Submitted:
02 January 2024
Posted:
03 January 2024
You are already at the latest version
Abstract
Keywords:
1. Current knowledge
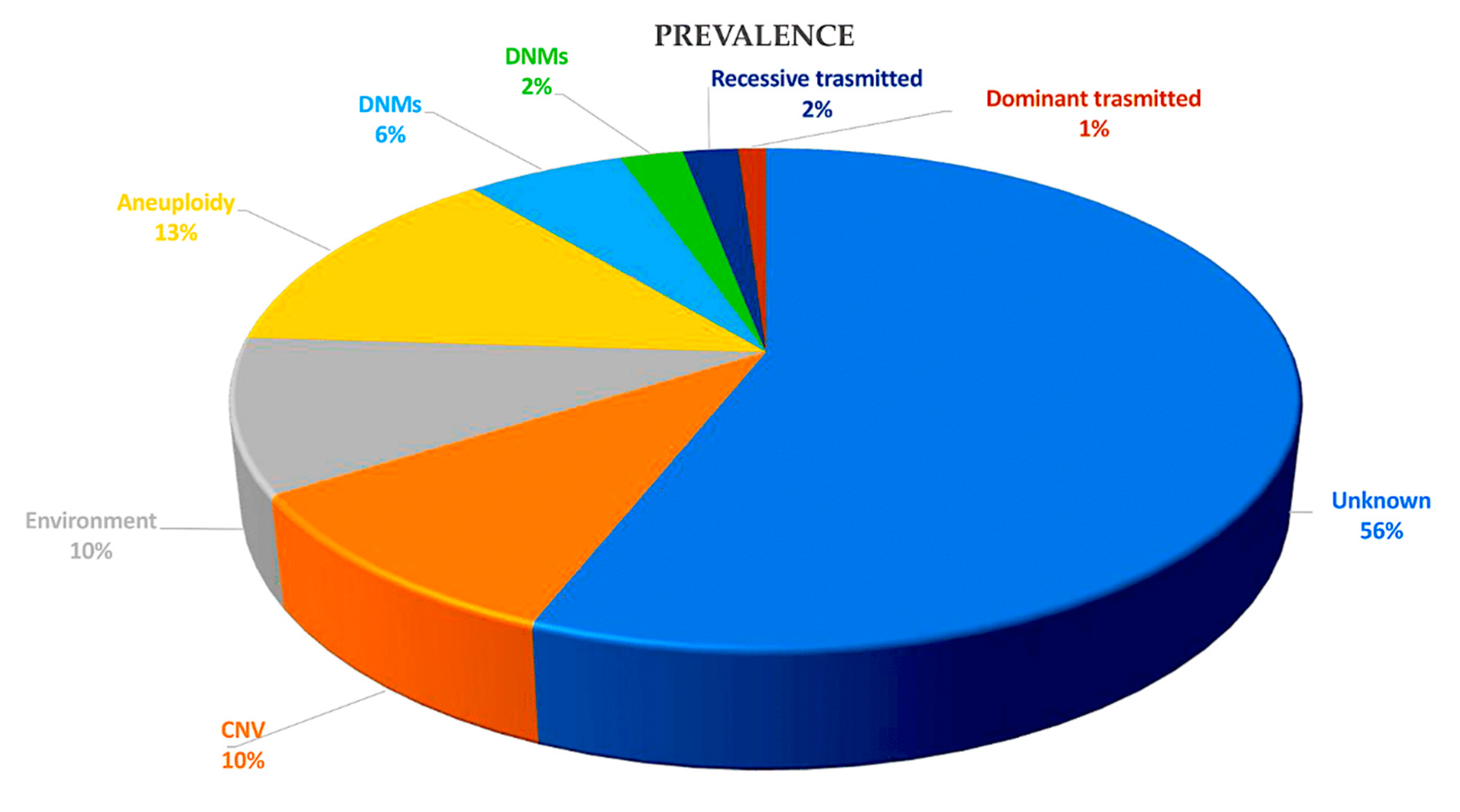
1.1. Genetic
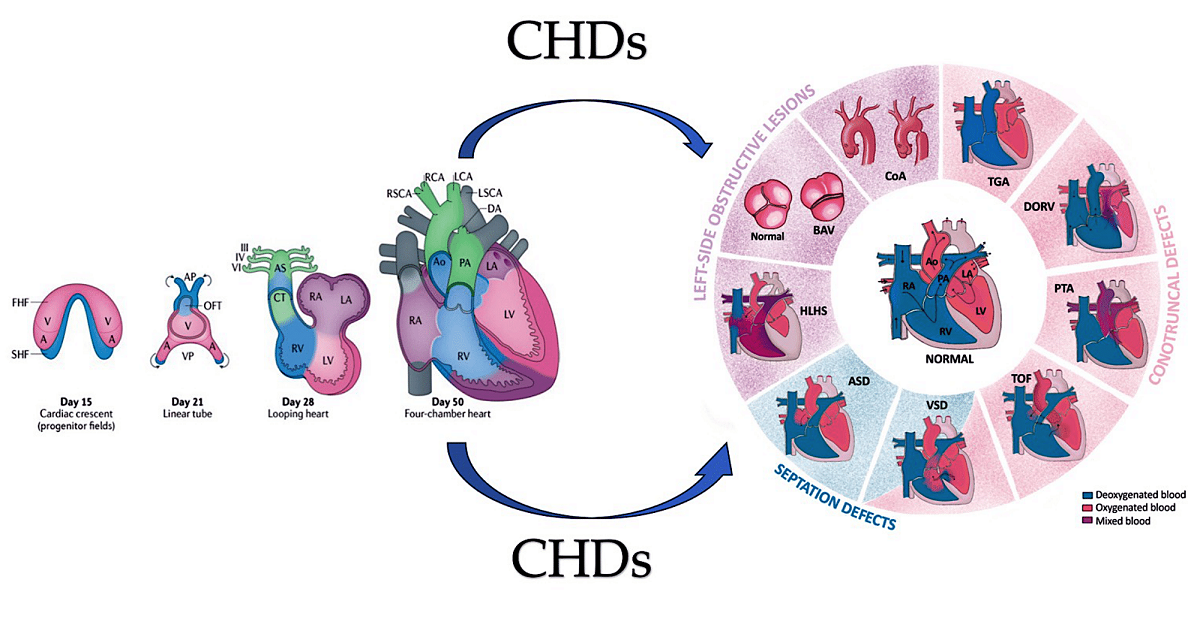
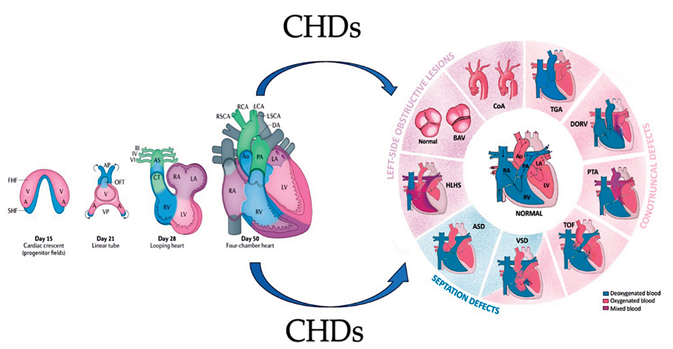
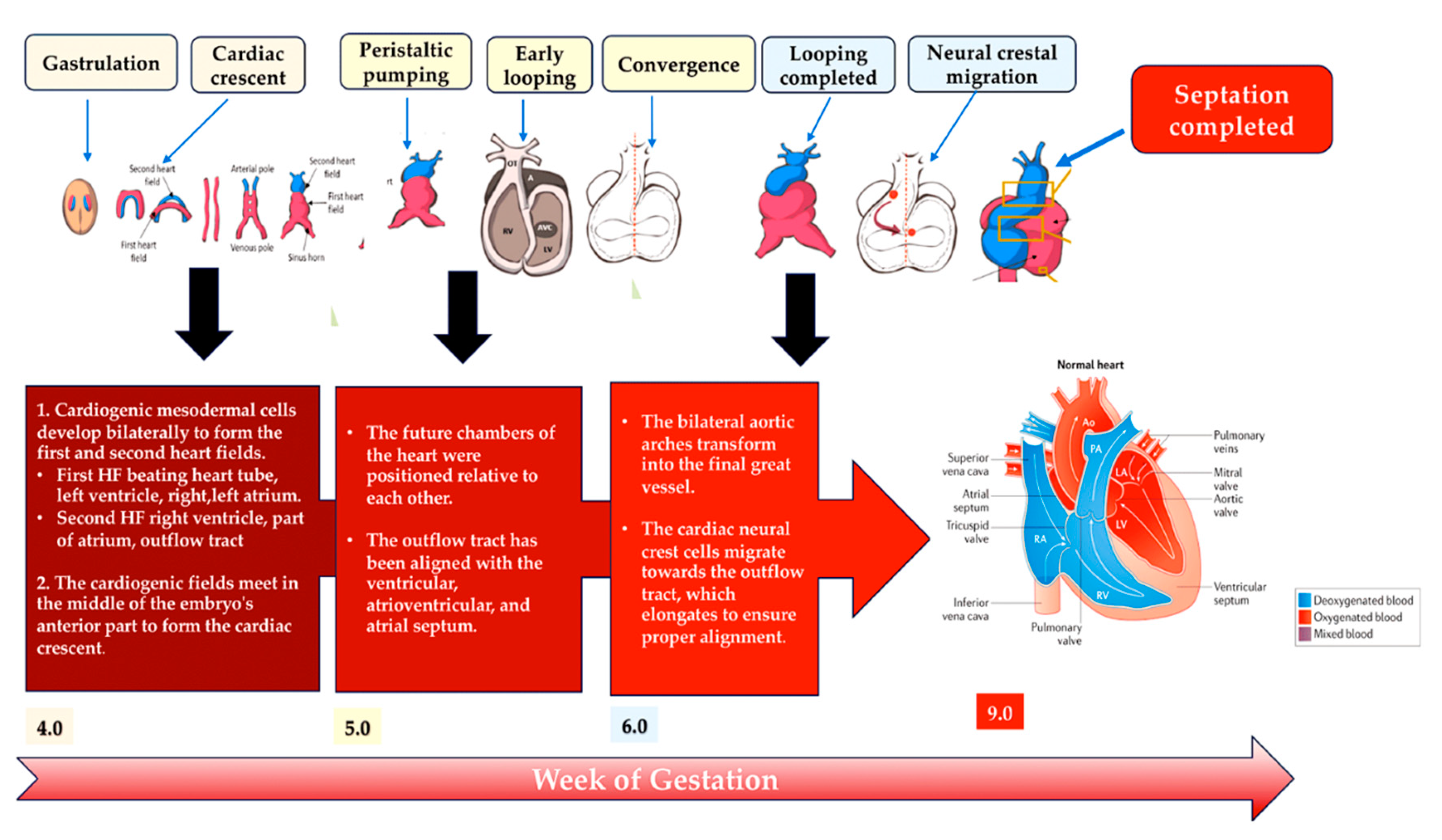
2. Development of heart
2.1. A look at the lines of progenitor cells and their morphogenesis.
2.2. Heart morphogenesis under genetic control
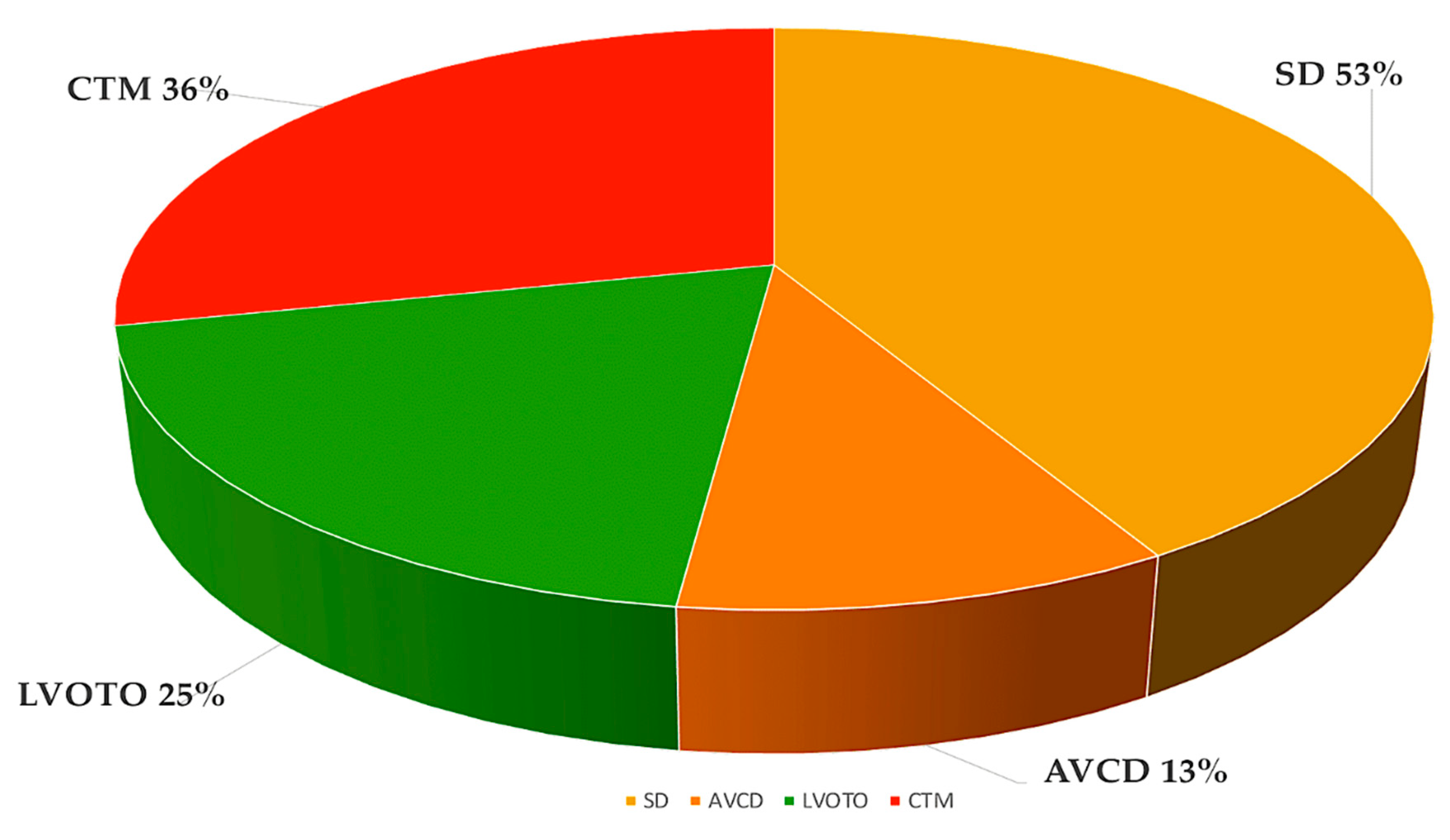
3. Interfering with Human Cardiac Development
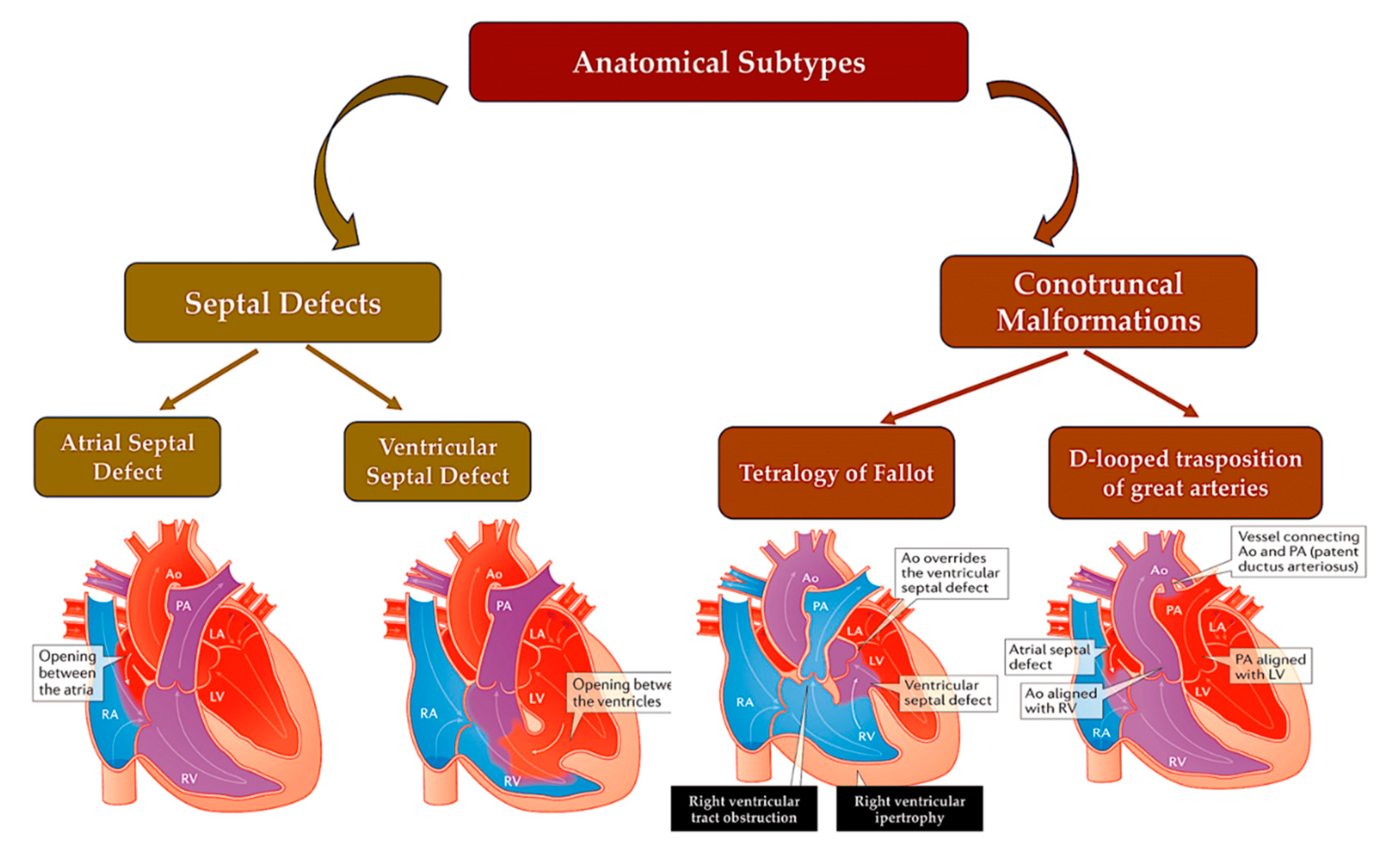
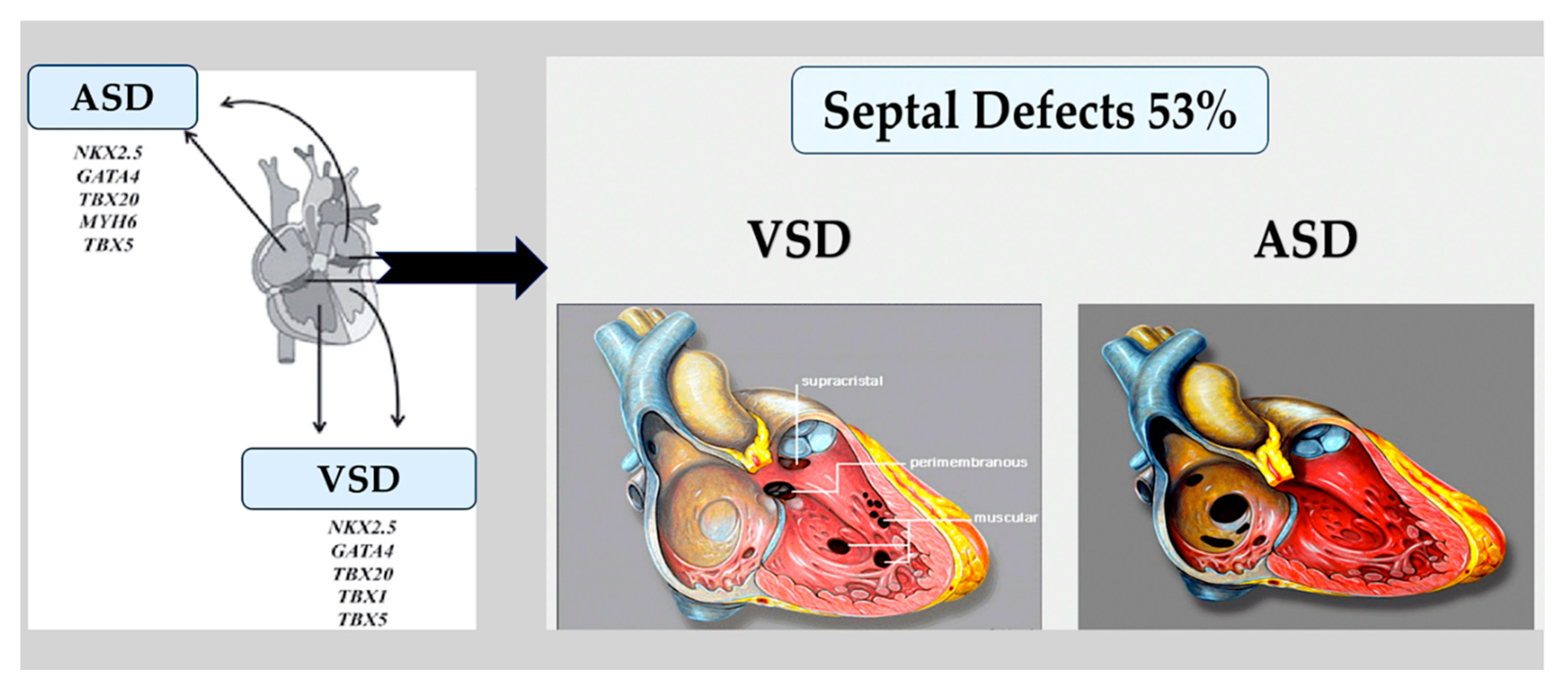
3.1. Atrial and ventricular septal defects.
3.2. Left-sided obstructive lesions.
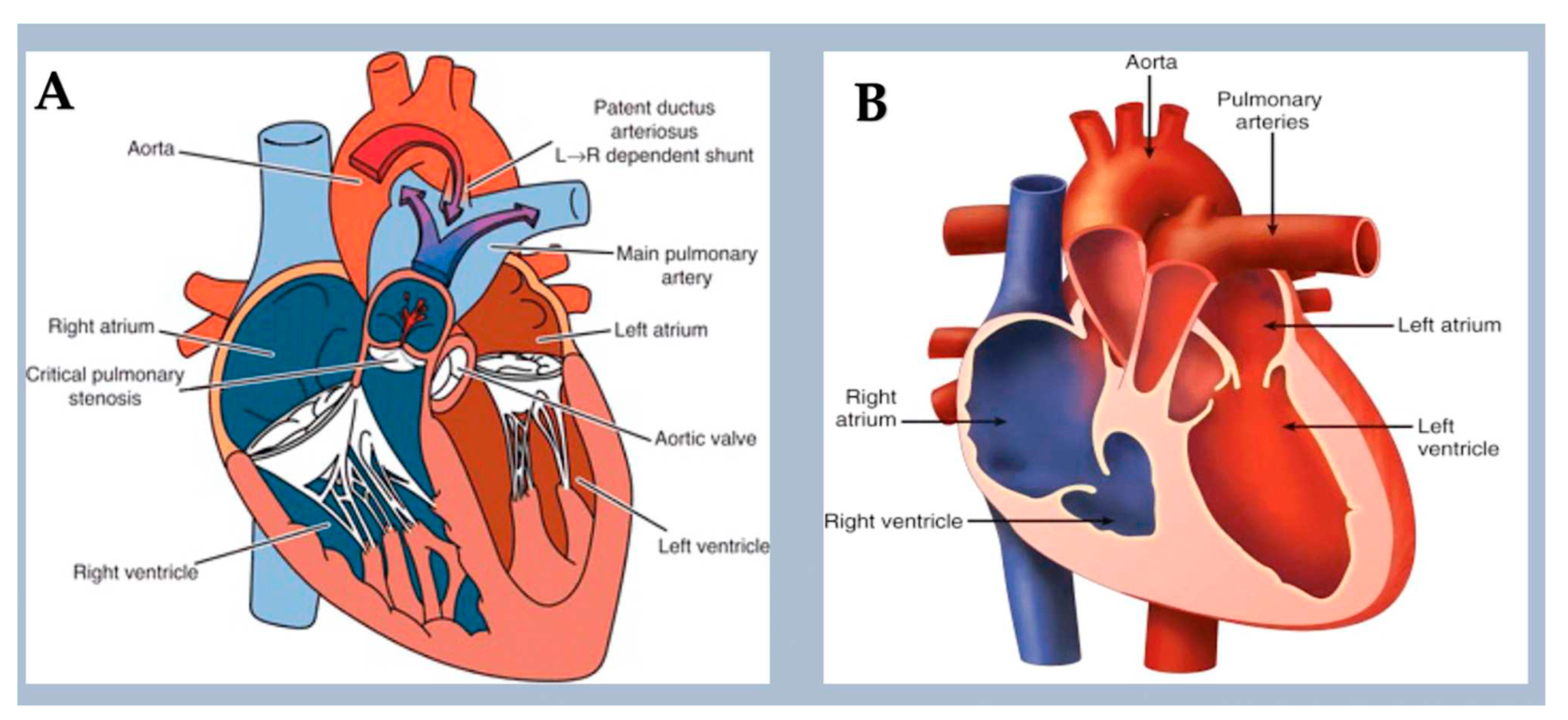
3.3. Right-sided obstructive lesions.
3.4. Defects in the left-right pattern.
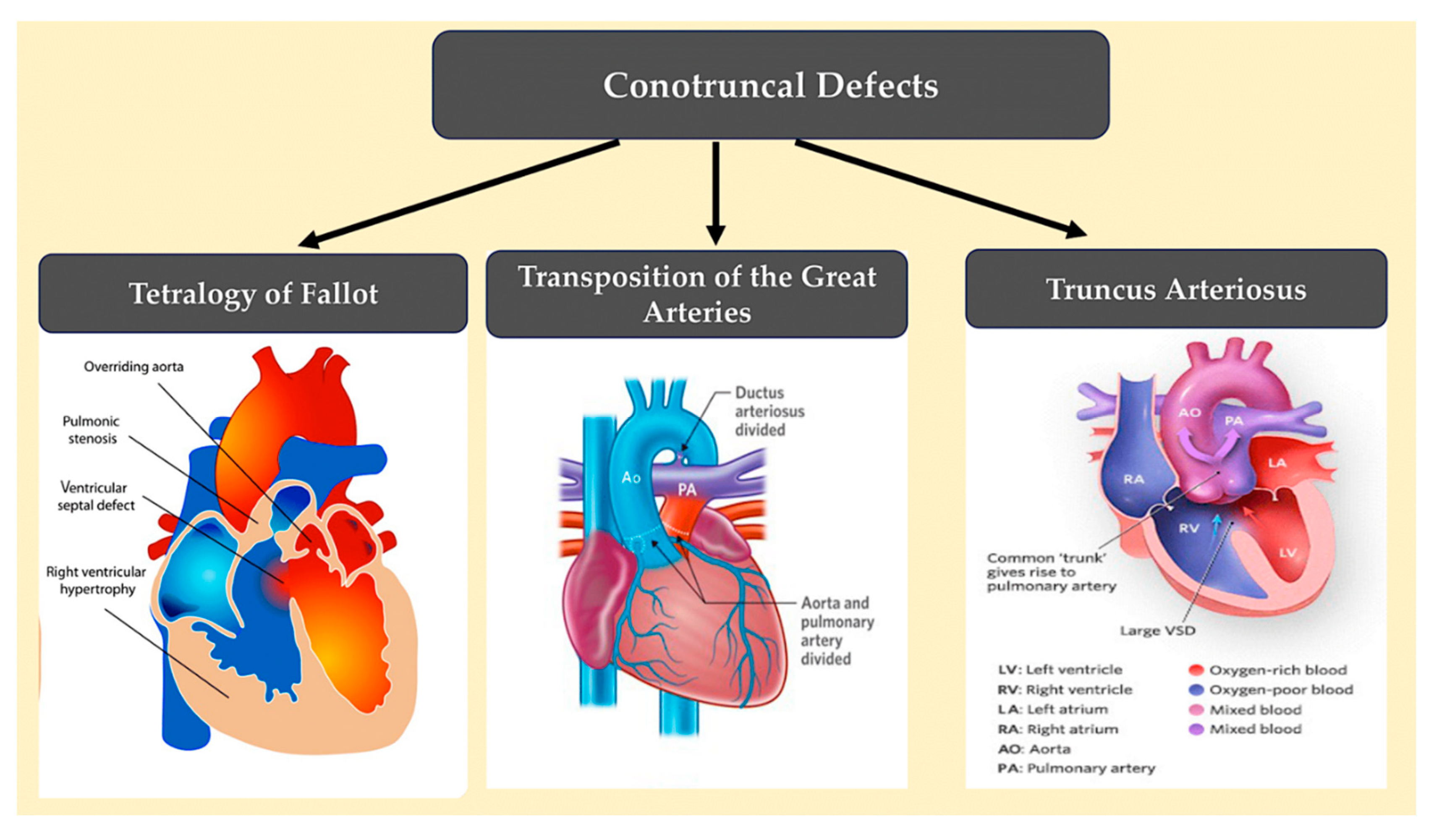
3.5. Conotruncal defects.
4. Cohort studies reveal new CHD genes.
4.1. CHD definitive genes.
4.2. Genes linked with CHD that have deleterious variants in humans.
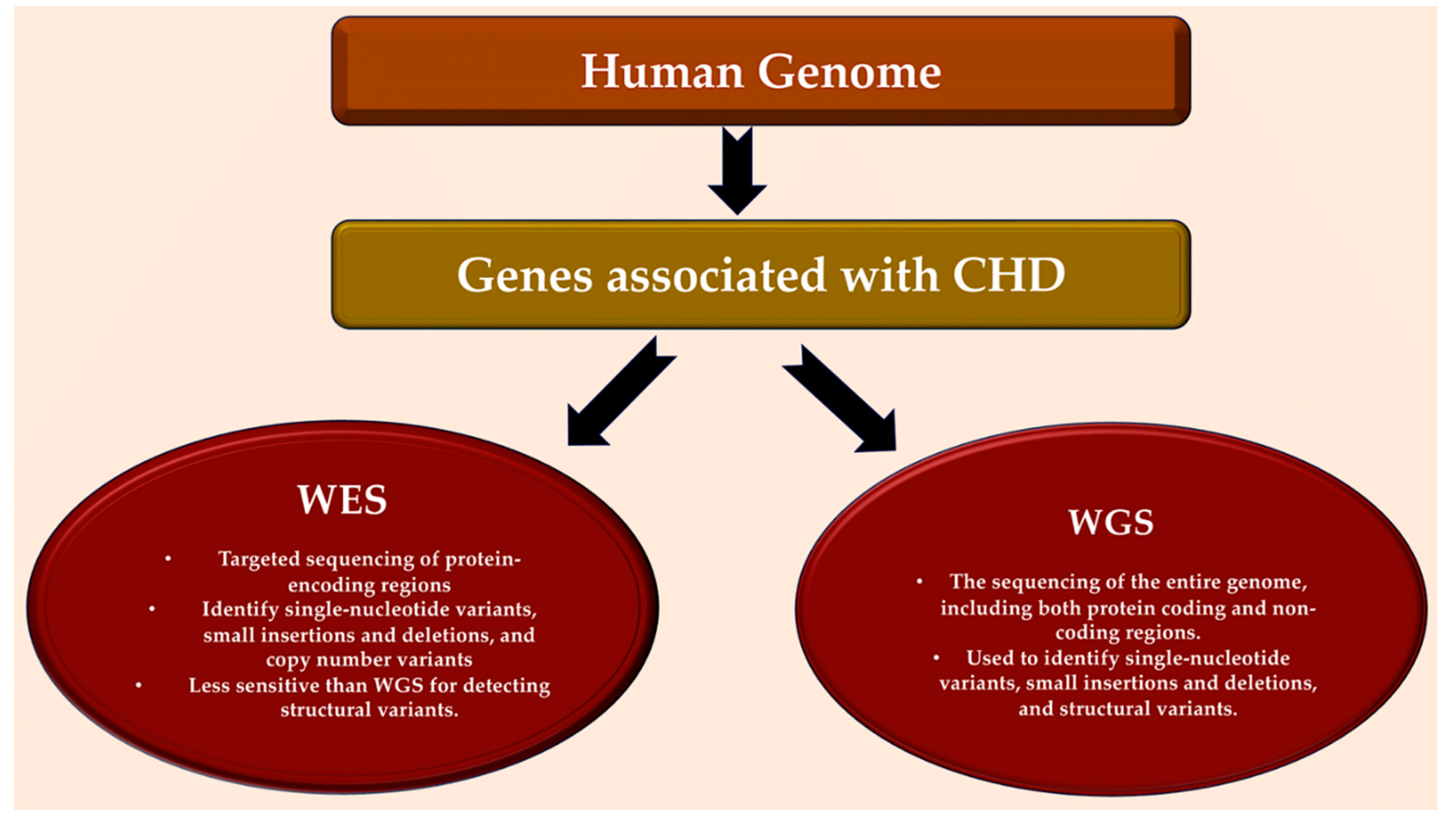
5. Whole-genome sequencing.
5.1. Variation in non-coding sequences through whole genome sequencing
5.2. Using whole genome sequencing to identify structural variation.
5.3. Future uses of genomics.
6. Deriving causality by integrating biology
6.1. Insights gained from single cell transcriptomics.
6.2. Findings from pluripotent stem cell models of congenital heart disease.
7. Moving genomics into the clinical setting
7.1. CHD in association with extracardiac abnormalities or neurodevelopmental disabilities.
7.2. Cancer risk in patients with congenital heart disease.
7.3. Forthcoming applications in the clinic.
8. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ibrahim S, Gaborit B, Lenoir M, Collod-Beroud G, Stefanovic S. Maternal Pre-Existing Diabetes: A Non-Inherited Risk Factor for Congenital Cardiopathies. Int J Mol Sci. 2023 Nov 13;24(22):16258. [CrossRef]
- Dilli D, Akduman H, Zenciroğlu A, Çetinkaya M, Okur N, Turan Ö, Özlü F, Çalkavur Ş, Demirel G, Koksal N, Çolak R, Örün UA, Öztürk E, Gül Ö, Tokel NK, Erdem S, Meşe T, Erdem A, Bostan ÖM, Polat TB, Taşar M, Hatemi AC, Doyurgan O, Özkan M, Avşar MK, Sarıosmanoğlu ON, Uğurlucan M, Sığnak IŞ, Başaran M. Neonatal Outcomes of Critical Congenital Heart Defects: A Multicenter Epidemiological Study of Turkish Neonatal Society: Neonatal Outcomes of CCHD. Pediatr Cardiol. 2023 Dec 28. [CrossRef]
- Hossin MZ, de la Cruz LF, McKay KA, Oberlander TF, Sandström A, Razaz N. Association of pre-existing maternal cardiovascular diseases with neurodevelopmental disorders in offspring: a cohort study in Sweden and British Columbia, Canada. Int J Epidemiol. 2023 Dec 27: dyad184. [CrossRef]
- Bakker MK, Bergman JEH, Krikov S, Amar E, Cocchi G, Cragan J, de Walle HEK, Gatt M, Groisman B, Liu S, Nembhard WN, Pierini A, Rissmann A, Chidambarathanu S, Sipek A Jr, Szabova E, Tagliabue G, Tucker D, Mastroiacovo P, Botto LD. Prenatal diagnosis and prevalence of critical congenital heart defects: an international retrospective cohort study. BMJ Open. 2019 Jul 2;9(7):e028139. [CrossRef]
- Hoffman JIE, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol. 2002 Jun 19;39(12):1890-900. [CrossRef]
- Reller MD, Strickland MJ, Riehle-Colarusso T, Mahle WT, Correa A Prevalence of congenital heart defects in metropolitan Atlanta. J Pediatr. 2008 Dec;153(6):807-13. [CrossRef]
- Spector LG, Menk JS, Knight JH, McCracken C, Thomas AS, Vinocur JM, Oster ME, St Louis JD, Moller JH, Kochilas L. Trends in Long-Term Mortality After Congenital Heart Surgery. J Am Coll Cardiol. 2018 May 29;71(21):2434-2446. [CrossRef]
- Egbe A, Lee S, Ho D, Uppu S, Srivastava S. Prevalence of congenital anomalies in newborns with congenital heart disease diagnosis. Ann Pediatr Cardiol. 2014 May;7(2):86-91. [CrossRef]
- Wang H, Lin X, Lyu G, He S, Dong B, Yang Y. Chromosomal abnormalities in fetuses with congenital heart disease: a meta-analysis. Arch Gynecol Obstet. 2023 Sep;308(3):797-811. [CrossRef]
- Diniz BL, Deconte D, Gadelha KA, Glaeser AB, Guaraná BB, de Moura AÁ, Rosa RFM, Zen PRG. Congenital Heart Defects and 22q11.2 Deletion Syndrome: A 20-Year Update and New Insights to Aid Clinical Diagnosis.
- J Pediatr Genet. 2023 Feb 17;12(2):113-122. [CrossRef]
- Thienpont B, Mertens L, de Ravel T, Eyskens B, Boshoff D, Maas N, Fryns JP, Gewillig M, Vermeesch JR, Devriendt K. Submicroscopic chromosomal imbalances detected by array-CGH are a frequent cause of congenital heart defects in selected patients. Eur Heart J. 2007 Nov;28(22):2778-84. [CrossRef]
- Wilson DI, Cross IE, Goodship JA, Brown J, Scambler PJ, Bain HH, Taylor JF, Walsh K, Bankier A, Burn J, et al. A prospective cytogenetic study of 36 cases of DiGeorge syndrome. Am J Hum Genet. 1992 Nov;51(5):957-63.
- Sgardioli IC, Vieira TP, Simioni M, Monteiro FP, Gil-da-Silva-Lopes VL 22q11.2 Deletion Syndrome: Laboratory Diagnosis and TBX1 and FGF8 Mutation Screening. J Pediatr Genet. 2015 Mar;4(1):17-22. [CrossRef]
- Agergaard P, Olesen C, Østergaard JR, Christiansen M, Sørensen KM The prevalence of chromosome 22q11.2 deletions in 2,478 children with cardiovascular malformations. A population-based study. Am J Med Genet A. 2012 Mar;158A(3):498-508. [CrossRef]
- Agergaard P, Hebert A, Sørensen KM, Østergaard JR, Olesen C. Can clinical assessment detect 22q11.2 deletions in patients with cardiac malformations? A review. Eur J Med Genet. 2011 Jan-Feb;54(1):3-8. [CrossRef]
- Peyvandi S, Lupo PJ, Garbarini J, Woyciechowski S, Edman S, Emanuel BS, Mitchell LE, Goldmuntz E.. 22q11.2 deletions in patients with conotruncal defects: data from 1,610 consecutive cases. Pediatr Cardiol. 2013 Oct;34(7):1687-94. [CrossRef]
- Dehghan B, Sabri MR, Ahmadi A, Ghaderian M, Mahdavi C, Ramezani Nejad D, Sattari M. Identifying the Factors Affecting the Incidence of Congenital Heart Disease Using Support Vector Machine and Particle Swarm Optimization. Adv Biomed Res. 2023 May 19;12:130. [CrossRef]
- Pierpont ME, Basson CT, Benson DW Jr, Gelb BD, Giglia TM, Goldmuntz E, McGee G, Sable CA, Srivastava D, Webb CL; American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young. Genetic basis for congenital heart defects: current knowledge: a scientific statement from the American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation. 2007 Jun 12;115(23):3015-38. [CrossRef]
- Goldmuntz, E. 22q11.2 deletion syndrome and congenital heart disease. Am J Med Genet C Semin Med Genet. 2020 Mar;184(1):64-72. [CrossRef]
- Fahed AC, Gelb BD, Seidman JG, Seidman CE Genetics of congenital heart disease: the glass half empty. Circ Res. 2013 Feb 15;112(4):707-20. [CrossRef]
- LaHaye S, Corsmeier D, Basu M, Bowman JL, Fitzgerald-Butt S, Zender G, Bosse K, McBride KL, White P, Garg V. Utilization of Whole Exome Sequencing to Identify Causative Mutations in Familial Congenital Heart Disease. Circ Cardiovasc Genet. 2016 Aug;9(4):320-9. [CrossRef]
- Jin SC, Homsy J, Zaidi S, Lu Q, Morton S, DePalma SR, Zeng X, Qi H, Chang W, Sierant MC, Hung WC, Haider S, Zhang J, Knight J, Bjornson RD, Castaldi C, Tikhonoa IR, Bilguvar K, Mane SM, Sanders SJ, Mital S, Russell MW, Gaynor JW, Deanfield J, Giardini A, Porter GA Jr, Srivastava D, Lo, et al.. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet. 2017 Nov;49(11):1593-1601. [CrossRef]
- Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewski KJ, DePalma SR, McKean D, Wakimoto H, Gorham J, Jin SC, Deanfield J, Giardini A, Porter GA Jr, Kim R, Bilguvar K, López-Giráldez F, Tikhonova I, Mane S, Romano-Adesman A, Qi H, Vardarajan B, Ma L, Daly M, Roberts AE, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015 Dec 4;350(6265):1262-6. [CrossRef]
- Deciphering Developmental Disorders Study. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017 Feb 23;542(7642):433-438. [CrossRef]
- Sifrim A, Hitz MP, Wilsdon A, Breckpot J, Turki SH, Thienpont B, McRae J, Fitzgerald TW, Singh T, Swaminathan GJ, Prigmore E, Rajan D, Abdul-Khaliq H, Banka S, Bauer UM, Bentham J, Berger F, Bhattacharya S, Bu'Lock F, Canham N, Colgiu IG, Cosgrove C, Cox H, Daehnert I, Daly A, Danesh J, Fryer A, Gewillig M, Hobson E, Hoff K, Homfray T; INTERVAL Study; Kahlert AK, Ketley A, Kramer HH, Lachlan K, Lampe AK, Louw JJ, Manickara AK, Manase D, McCarthy KP, Metcalfe K, Moore C, Newbury-Ecob R, Omer SO, Ouwehand WH, Park SM, Parker MJ, Pickardt T, Pollard MO, Robert L, Roberts DJ, Sambrook J, Setchfield K, Stiller B, Thornborough C, Toka O, Watkins H, Williams D, Wright M, Mital S, Daubeney PE, Keavney B, Goodship J; UK10K Consortium; Abu-Sulaiman RM, Klaassen S, Wright CF, Firth HV, Barrett JC, Devriendt K, FitzPatrick DR, Brook JD; Deciphering Developmental Disorders Study; Hurles ME. Distinct genetic architectures for syndromic and nonsyndromic congenital heart defects identified by exome sequencing. Nat Genet. 2016 Sep;48(9):1060-5. [CrossRef]
- Shan W, Yuanqing X, Jing Z, Xi W, Huifeng G, Yi W. Risk factor analysis for adverse prognosis of the fetal ventricular septal defect (VSD). BMC Pregnancy Childbirth. 2023 Sep 21;23(1):683. [CrossRef]
- International Society of Ultrasound in Obstetrics & Gynecology. Cardiac screening examination of the fetus: guidelines for performing the ‘basic’ and ‘extended basic’ cardiac scan. Ultrasound Obstet Gynecol2006 Jan;27(1):107-113.
- Meilhac SM, Buckingham ME The deployment of cell lineages that form the mammalian heart. Nat Rev Cardiol. 2018 Nov;15(11):705-724. [CrossRef]
- Kathiriya IS, Nora EP, Bruneau BG Investigating the transcriptional control of cardiovascular development. Circ Res. 2015 Feb 13;116(4):700-14. [CrossRef]
- Maas RGC, van den Dolder FW, Yuan Q, van der Velden J, Wu SM, Sluijter JPG, Buikema JW. Harnessing developmental cues for cardiomyocyte production. Development. 2023 Aug 1;150(15):dev201483. [CrossRef]
- Günthel M, Barnett P, Christoffels VM Development, proliferation, and growth of the mammalian heart. Mol Ther. 2018 Jul 5;26(7):1599-1609. [CrossRef]
- Cui M, Wang Z, Bassel-Duby R & Olson EN Genetic and epigenetic regulation of cardiomyocytes in development, regeneration and disease. Development 145, dev171983 (2018).
- Chien KR, Domian IJ, Parker KK. Cardiogenesis and the complex biology of regenerative cardiovascular medicine.Science. 2008 Dec 5;322(5907):1494-7. [CrossRef]
- Jain R, Epstein JA Competent for commitment: you’ve got to have heart! Genes Dev. 2018 Jan 1;32(1):4-13. [CrossRef]
- Zaffran S, Kelly RG, Meilhac SM, Buckingham ME, Brown NA. Right ventricular myocardium derives from the anterior heart field. Circ Res. 2004 Aug 6;95(3):261-8. [CrossRef]
- Zaidi, S.; Choi, M.; Wakimoto, H.; Ma, L.; Jiang, J.; Overton, J.D.; Romano-Adesman, A.; Bjornson, R.D.; Breitbart, R.E.; Brown, K.K.; et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature 2013, 498, 220–223. [Google Scholar] [CrossRef]
- Hedermann, G.; Hedley, P.L.; Thagaard, I.N.; Krebs, L.; Ekelund, C.K.; Sørensen, T.I.A.; Christiansen, M. Maternal obesity and metabolic disorders associate with congenital heart defects in the offspring: A systematic review. PLoS ONE 2021, 16, e0252343. [Google Scholar] [CrossRef] [PubMed]
- Cavadino, A.; Sandberg, L.; Öhman, I.; Bergvall, T.; Star, K.; Dolk, H.; Loane, M.; Addor, M.C.; Barisic, I.; Cavero-Carbonell, C.; et al. Signal Detection in EUROmediCAT: Identification and Evaluation of Medication-Congenital Anomaly Associations and Use of VigiBase as a Complementary Source of Reference. Drug Saf. 2021, 44, 765–785. [Google Scholar] [CrossRef] [PubMed]
- Kalisch-Smith, J.I.; Ved, N.; Sparrow, D.B. Environmental Risk Factors for Congenital Heart Disease. Cold Spring Harb. Perspect. Biol. 2020, 12, a037234. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, K.J.; Correa, A.; Feinstein, J.A.; Botto, L.; Britt, A.E.; Daniels, S.R.; Elixson, M.; Warnes, C.A.; Webb, C.L. American Heart Association Council on Cardiovascular Disease in the Y: Noninherited risk factors and congenital cardiovascular defects: Current knowledge: A scientific statement from the American Heart Association Council on Cardiovascular Disease in the Young: Endorsed by the American Academy of Pediatrics. Circulation 2007, 115, 2995–3014. [Google Scholar] [PubMed]
- Hutson MR, Kirby ML Model systems for the study of heart development and disease. Semin Cell Dev Biol. 2007 Feb;18(1):101-10. [CrossRef]
- de la Pompa JL, Timmerman LA, Takimoto H, Yoshida H, Elia AJ, Samper E, Potter J, Wakeham A, Marengere L, Langille BL, Crabtree GR, Mak TW. Role of the NF-ATc transcription factor in morphogenesis of cardiac valves and septum.Nature. 1998 Mar 12;392(6672):182-6. [CrossRef]
- Lin C-J, Lin C-Y, Chen C-H, Zhou B, Chang C-P Partitioning the heart: mechanisms of cardiac septation and valve development. Development. 2012 Sep;139(18):3277-99. [CrossRef]
- Wu B, Wu B, Benkaci S, Shi L, Lu P, Park T, Morrow BE, Wang Y, Zhou B. Crk and Crkl Are Required in the Endocardial Lineage for Heart Valve Development. J Am Heart Assoc. 2023 Sep 19;12(18):e029683. [CrossRef]
- Christoffels VM, Habets PE, Franco D, Campione M, de Jong F, Lamers WH, Bao ZZ, Palmer S, Biben C, Harvey RP, Moorman AF. Chamber formation and morphogenesis in the developing mammalian heart. Dev Biol. 2000 Jul 15;223(2):266-78. [CrossRef]
- Alsan BH, Schultheiss TM Regulation of avian cardiogenesis by Fgf8 signaling. Development. 2002 Apr;129(8): 1935-43. [CrossRef]
- Astrof S, Arriagada C, Saijoh Y, Francou A, Kelly RG, Moon A. Aberrant differentiation of second heart field mesoderm prefigures cellular defects in the outflow tract in response to loss of FGF8. Dev Biol. 2023 Jul;499:10-21. [CrossRef]
- Ivanovitch K, Soro-Barrio P, Chakravarty P, Jones RA, Bell DM, Mousavy Gharavy SN, Stamataki D, Delile J, Smith JC, Briscoe J. Ventricular, atrial, and outflow tract heart progenitors arise from spatially and molecularly distinct regions of the primitive streak. PLoS Biol. 2021 May 17;19(5):e3001200. [CrossRef]
- Itoh N, Ohta H, Nakayama Y & Konishi M Roles of FGF signals in heart development, health, and disease. Front. Cell Dev. Biol. 4, 110 (2016).
- de Pater E, Ciampricotti M, Priller F, Veerkamp J, Strate I, Smith K, Lagendijk AK, Schilling TF, Herzog W, Abdelilah-Seyfried S, Hammerschmidt M, Bakkers J. Bmp signaling exerts opposite effects on cardiac differentiation. Circ Res. 2012 Feb 17;110(4):578-87. [CrossRef]
- Targoff KL, Colombo S, George V, Schell T, Kim SH, Solnica-Krezel L, Yelon D. Nkx genes are essential for maintenance of ventricular identity.Development. 2013 Oct;140(20):4203-13. [CrossRef]
- Nelson DO, Lalit PA, Biermann M, Markandeya YS, Capes DL, Addesso L, Patel G, Han T, John MC, Powers PA, Downs KM, Kamp TJ, Lyons GE. Irx4 Marks a Multipotent, Ventricular-Specific Progenitor Cell. Stem Cells. 2016 Dec;34(12):2875-2888. [CrossRef]
- Goldfracht I, Protze S, Shiti A, Setter N, Gruber A, Shaheen N, Nartiss Y, Keller G, Gepstein L. Generating ring-shaped engineered heart tissues from ventricular and atrial human pluripotent stem cell-derived cardiomyocytes.
- Nat Commun. 2020 Jan 7;11(1):75. [CrossRef]
- Giacomelli E, Bellin M, Orlova VV, Mummery CL. Co-Differentiation of Human Pluripotent Stem Cells-Derived Cardiomyocytes and Endothelial Cells from Cardiac Mesoderm Provides a Three-Dimensional Model of Cardiac Microtissue. Curr Protoc Hum Genet. 2017 Oct 18; 95:21.9.1-21.9.22. [CrossRef]
- Cheng Z et al. Two novel mutations of the IRX4 gene in patients with congenital heart disease. Hum. Genet. 130, 657–662 (2011).
- de Soysa TY et al. Single-cell analysis of cardiogenesis reveals basis for organ-level developmental defects. Nature 572, 120–124 (2019).
- Chen YH, Ishii M, Sun J, Sucov HM & Maxson RE Msx1 and Msx2 regulate survival of secondary heart field precursors and post-migratory proliferation of cardiac neural crest in the outflow tract. Dev. Biol. 308, 421–437 (2007).
- Sharma A et al. GATA6 mutations in hiPSCs inform mechanisms for maldevelopment of the heart, pancreas, and diaphragm. eLife 9, e53278 (2020).
- Uribe V et al. Arid3b is essential for second heart field cell deployment and heart patterning. Development 141,4168–4181 (2014).
- Wang Z, Wang DZ, Hockemeyer D, McAnally J, Nordheim A, Olson EN. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature. 2004 Mar 11;428(6979):185-9. [CrossRef]
- Felker A, Prummel KD, Merks AM, Mickoleit M, Brombacher EC, Huisken J, Panáková D, Mosimann C. Continuous addition of progenitors forms the cardiac ventricle in zebrafish. Nat Commun. 2018 May 21;9(1):2001. [CrossRef]
- Sánchez-Iranzo H, Galardi-Castilla M, Minguillón C, Sanz-Morejón A, González-Rosa JM, Felker A, Ernst A, Guzmán-Martínez G, Mosimann C, Mercader N. Tbx5a lineage tracing shows cardiomyocyte plasticity during zebrafish heart regeneration. Nat Commun. 2018 Jan 30;9(1):428. [CrossRef]
- Jiang X, Choudhary B, Merki E, Chien KR, Maxson RE, Sucov HM.Normal fate and altered function of the cardiac neural crest cell lineage in retinoic acid receptor mutant embryos. Mech Dev. 2002 Sep;117(1–2):115-22. [CrossRef]
- Zhou C, Häneke T, Rohner E, Sohlmér J, Kameneva P, Artemov A, Adameyko I, Sahara M. STRA6 is essential for induction of vascular smooth muscle lineages in human embryonic cardiac outflow tract development. Cardiovasc Res. 2023 May 22;119(5):1202-1217. [CrossRef]
- Sanchez J, Miyake R, Cheng A, Liu T, Iseki S, Kume T. Conditional inactivation of Foxc1 and Foxc2 in neural crest cells leads to cardiac abnormalities.Genesis. 2020 Jul;58(7):e23364. [CrossRef]
- Kodo K, Shibata S, Miyagawa-Tomita S, Ong SG, Takahashi H, Kume T, Okano H, Matsuoka R, Yamagishi H. Regulation of Sema3c and the interaction between cardiac neural crest and second heart field during outflow tract development. Sci Rep. 2017 Jul 28;7(1):6771. [CrossRef]
- Greulich F, Rudat C, Kispert A. Mechanisms of T-box gene function in the developing heart. Cardiovasc Res. 2011 Jul 15;91(2):212-22. [CrossRef]
- MacGrogan D, Nus M, de la Pompa JL Notch signaling in cardiac development and disease.Curr Top Dev Biol. 2010;92:333-65. [CrossRef]
- Stankunas K, Ma GK, Kuhnert FJ, Kuo CJ, Chang CP. VEGF signaling has distinct spatiotemporal roles during heart valve development.Dev Biol. 2010 Nov 15;347(2):325-36. [CrossRef]
- Alvandi Z, Bischoff J. Endothelial-Mesenchymal Transition in Cardiovascular Disease.Arterioscler Thromb Vasc Biol. 2021 Sep;41(9):2357-2369. [CrossRef]
- Singh N, Trivedi CM, Lu M, Mullican SE, Lazar MA, Epstein JA. Histone deacetylase 3 regulates smooth muscle differentiation in neural crest cells and development of the cardiac outflow tract. Circ Res. 2011 Nov 11;109(11):1240-9. [CrossRef]
- Kelly RG, Papaioannou VE. Visualization of outflow tract development in the absence of Tbx1 using an FgF10 enhancer trap transgene.Dev Dyn. 2007 Mar;236(3):821-8. [CrossRef]
- Peng T, Tian Y, Boogerd CJ, Lu MM, Kadzik RS, Stewart KM, Evans SM, Morrisey E. Coordination of heart and lung co-development by a multipotent cardiopulmonary progenitor. Nature. 2013 Aug 29;500(7464):589-92. [CrossRef]
- Liu X, Chen W, Li W, Li Y, Priest JR, Zhou B, Wang J, Zhou Z. Single-Cell RNA-seq of the developing cardiac outflow tract reveals convergent development of the vascular smooth muscle cells. Cell Rep. 2019 Jul 30;28(5):1346-1361.e4. [CrossRef]
- Forrest K, Barricella AC, Pohar SA, Hinman AM, Amack JD. Understanding laterality disorders and the left-right organizer: Insights from zebrafish. Front Cell Dev Biol. 2022 Dec 23;10:1035513. [CrossRef]
- Lopes Floro K, Artap ST, Preis JI, Fatkin D, Chapman G, Furtado MB, Harvey RP, Hamada H, Sparrow DB, Dunwoodie S. Loss of Cited2 causes congenital heart disease by perturbing left-right patterning of the body axis. Hum Mol Genet. 2011 Mar 15;20(6):1097-110. [CrossRef]
- Bellchambers HM, Ware SM. ZIC3 in Heterotaxy. Adv Exp Med Biol. 2018;1046:301-327. [CrossRef]
- Levin, M. Left-right asymmetry in vertebrate embryogenesis. Bioessays. 1997 Apr;19(4):287-96. [CrossRef]
- Chang H, Zwijsen A, Vogel H, Huylebroeck D, Matzuk MM. Smad5 is essential for left-right asymmetry in mice Dev Biol. 2000 Mar 1;219(1):71-8. [CrossRef]
- Kumar S, Donofrio M, Frank L, He D, Jonas R. Complete atrioventricular canal with guarded primum septal defect.Pediatr Cardiol. 2011 Apr;32(4):503-5. [CrossRef]
- Paladini D, Tartaglione A, Agangi A, Teodoro A, Forleo F, Borghese A, Martinelli P. The association between congenital heart disease and Down syndrome in prenatal life. Ultrasound Obstet Gynecol. 2000 Feb;15(2):104-8. [CrossRef]
- Santoro M, Coi A, Spadoni I, Bianchi F, Pierini A. Sex differences for major congenital heart defects in Down Syndrome: A population based study. Eur J Med Genet. 2018 Sep;61(9):546-550. [CrossRef]
- Pelleri MC, Locatelli C, Mattina T, Bonaglia MC, Piazza F, Magini P, Antonaros F, Ramacieri G, Vione B, Vitale L, Seri M, Strippoli P, Cocchi G, Piovesan A, Caracausi M. Partial trisomy 21 with or without highly restricted Down syndrome critical region (HR-DSCR): report of two new cases and reanalysis of the genotype-phenotype association. BMC Med Genomics. 2022 Dec 21;15(1):266. [CrossRef]
- Ang Y-S, Rivas RN, Ribeiro AJS, Srivas R, Rivera J, Stone NR, Pratt K, Mohamed TMA, Fu JD, Spencer CI, Tippens ND, Li M, Narasimha A, Radzinsky E, Moon-Grady AJ, Yu H, Pruitt BL, Snyder MP, Srivastava D. Disease model of GATA4 mutation reveals transcription factor cooperativity in human cardiogenesis. Cell. 2016 Dec 15;167(7):1734-1749.e22. [CrossRef]
- Misra C, Sachan N, McNally CR, Koenig SN, Nichols HA, Guggilam A, Lucchesi PA, Pu WT, Srivastava D, Garg V. Congenital heart disease-causing Gata4 mutation displays functional deficits in vivo. PLoS Genet. 2012;8(5):e1002690. [CrossRef]
- Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS, Hirayama-Yamada K, Joo K, Matsuoka R, Cohen JC, Srivastava D. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003 Jul 24;424(6947):443-7. [CrossRef]
- Brown CO 3rd, Chi X, Garcia-Gras E, Shirai M, Feng XH, Schwartz RJ. The cardiac determination factor, Nkx2-5, is activated by mutual cofactors GATA-4 and Smad1/4 via a novel upstream enhancer. J Biol Chem. 2004 Mar 12;279(11):10659-69. [CrossRef]
- Vecoli C, Pulignani S, Foffa I, Andreassi MG. Congenital heart disease: the crossroads of genetics, epigenetics and environment. Curr Genomics. 2014 Oct;15(5):390-9. [CrossRef]
- Kirklin/Barratt-Boyes Cardiac Surgery 4th Edition by James K Kirklin MD (Author), Eugene H. Blackstone MD.
- McBride KL Pignatelli R, Lewin M, Ho T, Fernbach S, Menesses A, Lam W, Leal SM, Kaplan N, Schliekelman P, Towbin JA, Belmont JW. Inheritance analysis of congenital left ventricular outflow tract obstruction malformations: segregation, multiplex relative risk, and heritability. Am J Med Genet A. 2005 Apr 15;134A(2):180-6. [CrossRef]
- Nappi F, Giacinto O, Lusini M, Garo M, Caponio C, Nenna A, Nappi P, Rousseau J, Spadaccio C, Chello M. Patients with Bicuspid Aortopathy and Aortic Dilatation. J Clin Med. 2022 Oct 11;11(20):6002. [CrossRef]
- Agasthi P, Pujari SH, Tseng A, Graziano JN, Marcotte F, Majdalany D, Mookadam F, Hagler DJ, Arsanjani R. Management of adults with coarctation of aorta. World J Cardiol. 2020 May 26;12(5):167-191. [CrossRef]
- Silberbach M et al. Cardiovascular health in Turner syndrome: a scientific statement from the American Heart Association. Circulation. Genomic Precis. Med. 11, e000048 (2018).
- Lara DA, Ethen MK, Canfield MA, Nembhard WN & Morris SA A population-based analysis of mortality in patients with Turner syndrome and hypoplastic left heart syndrome using the Texas Birth Defects Registry. Congenit. Heart Dis. 12, 105–112 (2017).
- Bouayed Abdelmoula N, Abdelmoula B, Smaoui W, Trabelsi I, Louati R, Aloulou S, Aloulou W, Abid F, Kammoun S, Trigui K, Bedoui O, Denguir H, Mallek S, Ben Aziza M, Dammak J, Kaabi O, Abdellaoui N, Turki F, Kaabi A, Kamoun W, Jabeur J, Ltaif W, Chaker K, Fourati H, M'rabet S, Ben Ameur H, Gouia N, Mhiri MN, Rebai T. Left-sided congenital heart lesions in mosaic Turner syndrome. Mol Genet Genomics. 2018 Apr;293(2):495-501. [CrossRef]
- Ribé L, Shihadeh FD, Afifi RO, Estrera AL, Prakash SK. Outcomes of cardiothoracic surgery in women with Turner syndrome. Ann Cardiothorac Surg. 2023 Nov 27;12(6):569-576. [CrossRef]
- Prakash SK, Bondy CA, Maslen CL, Silberbach M, Lin AE, Perrone L, Limongelli G, Michelena HI, Bossone E, Citro R; BAVCon Investigators, GenTAC Registry Investigators; Lemaire SA, Body SC, Milewicz DM. Autosomal and X chromosome structural variants are associated with congenital heart defects in Turner syndrome: The NHLBI GenTAC registry. Am J Med Genet A. 2016 Dec;170(12):3157-3164. [CrossRef]
- Wenger SL, Grossfeld PD, Siu BL, Coad JE, Keller FG, Hummel M. Molecular characterization of an 11q interstitial deletion in a patient with the clinical features of Jacobsen syndrome. Am J Med Genet A. 2006 Apr 1;140(7):704-8. [CrossRef]
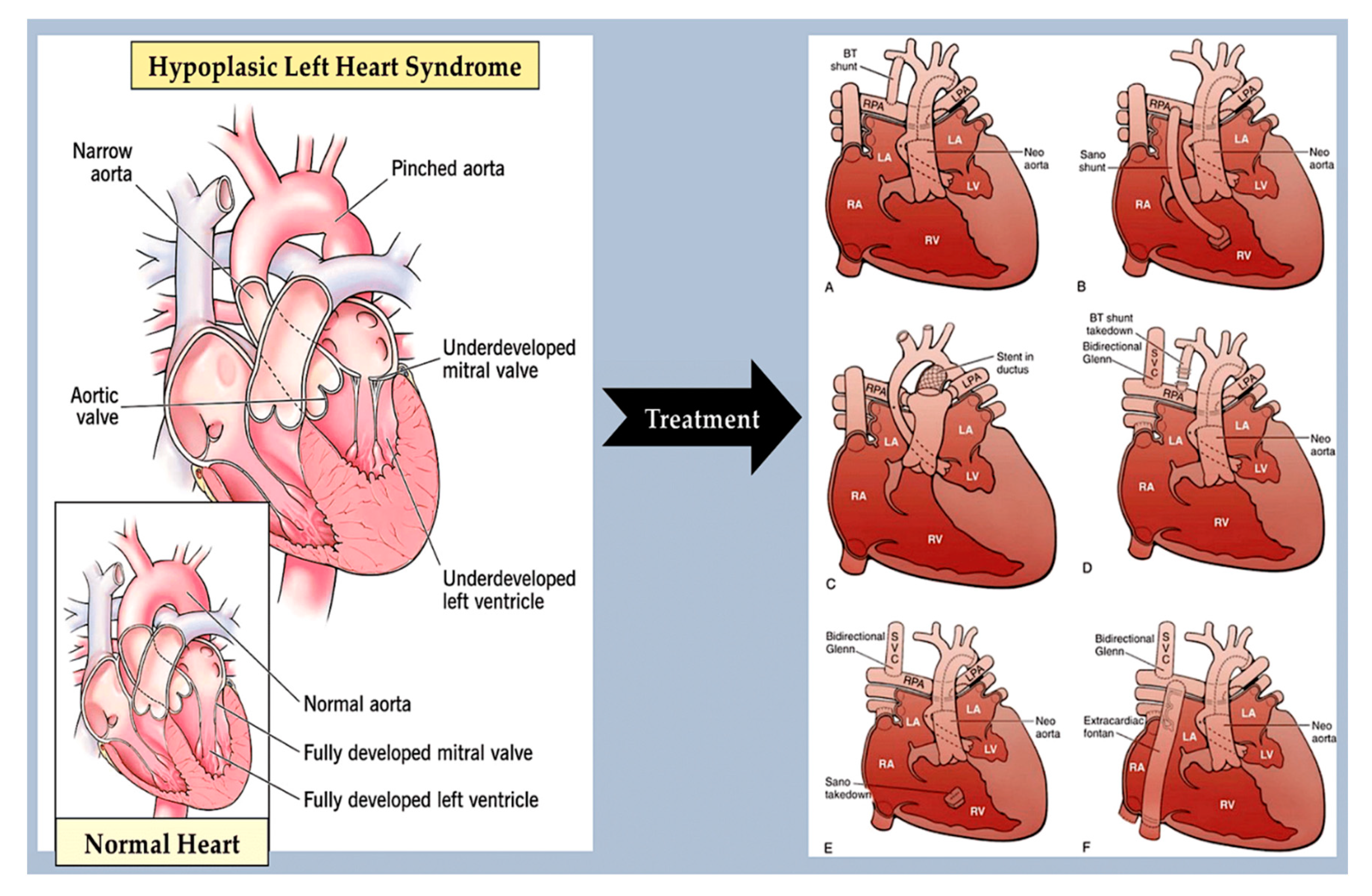
- Crucean A, Alqahtani A, Barron DJ, Brawn WJ, Richardson RV, O'Sullivan J, Anderson RH, Henderson DJ, Chaudhry B. Re-evaluation of hypoplastic left heart syndrome from a developmental and morphological perspective. Orphanet J Rare Dis. 2017 Aug 10;12(1):138. [CrossRef]
- Miao Y, Tian L, Martin M, Paige SL, Galdos FX, Li J, Klein A, Zhang H, Ma N, Wei Y, Stewart M, Lee S, Moonen JR, Zhang B, Grossfeld P, Mital S, Chitayat D, Wu JC, Rabinovitch M, Nelson TJ, Nie S, Wu SM, Gu M.. Intrinsic endocardial defects contribute to hypoplastic left heart syndrome. Cell Stem Cell. 2020 Oct 1;27(4):574-589.e8. [CrossRef]
- Jiang Y, Habibollah S, Tilgner K, Collin J, Barta T, Al-Aama JY, Tesarov L, Hussain R, Trafford AW, Kirkwood G, Sernagor E, Eleftheriou CG, Przyborski S, Stojković M, Lako M, Keavney B, Armstrong LAn induced pluripotent stem cell model of hypoplastic left heart syndrome (HLHS) reveals multiple expression and functional differences in HLHS-derived cardiac myocytes. Stem Cells Transl Med. 2014 Apr;3(4):416-23. [CrossRef]
- Shi LM, Tao JW, Qiu XB, Wang J, Yuan F, Xu L, Liu H, Li RG, Xu YJ, Wang Q, Zheng HZ, Li X, Wang XZ, Zhang M, Qu XK, Yang YQ.. GATA5 loss-of-function mutations associated with congenital bicuspid aortic valve. Int J Mol Med. 2014 May;33(5):1219-26. [CrossRef]
- Bonachea EM et al. Rare GATA5 sequence variants identified in individuals with bicuspid aortic valve. Pediatr. Res. 76, 211–216 (2014).
- Huang M, Akerberg AA, Zhang X, Yoon H, Joshi S, Hallinan C, Nguyen C, Pu WT, Haigis MC, Burns CG, Burns CE. Intrinsic myocardial defects underlie an Rbfox-deficient zebrafish model of hypoplastic left heart syndrome. Nat Commun. 2022 Oct 5;13(1):5877. [CrossRef]
- Xu YJ, Di RM, Qiao Q, Li XM, Huang RT, Xue S, Liu XY, Wang J, Yang YQ. GATA6 loss-of-function mutation contributes to congenital bicuspid aortic valve. Gene. 2018 Jul 15; 663:115-120. [CrossRef]
- Theis JL, Hu JJ, Sundsbak RS, Evans JM, Bamlet WR, Qureshi MY, O'Leary PW, Olson TM. Genetic Association Between Hypoplastic Left Heart Syndrome and Cardiomyopathies. Circ Genom Precis Med. 2021 Feb;14(1):e003126. [CrossRef]
- Theis JL, Zimmermann MT, Evans JM, Eckloff BW, Wieben ED, Qureshi MY, O'Leary PW, Olson TM. Recessive MYH6 mutations in hypoplastic left heart with reduced ejection fraction. Circ Cardiovasc Genet. 2015 Aug;8(4):564-71. [CrossRef]
- Berg C, Lachmann R, Kaiser C, Kozlowski P, Stressig R, Schneider M, Asfour B, Herberg U, Breuer J, Gembruch U, Geipel A. Prenatal diagnosis of tricuspid atresia: intrauterine course and outcome. Ultrasound Obstet Gynecol. 2010 Feb;35(2):183-90. [CrossRef]
- Sarkozy A, Conti E, D'Agostino R, Digilio MC, Formigari R, Picchio F, Marino B, Pizzuti A, Dallapiccola B. ZFPM2/FOG2 and HEY2 genes analysis in nonsyndromic tricuspid atresia. Am J Med Genet A. 2005 Feb 15;133A(1):68-70. [CrossRef]
- Pierpont ME, Digilio MC. Cardiovascular disease in Noonan syndrome. Curr Opin Pediatr. 2018 Oct;30(5):601-608. [CrossRef]
- Lee ST, Ki CS, Lee HJ. Mutation analysis of the genes involved in the Ras-mitogen-activated protein kinase (MAPK) pathway in Korean patients with Noonan syndrome. Clin Genet. 2007 Aug;72(2):150-5. [CrossRef]
- Pandit B, Sarkozy A, Pennacchio LA, Carta C, Oishi K, Martinelli S, Pogna EA, Schackwitz W, Ustaszewska A, Landstrom A, Bos JM, Ommen SR, Esposito G, Lepri F, Faul C, Mundel P, López Siguero JP, Tenconi R, Selicorni A, Rossi C, Mazzanti L, Torrente I, Marino B, Digilio MC, Zampino G, Ackerman MJ, Dallapiccola B, Tartaglia M, Gelb BD. Gain-of-function RAF1 mutations cause Noonan and LEOPARD syndromes with hypertrophic cardiomyopathy. Nat Genet. 2007 Aug;39(8):1007-12. [CrossRef]
- Roberts A, Allanson J, Jadico SK, Kavamura MI, Noonan J, Opitz JM, Young T, Neri G. The cardiofaciocutaneous syndrome. J Med Genet. 2006 Nov;43(11):833-42. [CrossRef]
- Tidyman WE, Rauen KA. Noonan, Costello and cardio-facio-cutaneous syndromes: dysregulation of the Ras-MAPK pathway.Expert Rev Mol Med. 2008 Dec 9;10: e37. [CrossRef]
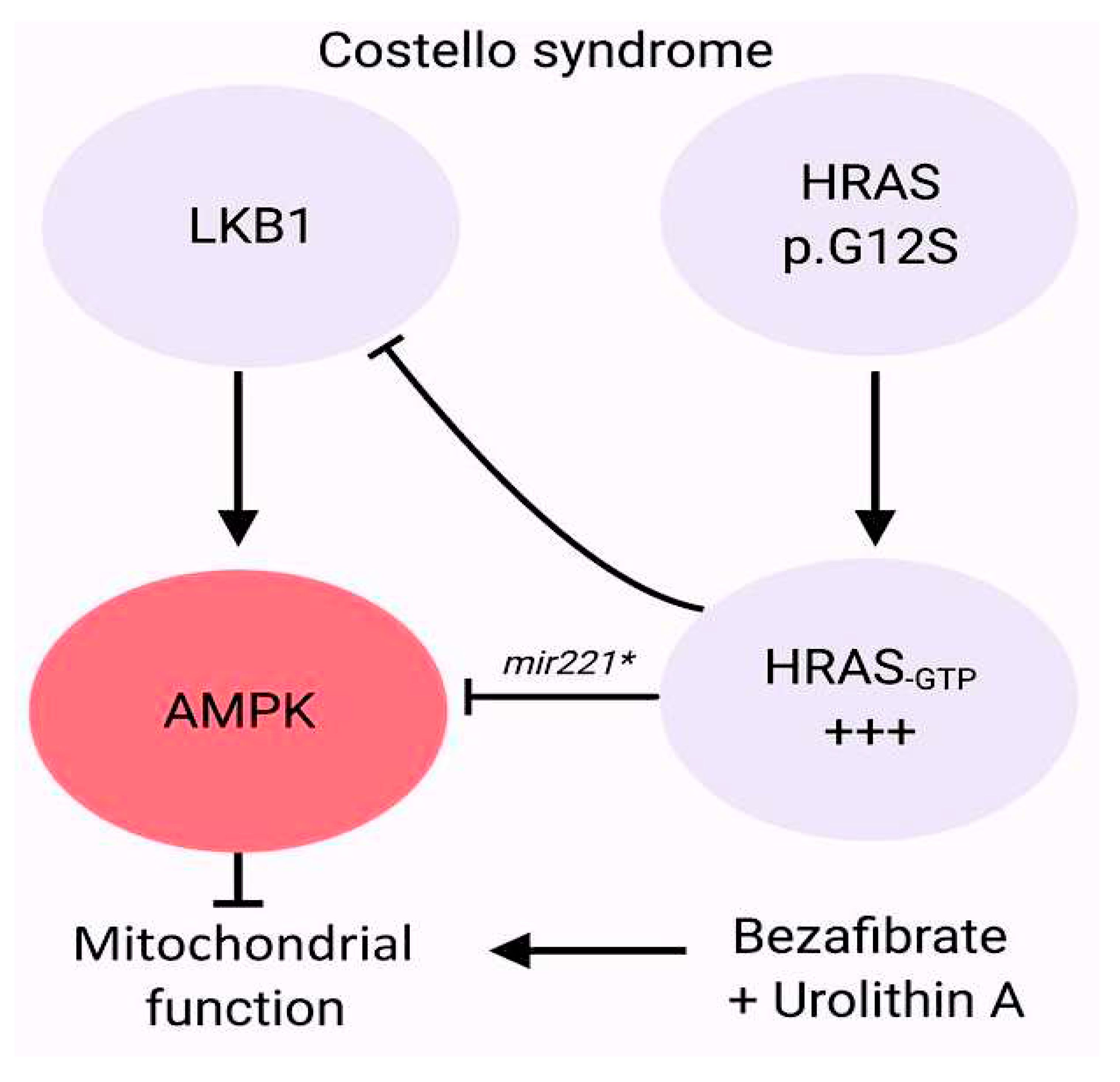
- Dard L, Hubert C, Esteves P, Blanchard W, Bou About G, Baldasseroni L, Dumon E, Angelini C, Delourme M, Guyonnet-Dupérat V, Claverol S, Fontenille L, Kissa K, Séguéla PE, Thambo JB, Nicolas L, Herault Y, Bellance N, Dias Amoedo N, Magdinier F, Sorg T, Lacombe D, Rossignol R. HRAS germline mutations impair LKB1/AMPK signaling and mitochondrial homeostasis in Costello syndrome models. J Clin Invest. 2022 Apr 15;132(8):e131053. [CrossRef]
- White JJ, Mazzeu JF, Hoischen A, Bayram Y, Withers M, Gezdirici A, Kimonis V, Steehouwer M, Jhangiani SN, Muzny DM, Gibbs RA; Baylor-Hopkins Center for Mendelian Genomics; van Bon BWM, Sutton VR, Lupski JR, Brunner HG, Carvalho CMB. DVL3 Alleles Resulting in a -1 Frameshift of the Last Exon Mediate Autosomal-Dominant Robinow Syndrome. Am J Hum Genet. 2016 Mar 3;98(3):553-561. [CrossRef]
- Rai A, Patil SJ, Srivastava P, Gaurishankar K, Phadke SR. Clinical and molecular characterization of four patients with Robinow syndrome from different families. Am J Med Genet A. 2021 Apr;185(4):1105-1112. [CrossRef]
- Hu R, Qiu Y, Li Y, Li J. A novel frameshift mutation of DVL1-induced Robinow syndrome: A case report and literature review. Mol Genet Genomic Med. 2022 Mar;10(3):e1886. [CrossRef]
- Danyel M, Kortüm F, Dathe K, Kutsche K, Horn D. Autosomal dominant Robinow syndrome associated with a novel DVL3 splice mutation. Am J Med Genet A. 2018 Apr;176(4):992-996. [CrossRef]
- Mašek J, Andersson ER. The developmental biology of genetic Notch disorders. Development. 2017 May 15;144(10):1743-1763. [CrossRef]
- Hofmann JJ, Briot A, Enciso J, Zovein AC, Ren S, Zhang ZW, Radtke F, Simons M, Wang Y, Iruela-Arispe ML Endothelial deletion of murine Jag1 leads to valve calcification and congenital heart defects associated with Alagille syndrome. Development. 2012 Dec 1;139(23):4449-60. [CrossRef]
- McCright B, Lozier J, Gridley T. A mouse model of Alagille syndrome: Notch2 as a genetic modifier of Jag1 haploinsufficiency. Development. 2002 Feb;129(4):1075-82. [CrossRef]
- Liu X, Chen W, Li W, Priest JR, Fu Y, Pang K, Ma B, Han B, Liu X, Hu S, Zhou Z.. Exome-based case-control analysis highlights the pathogenic role of ciliary genes in transposition of the great arteries. Circ Res. 2020 Mar 27;126(7):811-821. [CrossRef]
- Blue GM, Mekel M, Das D, Troup M, Rath E, Ip E, Gudkov M, Perumal G, Harvey RP, Sholler GF, Gecz J, Kirk EP, Liu J, Giannoulatou E, Hong H, Dunwoodie SL, Winlaw DS. Whole genome sequencing in transposition of the great arteries and associations with clinically relevant heart, brain and laterality genes. Am Heart J. 2022 Feb;244:1-13. [CrossRef]
- Li AH, Hanchard NA, Azamian M, D'Alessandro LCA, Coban-Akdemir Z, Lopez KN, Hall NJ, Dickerson H, Nicosia A, Fernbach S, Boone PM, Gambin T, Karaca E, Gu S, Yuan B,. Genetic architecture of laterality defects revealed by whole exome sequencing. Eur J Hum Genet. 2019 Apr;27(4):563-573. [CrossRef]
- Yi T, Sun H, Fu Y, Hao X, Sun L, Zhang Y, Han J, Gu X, Liu X, Guo Y, Wang X, Zhou X, Zhang S, Yang Q, Fan J, He Y. Genetic and Clinical Features of Heterotaxy in a Prenatal Cohort. Front Genet. 2022 Apr 19; 13:818241. [CrossRef]
- Papanayotou C, Collignon J.Philos Trans R Soc Lond B Activin/Nodal signalling before implantation: setting the stage for embryo patterning. Biol Sci. 2014 Dec 5;369(1657):20130539. [CrossRef]
- Mohapatra B, Casey B, Li H, Ho-Dawson T, Smith L, Fernbach SD, Molinari L, Niesh SR, Jefferies JL, Craigen WJ, Towbin JA, Belmont JW, Ware SM.. Identification and functional characterization of NODAL rare variants in heterotaxy and isolated cardiovascular malformations. Hum Mol Genet. 2009 Mar 1;18(5):861-71. [CrossRef]
- Bedard JE, Haaning AM, Ware SM. Identification of a novel ZIC3 isoform and mutation screening in patients with heterotaxy and congenital heart disease. PLoS One. 2011;6(8):e23755. [CrossRef]
- D'Alessandro LC, Latney BC, Paluru PC, Goldmuntz E. The phenotypic spectrum of ZIC3 mutations includes isolated d-transposition of the great arteries and double outlet right ventricle. Am J Med Genet A. 2013 Apr;161A(4):792-802. [CrossRef]
- Zhao Y, Wang Y, Shi L, McDonald-McGinn DM, Crowley TB, McGinn DE, Tran OT, Miller D, Lin JR, Zackai E, Johnston HR, Chow EWC, Vorstman JAS, Vingerhoets C, van Amelsvoort T, Gothelf D, Swillen A, Breckpot J, Vermeesch JR, Eliez S, Schneider M, van den Bree MBM, Owen MJ, Kates WR, Repetto GM,et al. Chromatin regulators in the TBX1 network confer risk for conotruncal heart defects in 22q11.2DS. NPJ Genom Med. 2023 Jul 18;8(1):17. [CrossRef]
- Khositseth A, Tocharoentanaphol C, Khowsathit P, Ruangdaraganon N. Chromosome 22q11 deletions in patients with conotruncal heart defects. Pediatr Cardiol. 2005 Sep-Oct;26(5):570-3.
- Ziolkowska L, Kawalec W, Turska-Kmiec A, Krajewska-Walasek M, Brzezinska-Rajszys G, Daszkowska J, Maruszewski B, Burczynski P. Chromosome 22q11.2 microdeletion in children with conotruncal heart defects: frequency, associated cardiovascular anomalies, and outcome following cardiac surgery. Eur J Pediatr. 2008 Oct;167(10):1135-40. [CrossRef]
- Carotti A, Digilio MC, Piacentini G, Saffirio C, Di Donato RM, Marino B. Cardiac defects and results of cardiac surgery in 22q11.2 deletion syndrome. Dev Disabil Res Rev. 2008;14(1):35-42. [CrossRef]
- Corsten-Janssen N, du Marchie Sarvaas GJ, Kerstjens-Frederikse WS, Hoefsloot LH, van Beynum IM, Kapusta L, van Ravenswaaij-Arts CM. CHD7 mutations are not a major cause of atrioventricular septal and conotruncal heart defects. Am J Med Genet A. 2014 Dec;164A(12):3003-9. [CrossRef]
- Corsten-Janssen N, Scambler PJ. Clinical and molecular effects of CHD7 in the heart. Am J Med Genet C Semin Med Genet. 2017 Dec;175(4):487-495. [CrossRef]
- Jongmans MC, Admiraal RJ, van der Donk KP, Vissers LE, Baas AF, Kapusta L, van Hagen JM, Donnai D, de Ravel TJ, Veltman JA, Geurts van Kessel A, De Vries BB, Brunner HG, Hoefsloot LH, van Ravenswaaij CM. CHARGE syndrome: the phenotypic spectrum of mutations in the CHD7 gene. J Med Genet. 2006 Apr;43(4):306-14. [CrossRef]
- Meisner JK, Martin DM. Congenital heart defects in CHARGE: The molecular role of CHD7 and effects on cardiac phenotype and clinical outcomes. Am J Med Genet C Semin Med Genet. 2020 Mar;184(1):81-89. [CrossRef]
- Zentner GE, Layman WS, Martin DM, Scacheri PC. Molecular and phenotypic aspects of CHD7 mutation in CHARGE syndrome. Am J Med Genet A. 2010 Mar;152A (3):674-86. [CrossRef]
- Rozas MF, Benavides F, León L, Repetto GM Association between phenotype and deletion size in 22q11.2 microdeletion syndrome: systematic review and meta-analysis. Orphanet J Rare Dis. 2019 Aug 9;14(1):195. [CrossRef]
- Zhao Y, Diacou A, Johnston HR, Musfee FI, McDonald-McGinn DM, McGinn D, Crowley TB, Repetto GM, Swillen A, Breckpot J, Vermeesch JR, Kates WR, Digilio MC, Unolt M, Marino B, Pontillo M, Armando M, Di Fabio F, Vicari S, van den Bree M, Moss H, Owen MJ, Murphy KC, Murphy CM, Murphy D, Schoch K, Shashi V, Tassone F, Simon TJ, Shprintzen RJ, Campbell L. Complete sequence of the 22q11.2 allele in 1,053 subjects with 22q11.2 deletion syndrome reveals modifiers of conotruncal heart defects. Am J Hum Genet. 2020 Jan 2;106(1):26-40. [CrossRef]
- He GW, Maslen CL, Chen HX, Hou HT, Bai XY, Wang XL, Liu XC, Lu WL, Chen XX, Chen WD, Xing QS, Wu Q, Wang J, Yang Q. Identification of novel rare copy number variants associated with sporadic tetralogy of Fallot and clinical implications.Clin Genet. 2022 Nov;102(5):391-403. [CrossRef]
- Mefford HC, Sharp AJ, Baker C, Itsara A, Jiang Z, Buysse K, Huang S, Maloney VK, Crolla JA, Baralle D, Collins A, Mercer C, Norga K, de Ravel T, Devriendt K, Bongers EM, de Leeuw N, Reardon W, Gimelli S, Bena F, Hennekam RC, Male A, Gaunt L, Clayton-Smith J, Simonic I, Park SM, Mehta SG, Nik-Zainal S, Woods CG, Firth HV, Parkin G, Fichera M, Reitano S, Lo Giudice M, Li KE, Casuga I, Broomer A, Conrad B, Schwerzmann M, Räber L, Gallati S, Striano P, Coppola A, et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008 Oct 16;359(16):1685-99. [CrossRef]
- Ceylan AC, Sahin I, Erdem HB, Kayhan G, Simsek-Kiper PO, Utine GE, Percin F, Boduroglu K, Alikasifoglu M.J An eight-case 1q21 region series: novel aberrations and clinical variability with new features. Intellect Disabil Res. 2019 Jun;63(6):548-557. [CrossRef]
- Reuter MS, Chaturvedi RR, Jobling RK, Pellecchia G, Hamdan O, Sung WWL, Nalpathamkalam T, Attaluri P, Silversides CK, Wald RM, Marshall CR, Williams SG, Keavney BD, Thiruvahindrapuram B, Scherer SW, Bassett AS. Clinical Genetic Risk Variants Inform a Functional Protein Interaction Network for Tetralogy of Fallot. Circ Genom Precis Med. 2021 Aug;14(4):e003410. [CrossRef]
- Page DJ, Miossec MJ, Williams SG, Monaghan RM, Fotiou E, Cordell HJ, Sutcliffe L, Topf A, Bourgey M, Bourque G, Eveleigh R, Dunwoodie SL, Winlaw DS, Bhattacharya S, Breckpot J, Devriendt K, Gewillig M, Brook JD, Setchfield KJ, Bu'Lock FA, O'Sullivan J, Stuart G,. Whole exome sequencing reveals the major genetic contributors to nonsyndromic tetralogy of Fallot. Circ Res. 2019 Feb 15;124(4):553-563. [CrossRef]
- Reuter MS et al. Haploinsufficiency of vascular endothelial growth factor related signaling genes is.
- associated with tetralogy of Fallot. Genet. Med. 21, 1001–1007 (2019).
- Škorić-Milosavljević D, Lahrouchi N, Bosada FM, Dombrowsky G, Williams SG, Lesurf R, Tjong FVY, Walsh R, El Bouchikhi I, Breckpot J, Audain E, Ilgun A, Beekman L, Ratbi I, Strong A, Muenke M, Heide S, Muir AM, Hababa M, Cross L, Zhou D, Pastinen T; et al. Rare variants in KDR, encoding VEGF Receptor 2, are associated with tetralogy of Fallot. Genet Med. 2021 Oct;23(10):1952-1960. [CrossRef]
- Tan ZP, Huang C, Xu ZB, Yang JF, Yang YF. Novel ZFPM2/FOG2 variants in patients with double outlet right ventricle. Clin Genet. 2012 Nov;82(5):466-71. [CrossRef]
- Huang X, Niu W, Zhang Z, Zhou C, Xu Z, Liu J, Su Z, Ding W, Zhang H. Identification of novel significant variants of ZFPM2/FOG2 in non-syndromic Tetralogy of Fallot and double outlet right ventricle in a Chinese Han population. Mol Biol Rep. 2014;41(4):2671-7. [CrossRef]
- Su W, Zhu P, Wang R, Wu Q, Wang M, Zhang X, Mei L, Tang J, Kumar M, Wang X, Su L, Dong N. Congenital heart diseases and their association with the variant distribution features on susceptibility genes. Clin Genet. 2017 Mar;91(3):349-354. [CrossRef]
- Yang YQ, Gharibeh L, Li RG, Xin YF, Wang J, Liu ZM, Qiu XB, Xu YJ, Xu L, Qu XK, Liu X, Fang WY, Huang RT, Xue S, Nemer G. GATA4 loss-of-function mutations underlie familial tetralogy of fallot. Hum Mutat. 2013 Dec ;34(12):1662-71. [CrossRef]
- Kodo K, Nishizawa T, Furutani M, Arai S, Yamamura E, Joo K, Takahashi T, Matsuoka R, Yamagishi H. GATA6 mutations cause human cardiac outflow tract defects by disrupting semaphorinplexin signaling. Proc Natl Acad Sci U S A. 2009 Aug 18;106(33):13933-8. [CrossRef]
- Lahaye S, Corsmeier D, Basu M, Bowman JL, Fitzgerald-Butt S, Zender G, Bosse K, McBride KL, White P, Garg V. Utilization of whole exome sequencing to identify causative mutations in familial congenital heart disease. Circ Cardiovasc Genet. 2016 Aug;9(4):320-9. [CrossRef]
- Hoang TT, Goldmuntz E, Roberts AE, Chung WK, Kline JK, Deanfield JE, Giardini A, Aleman A, Gelb BD, Mac Neal M, Porter GA Jr, Kim R, Brueckner M, Lifton RP, Edman S, Woyciechowski S, Mitchell LE, Agopian AJ. The congenital heart disease genetic network study: cohort description. PLoS One. 2018 Jan 19;13(1):e0191319. [CrossRef]
- Preuss C, Capredon M, Wünnemann F, Chetaille P, Prince A, Godard B, Leclerc S, Sobreira N, Ling H, Awadalla P, Thibeault M, Khairy P; MIBAVA Leducq consortium; Samuels ME, Andelfinger G Family based whole exome sequencing reveals the multifaceted role of Notch signaling in congenital heart disease. PLoS Genet. 2016 Oct 19;12(10):e1006335. [CrossRef]
- Sevim Bayrak C, Zhang P, Tristani-Firouzi M, Gelb BD, Itan Y. De novo variants in exomes of congenital heart disease patients identify risk genes and pathways. Genome Med. 2020 Jan 15;12(1):9.
- Morton SU, Shimamura A, Newburger PE, Opotowsky AR, Quiat D, Pereira AC, Jin SC, Gurvitz M, Brueckner M, Chung WK, Shen Y, Bernstein D, Gelb BD, Giardini A, Goldmuntz E, Kim RW, Lifton RP, Porter GA Jr, Srivastava D, Tristani-Firouzi M, Newburger JW, Seidman JG, Seidman CE. Association of damaging variants in genes with increased cancer risk among patients with congenital heart disease. JAMA Cardiol. 2021 Apr 1;6(4):457-462. [CrossRef]
- Park JE, Park JS, Jang SY, Park SH, Kim JW, Ki CS, Kim DK. A novel SMAD6 variant in a patient with severely calcified bicuspid aortic valve and thoracic aortic aneurysm. Mol Genet Genomic Med. 2019 May;7(5):e620. doi10.1002/mgg3.620.
- Raya A, Kawakami Y, Rodriguez-Esteban C, Buscher D, Koth CM, Itoh T, Morita M, Raya RM, Dubova I, Bessa JG, de la Pompa JL, Izpisua Belmonte JC. Notch activity induces Nodal expression and mediates the establishment of left-right asymmetry in vertebrate embryos.Genes Dev. 2003 May 15;17(10):1213-8. [CrossRef]
- Galvin KM, Donovan MJ, Lynch CA, Meyer RI, Paul RJ, Lorenz JN, Fairchild-Huntress V, Dixon KL, Dunmore JH, Gimbrone MA Jr, Falb D, Huszar D. A role for Smad6 in development and homeostasis of the cardiovascular system. Nat. Genet. Nat Genet. 2000 Feb;24(2):171-4. [CrossRef]
- McKean DM, Homsy J, Wakimoto H, Patel N, Gorham J, DePalma SR, Ware JS, Zaidi S, Ma W, Patel N, Lifton RP, Chung WK, Kim R, Shen Y, Brueckner M, Goldmuntz E, Sharp AJ, Seidman CE, Gelb BD, Seidman JG. Loss of RNA expression and allele-specific expression associated with congenital heart disease. Nat Commun. 2016 Sep 27; 7:12824. [CrossRef]
- Brault V, Nguyen TL, Flores-Gutiérrez J, Iacono G, Birling MC, Lalanne V, Meziane H, Manousopoulou A, Pavlovic G, Lindner L, Selloum M, Sorg T, Yu E, Garbis SD, Hérault Y. Dyrk1a gene dosage in glutamatergic neurons has key effects in cognitive deficits observed in mouse models of MRD7 and Down syndrome. PLoS Genet. 2021 Sep 29;17(9):e1009777. [CrossRef]
- Feki A, Hibaoui Y. DYRK1A Protein, A Promising Therapeutic Target to Improve Cognitive Deficits in Down Syndrome.
- Brain Sci. 2018 Oct 16;8(10):187. [CrossRef]
- Vandeweyer G, Helsmoortel C, Van Dijck A, Vulto-van Silfhout AT, Coe BP, Bernier R, Gerdts J, Rooms L, van den Ende J, Bakshi M, Wilson M, Nordgren A, Hendon LG, Abdulrahman OA, Romano C, de Vries BB, Kleefstra T, Eichler EE, Van der Aa N, Kooy RF. The transcriptional regulator ADNP links the BAF (SWI/SNF) complexes with autism. Am J Med Genet C Semin Med Genet. 2014 Sep ;166C (3):315-26. [CrossRef]
- Ockeloen CW, Willemsen MH, de Munnik S, van Bon BW, de Leeuw N, Verrips A, Kant SG, Jones EA, Brunner HG, van Loon RL, Smeets EE, van Haelst MM, van Haaften G, Nordgren A, Malmgren H, Grigelioniene G, Vermeer S, Louro P, Ramos L, Maal TJ, van Heumen CC, Yntema HG, Carels CE, Kleefstra T. Further delineation of the KBG syndrome phenotype caused by ANKRD11 aberrations. Eur J Hum Genet. 2015 Sep ;23(9):1176-85. [CrossRef]
- Hamilton MJ, Caswell RC, Canham N, Cole T, Firth HV, Foulds N, Heimdal K, Hobson E, Houge G, Joss S, Kumar D, Lampe AK, Maystadt I, McKay V, Metcalfe K, Newbury-Ecob R, Park SM, Robert L, Rustad CF, Wakeling E, Wilkie AOM, Study TDDD, Twigg SRF, Suri M. Heterozygous mutations affecting the protein kinase domain of CDK13 cause a syndromic form of developmental delay and intellectual disability. J Med Genet. 2018 Jan;55(1):28-38. [CrossRef]
- van den Akker WMR, Brummelman I, Martis LM, Timmermans RN, Pfundt R, Kleefstra T, Willemsen MH, Gerkes EH, Herkert JC, van Essen AJ, Rump P, Vansenne F, Terhal PA, van Haelst MM, Cristian I, Turner CE, Cho MT, Begtrup A, Willaert R, Fassi E, van Gassen KLI, Stegmann APA, de Vries BBA, Schuurs-Hoeijmakers JHM. De novo variants in CDK13 associated with syndromic ID/DD: Molecular and clinical delineation of 15 individuals and a further review. Clin Genet. 2018 May;93(5):1000-1007. [CrossRef]
- Beal B, Hayes I, McGaughran J, Amor DJ, Miteff C, Jackson V, van Reyk O, Subramanian G, Hildebrand MS, Morgan AT, Goel H. Expansion of phenotype of DDX3X syndrome: six new cases. Clin Dysmorphol. 2019 Oct;28(4):169-174. [CrossRef]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M,. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016 Aug 18;536(7616):285-91. [CrossRef]
- Belkadi A, Bolze A, Itan Y, Cobat A, Vincent QB, Antipenko A, Shang L, Boisson B, Casanova JL, Abel L Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants.
- Proc Natl Acad Sci U S, A. 2015 Apr 28;112(17):5473-8. [CrossRef]
- Hsieh A, Morton SU, Willcox JAL, Gorham JM, Tai AC, Qi H, DePalma S, McKean D, Griffin E, Manheimer KB, Bernstein D, Kim RW, Newburger JW, Porter GA Jr, Srivastava D, Tristani-Firouzi M, Brueckner M, Lifton RP, Goldmuntz E, Gelb BD, Chung WK, Seidman CE, Seidman JG, Shen Y. EM-mosaic detects mosaic point mutations that contribute to congenital heart disease. Genome Med. 2020 Apr 29;12(1):42. [CrossRef]
- Manheimer KB, Richter F, Edelmann LJ, D'Souza SL, Shi L, Shen Y, Homsy J, Boskovski MT, Tai AC, Gorham J, Yasso C, Goldmuntz E, Brueckner M, Lifton RP, Chung WK, Seidman CE, Seidman JG, Gelb BD. Robust identification of mosaic variants in congenital heart disease. Hum Genet. 2018 Feb;137(2):183-193. [CrossRef]
- Wei W, Keogh MJ, Aryaman J, Golder Z, Kullar PJ, Wilson I, Talbot K, Turner MR, McKenzie CA, Troakes C, Attems J, Smith C, Sarraj SA, Morris CM, Ansorge O, Jones NS, Ironside JW, Chinnery PF. Frequency and signature of somatic variants in 1461 human brain exomes. Genet Med. 2019 Apr;21(4):904-912. [CrossRef]
- .Zech M, Jech R, Boesch S, Škorvánek M, Weber S, Wagner M, Zhao C, Jochim A, Necpál J, Dincer Y, Vill K, Distelmaier F, Stoklosa M, Krenn M, Grunwald S, Bock-Bierbaum T, Fečíková A, Havránková P, Roth J, Příhodová I, Adamovičová M, Ulmanová O, Bechyně K, Danhofer P, Veselý B, Haň V, Pavelekova P, Gdovinová Z, Mantel T, Meindl T, Sitzberger A, Schröder S, Blaschek A, Roser T, Bonfert MV, et al. Monogenic variants in dystonia: an exome-wide sequencing study Lancet Neurol. 2020 Nov;19(11):908-918. [CrossRef]
- Gardner EJ, Sifrim A, Lindsay SJ, Prigmore E, Rajan D, Danecek P, Gallone G, Eberhardt RY, Martin HC, Wright CF, FitzPatrick DR, Firth HV, Hurles ME. Detecting cryptic clinically relevant structural variation in exome-sequencing data increases diagnostic yield for developmental disorders. Am J Hum Genet. 2021 Nov 4;108(11):2186-2194. [CrossRef]
- Wright CF, Fitzgerald TW, Jones WD, Clayton S, McRae JF, van Kogelenberg M, King DA, Ambridge K, Barrett DM, Bayzetinova T, Bevan AP, Bragin E, Chatzimichali EA, Gribble S, Jones P, Krishnappa N, Mason LE, Miller R, Morley KI, Parthiban V, Prigmore E, Rajan D, Sifrim A, Swaminathan GJ, Tivey AR, Middleton A, Parker M, Carter NP, Barrett JC, Hurles ME, FitzPatrick DR, Firth HV; DDD study. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet. 2015 Apr 4;385(9975):1305-14. [CrossRef]
- Padhi EM, Hayeck TJ, Cheng Z, Chatterjee S, Mannion BJ, Byrska-Bishop M, Willems M, Pinson L, Redon S, Benech C, Uguen K, Audebert-Bellanger S, Le Marechal C, Férec C, Efthymiou S, Rahman F, Maqbool S, Maroofian R, Houlden H, Musunuri R, Narzisi G, Abhyankar A, Hunter RD, Akiyama J, Fries LE, Ng JK, Mehinovic E, Stong N, Allen AS, Dickel DE, Bernier RA, Gorkin DU, Pennacchio LA, Zody MC, Turner TN. Coding and noncoding variants in EBF3 are involved in HADDS and simplex autism. Hum Genomics. 2021 Jul 13;15(1):44. [CrossRef]
- Choy MK, Javierre BM, Williams SG, Baross SL, Liu Y, Wingett SW, Akbarov A, Wallace C, Freire-Pritchett P, Rugg-Gunn PJ, Spivakov M, Fraser P, Keavney BD. Promoter interactome of human embryonic stem cell-derived cardiomyocytes connects GWAS regions to cardiac gene networks. Nat Commun. 2018 Jun 28;9(1):2526. [CrossRef]
- Hoelscher SC, Stich T, Diehm A, Lahm H, Dreßen M, Zhang Z, Neb I, Aherrahrou Z, Erdmann J, Schunkert H, Santamaria G, Cuda G, Gilsbach R, Hein L, Lange R, Hassel D, Krane M, Doppler SA. miR-128a Acts as a Regulator in Cardiac Development by Modulating Differentiation of Cardiac Progenitor Cell Populations. Int J Mol Sci. 2020 Feb 10;21(3):1158. [CrossRef]
- Kheradpour P, Ernst J, Melnikov A, Rogov P, Wang L, Zhang X, Alston J, Mikkelsen TS, Kellis M. Systematic dissection of regulatory motifs in 2000 predicted human enhancers using a massively parallel reporter assay. Genome Res. 2013 May;23(5):800-11. [CrossRef]
- Akerberg BN, Gu F, VanDusen NJ, Zhang X, Dong R, Li K, Zhang B, Zhou B, Sethi I, Ma Q, Wasson L, Wen T, Liu J, Dong K, Conlon FL, Zhou J, Yuan GC, Zhou P, Pu WT. A reference map of murine cardiac transcription factor chromatin occupancy identifies dynamic and conserved enhancers. Nat Commun. 2019 Oct 28;10(1):4907. [CrossRef]
- Vanoudenhove J, Yankee TN, Wilderman, Cotney J Epigenomic and transcriptomic dynamics during human heart organogenesis. Circ Res. 2020 Oct 9;127(9):e184-e209. [CrossRef]
- Hussein IR, Bader RS, Chaudhary AG, Bassiouni R, Alquaiti M, Ashgan F, Schulten HJ, Al Qahtani MH. dentification of De Novo and Rare Inherited Copy Number Variants in Children with Syndromic Congenital Heart Defects Pediatr Cardiol. 2018 Jun;39(5):924-940. [CrossRef]
- van Ouwerkerk AF, Bosada FM, van Duijvenboden K, Houweling AC, Scholman KT, Wakker V, Allaart CP, Uhm JS, Mathijssen IB, Baartscheer T, Postma AV, Barnett P, Verkerk AO, Boukens BJ, Christoffels VM. Patient-Specific TBX5-G125R Variant Induces Profound Transcriptional Deregulation and Atrial Dysfunction. Circulation. 2022 Feb 22;145(8):606-619. [CrossRef]
- Richter F, Morton SU, Kim SW, Kitaygorodsky A, Wasson LK, Chen KM, Zhou J, Qi H, Patel N, DePalma SR, Parfenov M, Homsy J, Gorham JM, Manheimer KB, Velinder M, Farrell A, Marth G, Schadt EE, Kaltman JR, Newburger JW, Giardini A, Goldmuntz E, Brueckner M, et al. Genomic analyses implicate noncoding denovo variants in congenital heart disease. Nat Genet. 2020 Aug;52(8):769-777. [CrossRef]
- Morton SU, Pereira AC, Quiat D, Richter F, Kitaygorodsky A, Hagen J, Bernstein D, Brueckner M, Goldmuntz E, Kim RW, Lifton RP, Porter GA Jr, Tristani-Firouzi M, Chung WK, Roberts A, Gelb BD, Shen Y, Newburger JW, Seidman JG, Seidman CE. Genome-Wide De Novo Variants in Congenital Heart Disease Are Not Associated With Maternal Diabetes or Obesity. Circ Genom Precis Med. 2022 Apr;15(2):e003500. [CrossRef]
- Wang T, Zhao PA, Eichler EE. Rare variants and the oligogenic architecture of autism. Trends Genet. 2022 Sep;38(9):895-903. [CrossRef]
- Karczewski KJ et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581,434–443 (2020).
- Collins RL, Brand H, Karczewski KJ, Zhao X, Alföldi J, Francioli LC, Khera AV, Lowther C, Gauthier LD, Wang H, Watts NA, Solomonson M, O'Donnell-Luria A, Baumann A, Munshi R, Walker M, Whelan CW, Huang Y, Brookings T, Sharpe T, Stone MR, Valkanas E, Fu J, Tiao G, Laricchia KM, Ruano-Rubio V, Stevens C, Gupta N, Cusick C, Margolin L; Genome Aggregation Database Production Team; Genome Aggregation Database Consortium; Taylor KD, Lin HJ, Rich SS, Post WS, Chen YI, Rotter JI, Nusbaum C, Philippakis A, Lander E, Gabriel S, Neale BM, Kathiresan S, Daly MJ, Banks E, MacArthur DG, Talkowski ME.A structural variation reference for medical and population genetics. Nature. 2020 May;581(7809):444-451. [CrossRef]
- Hureaux M, Guterman S, Hervé B, Till M, Jaillard S, Redon S, Valduga M, Coutton C, Missirian C, Prieur F, Simon-Bouy B, Beneteau C, Kuentz P, Rooryck C, Gruchy N, Marle N, Plutino M, Tosca L, Dupont C, Puechberty J, Schluth-Bolard C, Salomon L, Sanlaville D, Malan V, Vialard F. Chromosomal microarray analysis in fetuses with an isolated congenital heart defect: A retrospective, nationwide, multicenter study in France. Prenat Diagn. 2019 May;39(6):464-470. [CrossRef]
- Hatim O, Pavlinov I, Xu M, Linask K, Beers J, Liu C, Baumgärtel K, Gilbert M, Spinner N, Chen C, Zou J, Zheng W. Generation of an Alagille syndrome (ALGS) patient-derived induced pluripotent stem cell line (TRNDi032-A) carrying a heterozygous mutation (p.Cys682Leufs*7) in the JAG1 gene. Stem Cell Res. 2023 Dec; 73:103231. [CrossRef]
- Legoff L, D’Cruz SC, Tevosian S, Primig M, Smagulova F Transgenerational inheritance of environmentally induced epigenetic alterations during mammalian development. Cells. 2019 Dec 3;8(12):1559. [CrossRef]
- Barua S, Junaid MA Lifestyle, pregnancy and epigenetic effects. Epigenomics. 2015;7(1):85-102. [CrossRef]
- Yokouchi-Konishi T, Yoshimatsu J, Sawada M, Shionoiri T, Nakanishi A, Horiuchi C, Tsuritani M, Iwanaga N, Kamiya CA, Neki R, Miyake A, Kurosaki K, Shiraishi I.. Recurrent congenital heart diseases among neonates born to mothers with congenital heart diseases. Pediatr Cardiol. 2019 Apr;40(4):865-870. [CrossRef]
- Ellesøe SG, Workman CT, Bouvagnet P, Loffredo CA, McBride KL, Hinton RB, van Engelen K, Gertsen EC, Mulder BJM, Postma AV, Anderson RH, Hjortdal VE, Brunak S, Larsen LA. Familial co-occurrence of congenital heart defects follows distinct patterns. Eur Heart J. 2018 Mar 21;39(12):1015-1022. [CrossRef]
- Øyen N, Poulsen G, Boyd HA, Wohlfahrt J, Jensen PK, Melbye M. Recurrence of congenital heart defects in families. Circulation. 2009 Jul 28;120(4):295-301. [CrossRef]
- Argelaguet R, Clark SJ, Mohammed H, Stapel LC, Krueger C, Kapourani CA, Imaz-Rosshandler I, Lohoff T, Xiang Y, Hanna CW, Smallwood S, Ibarra-Soria X, Buettner F, Sanguinetti G, Xie W, Krueger F, Göttgens B, Rugg-Gunn PJ, Kelsey G, Dean W, Nichols J, Stegle O, Marioni JC, Reik W. Multi-omics profiling of mouse gastrulation at single-cell resolution. Nature. 2019 Dec;576(7787):487-491. [CrossRef]
- Tucker NR, Chaffin M, Fleming SJ, Hall AW, Parsons VA, Bedi KC Jr, Akkad AD, Herndon CN, Arduini A, Papangeli I, Roselli C, Aguet F, Choi SH, Ardlie KG, Babadi M, Margulies KB, Stegmann CM, Ellinor PT. Transcriptional and Cellular Diversity of the Human Heart. Circulation. 2020 Aug 4;142(5):466-482. [CrossRef]
- Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, Trapnell C, Shendure J. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019 Feb;566(7745):496-502. [CrossRef]
- Cui Y, Zheng Y, Liu X, Yan L, Fan X, Yong J, Hu Y, Dong J, Li Q, Wu X, Gao S, Li J, Wen L, Qiao J, Tang F.. Single-cell transcriptome analysis maps the developmental track of the human heart. Cell Rep. 2019 Feb 12;26(7):1934-1950.e5. [CrossRef]
- Lescroart F, Wang X, Lin X, Swedlund B, Gargouri S, Sànchez-Dànes A, Moignard V, Dubois C, Paulissen C, Kinston S, Göttgens B, Blanpain C.. Defining the earliest step of cardiovascular lineage segregation by single‐cell.
- RNA-seq. Science. 2018 Mar 9;359(6380):1177-1181. [CrossRef]
- DeLaughter DM, Bick AG, Wakimoto H, McKean D, Gorham JM, Kathiriya IS, Hinson JT, Homsy J, Gray J, Pu W, Bruneau BG, Seidman JG, Seidman CE. Single-cell resolution of temporal gene expression during heart development. Dev Cell. 2016 Nov 21;39(4):480-490. [CrossRef]
- Pijuan-Sala B, Griffiths JA, Guibentif C, Hiscock TW, Jawaid W, Calero-Nieto FJ, Mulas C, Ibarra-Soria X, Tyser RCV, Ho DLL, Reik W, Srinivas S, Simons BD, Nichols J, Marioni JC, Göttgens B. A single-cell molecular map of mouse gastrulation and early organogenesis. Nature. 2019 Feb;566(7745):490-495. [CrossRef]
- Ulirsch JC, Verboon JM, Kazerounian S, Guo MH, Yuan D, Ludwig LS, Handsaker RE, Abdulhay NJ, Fiorini C, Genovese G, Lim ET, Cheng A, Cummings BB, Chao KR, Beggs AH, Genetti CA, Sieff CA, Newburger PE, Niewiadomska E, Matysiak M, Vlachos A, Lipton JM, Atsidaftos E. The genetic landscape of Diamond-Blackfan anemia. Am J Hum Genet. 2018 Dec 6;103(6):930-947. [CrossRef]
- Bramel EE, Creamer TJ, Saqib M, Camejo Nunez WA, Bagirzadeh R, Roker LA, Goff LA, MacFarlane EG. Postnatal Smad3 Inactivation in Murine Smooth Muscle Cells Elicits a Temporally and Regionally Distinct Transcriptional Response. Front Cardiovasc Med. 2022 Apr 8;9:826495. [CrossRef]
- Bissoli I, D'Adamo S, Pignatti C, Agnetti G, Flamigni F, Cetrullo S. Induced pluripotent stem cell-based models: Are we ready for that heart in a dish? Front Cell Dev Biol. 2023 Jan 19; 11:1129263. [CrossRef]
- Baumann, K. Achieving pluripotency. Nat Rev Mol Cell Biol. 2010 Oct;11(10):677. [CrossRef]
- Zhang J, Tao R, Campbell KF, Carvalho JL, Ruiz EC, Kim GC, Schmuck EG, Raval AN, da Rocha AM, Herron TJ, Jalife J, Thomson JA, Kamp TJ.Nat Commun. 2019 May 20;10(1):2238. [CrossRef]
- Liu JA, Cheung M. Neural crest stem cells and their potential therapeutic applications. Dev Biol. 2016 Nov 15;419(2):199-216. [CrossRef]
- Neri T, Hiriart E, van Vliet PP, Faure E, Norris RA, Farhat B, Jagla B, Lefrancois J, Sugi Y, Moore-Morris T, Zaffran S, Faustino RS, Zambon AC, Desvignes JP, Salgado D, Levine RA, de la Pompa JL, Terzic A, Evans SM, Markwald R, Pucéat M.Nat Commun. 2019 Apr 26;10(1):1929. [CrossRef]
- Kathiriya IS, Rao KS, Iacono G, Devine WP, Blair AP, Hota SK, Lai MH, Garay BI, Thomas R, Gong HZ, Wasson LK, Goyal P, Sukonnik T, Hu KM, Akgun GA, Bernard LD, Akerberg BN, Gu F, Li K, Speir ML, Haeussler M, Pu WT, Stuart JM, Seidman CE, Seidman JG, Heyn H, Bruneau BG. Dev Cell. 2021 Feb 8;56(3):292-309.e9. [CrossRef]
- Rydzanicz M, Zwoliński P, Gasperowicz P, Pollak A, Kostrzewa G, Walczak A, Konarzewska M, Płoski R. A recurrent de novo variant supports KCNC2 involvement in the pathogenesis of developmental and epileptic encephalopathy. Am J Med Genet A. 2021 Nov;185(11):3384-3389. [CrossRef]
- Pierpont ME, Brueckner M, Chung WK, Garg V, Lacro RV, McGuire AL, Mital S, Priest JR, Pu WT, Roberts A, Ware SM, Gelb BD, Russell MW; American Heart Association Council on Cardiovascular Disease in the Young; Council on Cardiovascular and Stroke Nursing; and Council on Genomic and Precision Medicine. Genetic basis for congenital heart disease: revisited: a scientific statement from the American Heart Association. Circulation. 2018 Nov 20;138(21):e653-e711. [CrossRef]
- DiStefano MT, Hemphill SE, Oza AM, Siegert RK, Grant AR, Hughes MY, Cushman BJ, Azaiez H, Booth KT, Chapin A, Duzkale H, Matsunaga T, Shen J, Zhang W, Kenna M, Schimmenti LA, Tekin M, Rehm HL, Tayoun ANA, Amr SS; ClinGen Hearing Loss Clinical Domain Working Group. ClinGen expert clinical validity curation of 164 hearing loss gene-disease pairs. Genet Med. 2019 Oct;21(10):2239-2247. [CrossRef]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015 May;17(5):405-24. [CrossRef]
- Boycott KM, Azzariti DR, Hamosh A, Rehm HL. Seven years since the launch of the Matchmaker Exchange: The evolution of genomic matchmaking. Hum Mutat. 2022 Jun;43(6):659-667. [CrossRef]
- Patel RY, Shah N, Jackson AR, Ghosh R, Pawliczek P, Paithankar S, Baker A, Riehle K, Chen H, Milosavljevic S, Bizon C, Rynearson S, Nelson T, Jarvik GP, Rehm HL, Harrison SM, Azzariti D, Powell B, Babb L, Plon SE, Milosavljevic A; ClinGen Resource. ClinGen Pathogenicity Calculator: a configurable system for assessing pathogenicity of genetic variants. Genome Med. 2017 Jan 12;9(1):3. [CrossRef]
- Yu Y, Lei W, Yang J, Wei YC, Zhao ZL, Zhao ZA, Hu S. Functional mutant GATA4 identification and potential application in preimplantation diagnosis of congenital heart. Gene. 2018 Jan 30; 641:349-354. [CrossRef]
- Boskovski MT, Homsy J, Nathan M, Sleeper LA, Morton S, Manheimer KB, Tai A, Gorham J, Lewis M, Swartz M, Alfieris GM, Bacha EA, Karimi M, Meyer D, Nguyen K, Bernstein D, Romano-Adesman A, Porter GA Jr, Goldmuntz E, Chung WK, Srivastava D, et al. De novo damaging variants, clinical phenotypes and post-operative outcomes in congenital heart disease. Circ Genom Precis Med. 2020 Aug;13(4):e002836. [CrossRef]
- Zomer AC, Ionescu-Ittu R, Vaartjes I, Pilote L, Mackie AS, Therrien J, Langemeijer MM, Grobbee DE, Mulder BJ, Marelli AJ. Sex differences in hospital mortality in adults with congenital heart disease: the impact of reproductive health.J Am Coll Cardiol. 2013 Jul 2;62(1):58-67. [CrossRef]
- Gurvitz M et al. Prevalence of cancer in adults with congenital heart disease compared with the general population. Am. J. Cardiol. 118, 1742–1750 (2016).
- Mandalenakis Z, Karazisi C, Skoglund K, Rosengren A, Lappas G, Eriksson P, Dellborg M. Risk of cancer among children and young adults with congenital heart disease compared with healthy controls. JAMA Netw Open. 2019 Jul 3;2(7):e196762. [CrossRef]
- Cohen S, Gurvitz MZ, Beauséjour-Ladouceur V, Lawler PR, Therrien J, Marelli AJ. Cancer Risk in Congenital Heart Disease-What Is the Evidence? Can J Cardiol. 2019 Dec;35(12):1750-1761. [CrossRef]
- Lim ET, Uddin M, De Rubeis S, Chan Y, Kamumbu AS, Zhang X, D'Gama AM, Kim SN, Hill RS, Goldberg AP, Poultney C, Minshew NJ, Kushima I, Aleksic B, Ozaki N, Parellada M, Arango C, Penzol MJ, Carracedo A, Kolevzon A, Hultman CM, Weiss LA, Fromer M, Chiocchetti AG, Freitag CM; Autism Sequencing Consortium; Church GM, Scherer SW, Buxbaum JD, Walsh CA. Rates, distribution and implications of postzygotic mosaic mutations in autism spectrum disorder. Nat Neurosci. 2017 Sep ;20(9):1217-1224. [CrossRef]
- Freed D, Pevsner J. The Contribution of Mosaic Variants to Autism Spectrum Disorder. PLoS Genet. 2016 Sep 15;12(9):e1006245. [CrossRef]
- Dou Y, Yang X, Li Z, Wang S, Zhang Z, Ye AY, Yan L, Yang C, Wu Q, Li J, Zhao B, Huang AY, Wei L. Postzygotic single-nucleotide mosaicisms contribute to the etiology of autism spectrum disorder and autistic traits and the origin of mutations. Hum Mutat. 2017 Aug;38(8):1002-1013. [CrossRef]
- Yu X, Tao Y, Liu X, Yu F, Jiang C, Xiao Y, Zhang H, He Y, Ye L, Wang Y, Zhou C, Wang J, Jiang Z, Hong H. The implication of chromosomal abnormalities in the surgical outcomes of Chinese pediatric patients with congenital heart disease. Front Cardiovasc Med. 2023 May 24;10:1164577. [CrossRef]
- Putotto C, Pugnaloni F, Unolt M, Maiolo S, Trezzi M, Digilio MC, Cirillo A, Limongelli G, Marino B, Calcagni G, Versacci P. 22q11.2 Deletion Syndrome: Impact of Genetics in the Treatment of Conotruncal Heart Defects. Children (Basel). 2022 May 25;9(6):772. [CrossRef]
- Mercer-Rosa L, Pinto N, Yang W, Tanel R, Goldmuntz E 22q11.2 deletion syndrome is associated with perioperative outcome in tetralogy of Fallot. J Thorac Cardiovasc Surg. 2013 Oct;146(4):868-73. [CrossRef]
- O’Byrne ML, O'Byrne ML, Yang W, Mercer-Rosa L, Parnell AS, Oster ME, Levenbrown Y, Tanel RE, Goldmuntz E.22q11.2 deletion syndrome is associated with increased perioperative events and more complicated postoperative course in infants undergoing infant operative correction of truncus arteriosus communis or interrupted aortic arch. J Thorac Cardiovasc Surg. 2014 Oct;148(4):1597-605. [CrossRef]
- Kim DS, Kim JH, Burt AA, Crosslin DR, Burnham N, Kim CE, McDonald-McGinn DM, Zackai EH, Nicolson SC, Spray TL, Stanaway IB, Nickerson DA, Heagerty PJ, Hakonarson H, Gaynor JW, Jarvik GP.. Burden of potentially pathologic copy number variants is higher in children with isolated congenital heart disease and significantly impairs covariate-adjusted transplant-free survival. J Thorac Cardiovasc Surg. 2016 Apr;151(4):1147-51.e4. [CrossRef]
- Bai W, Suzuki H, Huang J, Francis C, Wang S, Tarroni G, Guitton F, Aung N, Fung K, Petersen SE, Piechnik SK, Neubauer S, Evangelou E, Dehghan A, O'Regan DP, Wilkins MR, Guo Y, Matthews PM, Rueckert D. A population-based phenome-wide association study of cardiac and aortic structure and function. Nat Med. 2020 Oct;26(10):1654-1662. [CrossRef]
- Cohen S, Liu A, Gurvitz M, Guo L, Therrien J, Laprise C, Kaufman JS, Abrahamowicz M, Marelli AJ. Exposure to Low-Dose Ionizing Radiation From Cardiac Procedures and Malignancy Risk in Adults With Congenital Heart Disease. Circulation. 2018 Mar 27;137(13):1334-1345. [CrossRef]
- Danieli C, Cohen S, Liu A, Pilote L, Guo L, Beauchamp ME, Marelli AJ, Abrahamowicz M. Flexible Modeling of the Association Between Cumulative Exposure to Low-Dose Ionizing Radiation From Cardiac Procedures and Risk of Cancer in Adults With Congenital Heart Disease. Am J Epidemiol. 2019 Aug 1;188(8):1552-1562. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




