Stoppa-Lyonnet, D.; Durr, A.; Humbert, S. CAG repeat size in Huntingtin alleles is associated with cancer prognosis. Eur. J. Hum. Genet. 2016, 24, 1310–1315. [Google Scholar] [CrossRef]
- Malla, B.; Guo, X.; Senger, G.; Chasapopoulou, Z.; Yildirim, F. A systematic review of transcriptional dysregulation in huntington’s disease studied by RNA sequencing. Front. Genet. 2021, 12, 751033. [Google Scholar] [CrossRef]
- Yalçin, M.; El-Athman, R.; Ouk, K.; Priller, J.; Relógio, A. Analysis of the Circadian Regulation of Cancer Hallmarks by a Cross-Platform Study of Colorectal Cancer Time-Series Data Reveals an Association with Genes Involved in Huntington’s Disease. Cancers (Basel) 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Leavitt, B.R.; Kordasiewicz, H.B.; Schobel, S.A. Huntingtin-Lowering Therapies for Huntington Disease: A Review of the Evidence of Potential Benefits and Risks. JAMA Neurol. 2020, 77, 764–772. [Google Scholar] [CrossRef] [PubMed]
- Kryston, T.B.; Georgiev, A.B.; Pissis, P.; Georgakilas, A.G. Role of oxidative stress and DNA damage in human carcinogenesis. Mutat. Res. 2011, 711, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Suwaki, N.; Klare, K.; Tarsounas, M. RAD51 paralogs: roles in DNA damage signalling, recombinational repair and tumorigenesis. Semin. Cell Dev. Biol. 2011, 22, 898–905. [Google Scholar] [CrossRef] [PubMed]
- Impaired DNA Damage Repair as a Common Feature of Neurodegenerati...: Ingenta Connect. Available online: https://www.ingentaconnect.com/content/ben/cmm/2015/00000015/00000002/art00003 (accessed on 14 December 2023).
- Gupta, S.; You, P.; SenGupta, T.; Nilsen, H.; Sharma, K. Crosstalk between Different DNA Repair Pathways Contributes to Neurodegenerative Diseases. Biology (Basel) 2021, 10. [Google Scholar] [CrossRef]
- van der Meer, L.B.; van Duijn, E.; Wolterbeek, R.; Tibben, A. Adverse childhood experiences of persons at risk for Huntington’s disease or BRCA1/2 hereditary breast/ovarian cancer. Clin. Genet. 2012, 81, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Montalto, F.I.; De Amicis, F. Cyclin D1 in cancer: A molecular connection for cell cycle control, adhesion and invasion in tumor and stroma. Cells 2020, 9. [Google Scholar] [CrossRef]
- Monti, S.; Chapuy, B.; Takeyama, K.; Rodig, S.J.; Hao, Y.; Yeda, K.T.; Inguilizian, H.; Mermel, C.; Currie, T.; Dogan, A.; Kutok, J.L.; Beroukhim, R.; Neuberg, D.; Habermann, T.M.; Getz, G.; Kung, A.L.; Golub, T.R.; Shipp, M.A. Integrative analysis reveals an outcome-associated and targetable pattern of p53 and cell cycle deregulation in diffuse large B cell lymphoma. Cancer Cell 2012, 22, 359–372. [Google Scholar] [CrossRef]
- Cohen, T.J.; Lee, V.M.Y.; Trojanowski, J.Q. TDP-43 functions and pathogenic mechanisms implicated in TDP-43 proteinopathies. Trends Mol. Med. 2011, 17, 659–667. [Google Scholar] [CrossRef]
- Alqurashi, Y.E.; Al-Hetty, H.R.A.K.; Ramaiah, P.; Fazaa, A.H.; Jalil, A.T.; Alsaikhan, F.; Gupta, J.; Ramírez-Coronel, A.A.; Tayyib, N.A.; Peng, H. Harnessing function of EMT in hepatocellular carcinoma: From biological view to nanotechnological standpoint. Environ. Res. 2023, 227, 115683. [Google Scholar] [CrossRef]
- Deng, K.; Yao, J.; Huang, J.; Ding, Y.; Zuo, J. Abnormal alternative splicing promotes tumor resistance in targeted therapy and immunotherapy. Transl. Oncol. 2021, 14, 101077. [Google Scholar] [CrossRef] [PubMed]
- Barzilai, A.; Rotman, G.; Shiloh, Y. ATM deficiency and oxidative stress: a new dimension of defective response to DNA damage. DNA Repair (Amst) 2002, 1, 3–25. [Google Scholar] [CrossRef]
- Yan, S.; Sorrell, M.; Berman, Z. Functional interplay between ATM/ATR-mediated DNA damage response and DNA repair pathways in oxidative stress. Cell. Mol. Life Sci. 2014, 71, 3951–3967. [Google Scholar] [CrossRef] [PubMed]
- Kazanets, A.; Shorstova, T.; Hilmi, K.; Marques, M.; Witcher, M. Epigenetic silencing of tumor suppressor genes: Paradigms, puzzles, and potential. Biochim. Biophys. Acta 2016, 1865, 275–288. [Google Scholar] [CrossRef] [PubMed]
| GENE | FUNCTION | ROLE IN NEURODEGENERATION | ROLE IN CANCER |
|---|---|---|---|
|
LRRK2 |
LRRK2 regulates various cellular processes, including cell signaling, vesicle trafficking, and autophagy | The LRRK2 gene is associated with familial and sporadic PD forms. Mutations in this gene have been identified as significant genetic contributor to PD and development, affecting cellular processes such as protein degradation and mitochondrial function in neurons. |
Involve alterations in cell proliferation, survival pathways, or immune responses for instance, the impact of LRRK2 mutations on cellular processes like autophagy, inflammation, or DNA repair mechanisms might contribute to the pathogenesis of PD and cancer |
|
PINK1 |
Critical role in maintaining mitochondrial function, regulating mitochondrial quality control, and initiating the clearance of damaged mitochondria through a process called mitophagy. Plays a crucial role in mitochondrial quality control and the regulation of cell death pathways. |
Mutations in the PINK1 gene are associated with certain cases of PD, leading to mitochondrial dysfunction and impaired removal of damaged mitochondria, contributing to neuronal degeneration | Studies exploring the broader roles of PINK1 have indicated its involvement in cellular processes beyond PD, including aspects related to cancer biology Initially linked to cancer biology through its modulation by the tumor suppressor PTEN in a cancer cell model, PINK1 gained prominence as it exhibited robust expression in highly metastatic melanoma and colon carcinoma mouse cancer cell lines |
|
SNCA |
Encodes αS |
Mutations or duplications in the SNCA gene have been linked to familial forms of PD | It's been observed that elevated levels of αS could influence tumor growth and metastasis in particular cancer types, including breast cancer and colorectal cancer.. |
| PARK2 |
Parkin, a protein encoded by the PARK2 gene, is vital in removing damaged or unnecessary proteins within cells, a mechanism called ubiquitination | Mutations in the PARK2 gene are linked to familial forms of PD, impairing Parkin's function and accumulating toxic proteins contributing to neuronal degeneration | Some research has explored its involvement in cellular processes related to tumor suppression and apoptosis, indicating a possible connection to cancer biology |
|
PARK 7 |
Parkin-7 (PARK7), also known as DJ-1, protects cells from oxidative stress, maintains mitochondrial function, and regulates cellular pathways associated with cell survival | Mutations in the PARK7 gene are implicated in familial forms of PD, compromising cellular defenses against oxidative damage and contributing to neuronal degeneration. | Involvement in modulating cell proliferation, apoptosis, and tumor progression. While these genes primarily feature in the context of Parkinson's disease, their broader implications or potential involvement in aspects of cancer biology are areas of ongoing investigation. |
| GENE | FUNCTION | ROLE IN NEURODEGENERATION | ROLE IN CANCER |
|---|---|---|---|
|
APOE |
Encodes a protein involved in lipid metabolism and transportation, playing a crucial role in regulating cholesterol levels in the body. |
Specific variants of the APOE gene, notably the APOE ε4 allele, are known to significantly elevate the risk and influence the age of onset of the disease. |
Alterations in the HTT gene could impact cell survival, DNA repair mechanisms, or cellular processes relevant to tumorigenesis. |
| TP53 | A critical tumor suppressor gene, is commonly mutated in various cancers. | Mutations of TP53 have been observed in the brains of individuals with AD, | Tumor suppressor genes |
| PSEN | Play pivotal roles in various cellular functions, including processing certain proteins like APP. | Mutations in the PSEN1 and PSEN2 genes are strongly linked to early-onset familial AD, promoting the accumulation of amyloid-β peptides, | Studies suggest potential roles of PSENs in regulating cell proliferation, apoptosis, and cellular signaling pathways relevant to tumorigenesis |
| BACE1 | Critical enzyme that produces Aβ peptides | BACE1 cleaves APP to generate these Aβ fragments, contributing to the neurotoxicity seen in AD | Potential implications of BACE1 in cancer biology, highlighting its involvement in regulating cell proliferation, migration, and tumor growth |
| GENE | FUNCTION | ROLE IN NEURODEGENERATION | ROLE IN CANCER |
|---|---|---|---|
|
C9orf72 |
Crucial role in cellular functions, including vesicle trafficking, autophagy, and RNA metabolism |
Expansions of the hexanucleotide repeat in the C9orf72 gene are ALS's most common genetic cause. These expansions lead to toxic RNA and protein aggregates contributing to neuronal degeneration in ALS | Studies have identified associations between C9orf72 mutations and an increased risk of developing various malignancies, including brain tumors and certain types of lymphoma |
|
FUS |
Is involved in various cellular functions, including RNA processing, transport, and DNA repair. |
Mutations in the FUS gene are linked to familial and sporadic cases of ALS, where aberrant FUS proteins form toxic aggregates contributing to neuronal damage |
Aberrant FUS aggregation and pathology have also been associated with certain cancers, albeit in a relatively limited context compared to its involvement in ALS |
|
SOD1 |
Enzyme that is crucial in neutralizing harmful free radicals in cells by converting superoxide radicals into less toxic molecules | Mutations in the SOD1 gene are associated with familial cases of ALS, where altered SOD1 proteins contribute to motor neuron degeneration | Potential role of SOD1 in modulating oxidative stress, inflammation, and cell survival pathways that might have implications in specific cancers |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





