Submitted:
29 December 2023
Posted:
04 January 2024
You are already at the latest version
Abstract
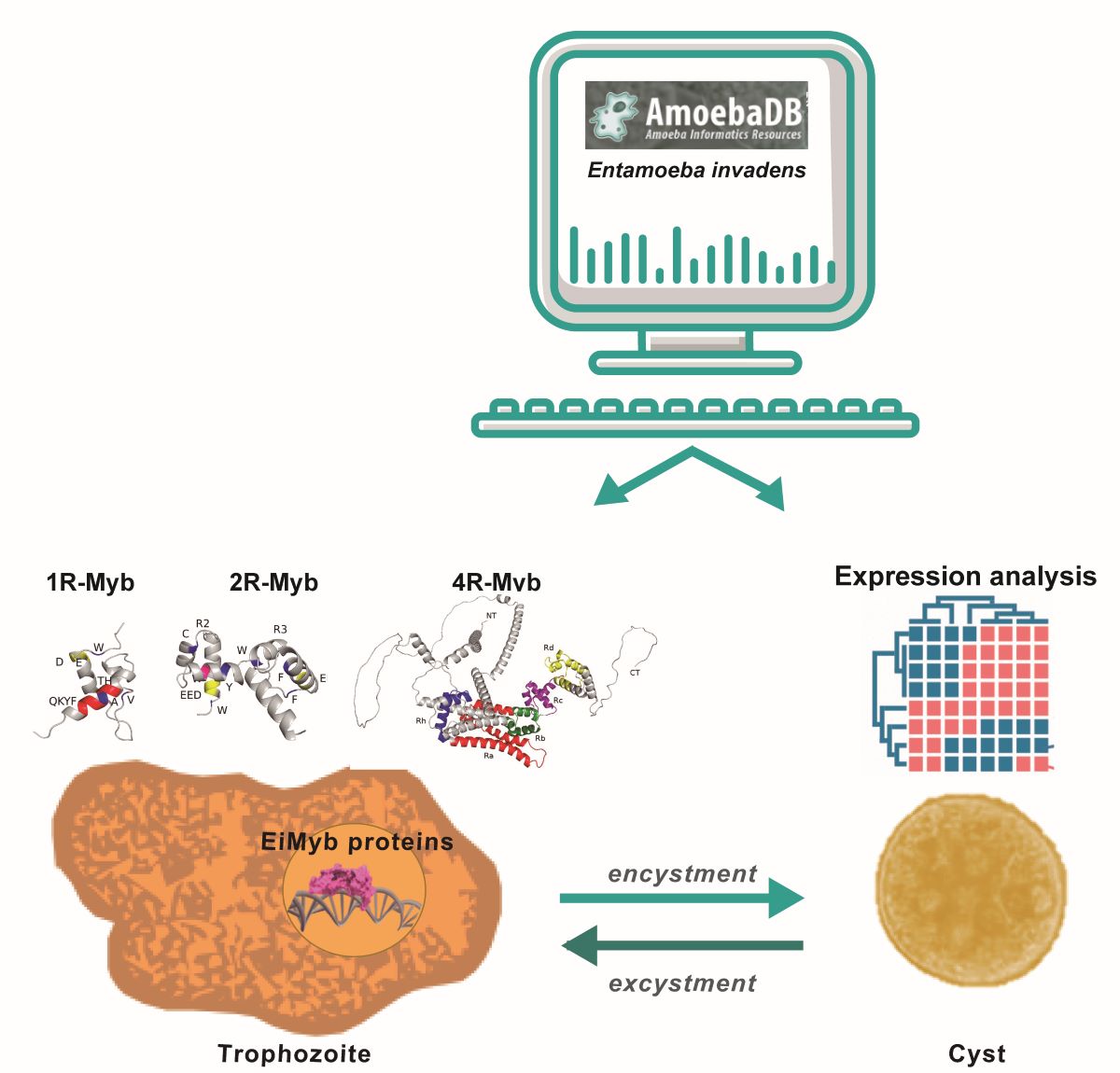
Keywords:
1. Introduction
2. Materials and Methods
2.1. Genomic data and identification of EiMyb-encoding proteins in E. invadens
2.2. EiMyb protein classification
2.3. Multiple Sequence Alignment and Phylogenetic Analysis of EiMyb Proteins
2.4. Amino acid sequence analysis of EiMyb proteins
2.5. eimyb gene expression analysis in E. invadens
2.6. Identification of Myb recognition elements (MRE) in the promoter regions of E. invadens genes
2.7. Analysis of enriched gene ontologies
3. Results and Discussion
3.1. Myb proteins in E. invadens
3.2. 1R-MYB subfamily in E. invadens
3.3. 2R-MYB subfamily in E. invadens
3.4. 4R MYB-DBD protein
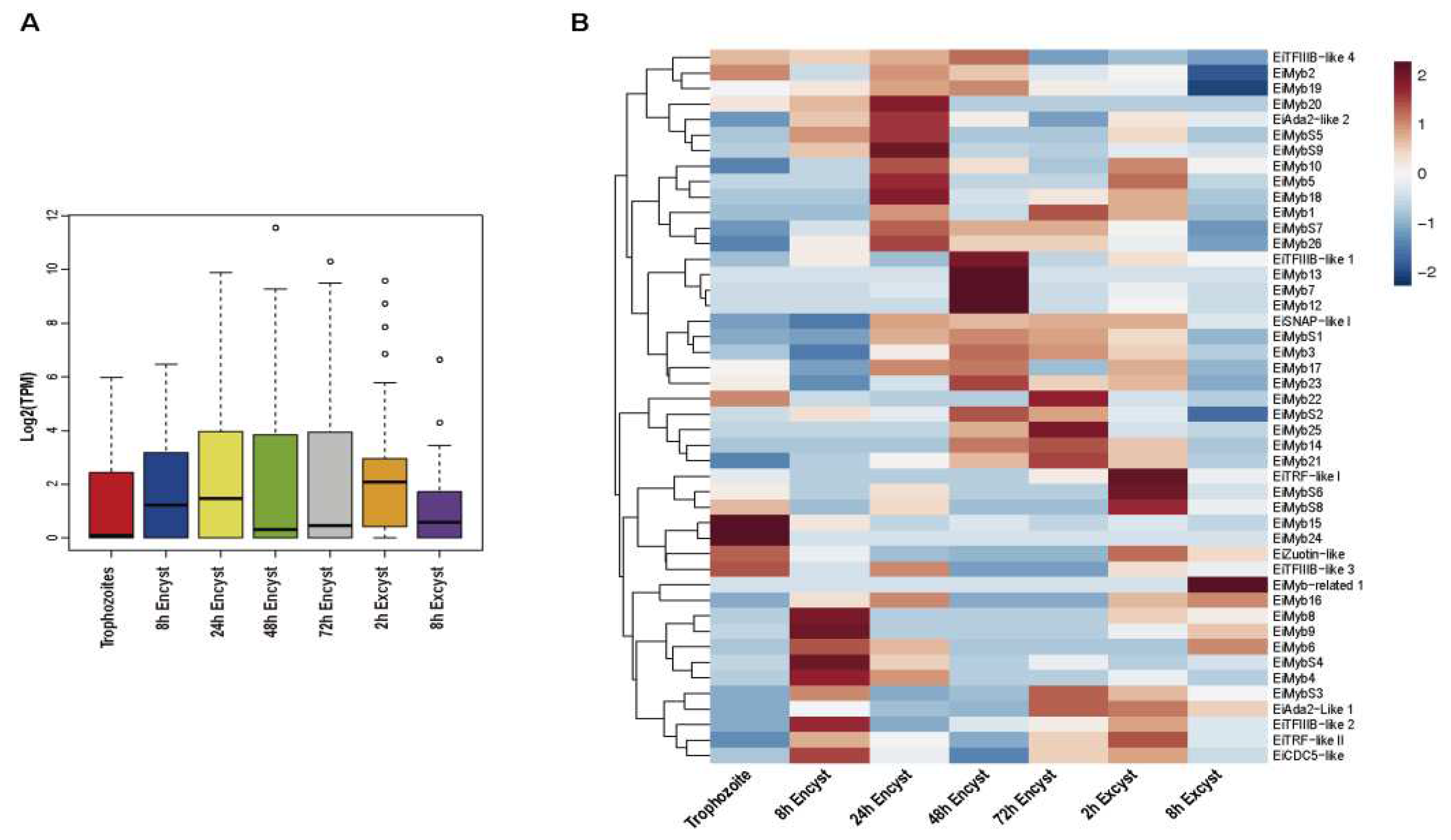
3.5. Expression analysis of the eimyb genes during trophozoite differentiation
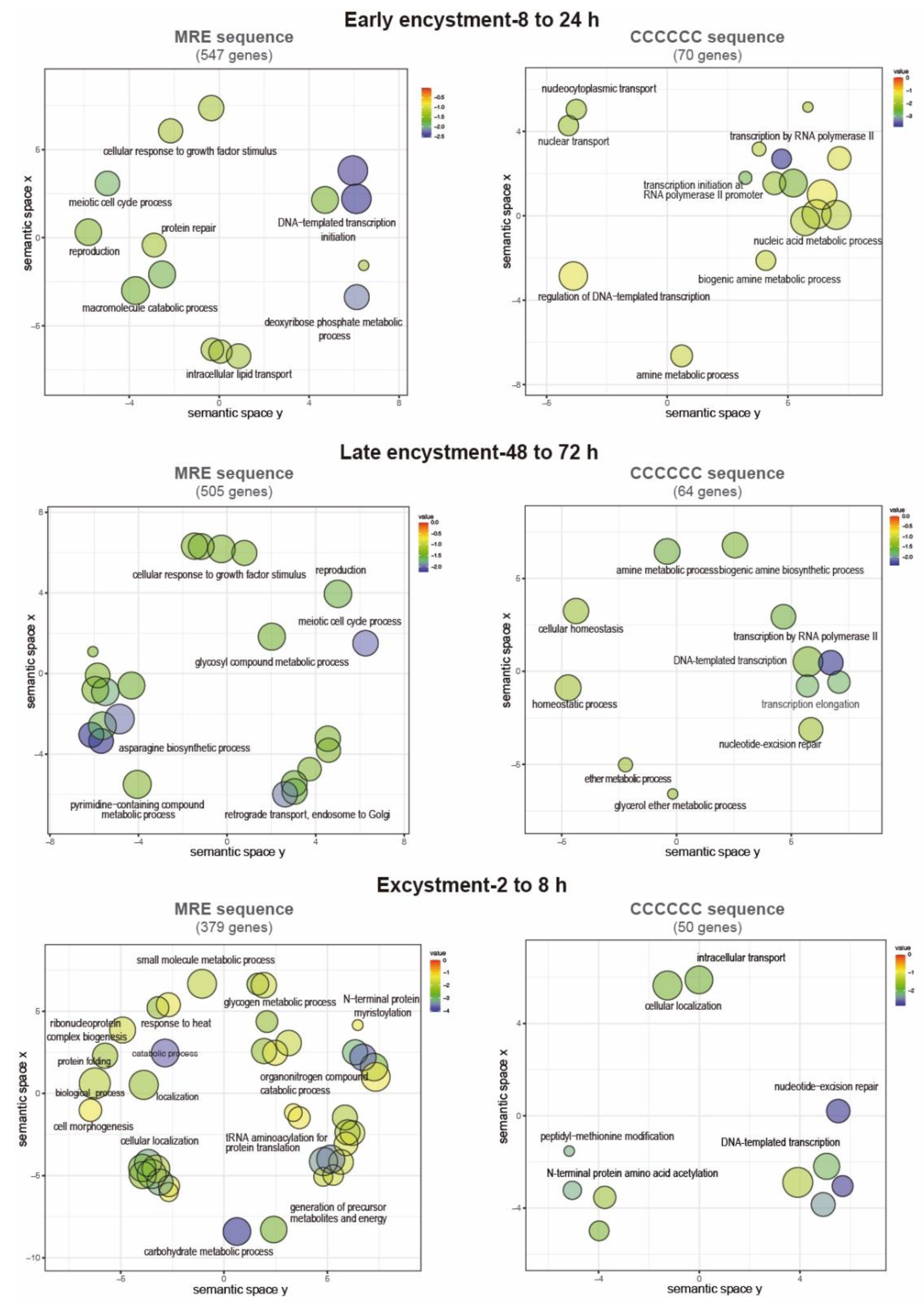
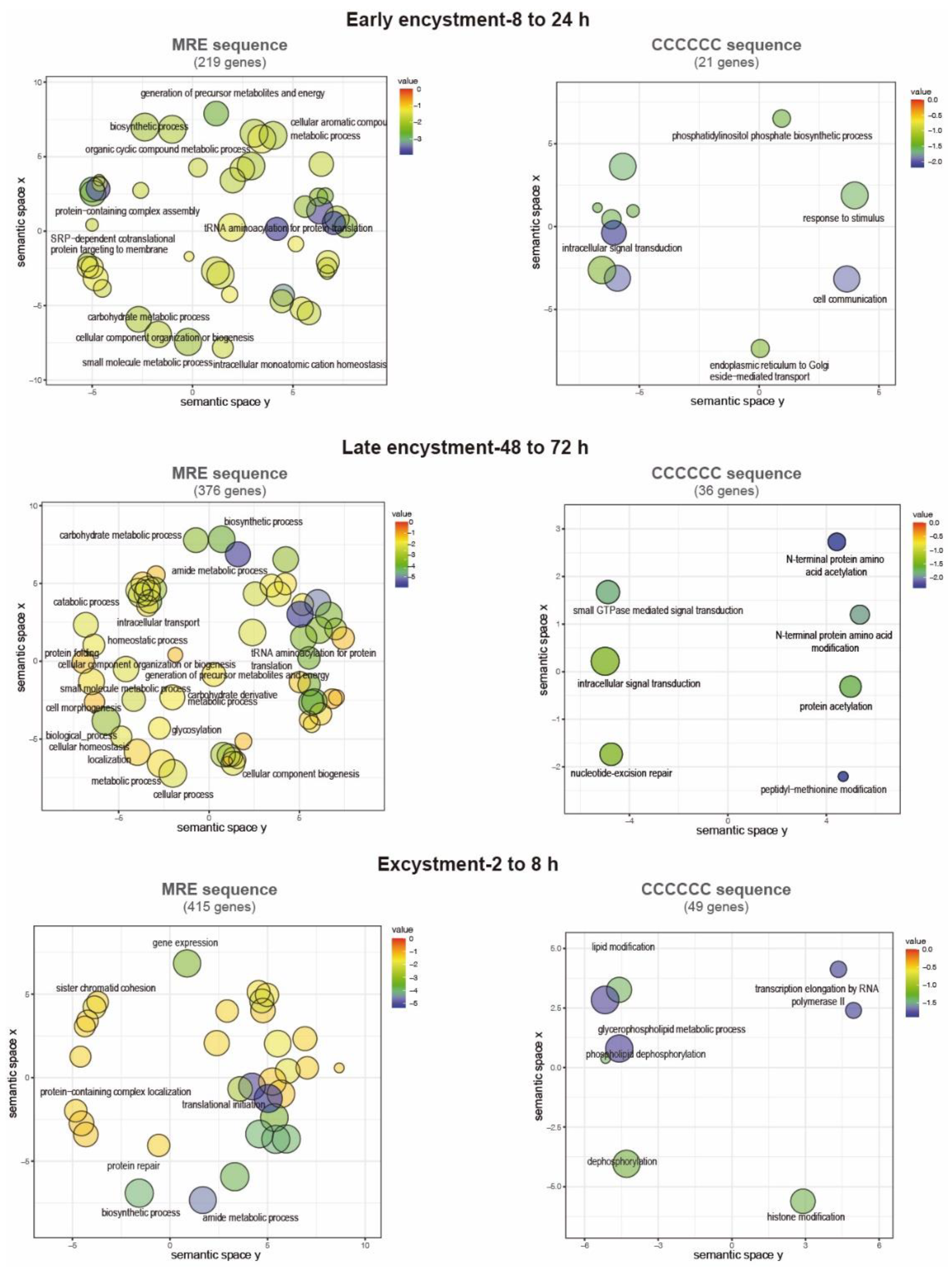
3.6. Presence of the Myb recognition element (MRE) and the C-rich sequence in E. invadens gene promoters
3.7. Functions of the putative EiMyb target genes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shirley, D.-A.T.; Farr, L.; Watanabe, K.; Moonah, S. A Review of the Global Burden, New Diagnostics, and Current Therapeutics for Amebiasis. Open Forum Infect Dis 2018, 5. [Google Scholar] [CrossRef] [PubMed]
- Haque, R.; Huston, C.D.; Hughes, M.; Houpt, E.; Petri, W.A. Amebiasis. New England Journal of Medicine 2003, 348, 1565–1573. [Google Scholar] [CrossRef] [PubMed]
- Eichinger, D. Encystation in Parasitic Protozoa. Curr Opin Microbiol 2001, 4, 421–426. [Google Scholar] [CrossRef]
- Eichinger, D. Encystation of Entamoeba Parasites. BioEssays 1997, 19, 633–639. [Google Scholar] [CrossRef] [PubMed]
- Wesel, J.; Shuman, J.; Bastuzel, I.; Dickerson, J.; Ingram-Smith, C. Encystation of Entamoeba Histolytica in Axenic Culture. Microorganisms 2021, 9, 873. [Google Scholar] [CrossRef]
- Ehrenkaufer, G.M.; Weedall, G.D.; Williams, D.; Lorenzi, H.A.; Caler, E.; Hall, N.; Singh, U. The Genome and Transcriptome of the Enteric Parasite Entamoeba Invadens, a Model for Encystation. Genome Biol 2013, 14, R77. [Google Scholar] [CrossRef]
- Wilson, I.W.; Weedall, G.D.; Lorenzi, H.; Howcroft, T.; Hon, C.-C.; Deloger, M.; Guillén, N.; Paterson, S.; Clark, C.G.; Hall, N. Genetic Diversity and Gene Family Expansions in Members of the Genus Entamoeba. Genome Biol Evol 2019, 11, 688–705. [Google Scholar] [CrossRef]
- Manna, D.; Lozano-Amado, D.; Ehrenkaufer, G.; Singh, U. The NAD+ Responsive Transcription Factor ERM-BP Functions Downstream of Cellular Aggregation and Is an Early Regulator of Development and Heat Shock Response in Entamoeba. Front Cell Infect Microbiol 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Manna, D.; Ehrenkaufer, G.M.; Singh, U. Regulation of Gene Expression in the Protozoan Parasite Entamoeba Invadens: Identification of Core Promoter Elements and Promoters with Stage-Specific Expression Patterns. Int J Parasitol 2014, 44, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Singh, U.; Ehrenkaufer, G. Recent Insights into Entamoeba Development: Identification of Transcriptional Networks Associated with Stage Conversion. Int J Parasitol 2009, 39 1, 41–47. [Google Scholar] [CrossRef]
- Manna, D.; Singh, U. Nuclear Factor Y (NF-Y) Modulates Encystation in Entamoeba via Stage-Specific Expression of the NF-YB and NF-YC Subunits. mBio 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Meenakshi; Balbhim, S.S.; Sarkar, S.; Vasudevan, M.; Ghosh, S.K. Three-amino Acid Loop Extension Homeodomain Proteins Regulate Stress Responses and Encystation in Entamoeba. Mol Microbiol. 2023; 120, 276–297. [CrossRef]
- De Cádiz, A.E.; Jeelani, G.; Nakada-Tsukui, K.; Caler, E.; Nozaki, T. Transcriptome Analysis of Encystation in Entamoeba Invadens. PLoS One 2013, 8, e74840. [Google Scholar] [CrossRef] [PubMed]
- Bilaud, T.; Koering, C.E.; Binet-Brasselet, E.; Ancelin, K.; Pollice, A.; Gasser, S.M.; Gilson, E. The Telobox, a Myb-Related Telomeric DNA Binding Motif Found in Proteins from Yeast, Plants and Human. Nucleic Acids Res 1996, 24, 1294–1303. [Google Scholar] [CrossRef] [PubMed]
- Lipsick, JS. One Billion Years of Myb. Oncogene 1996, 13, 223–35. [Google Scholar]
- Burns, C.G.; Ohi, R.; Krainer, A.R.; Gould, K.L. Evidence That Myb-Related CDC5 Proteins Are Required for Pre-MRNA Splicing. Proceedings of the National Academy of Sciences 1999, 96, 13789–13794. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB Transcription Factors in Arabidopsis. Trends Plant Sci 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Collier, S.E.; Voehler, M.; Peng, D.; Ohi, R.; Gould, K.L.; Reiter, N.J.; Ohi, M.D. Structural and Functional Insights into the N-Terminus of Schizosaccharomyces Pombe Cdc5. Biochemistry 2014, 53, 6439–6451. [Google Scholar] [CrossRef] [PubMed]
- Ogata, K.; Hojo, H.; Aimoto, S.; Nakai, T.; Nakamura, H.; Sarai, A.; Ishii, S.; Nishimura, Y. Solution Structure of a DNA-Binding Unit of Myb: A Helix-Turn-Helix-Related Motif with Conserved Tryptophans Forming a Hydrophobic Core. Proceedings of the National Academy of Sciences 1992, 89, 6428–6432. [Google Scholar] [CrossRef]
- Otsuka, H.; Van Haastert, P.J.M. A Novel Myb Homolog Initiates Dictyostelium Development by Induction of Adenylyl Cyclase Expression. Genes Dev 1998, 12, 1738–1748. [Google Scholar] [CrossRef] [PubMed]
- Tsujioka, M.; Zhukovskaya, N.; Yamada, Y.; Fukuzawa, M.; Ross, S.; Williams, J.G. Dictyostelium Myb Transcription Factors Function at Culmination as Activators of Ancillary Stalk Differentiation. Eukaryot Cell 2007, 6, 568–570. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Yang, T.; Yang, H.; Li, Z.; Qin, P.; Zhang, X.; Liang, X.; Li, J.; Chen, Q. Identification of Myb Genes in Euplotes Aediculatus May Indicate an Early Evolutionary Process. Gene 2013, 530, 266–272. [Google Scholar] [CrossRef]
- Alonso, A.M.; Schcolnicov, N.; Diambra, L.; Cóceres, V.M. In-Depth Comparative Analysis of Tritrichomonas Foetus Transcriptomics Reveals Novel Genes Linked with Adaptation to Feline Host. Sci Rep 2022, 12, 10057. [Google Scholar] [CrossRef]
- da Silva, M.S.; Perez, A.M.; da Silveira, R. de C.V; de Moraes, C.E.; Siqueira-Neto, J.L.; Freitas-Junior, L.H.; Cano, M.I.N. The Leishmania Amazonensis TRF (TTAGGG Repeat-Binding Factor) Homologue Binds and Co-Localizes with Telomeres. BMC Microbiol, 2010; 10, 136. [Google Scholar] [CrossRef]
- Li, B.; Espinal, A.; Cross, G.A.M. Trypanosome Telomeres Are Protected by a Homologue of Mammalian TRF2. Mol Cell Biol 2005, 25, 5011–5021. [Google Scholar] [CrossRef] [PubMed]
- Gissot, M.; Briquet, S.; Refour, P.; Boschet, C.; Vaquero, C. PfMyb1, a Plasmodium Falciparum Transcription Factor, Is Required for Intra-Erythrocytic Growth and Controls Key Genes for Cell Cycle Regulation. J Mol Biol 2005, 346, 29–42. [Google Scholar] [CrossRef] [PubMed]
- Alzan, H.F.; Knowles, D.P.; Suarez, C.E. Comparative Bioinformatics Analysis of Transcription Factor Genes Indicates Conservation of Key Regulatory Domains among Babesia Bovis, Babesia Microti, and Theileria Equi. PLoS Negl Trop Dis 2016, 10, e0004983. [Google Scholar] [CrossRef] [PubMed]
- Meneses, E.; Cárdenas, H.; Zárate, S.; Brieba, L.G.; Orozco, E.; López-Camarillo, C.; Azuara-Liceaga, E. The R2R3 Myb Protein Family in Entamoeba Histolytica. Gene 2010, 455, 32–42. [Google Scholar] [CrossRef]
- Hsu, H.-M.; Ong, S.-J.; Lee, M.-C.; Tai, J.-H. Transcriptional Regulation of an Iron-Inducible Gene by Differential and Alternate Promoter Entries of Multiple Myb Proteins in the Protozoan Parasite Trichomonas Vaginalis. Eukaryot Cell 2009, 8, 362–372. [Google Scholar] [CrossRef] [PubMed]
- Ong, S.-J.; Hsu, H.-M.; Liu, H.-W.; Chu, C.-H.; Tai, J.-H. Multifarious Transcriptional Regulation of Adhesion Protein Gene Ap65 - 1 by a Novel Myb1 Protein in the Protozoan Parasite Trichomonas Vaginalis. Eukaryot Cell 2006, 5, 391–399. [Google Scholar] [CrossRef]
- Huang, Y.-C.; Su, L.-H.; Lee, G.A.; Chiu, P.-W.; Cho, C.-C.; Wu, J.-Y.; Sun, C.-H. Regulation of Cyst Wall Protein Promoters by Myb2 in Giardia Lamblia. Journal of Biological Chemistry 2008, 283, 31021–31029. [Google Scholar] [CrossRef]
- Sun, C.-H.; Palm, D.; McArthur, A.G.; Svärd, S.G.; Gillin, F.D. A Novel Myb-Related Protein Involved in Transcriptional Activation of Encystation Genes in Giardia Lamblia. Mol Microbiol 2002, 46, 971–984. [Google Scholar] [CrossRef]
- Yang, H.; Chung, H.J.; Yong, T.; Lee, B.H.; Park, S. Identification of an Encystation-Specific Transcription Factor, Myb Protein in Giardia Lamblia. Mol Biochem Parasitol 2003, 128, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Waldman, B.S.; Schwarz, D.; Wadsworth, M.H.; Saeij, J.P.; Shalek, A.K.; Lourido, S. Identification of a Master Regulator of Differentiation in Toxoplasma. Cell 2020, 180, 359–372e16. [Google Scholar] [CrossRef]
- Ehrenkaufer, G.M.; Hackney, J.A.; Singh, U. A Developmentally Regulated Myb Domain Protein Regulates Expression of a Subset of Stage-Specific Genes in Entamoeba Histolytica. Cell Microbiol 2009, 11, 898–910. [Google Scholar] [CrossRef] [PubMed]
- Aurrecoechea, C.; Barreto, A.; Brestelli, J.; Brunk, B.P.; Caler, E. V.; Fischer, S.; Gajria, B.; Gao, X.; Gingle, A.; Grant, G.; et al. AmoebaDB and MicrosporidiaDB: Functional Genomic Resources for Amoebozoa and Microsporidia Species. Nucleic Acids Res 2011, 39, D612–D619. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Jarreta, J.; Amos, B.; Aurrecoechea, C.; Bah, S.; Barba, M.; Barreto, A.; Basenko, E.Y.; Belnap, R.; Blevins, A.; Böhme, U.; et al. VEuPathDB: The Eukaryotic Pathogen, Vector and Host Bioinformatics Resource Center in 2023. Nucleic Acids Res 2023. [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A Sequence Logo Generator: Figure 1. Genome Res 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.-F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny Fr: Robust Phylogenetic Analysis for the Non-Specialist. Nucleic Acids Res, 2008; 36, W465–W469. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Wen, L.; Gao, X.; Jin, C.; Xue, Y.; Yao, X. DOG 10: Illustrator of Protein Domain Structures. Cell Res. 2009, 19, 271–273. [Google Scholar] [CrossRef]
- Bailey, T.L. STREME: Accurate and Versatile Sequence Motif Discovery. Bioinformatics 2021, 37, 2834–2840. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS One 2011, 6, e21800. [Google Scholar] [CrossRef]
- Broccoli, D.; Smogorzewska, A.; Chong, L.; de Lange, T. Human Telomeres Contain Two Distinct Myb–Related Proteins, TRF1 and TRF2. Nat Genet 1997, 17, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Alabadı́, D.; Oyama, T.; Yanovsky, M.J.; Harmon, F.G.; Más, P.; Kay, S.A. Reciprocal Regulation Between TOC1 and LHY / CCA1 Within the Arabidopsis Circadian Clock. Science (1979) 2001, 293, 880–883. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, A.; Kudapa, H.; Pazhamala, L.T.; Garg, V.; Varshney, R.K. Gene Expression and Yeast Two-Hybrid Studies of 1R-MYB Transcription Factor Mediating Drought Stress Response in Chickpea (Cicer Arietinum L.). Front Plant Sci 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Aasland R, S.A.G.T. The SANT Domain: A Putative DNA-Binding Domain in the SWI-SNF and ADA Complexes, the Transcriptional Co-Repressor N-CoR and TFIIIB. . Trends Biochem Sci. 1996, 21, 87–95. [Google Scholar] [CrossRef]
- Boyer, L.A.; Latek, R.R.; Peterson, C.L. The SANT Domain: A Unique Histone-Tail-Binding Module? Nat Rev Mol Cell Biol 2004, 5, 158–163. [Google Scholar] [CrossRef]
- Carre, I.A. MYB Transcription Factors in the Arabidopsis Circadian Clock. J Exp Bot 2002, 53, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Schaffer, R.; Ramsay, N.; Samach, A.; Corden, S.; Putterill, J.; Carré, I.A.; Coupland, G. The Late Elongated Hypocotyl Mutation of Arabidopsis Disrupts Circadian Rhythms and the Photoperiodic Control of Flowering. Cell 1998, 93, 1219–1229. [Google Scholar] [CrossRef] [PubMed]
- Rendón-Gandarilla, F.J.; Álvarez-Hernández, V.; Castañeda-Ortiz, E.J.; Cárdenas-Hernández, H.; Cárdenas-Guerra, R.E.; Valdés, J.; Betanzos, A.; Chávez-Munguía, B.; Lagunes-Guillen, A.; Orozco, E.; et al. Telomeric Repeat-Binding Factor Homologs in Entamoeba Histolytica: New Clues for Telomeric Research. Front Cell Infect Microbiol 2018, 8. [Google Scholar] [CrossRef]
- Fan, Q.; An, L.; Cui, L. PfADA2, a Plasmodium Falciparum Homologue of the Transcriptional Coactivator ADA2 and Its in Vivo Association with the Histone Acetyltransferase PfGCN5. Gene 2004, 336, 251–261. [Google Scholar] [CrossRef]
- Shoji, W.; Inoue, T.; Yamamoto, T.; Obinata, M. MIDA1, a Protein Associated with Id, Regulates Cell Growth. Journal of Biological Chemistry 1995, 270, 24818–24825. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Lockshin, C.; Herbert, A.; Winter, E.; Rich, A. Zuotin, a Putative Z-DNA Binding Protein in Saccharomyces Cerevisiae. EMBO J 1992, 11, 3787–3796. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, M.L.; Reinbolt, J.; Gangloff, J.; Dirheimer, G.; Wilhelm, F.X. Transfer RNA Binding Protein in the Nucleus of Saccharomyces Cerevisiae. FEBS Lett 1994, 349, 260–264. [Google Scholar] [CrossRef] [PubMed]
- Yan, W. Zuotin, a Ribosome-Associated DnaJmolecular Chaperone. EMBO J 1998, 17, 4809–4817. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, O.K.; Sharma, R.; Tomiczek, B.; Lee, W.; Tonelli, M.; Cornilescu, G.; Stolarska, M.; Nierzwicki, L.; Czub, J.; Markley, J.L.; et al. Structure and Evolution of the 4-Helix Bundle Domain of Zuotin, a J-Domain Protein Co-Chaperone of Hsp70. PLoS One 2019, 14, e0217098. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.-L.; Wu, C.-C.; Lee, J.-C.; Chen, H.-T. A Region of Bdp1 Necessary for Transcription Initiation That Is Located within the RNA Polymerase III Active Site Cleft. Mol Cell Biol 2015, 35, 2831–2840. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.-K.; Rao, G.-Y. Insights into the Diversification and Evolution of R2R3-MYB Transcription Factors in Plants. Plant Physiol 2020, 183, 637–655. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Kumar, N.; Dwivedi, A.K.; Sharma, R.; Sharma, M.K. Phylogenomic Analysis of R2R3 MYB Transcription Factors in Sorghum and Their Role in Conditioning Biofuel Syndrome. Curr Genomics 2020, 21, 138–154. [Google Scholar] [CrossRef]
- Millard, P.S.; Kragelund, B.B.; Burow, M. R2R3 MYB Transcription Factors – Functions Outside the DNA-Binding Domain. Trends Plant Sci 2019, 24, 934–946. [Google Scholar] [CrossRef] [PubMed]
- Guehmann, S.; Vorbrueggen, G.; Kalkbrenner, F.; Moelling, K. Reduction of a Conserved Cys Is Essential for Myb DNA-Binding. Nucleic Acids Res 1992, 20, 2279–2286. [Google Scholar] [CrossRef]
- Hirayama, T.; Shinozaki, K. A Cdc5+ Homolog of a Higher Plant, Arabidopsis Thaliana. Proceedings of the National Academy of Sciences 1996, 93, 13371–13376. [Google Scholar] [CrossRef] [PubMed]
- Thiedig, K.; Weisshaar, B.; Stracke, R. Functional and Evolutionary Analysis of the Arabidopsi s 4R-MYB Protein SNAPc4 as Part of the SNAP Complex. Plant Physiol 2021, 185, 1002–1020. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Li, X.; Hou, X.; Cao, S.; Cao, W.; Zhang, Y.; Song, J.; Wang, M.; Wang, H.; Yan, X.; et al. Structural Basis of Human SNAPc Recognizing Proximal Sequence Element of SnRNA Promoter. Nat Commun 2022, 13, 6871. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.W.; Henry, R.W.; Ma, B.; Kobayashi, R.; Klages, N.; Matthias, P.; Strubin, M.; Hernandez, N. The Large Subunit of Basal Transcription Factor SNAP c Is a Myb Domain Protein That Interacts with Oct-1. Mol Cell Biol 1998, 18, 368–377. [Google Scholar] [CrossRef] [PubMed]
- Henry, R.W.; Mittal, V.; Ma, B.; Kobayashi, R.; Hernandez, N. SNAP19 Mediates the Assembly of a Functional Core Promoter Complex (SNAP c ) Shared by RNA Polymerases II and III. Genes Dev 1998, 12, 2664–2672. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, D.; Ghosh, S.K. Cellular Events of Multinucleated Giant Cells Formation During the Encystation of Entamoeba Invadens. Front Cell Infect Microbiol 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Reiner, D.S.; McCaffery, M.; Gillin, F.D. Sorting of Cyst Wall Proteins to a Regulated Secretory Pathway during Differentiation of the Primitive Eukaryote, Giardia Lamblia. Eur J Cell Biol 1990, 53, 142–153. [Google Scholar]
- Galán-Vásquez, E.; Gómez-García, M. del C; Pérez-Rueda, E. A Landscape of Gene Regulation in the Parasitic Amoebozoa Entamoeba Spp. PLoS One 2022, 17, e0271640. [Google Scholar] [CrossRef]

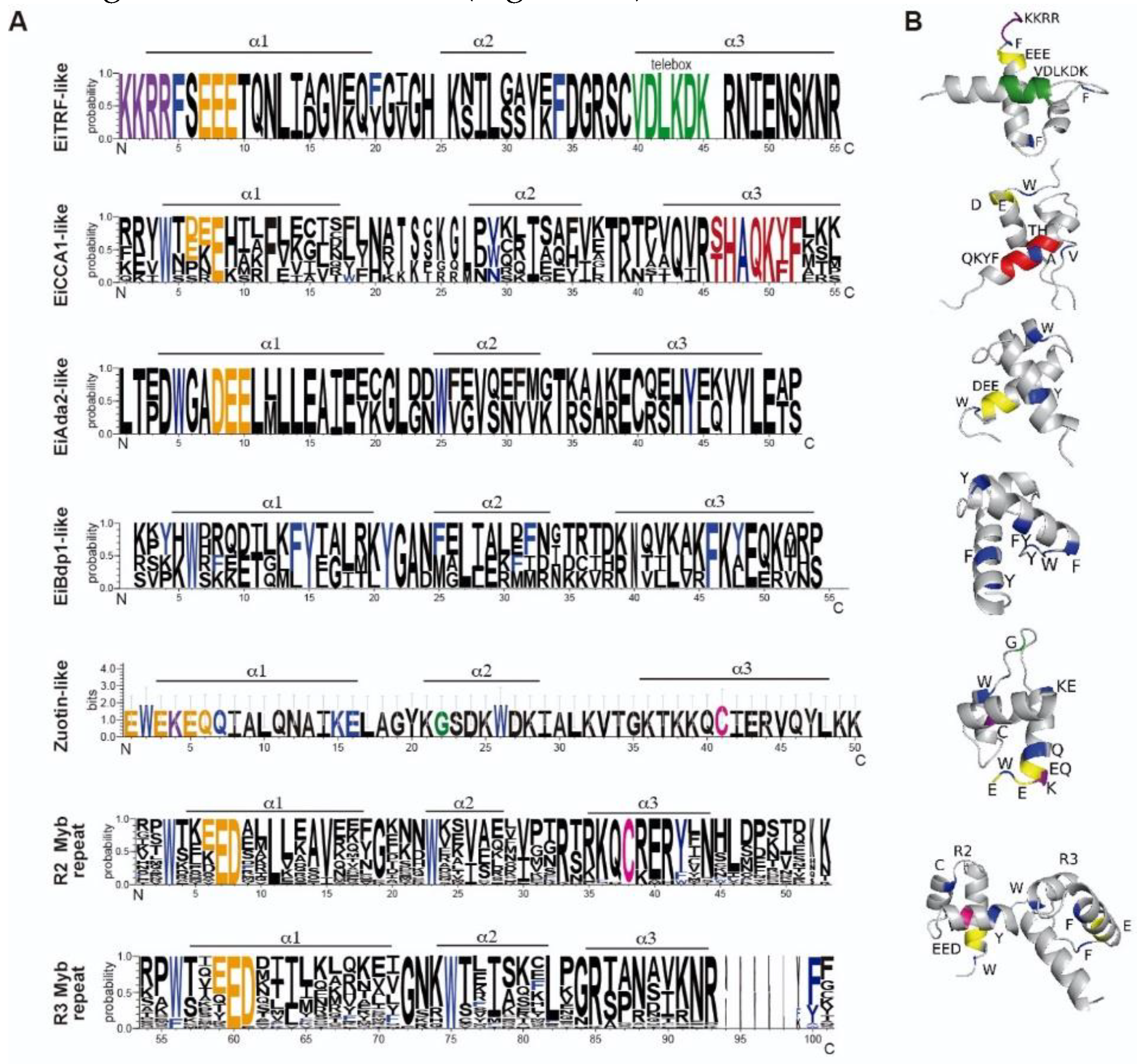
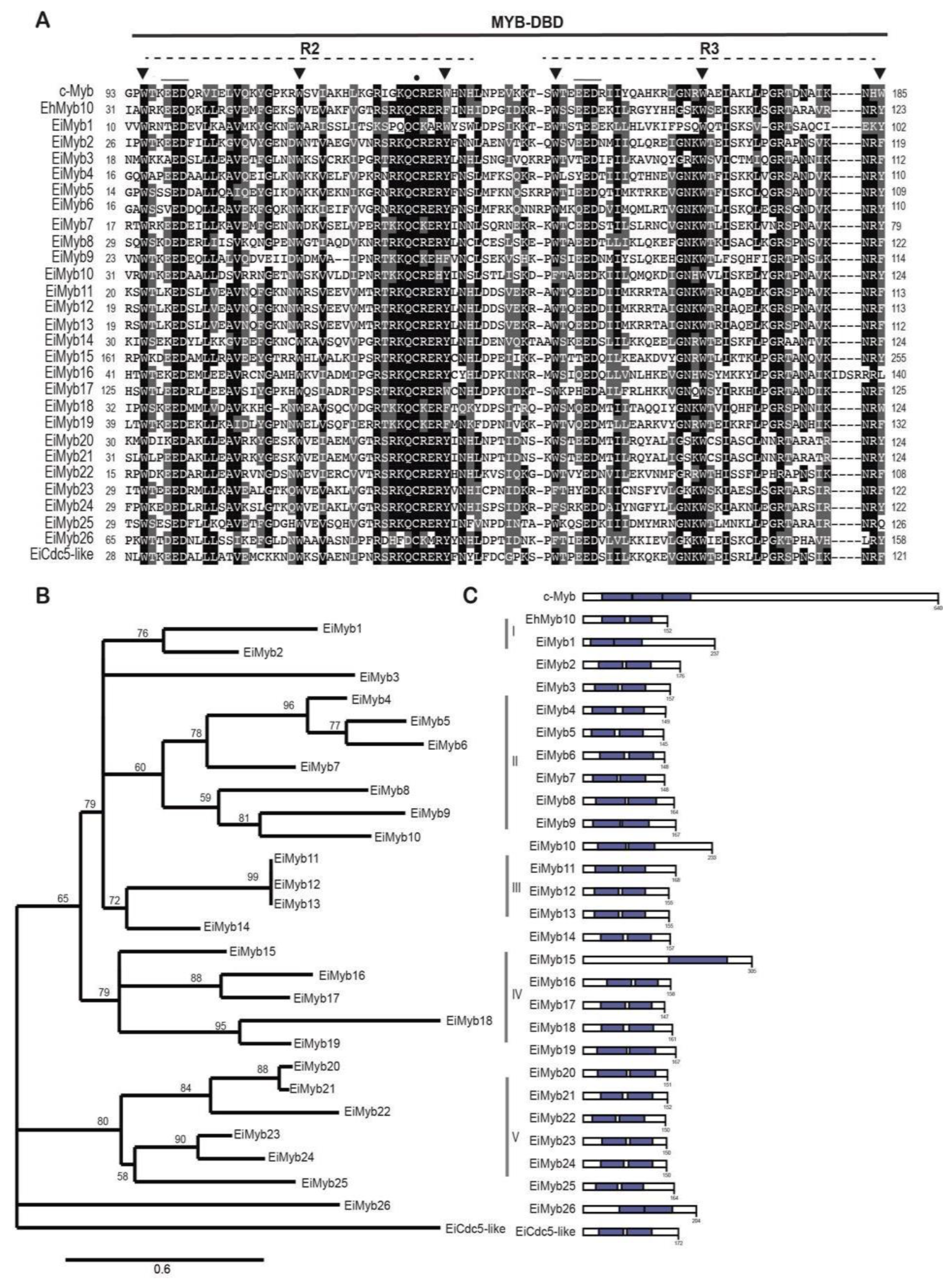
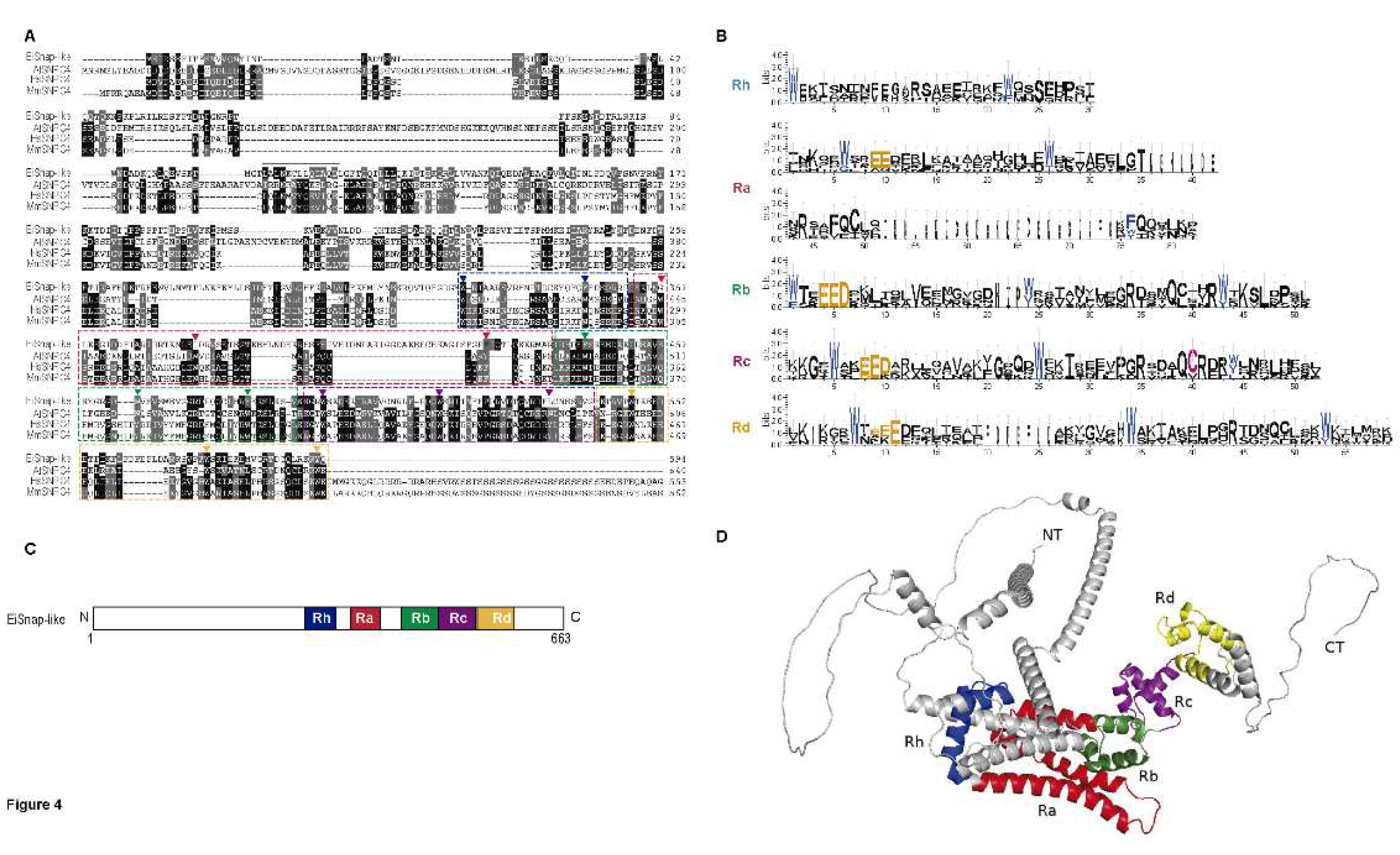
| Myb subfamily | Groups | Number of members |
| 1R-MYB | SHAQKYF (CCA1-like ) | 9 |
| Bdp1-like | 4 | |
| TRF-like | 2 | |
| Ada2-like (Transcriptional adapter putative-Ada2) | 2 | |
| Myb-related | 1 | |
| Zuotin-like | 1 | |
| 2R-MYB | Myb transcription factors | 13 |
| Trichome differentiation protein GL1 related | 6 | |
| Werewolf transcription factors related | 3 | |
| Hypothetical proteins | 3 | |
| R2R3-Myb transcription factors | 2 | |
| 4R-MYB | Snap190 | 1 |
| Total | 47 |
| 1R-MYBS | ||||||||||
| Group | Gene ID |
Gene (pb) |
mRNA |
Protein name | DBD-MYB | InterProScan domains | CD-Search domains |
H. sapiens c-Myb |
A. thaliana | E. histolytica |
| TRF-like |
EIN_023650 |
1290 | 1290 |
EiTRF-like I |
348-392 |
Telomeric Repeat Binding Factor 1// TM Helix |
SANT_TRF/SANT Superfamily |
29.07% 7e-12 29.90% 3e-17 (TERF1) |
48.08% 3e-13 (CAD531509.1) |
49.89% 5e-133 (EHI_148140) |
|
EIN_079420 |
1404 | 1404 |
EiTRF-like II |
378-422 |
Telomeric Repeat Binding Factor 1// TM Helix |
SANT_TRF/SANT Superfamily |
26.32% 4e-14 35.87 8e-17 (TERF1) |
21.54% 2e-15 (OAP03200.1) |
55.13% 5e-164 (EHI_001110) |
|
| CCA-like (SHAQKYF) | EIN_086260 |
540 |
540 |
EiMybS1 |
48-92 |
Myb-DNA Binding/SANT Superfamily | Myb-DNA Binding/SANT Superfamily/RSC8 Chromatin remodeling | 22.95% 4e-14 |
64.71% 6e-25 (AAF23291.1) |
63.10% 2e-45 (EHI_092160) |
| EIN_087120 |
537 |
537 |
EiMybS2 |
46-90 |
Myb-DNA Binding/SANT Superfamily | Myb-DNA Binding/SANT Superfamily | 21.92% 1e-14 |
45.37% 6e-22 (AAF81310.1) |
52.43% 2e-35 (EHI_092160) |
|
| EIN_031250* |
601 |
534 |
EiMybS3 |
41-85 |
Myb-DNA Binding/SANT Superfamily | Myb-DNA Binding/SANT Superfamily | 25.45% 2e-14 |
58.06% 6e-20 (OAP07468.1) |
54.55% 4e-35 (EHI_136420) |
|
| EIN_095950 |
519 |
519 |
EiMybS4 |
83-133 |
Myb-DNA Binding/SANT Superfamily | SANT Superfamily |
16.13% 1e-11 |
20.83% 3e-12 (CAA0383923.1) |
38.22% 1e-18 (EHI_155580) |
|
| EIN_224050 |
516 |
516 |
EiMybS5 |
83-133 |
Myb-DNA Binding/SANT Superfamily | SANT Superfamily |
25.64% 2e-09 |
27.78% 7e-12 (NP_00107786.1) |
39.10% 6e-17 (EHI_155580) |
|
| EIN_020720 |
438 |
438 |
EiMybS6 |
56-106 |
Myb-DNA Binding/SANT Superfamily | SANT Superfamily |
- |
24.29% 2e-12 (CAA0367555.1) |
62.02% 5e-36 (EHI_051440) |
|
| EIN_469690 |
399 |
399 |
EiMybS7 |
41-91 |
Myb-DNA Binding/SANT Superfamily | SANT Superfamily |
- |
21.51% 4e-16 (AAM63125.1) |
59.84% 5e-38 (EHI_051440) |
|
| EIN_081930 |
408 |
408 |
EiMybS8 |
45-95 |
Myb-DNA Binding/SANT Superfamily | SANT Superfamily |
- |
29.63% 5e-14 (CAA0198797.1) |
51.88% 1e-33 (EHI_013340) |
|
| EIN_407300 |
546 |
546 |
EiMybS9 |
88-138 |
Myb-DNA Binding/SANT Superfamily | SANT Superfamily |
23.64 % 3e-11 |
19.77% 5e-12 (NP_001330337.1) |
46.92% 6e-19 (EHI_038640) |
|
| Ada2-like | EIN_359680* |
1229 |
993 |
EiAda2-Like 1 |
66-108 |
ADA2-like ZZ |
Histone acetyltransferase complex SAGA/ADA, subunit ADA2 [Chromatin structure and dynamics] | 18.75% 3e-13 |
30.59% 1e-62 (CAD5328117.1) |
64.85% 9e-146 (EHI_142140) |
|
EIN_390470 |
1032 |
1032 |
EiAda2-like 2 |
74-116 |
ADA2-like ZZ |
Histone acetyltransferase complex SAGA/ADA, subunit ADA2 [Chromatin structure and dynamics] | 20.83% 1e-09 |
34.38% 2e-60 (CAD5328118.1) |
46.86% 9e-92 (EHI_142140) |
|
| Myb-related | EIN_020090 |
423 |
423 |
EiMyb-related 1 |
47-91 |
Myb-DNA Binding/SANT Superfamily | Myb-DNA Binding/SANT Superfamily | 29.1% 4e-17 |
32% 4e-18 (NP_201038.1) |
25.00% 6e-19 (EHI_009930) |
|
Zuotin |
EIN_182440* |
1658 |
1596 |
EiZuotin-like |
472-521 |
DNAJ domain |
ZUO1 Superfamily / SANT Superfamily | 16.26% 2e-21 |
30.24% 2e-29 (AAG51437.1) |
52.35% 8e-62 (EHI_128200) |
|
Bdp1-like |
EIN_223710 |
366 | 366 | EiBdp1-like 1 |
45-111 |
Transcription factor TFIIIB component B’, Myb domain |
SANT/Myb-like DNA-binding domain-containing protein |
- 28.95% 0.017 TFIIIB |
31.58% 8e-05 (CAB43631.1) |
34.29% 2e-10 (EHI_074810) |
|
EIN_034860 |
363 | 363 |
EiBdp1-like 2 |
48-110 |
Transcription factor TFIIIB component B'', Myb domain |
SANT/Myb-like DNA-binding domain-containing protein |
- 30.88% 5e-20 TFIIIB |
25.69% 4e-20 (CAD5330371.1) |
ND |
|
|
EIN_314460 |
522 | 522 |
EiBdp1-like 3 |
84-126 |
Transcription factor TFIIIB component B'', Myb domain |
SANT/Myb-like DNA-binding domain-containing protein |
17.65% 6e-15 |
23.70% 5e-20 (CAD5330371.1) |
65.68% 3e-37 (EHI_009820) |
|
| EIN_096130 | 312 | 312 | EiBdp1-like 4 | 30-90 | Transcription factor TFIIIB component B'', Myb domain | BDP1 super family | - | 30.77% 3e-06 (CAB43631.1) |
40.59% 3e-12 (EHI_009820) |
|
| 2R-MYBS | ||||||||||
|
I |
EIN_284910 |
519 |
519 |
EiMyb1 |
28-121 |
Myb-DNA Binding/SANT Superfamily |
Transcription repressor MYB5; Provisional |
33.63% 6e-44 |
34.35% 1e-42 (NP_001330339.1) |
42.55% 2e-43 (EHI_063550) |
|
EIN_178740 |
531 |
531 |
EiMyb2 |
26-119 |
Myb-DNA Binding/SANT Superfamily |
Transcription repressor MYB5; Provisional |
30.89% 4e-46 |
33.33% 1e-43 (NP_190575.1) |
42.25% 1e-39 (EHI_063550) |
|
|
- |
EIN_047330 |
474 |
474 |
EiMyb3 |
18-112 |
Myb-DNA Binding/SANT Superfamily |
SANT DNA binding domain / Transcription repressor MYB5; Provisional |
38.39% 2e-40 |
31.45% 2e-42 (NP_190575.1) |
35.26% 1e-41 (EHI_063550) |
|
II |
EIN_206260 |
450 |
450 |
EiMyb4 |
16-110 |
Myb-DNA Binding/SANT Superfamily |
PLN03091 super family hypothetical protein; Provisional | 34.17% 6e-45 |
35.42% 1e-45 (NP_190575.1) |
34.09% 6e-41 (EHI_063550) |
|
EIN_169560 |
438 |
438 |
EiMyb5 |
14-109 |
Myb-DNA Binding/SANT Superfamily |
REB1 superfamily |
25.19% 1e-44 |
37.93% 5e-43 (NP_190575.1) |
37.40% 1e-37 (EHI_063550) |
|
|
EIN_168610 |
447 |
447 |
EiMyb6 |
16-110 |
Myb-DNA Binding/SANT Superfamily |
PLN03091 super family hypothetical protein; Provisional |
27.83% 1e-42 |
37.93% 5e-43 (NP_190575.1) |
39.69% 1e-37 (EHI_098070) |
|
|
EIN_207200 |
447 |
447 |
EiMyb7 |
17-79 |
Myb-DNA Binding/SANT Superfamily |
REB1 superfamily |
29.29% 4e-45 |
36.36% 1e-44 (NP_190575.1) |
42.34% 5e-41 (EHI_098070) |
|
|
EIN_022390 |
495 |
495 |
EiMyb8 |
29-122 |
Myb-DNA Binding/SANT Superfamily |
REB1 superfamily |
31.03% 5e-46 |
30.71% 4e-43 (NP_190575.1) |
42.31% 2e-39 (EHI_063550) |
|
|
EIN_080130 |
504 |
504 |
EiMyb9 |
23-114 |
Myb-DNA Binding/SANT Superfamily |
REB1 superfamily |
32.65% 4e-46 |
33.10% 1e-41 (NP_190575.1) |
38.40% 2e-40 (EHI_063550) |
|
|
EIN_276810* |
754 |
702 |
EiMyb10 |
31-124 |
Myb-DNA Binding/SANT Superfamily |
Transcription repressor MYB5; Provisional |
38.53% 4e-42 |
33.56% 6e-42 (NP_190575.1) |
31.48% 2e-38 (EHI_063550) |
|
|
III |
EIN_307410 |
507 |
507 |
EiMyb11 |
20-113 |
Myb-DNA Binding/SANT Superfamily |
Transcription repressor MYB5; Provisional |
30.58% 5e-46 |
31.72 7e-46 (VYS56784.1) |
47.47% 4e-39 (EHI_063550) |
|
EIN_307180 |
468 |
468 |
EiMyb12 |
19-113 |
Myb-DNA Binding/SANT Superfamily |
Transcription repressor MYB5; Provisional |
38.61% 5e-43 |
32.88% 5e-44 (NP_190575.1) |
40.27% 2e-41 (EHI_063550) |
|
|
EIN_308550 |
468 |
468 |
EiMyb13 |
19-112 |
Myb-DNA Binding/SANT Superfamily |
Transcription repressor MYB5; Provisional |
38.53% 1e-43 |
34.04% 4e-43 (NP_190575.1) |
40.29% 1e-40 (EHI_063550) |
|
|
- |
EIN_095310 |
474 | 474 |
EiMyb14 |
30-124 |
Myb-DNA Binding/SANT Superfamily |
PLN03091 super family hypothetical protein; Provisional |
37.07% 3e-40 |
33.87% 1e-40 (NP_195443.1) |
51.52% 2e-40 (EHI_153350 |
|
IV |
EIN_399710* |
1119 |
918 |
EiMyb15 |
161-255 |
Myb-DNA Binding/SANT Superfamily |
REB1 superfamily |
35.66% 2e-43 |
34.27% 2e-42 (NP_195443.1) |
68.29% 1e-48 (EHI_098070) |
|
EIN_490880 |
477 |
477 |
EiMyb16 |
41-140 |
Myb-DNA Binding/SANT Superfamily |
REB1 superfamily |
38.89% 2e-41 |
35.83% 1e-42 (NP_195443.1) |
39.09% 3e-39 (EHI_063550) |
|
|
EIN_310240 |
444 |
444 |
EiMyb17 |
31-125 |
Myb-DNA Binding/SANT Superfamily |
REB1 superfamily |
45.05% 4e-44 |
36.36% 3e-44 (NP_195443.1) |
44.25% 9e-40 (EHI_098070) |
|
|
EIN_425380 |
486 |
486 |
EiMyb18 |
32-124 |
Myb-DNA Binding/SANT Superfamily |
Transcription repressor MYB5; Provisional |
28.93% 1e-43 |
33.05% 6e-42 (OAO92063.1) |
44.14% 4e-35 (EHI_098070) |
|
|
EIN_046410 |
504 |
504 |
EiMyb19 |
39-132 |
Myb-DNA Binding/SANT Superfamily |
REB1 superfamily |
34.58% 2e-43 |
34.91% 1e-41 (OAO92063.1) |
51.25% 3e-47 (EHI_063550) |
|
|
V |
EIN_183110 |
456 |
456 |
EiMyb20 |
31-124 |
Myb-DNA Binding/SANT Superfamily |
PLN03091 super family hypothetical protein; Provisional |
33.07% 4e-46 |
37.01% 3e-46 (VYS56784.1) |
38.84% 1e-38 (EHI_098070) |
|
EIN_183730 |
459 |
459 |
EiMyb21 |
31-124 |
Myb-DNA Binding/SANT Superfamily |
PLN03091 super family hypothetical protein; Provisional |
38.10% 2e-42 |
33.08% 2e-42 (CAA0401764.1) |
64.10% 2e-39 (EHI_130060) |
|
|
EIN_169190 |
453 |
453 |
EiMyb22 |
15-108 |
Myb-DNA Binding/SANT Superfamily |
Transcription repressor MYB5; Provisional |
29.82% 2e-41 |
35.77% 2e-41 (AAS58517.1) |
46.48% 7e-39 (EHI_168310) |
|
|
EIN_359630 |
453 |
453 |
EiMyb23 |
29-122 |
Myb-DNA Binding/SANT Superfamily |
PLN03091 super family hypothetical protein; Provisional |
35.45% 4e-43 |
39.05% 2e-42 (CAA0383923.1) |
57.58% 4e-44 (EHI_129790) |
|
|
EIN_379820 |
453 |
453 |
EiMyb24 |
29-122 |
Myb-DNA Binding/SANT Superfamily |
PLN03091 super family hypothetical protein; Provisional |
34.43% 5e-44 |
32.41% 1e-43 (VYS56784.1) |
53.62% 5e-42 (EHI_129790) |
|
|
- |
EIN_168860 |
495 |
495 |
EiMyb25 |
29-126 |
Myb-DNA Binding/SANT Superfamily |
PLN03091 super family hypothetical protein; Provisional |
29.13% 9e-46 |
33.93% 3e-42 (CAD5329766.1) |
46.23% 1e-32 (EHI_092700) |
|
- |
EIN_405040 |
615 |
615 |
EiMyb26 |
65-458 |
Myb-DNA Binding/SANT Superfamily |
REB1 superfamily |
28.23% 4e-40 |
30.86% 1e-43 (AAS58517.1) |
43.37% 5e-36 (EHI_053000) |
|
- |
EIN_248780 |
714 |
714 |
EiCDC5-like |
10-102 |
CDC5L_II |
22.78% 5e-39 58.71% 8e-41 (CDC5) |
53.45% 2e-55 (OAP18307.1 CDC5) |
78.48% 2e-22 (EHI_000550) |
|
| 4R-MYBS | ||||||||||
| SNAP-like |
EIN_267690 |
1992 | 1992 | EiSNAP-like I |
R1 436-599 |
SANT/Myb domain |
SANT/Myb domain |
28.22% 2e-11 39.08% 7e-08 (SNAP190) |
31.22% 3e-18 |
29.97% 1e-185 (EHI_130710) |
| Motif consensus sequence | Modified sequence | Motif containing genes* |
E-value |
STREME confirmed |
Stage related genes |
|
[T/C]AAC[G/T]G |
CAACTG | 2559 (21.31%) | 2.0 e-036 | 2541 (99.29%) | Trophozoite |
| DCAACTG | 815 (6.78%) | 1.3 e-011 | 807 (99.01%) | Encystation | |
| CAACTG | 838 (6.97%) | 5.5 e-011 | 834 (99.52%) | Excystation | |
|
[CA]CCCCC |
MCCCCC | 288 (2.39%) | 1.1 e-007 | 284 (98.6%) | Trophozoite |
| ACCCCCA | 99 (0.82%) | 6.8 e-003 | 97 (97.97%) | Encystation | |
| CCCCCC | 100 (0.83%) | 1.8 e-001 | 98 (98.0%) | Excystation |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





