Submitted:
29 December 2023
Posted:
03 January 2024
You are already at the latest version
Abstract
Keywords:
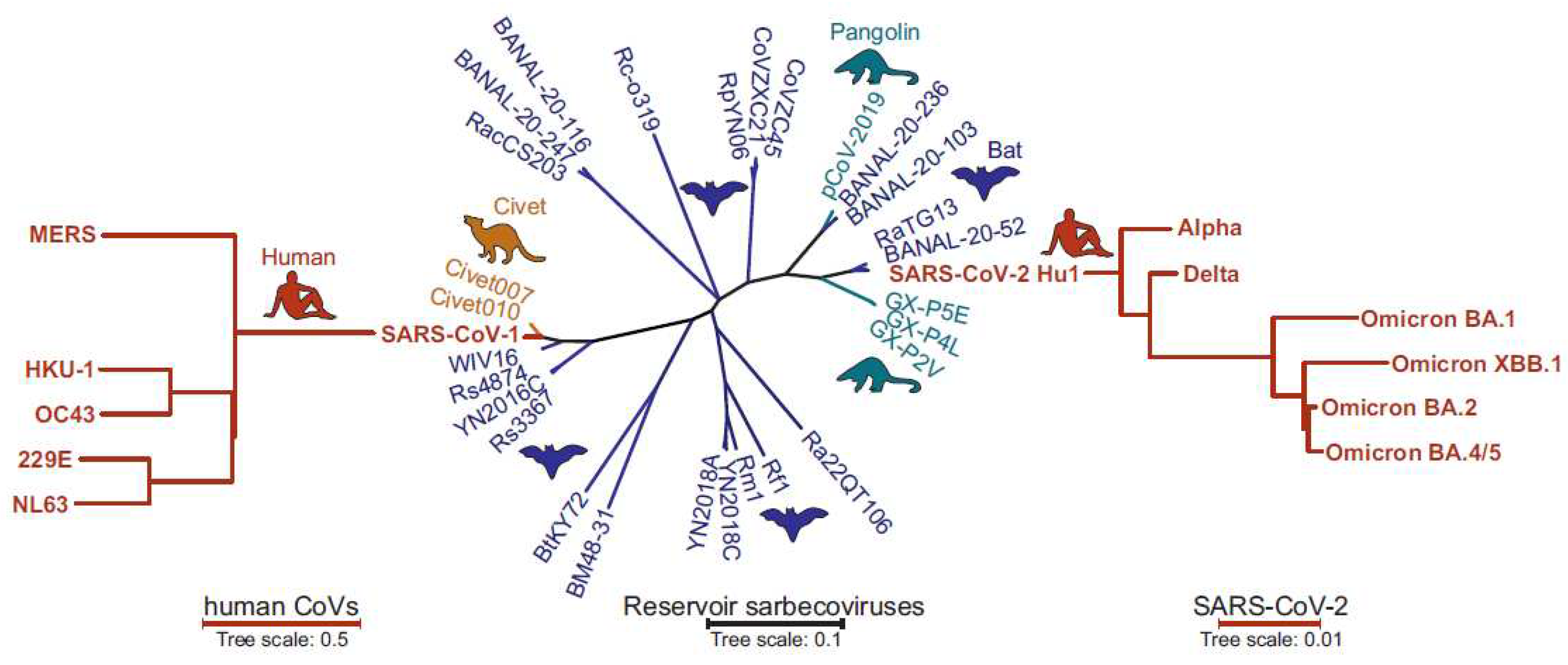
Introduction
Early features of the SARS-CoV-2 Spike protein
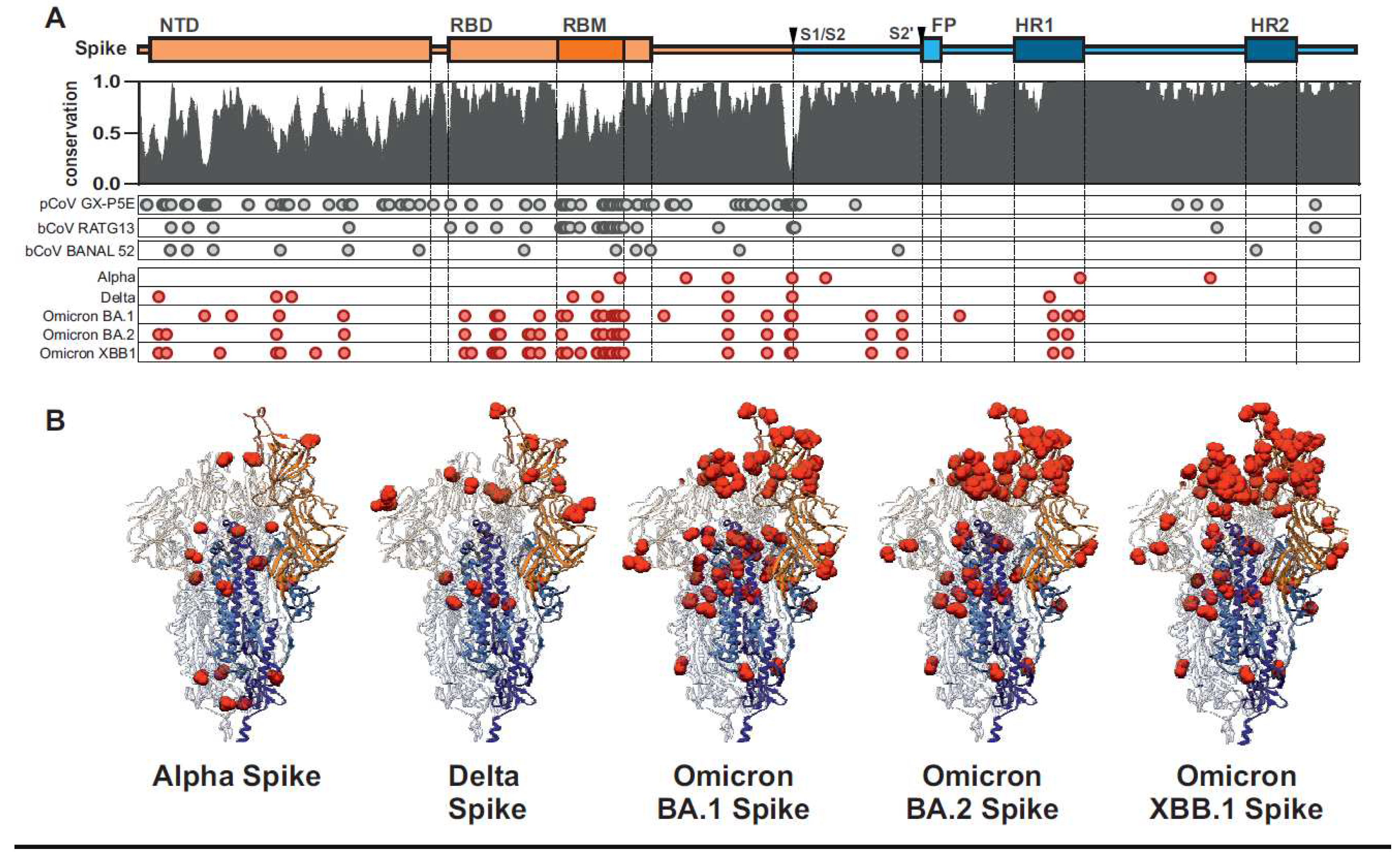
Initial human adaptation of SARS-CoV-2 Spike proteins
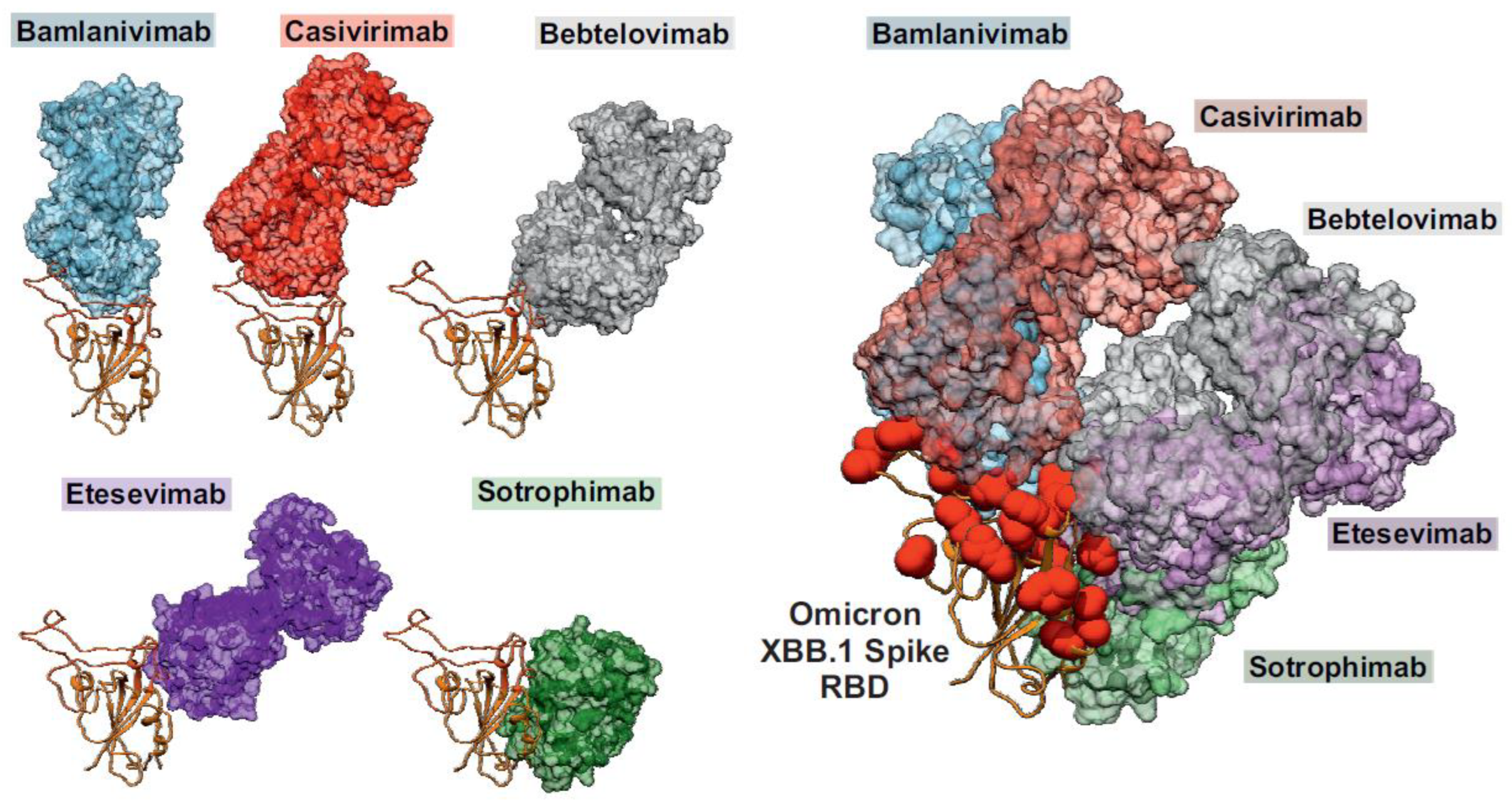
Evasion of adaptive immunity
Broadly acting vaccines or therapeutics targeting the SARS-CoV-2 Spike protein
Conclusions and future perspectives
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Banerjee, A., Kulcsar, K., Misra, V., Frieman, M., and Mossman, K. (2019). Bats and Coronaviruses. Viruses 11. [CrossRef]
- Mahdy, M.A.A., Younis, W., and Ewaida, Z. (2020). An Overview of SARS-CoV-2 and Animal Infection. Frontiers in Veterinary Science 7. [CrossRef]
- Dhama, K., Patel, S.K., Sharun, K., Pathak, M., Tiwari, R., Yatoo, M.I., Malik, Y.S., Sah, R., Rabaan, A.A., Panwar, P.K., et al. (2020). SARS-CoV-2 jumping the species barrier: Zoonotic lessons from SARS, MERS and recent advances to combat this pandemic virus. Travel Med Infect Dis 37. 101830. [CrossRef]
- Ye, Z.W., Yuan, S., Yuen, K.S., Fung, S.Y., Chan, C.P., and Jin, D.Y. (2020). Zoonotic origins of human coronaviruses. Int J Biol Sci. 16. [CrossRef]
- Corman, V.M., Muth, D., Niemeyer, D., and Drosten, C. (2018). Hosts and Sources of Endemic Human Coronaviruses. In Advances in Virus Research 100. [CrossRef]
- Cui, J., Li, F., and Shi, Z.L. (2019). Origin and evolution of pathogenic coronaviruses Nature Reviews Microbiology 17. [CrossRef]
- Ksiazek, T.G., Erdman, D., Goldsmith, C.S., Zaki, S.R., Peret, T., Emery, S., Tong, S., Urbani, C., Comer, J.A., Lim, W., et al. (2003). A novel coronavirus associated with severe acute respiratory syndrome. New England Journal of Medicine 348. 1953–1966. [CrossRef]
- Bermingham, A., Chand, M.A., Brown, C.S., Aarons, E., Tong, C., Langrish, C., Hoschler, K., Brown, K., Galiano, M., Myers, R., et al. (2012). Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Eurosurveillance 17. [CrossRef]
- Zhou, P., Yang, X.L., Wang, X.G., Hu, B., Zhang, L., Zhang, W., Si, H.R., Zhu, Y., Li, B., Huang, C.L., et al. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579. [CrossRef]
- Boni, M.F., Lemey, P., Jiang, X., Lam, T.T.Y., Perry, B.W., Castoe, T.A., Rambaut, A., and Robertson, D.L. (2020). Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nature Microbiology 5. [CrossRef]
- Worobey, M., Levy, J.I., Malpica Serrano, L., Crits-Christoph, A., Pekar, J.E., Goldstein, S.A., Rasmussen, A.L., Kraemer, M.U.G., Newman, C., Koopmans, M.P.G., et al. (2022). The Huanan Seafood Wholesale Market in Wuhan was the early epicenter of the COVID-19 pandemic. Science 377. [CrossRef]
- Hao, X., Cheng, S., Wu, D., Wu, T., Lin, X., and Wang, C. (2020). Reconstruction of the full transmission dynamics of COVID-19 in Wuhan. Nature 584. [CrossRef]
- Sanjuán, R., Nebot, M.R., Chirico, N., Mansky, L.M., and Belshaw, R. (2010). Viral mutation rates. J Virol 84. [CrossRef]
- De Maio, N., Walker, C.R., Turakhia, Y., Lanfear, R., Corbett-Detig, R., and Goldman, N. (2021). Mutation Rates and Selection on Synonymous Mutations in SARS-CoV-2. Genome Biol Evol 13. [CrossRef]
- Moeller, N.H., Shi, K., Demir, Ö., Belica, C., Banerjee, S., Yin, L., Durfee, C., Amaro, R.E., and Aihara, H. (2022). Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN. Proceedings of the National Academy of Sciences 119. [CrossRef]
- Eckerle, L.D., Becker, M.M., Halpin, R.A., Li, K., Venter, E., Lu, X., Scherbakova, S., Graham, R.L., Baric, R.S., Stockwell, T.B., et al. (2010). Infidelity of SARS-CoV Nsp14-Exonuclease Mutant Virus Replication Is Revealed by Complete Genome Sequencing. PLOS Pathogens 6. [CrossRef]
- Cao, C., Cai, Z., Xiao, X., Rao, J., Chen, J., Hu, N., Yang, M., Xing, X., Wang, Y., Li, M., et al. (2021). The architecture of the SARS-CoV-2 RNA genome inside virion. Nat Commun 12. [CrossRef]
- Jones, A.N., Mourão, A., Czarna, A., Matsuda, A., Fino, R., Pyrc, K., Sattler, M., and Popowicz, G.M. (2022). Characterization of SARS-CoV-2 replication complex elongation and proofreading activity. Sci Rep 12. [CrossRef]
- Markov, P.V., Ghafari, M., Beer, M., Lythgoe, K., Simmonds, P., Stilianakis, N.I., and Katzourakis, A. (2023). The evolution of SARS-CoV-2. Nat Rev Microbiol, 21. [CrossRef]
- Focosi, D., and Maggi, F. (2022). Recombination in Coronaviruses, with a Focus on SARS-CoV-2. Viruses 14. [CrossRef]
- Carabelli, A.M., Peacock, T.P., Thorne, L.G., Harvey, W.T., Hughes, J., de Silva, T.I., Peacock, S.J., Barclay, W.S., de Silva, T.I., Towers, G.J., et al. (2023). SARS-CoV-2 variant biology: immune escape, transmission and fitness. Nat Rev Microbiol 21. [CrossRef]
- Chakraborty, C., Sharma, A.R., Bhattacharya, M., and Lee, S.-S. (2022). A Detailed Overview of Immune Escape, Antibody Escape, Partial Vaccine Escape of SARS-CoV-2 and Their Emerging Variants With Escape Mutations. Front Immunol 13. [CrossRef]
- Pastorio, C., Noettger, S., Nchioua, R., Zech, F., Sparrer, K.M.J., and Kirchhoff, F. (2023). Impact of mutations defining SARS-CoV-2 Omicron subvariants BA.2.12.1 and BA.4/5 on Spike function and neutralization. iScience 26. [CrossRef]
- Zmasek, C.M., Lefkowitz, E.J., Niewiadomska, A., and Scheuermann, R.H. (2022). Genomic evolution of the Coronaviridae family. Virology 570. [CrossRef]
- Temmam, S., Vongphayloth, K., Baquero, E., Munier, S., Bonomi, M., Regnault, B., Douangboubpha, B., Karami, Y., Chrétien, D., Sanamxay, D., et al. (2022). Bat coronaviruses related to SARS-CoV-2 and infectious for human cells. Nature 604. [CrossRef]
- Ou, X., Liu, Y., Lei, X., Li, P., Mi, D., Ren, L., Guo, L., Guo, R., Chen, T., Hu, J., et al. (2020). Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun 11. [CrossRef]
- Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., Schiergens, T.S., Herrler, G., Wu, N.-H., Nitsche, A., et al. (2020). SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 181. [CrossRef]
- Hofmann, H., Pyrc, K., van der Hoek, L., Geier, M., Berkhout, B., and Pöhlmann, S. (2005). Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc Natl Acad Sci U S A 102. [CrossRef]
- Letko, M., Marzi, A., and Munster, V. (2020). Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nature Microbiology 5. [CrossRef]
- Zheng, M., Zhao, X., Zheng, S., Chen, D., Du, P., Li, X., Jiang, D., Guo, J.-T., Zeng, H., and Lin, H. (2020). Bat SARS-Like WIV1 coronavirus uses the ACE2 of multiple animal species as receptor and evades IFITM3 restriction via TMPRSS2 activation of membrane fusion. Emerging Microbes & Infections 9. [CrossRef]
- Wang, Q., Noettger, S., Xie, Q., Pastorio, C., Seidel, A., Müller, J.A., Jung, C., Jacob, T., Sparrer, K.M.J., Zech, F., et al. (2023). Determinants of species-specific utilization of ACE2 by human and animal coronaviruses. Commun Biol 6. [CrossRef]
- Zech, F., Schniertshauer, D., Jung, C., Herrmann, A., Cordsmeier, A., Xie, Q., Nchioua, R., Prelli Bozzo, C., Volcic, M., Koepke, L., et al. (2021). Spike residue 403 affects binding of coronavirus spikes to human ACE2. Nat Commun 12. [CrossRef]
- Lim, S., Zhang, M., and Chang, T.L. (2022). ACE2-Independent Alternative Receptors for SARS-CoV-2. Viruses 14. 2535. [CrossRef]
- Sasaki, M., Toba, S., Itakura, Y., Chambaro, H.M., Kishimoto, M., Tabata, K., Intaruck, K., Uemura, K., Sanaki, T., Sato, A., et al. (2021). SARS-CoV-2 Bearing a Mutation at the S1/S2 Cleavage Site Exhibits Attenuated Virulence and Confers Protective Immunity. mBio 12. e0141521. [CrossRef]
- Whittaker, G.R. (2021). SARS-CoV-2 spike and its adaptable furin cleavage site. The Lancet Microbe 2. [CrossRef]
- Peacock, T.P., Goldhill, D.H., Zhou, J., Baillon, L., Frise, R., Swann, O.C., Kugathasan, R., Penn, R., Brown, J.C., Sanchez-David, R.Y., et al. (2021). The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat Microbiol 6. [CrossRef]
- Örd, M., Faustova, I., and Loog, M. (2020). The sequence at Spike S1/S2 site enables cleavage by furin and phospho-regulation in SARS-CoV2 but not in SARS-CoV1 or MERS-CoV. Sci Rep 10. [CrossRef]
- Chan, Y.A., and Zhan, S.H. (2022). The Emergence of the Spike Furin Cleavage Site in SARS-CoV-2. Mol Biol Evol 39. [CrossRef]
- Coutard, B., Valle, C., de Lamballerie, X., Canard, B., Seidah, N.G., and Decroly, E. (2020). The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res 176. [CrossRef]
- Andersen, K.G., Rambaut, A., Lipkin, W.I., Holmes, E.C., and Garry, R.F. (2020). The proximal origin of SARS-CoV-2. Nature Medicine 26. [CrossRef]
- Zhang, T., Wu, Q., and Zhang, Z. (2020). Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak. Current Biology 30. [CrossRef]
- Xiong, Q., Cao, L., Ma, C., Tortorici, M.A., Liu, C., Si, J., Liu, P., Gu, M., Walls, A.C., Wang, C., et al. (2022). Close relatives of MERS-CoV in bats use ACE2 as their functional receptors. Nature 612. [CrossRef]
- Stout, A.E., Millet, J.K., Stanhope, M.J., and Whittaker, G.R. (2021). Furin cleavage sites in the spike proteins of bat and rodent coronaviruses: Implications for virus evolution and zoonotic transfer from rodent species. One Health 13. [CrossRef]
- Lubinski, B., Fernandes, M.H.V., Frazier, L., Tang, T., Daniel, S., Diel, D.G., Jaimes, J.A., and Whittaker, G.R. (2021). Functional evaluation of the P681H mutation on the proteolytic activation of the SARS-CoV-2 variant B.1.1.7 (Alpha) spike. iScience 25. [CrossRef]
- Vu, M.N., Alvarado, R.E., Morris, D.R., Lokugamage, K.G., Zhou, Y., Morgan, A.L., Estes, L.K., McLeland, A.M., Schindewolf, C., Plante, J.A., et al. (2023). Loss-of-function mutation in Omicron variants reduces spike protein expression and attenuates SARS-CoV-2 infection. bioRxiv, 2023.04.17.536926. [CrossRef]
- Schaefer, S.L., Jung, H., and Hummer, G. (2021). Binding of SARS-CoV-2 Fusion Peptide to Host Endosome and Plasma Membrane. J Phys Chem B 125. [CrossRef]
- Yu, S., Zheng, X., Zhou, B., Li, J., Chen, M., Deng, R., Wong, G., Lavillette, D., and Meng, G. (2022). SARS-CoV-2 spike engagement of ACE2 primes S2’ site cleavage and fusion initiation. Proc Natl Acad Sci U S A 119. [CrossRef]
- Hu, B., Chan, J.F.-W., Liu, H., Liu, Y., Chai, Y., Shi, J., Shuai, H., Hou, Y., Huang, X., Yuen, T.T.-T., et al. (2022). Spike mutations contributing to the altered entry preference of SARS-CoV-2 omicron BA.1 and BA.2. Emerging Microbes & Infections 11. [CrossRef]
- Meng, B., Abdullahi, A., Ferreira, I.A.T.M., Goonawardane, N., Saito, A., Kimura, I., Yamasoba, D., Gerber, P.P., Fatihi, S., Rathore, S., et al. (2022). Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature 603. [CrossRef]
- Qu, P., Evans, J.P., Kurhade, C., Zeng, C., Zheng, Y.-M., Xu, K., Shi, P.-Y., Xie, X., and Liu, S.-L. (2023). Determinants and Mechanisms of the Low Fusogenicity and High Dependence on Endosomal Entry of Omicron Subvariants. mBio 14. [CrossRef]
- Nchioua, R., Schundner, A., Kmiec, D., Prelli Bozzo, C., Zech, F., Koepke, L., Graf, A., Krebs, S., Blum, H., Frick, M., et al. (2022). SARS-CoV-2 Variants of Concern Hijack IFITM2 for Efficient Replication in Human Lung Cells. J Virol 96. [CrossRef]
- Prelli Bozzo, C., Nchioua, R., Volcic, M., Koepke, L., Krüger, J., Schütz, D., Heller, S., Stürzel, C.M., Kmiec, D., Conzelmann, C., et al. (2021). IFITM proteins promote SARS-CoV-2 infection and are targets for virus inhibition in vitro. Nat Commun 12. [CrossRef]
- Xie, Q., Bozzo, C.P., Eiben, L., Noettger, S., Kmiec, D., Nchioua, R., Niemeyer, D., Volcic, M., Lee, J.-H., Zech, F., et al. (2023). Endogenous IFITMs boost SARS-coronavirus 1 and 2 replication whereas overexpression inhibits infection by relocalizing ACE2. iScience 26. [CrossRef]
- Basile, A., Zannella, C., De Marco, M., Sanna, G., Franci, G., Galdiero, M., Manzin, A., De Laurenzi, V., Chetta, M., Rosati, A., et al. (2023). Spike-mediated viral membrane fusion is inhibited by a specific anti-IFITM2 monoclonal antibody. Antiviral Res 211. [CrossRef]
- Stewart, H., Palmulli, R., Johansen, K.H., McGovern, N., Shehata, O.M., Carnell, G.W., Jackson, H.K., Lee, J.S., Brown, J.C., Burgoyne, T., et al. (2023). Tetherin antagonism by SARS-CoV-2 ORF3a and spike protein enhances virus release. EMBO Rep. [CrossRef]
- Dangi, T., Palacio, N., Sanchez, S., Park, M., Class, J., Visvabharathy, L., Ciucci, T., Koralnik, I.J., Richner, J.M., and Penaloza-MacMaster, P. (2021). Cross-protective immunity following coronavirus vaccination and coronavirus infection. J Clin Invest. [CrossRef]
- Weissman, D., Alameh, M.G., de Silva, T., Collini, P., Hornsby, H., Brown, R., LaBranche, C.C., Edwards, R.J., Sutherland, L., Santra, S., et al. (2021). D614G Spike Mutation Increases SARS CoV-2 Susceptibility to Neutralization. Cell Host and Microbe 29. [CrossRef]
- Zhang, L., Jackson, C.B., Mou, H., Ojha, A., Peng H., Quinlan B.D., Rangarajan, E.S., Pan A., Vanderheiden A., Suthar M.S., Li W., Izard, T., Rader C., Farzan, M., and Choe, H. (2020). SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat Commun. 11. 10.1038/s41467-020-19808-4.
- Plante, J.A., Liu, Y., Liu, J., Xia, H., Johnson, B.A., Lokugamage, K.G., Zhang, X., Muruato, A.E., Zou, J., Fontes-Garfias, C.R., et al. (2021). Spike mutation D614G alters SARS-CoV-2 fitness. Nature 592. [CrossRef]
- Daniloski, Z., Jordan, T.X., Ilmain, J.K., Guo, X., Bhabha, G., TenOever, B.R., and Sanjana, N.E. (2021). The Spike D614G mutation increases SARS-CoV-2 infection of multiple human cell types. eLife 10. [CrossRef]
- Bhattacharya, M., Chatterjee, S., Sharma, A.R., Agoramoorthy, G., and Chakraborty, C. (2021). D614G mutation and SARS-CoV-2: impact on S-protein structure, function, infectivity, and immunity. Appl Microbiol Biotechnol 105. [CrossRef]
- Yurkovetskiy, L., Wang, X., Pascal, K.E., Tomkins-Tinch, C., Nyalile, T.P., Wang, Y., Baum, A., Diehl, W.E., Dauphin, A., Carbone, C., et al. (2020). Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell 183. [CrossRef]
- Korber, B., Fischer, W.M., Gnanakaran, S., Yoon, H., Theiler, J., Abfalterer, W., Hengartner, N., Giorgi, E.E., Bhattacharya, T., Foley, B., et al. (2020). Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 182. [CrossRef]
- Martin, D.P., Weaver, S., Tegally, H., San, J.E., Shank, S.D., Wilkinson, E., Lucaci, A.G., Giandhari, J., Naidoo, S., Pillay, Y., et al. (2021). The emergence and ongoing convergent evolution of the SARS-CoV-2 N501Y lineages. Cell 184. [CrossRef]
- Tian, F., Tong, B., Sun, L., Shi, S., Zheng, B., Wang, Z., Dong, X., and Zheng, P. (2021). N501Y mutation of spike protein in SARS-CoV-2 strengthens its binding to receptor ACE2. eLife 10. [CrossRef]
- Wright, E.S., Lakdawala, S.S., and Cooper, V.S. (2020). SARS-CoV-2 genome evolution exposes early human adaptations. bioRxiv. [CrossRef]
- Posani, E., Dilucca, M., Forcelloni, S., Pavlopoulou, A., Georgakilas, A.G., and Giansanti, A. (2022). Temporal evolution and adaptation of SARS-CoV-2 codon usage. Frontiers in Bioscience-Landmark 27. [CrossRef]
- Choe, H., and Farzan, M. (2021). How SARS-CoV-2 first adapted in humans: An early spike protein mutation promotes transmission and will shape the next vaccines. Science 372. [CrossRef]
- Tosta, E. (2021). The adaptation of SARS-CoV-2 to humans. Memórias do Instituto Oswaldo Cruz 116. [CrossRef]
- Cosar, B., Karagulleoglu, Z.Y., Unal, S., Ince, A.T., Uncuoglu, D.B., Tuncer, G., Kilinc, B.R., Ozkan, Y.E., Ozkoc, H.C., Demir, I.N., et al. (2022). SARS-CoV-2 Mutations and their Viral Variants. Cytokine & growth factor reviews 63. [CrossRef]
- Tao, K., Tzou, P.L., Nouhin, J., Gupta, R.K., de Oliveira, T., Kosakovsky Pond, S.L., Fera, D., and Shafer, R.W. (2021). The biological and clinical significance of emerging SARS-CoV-2 variants. Nat Rev Genet 22. [CrossRef]
- Jung, C., Kmiec, D., Koepke, L., Zech, F., Jacob, T., Sparrer, K.M.J., and Kirchhoff, F. (2022). Omicron: what makes the latest SARS-CoV-2 variant of concern so concerning? J Virol. [CrossRef]
- Nchioua, R., Diofano, F., Noettger, S., von Maltitz, P., Stenger, S., Zech, F., Münch, J., Sparrer, K.M.J., Just, S., and Kirchhoff, F. (2022). Strong attenuation of SARS-CoV-2 Omicron BA.1 and increased replication of the BA.5 subvariant in human cardiomyocytes. Sig Transduct Target Ther 7. [CrossRef]
- van Doremalen, N., Singh, M., Saturday, T.A., Yinda, C.K., Perez-Perez, L., Bohler, W.F., Weishampel, Z.A., Lewis, M., Schulz, J.E., Williamson, B.N., et al. (2022). SARS-CoV-2 Omicron BA.1 and BA.2 are attenuated in rhesus macaques as compared to Delta. bioRxiv. [CrossRef]
- Shuai, H., Chan, J.F.-W., Hu, B., Chai, Y., Yoon, C., Liu, H., Liu, Y., Shi, J., Zhu, T., Hu, J.-C., et al. (2023). The viral fitness and intrinsic pathogenicity of dominant SARS-CoV-2 Omicron sublineages BA.1, BA.2, and BA.5. eBioMedicine 95. [CrossRef]
- Pastorio, C., Zech, F., Noettger, S., Jung, C., Jacob, T., Sanderson, T., Sparrer, K.M.J., and Kirchhoff, F. (2022). Determinants of Spike infectivity, processing, and neutralization in SARS-CoV-2 Omicron subvariants BA.1 and BA.2. Cell Host & Microbe 30. [CrossRef]
- Hachmann, N.P., Miller, J., Collier, A.Y., Ventura, J.D., Yu, J., Rowe, M., Bondzie, E.A., Powers, O., Surve, N., Hall, K., et al. (2022). Neutralization Escape by SARS-CoV-2 Omicron Subvariants BA.2.12.1, BA.4, and BA.5. New England Journal of Medicine 387. [CrossRef]
- Wang, Q., Guo, Y., Iketani, S., Nair, M.S., Li, Z., Mohri, H., Wang, M., Yu, J., Bowen, A.D., Chang, J.Y., et al. (2022). Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature 608. [CrossRef]
- Wang, Q., Guo, Y., Iketani, S., Nair, M.S., Li, Z., Mohri, H., Wang, M., Yu, J., Bowen, A.D., Chang, J.Y., et al. (2022). Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature 608. [CrossRef]
- Park, Y.-J., Pinto, D., Walls, A.C., Liu, Z., De Marco, A., Benigni, F., Zatta, F., Silacci-Fregni, C., Bassi, J., Sprouse, K.R., et al. (2022). Imprinted antibody responses against SARS-CoV-2 Omicron sublineages. Science 378. [CrossRef]
- Cao, Y., Jian, F., Wang, J., Yu, Y., Song, W., Yisimayi, A., Wang, J., An, R., Chen, X., Zhang, N., et al. (2023). Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature 614. [CrossRef]
- Ito, J., Suzuki, R., Uriu, K., Itakura, Y., Zahradnik, J., Kimura, K.T., Deguchi, S., Wang, L., Lytras, S., Tamura, T., et al. (2023). Convergent evolution of SARS-CoV-2 Omicron subvariants leading to the emergence of BQ.1.1 variant. Nature Communications 2023 14. [CrossRef]
- Ao, D., He, X., Hong, W., and Wei, X. (2023). The rapid rise of SARS-CoV-2 Omicron subvariants with immune evasion properties: XBB.1.5 and BQ.1.1 subvariants. MedComm 4. [CrossRef]
- Hoffmann, M., Arora, P., Nehlmeier, I., Kempf, A., Cossmann, A., Schulz, S.R., Morillas Ramos, G., Manthey, L.A., Jäck, H.-M., Behrens, G.M.N., et al. (2023). Profound neutralization evasion and augmented host cell entry are hallmarks of the fast-spreading SARS-CoV-2 lineage XBB.1.5. Cell Mol Immunol 20. [CrossRef]
- Wang, Q., Iketani, S., Li, Z., Liu, L., Guo, Y., Huang, Y., Bowen, A.D., Liu, M., Wang, M., Yu, J., et al. (2023). Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell 186. [CrossRef]
- Yue, C., Song, W., Wang, L., Jian, F., Chen, X., Gao, F., Shen, Z., Wang, Y., Wang, X., and Cao, Y. (2023). ACE2 binding and antibody evasion in enhanced transmissibility of XBB.1.5. The Lancet Infectious Diseases 23. [CrossRef]
- Yang, S., Yu, Y., Xu, Y., Jian, F., Song, W., Yisimayi, A., Wang, P., Wang, J., Liu, J., Yu, L., et al. (2023). Fast evolution of SARS-CoV-2 BA.2·86 to JN.1 under heavy immune pressure. The Lancet Infectious Diseases. [CrossRef]
- Belik, M., Liedes, O., Vara, S., Haveri, A., Pöysti, S., Kolehmainen, P., Maljanen, S., Huttunen, M., Reinholm, A., Lundberg, R., et al. (2023). Persistent T cell-mediated immune responses against Omicron variants after the third COVID-19 mRNA vaccine dose. Front Immunol 14. [CrossRef]
- Arieta, C.M., Xie, Y.J., Rothenberg, D.A., Diao, H., Harjanto, D., Meda, S., Marquart, K., Koenitzer, B., Sciuto, T.E., Lobo, A., et al. (2023). The T-cell-directed vaccine BNT162b4 encoding conserved non-spike antigens protects animals from severe SARS-CoV-2 infection. Cell 186. [CrossRef]
- Cohen, L.E., Spiro, D.J., and Viboud, C. (2022). Projecting the SARS-CoV-2 transition from pandemicity to endemicity: Epidemiological and immunological considerations. PLOS Pathogens 18. [CrossRef]
- Biancolella, M., Colona, V.L., Mehrian-Shai, R., Watt, J.L., Luzzatto, L., Novelli, G., and Reichardt, J.K.V. (2022). COVID-19 2022 update: transition of the pandemic to the endemic phase. Human Genomics 16. [CrossRef]
- Saunders, K.O., Lee, E., Parks, R., Martinez, D.R., Li, D., Chen, H., Edwards, R.J., Gobeil, S., Barr, M., Mansouri, K., et al. (2021). Neutralizing antibody vaccine for pandemic and pre-emergent coronaviruses. Nature 594. [CrossRef]
- Planas, D., Saunders, N., Maes, P., Guivel-Benhassine, F., Planchais, C., Buchrieser, J., Bolland, W.-H., Porrot, F., Staropoli, I., Lemoine, F., et al. (2021). Considerable escape of SARS-CoV-2 variant Omicron to antibody neutralization. Nature 602. [CrossRef]
- Imai, M., Ito, M., Kiso, M., Yamayoshi, S., Uraki, R., Fukushi, S., Watanabe, S., Suzuki, T., Maeda, K., Sakai-Tagawa, Y., Iwatsuki-Horimoto, K., Halfmann, P.J., Kawaoka, Y. (2022). Efficacy of Antiviral Agents against Omicron Subvariants BQ.1.1 and XBB. N Engl J Med. 388. [CrossRef]
- Wang, P., Nair, M.S., Liu, L., Iketani, S., Luo, Y., Guo, Y., Wang, M., Yu, J., Zhang, B., Kwong, P.D., et al. (2021). Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 593. [CrossRef]
- Cao, Y., Wang, J., Jian, F., Xiao, T., Song, W., Yisimayi, A., Huang, W., Li, Q., Wang, P., An, R., et al. (2022). Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 602. [CrossRef]
- Liu, L., Iketani, S., Guo, Y., Chan, J.F.-W., Wang, M., Liu, L., Luo, Y., Chu, H., Huang, Y., Nair, M.S., et al. (2022). Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature 602. [CrossRef]
- Zhou, P., Song, G., Liu, H., Yuan, M., He, W., Beutler, N., Zhu, X., Tse, L.V., Martinez, D.R., Schäfer, A., et al. (2023). Broadly neutralizing anti-S2 antibodies protect against all three human betacoronaviruses that cause deadly disease. Immunity 56. [CrossRef]
- Chen, Y., Zhao, X., Zhou, H., Zhu, H., Jiang, S., and Wang, P. (2023). Broadly neutralizing antibodies to SARS-CoV-2 and other human coronaviruses. Nat Rev Immunol 23. [CrossRef]
- Karoyan, P., Vieillard, V., Gómez-Morales, L., Odile, E., Guihot, A., Luyt, C.-E., Denis, A., Grondin, P., and Lequin, O. (2021). Human ACE2 peptide-mimics block SARS-CoV-2 pulmonary cells infection. Commun Biol 4. [CrossRef]
- Larue, R.C., Xing, E., Kenney, A.D., Zhang, Y., Tuazon, J.A., Li, J., Yount, J.S., Li, P.-K., and Sharma, A. (2021). Rationally Designed ACE2-Derived Peptides Inhibit SARS-CoV-2. Bioconjug Chem 32. [CrossRef]
- Adhikary, P., Kandel, S., Mamani, U.-F., Mustafa, B., Hao, S., Qiu, J., Fetse, J., Liu, Y., Ibrahim, N.M., Li, Y., et al. (2021). Discovery of Small Anti-ACE2 Peptides to Inhibit SARS-CoV-2 Infectivity. Adv Ther (Weinh) 4. [CrossRef]
- Xia, S., Lan, Q., Zhu, Y., Wang, C., Xu, W., Li, Y., Wang, L., Jiao, F., Zhou, J., Hua, C., et al. (2021). Structural and functional basis for pan-CoV fusion inhibitors against SARS-CoV-2 and its variants with preclinical evaluation. Sig Transduct Target Ther 6. [CrossRef]
- Yu, D., Zhu, Y., Yan, H., Wu, T., Chong, H., and He, Y. (2021). Pan-coronavirus fusion inhibitors possess potent inhibitory activity against HIV-1, HIV-2, and simian immunodeficiency virus. Emerg Microbes Infect 10. [CrossRef]
- Xia, S., Wang, L., Jiao, F., Yu, X., Xu, W., Huang, Z., Li, X., Wang, Q., Zhu, Y., Man, Q., et al. (2023). SARS-CoV-2 Omicron subvariants exhibit distinct fusogenicity, but similar sensitivity, to pan-CoV fusion inhibitors. Emerg Microbes Infect 12. [CrossRef]
- Cheng, Y.-W., Chao, T.-L., Li, C.-L., Chiu, M.-F., Kao, H.-C., Wang, S.-H., Pang, Y.-H., Lin, C.-H., Tsai, Y.-M., Lee, W.-H., et al. (2020). Furin Inhibitors Block SARS-CoV-2 Spike Protein Cleavage to Suppress Virus Production and Cytopathic Effects. Cell Rep 33. [CrossRef]
- Müller, P., Maus, H., Hammerschmidt, S.J., Knaff, P., Mailänder, V., Schirmeister, T., and Kersten, C. (2021). Interfering with Host Proteases in SARS-CoV-2 Entry as a Promising Therapeutic Strategy. Curr Med Chem. [CrossRef]
- Wettstein, L., Immenschuh, P., Weil, T., Conzelmann, C., Almeida-Hernández, Y., Hoffmann, M., Kempf, A., Nehlmeier, I., Lotke, R., Petersen, M., et al. (2023). Native and activated antithrombin inhibits TMPRSS2 activity and SARS-CoV-2 infection. J Med Virol 95. [CrossRef]
- Polack, F.P., Thomas, S.J., Kitchin, N., Absalon, J., Gurtman, A., Lockhart, S., Perez, J.L., Pérez Marc, G., Moreira, E.D., Zerbini, C., et al. (2020). Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N Engl J Med 383. [CrossRef]
- Folegatti, P.M., Ewer, K.J., Aley, P.K., Angus, B., Becker, S., Belij-Rammerstorfer, S., Bellamy, D., Bibi, S., Bittaye, M., Clutterbuck, E.A., et al. (2020). Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet 396. [CrossRef]
- Xia, S., Zhang, Y., Wang, Y., Wang, H., Yang, Y., Gao, G.F., Tan, W., Wu, G., Xu, M., Lou, Z., et al. (2021). Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBIBP-CorV: a randomised, double-blind, placebo-controlled, phase 1/2 trial. Lancet Infect Dis 21. [CrossRef]
- Voysey, M., Costa Clemens, S.A., Madhi, S.A., Weckx, L.Y., Folegatti, P.M., Aley, P.K., Angus, B., Baillie, V.L., Barnabas, S.L., Bhorat, Q.E., et al. (2021). Single-dose administration and the influence of the timing of the booster dose on immunogenicity and efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine: a pooled analysis of four randomised trials. Lancet 397. [CrossRef]
- Lopez Bernal, J., Andrews, N., Gower, C., Gallagher, E., Simmons, R., Thelwall, S., Stowe, J., Tessier, E., Groves, N., Dabrera, G., et al. (2021). Effectiveness of Covid-19 Vaccines against the B.1.617.2 (Delta) Variant. N Engl J Med 385. [CrossRef]
- Tseng, H.F., Ackerson, B.K., Sy, L.S., Tubert, J.E., Luo, Y., Qiu, S., Lee, G.S., Bruxvoort, K.J., Ku, J.H., Florea, A., et al. (2023). mRNA-1273 bivalent (original and Omicron) COVID-19 vaccine effectiveness against COVID-19 outcomes in the United States. Nat Commun 14. [CrossRef]
- Martinez, D.R., fer, A.S., Gavitt, T.D., Mallory, M.L., Lee, E., Catanzaro, N.J., Chen, H., Gully, K., Scobey, T., Brown, A., et al. (2023) Vaccine-mediated protection against merbecovirus and sarbecovirus challenge in mice. bioRxiv. [CrossRef]
- Martinez, D.R., Schäfer, A., Gavitt, T.D., Mallory, M.L., Lee, E., Catanzaro, N.J., Chen, H., Gully, K., Scobey, T., Korategere, P., et al. (2023). Vaccine-mediated protection against Merbecovirus and Sarbecovirus challenge in mice. Cell reports 42. [CrossRef]
- Evans, T.S., Tan, C.W., Aung, O., Phyu, S., Lin, H., Coffey, L.L., Toe, A.T., Aung, P., Aung, T.H., Aung, N.T., et al. (2023). Exposure to diverse sarbecoviruses indicates frequent zoonotic spillover in human communities interacting with wildlife. International Journal of Infectious Diseases 131. [CrossRef]
- Cantoni, D., Mayora-Neto, M., Thakur, N., Elrefaey, A.M.E., Newman, J., Vishwanath, S., Nadesalingam, A., Chan, A., Smith, P., Castillo-Olivares, J., et al. (2022). Pseudotyped Bat Coronavirus RaTG13 is efficiently neutralised by convalescent sera from SARS-CoV-2 infected patients. Commun Biol 5. [CrossRef]
- Lawrenz, J., Xie, Q., Zech, F., Weil, T., Seidel, A., Krnavek, D., van der Hoek, L., Münch, J., Müller, J.A., and Kirchhoff, F. (2022). Severe Acute Respiratory Syndrome Coronavirus 2 Vaccination Boosts Neutralizing Activity Against Seasonal Human Coronaviruses. Clinical Infectious Diseases 75. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




