Submitted:
29 December 2023
Posted:
03 January 2024
Read the latest preprint version here
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Treatment
2.2. Immunofluorescence
2.3. Fluorescence in Situ Hybridization (FISH)
3. Results
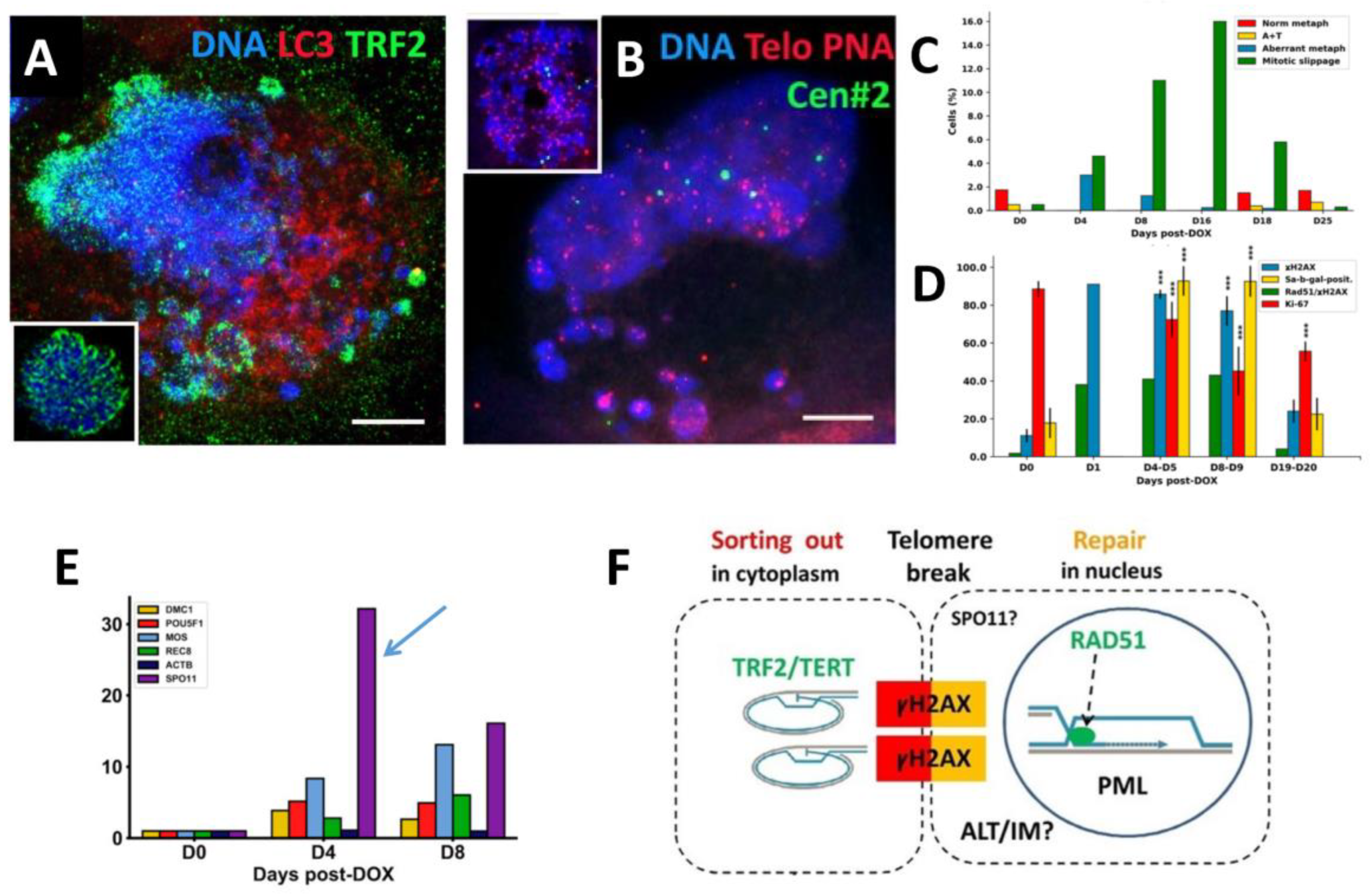
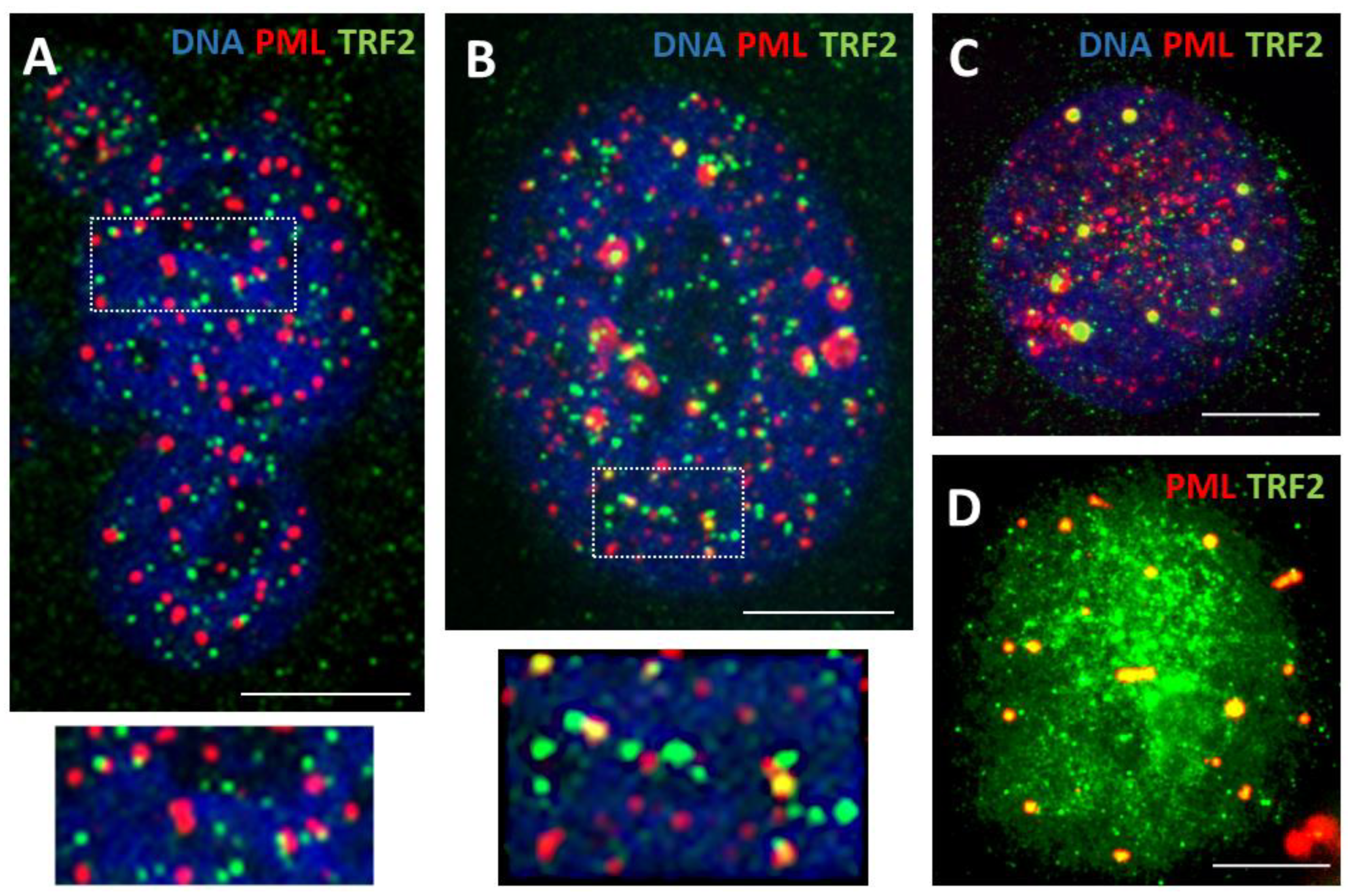
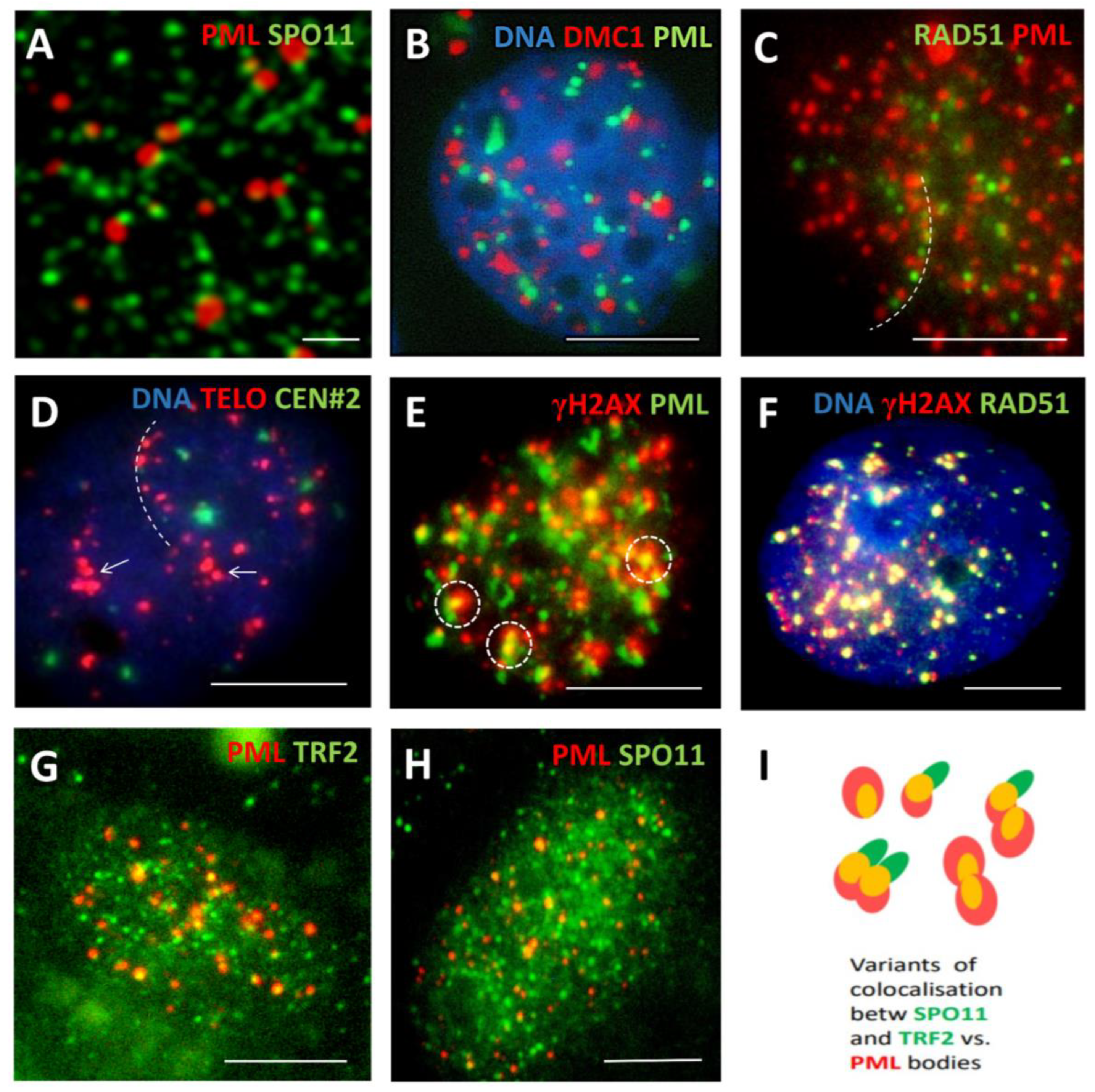
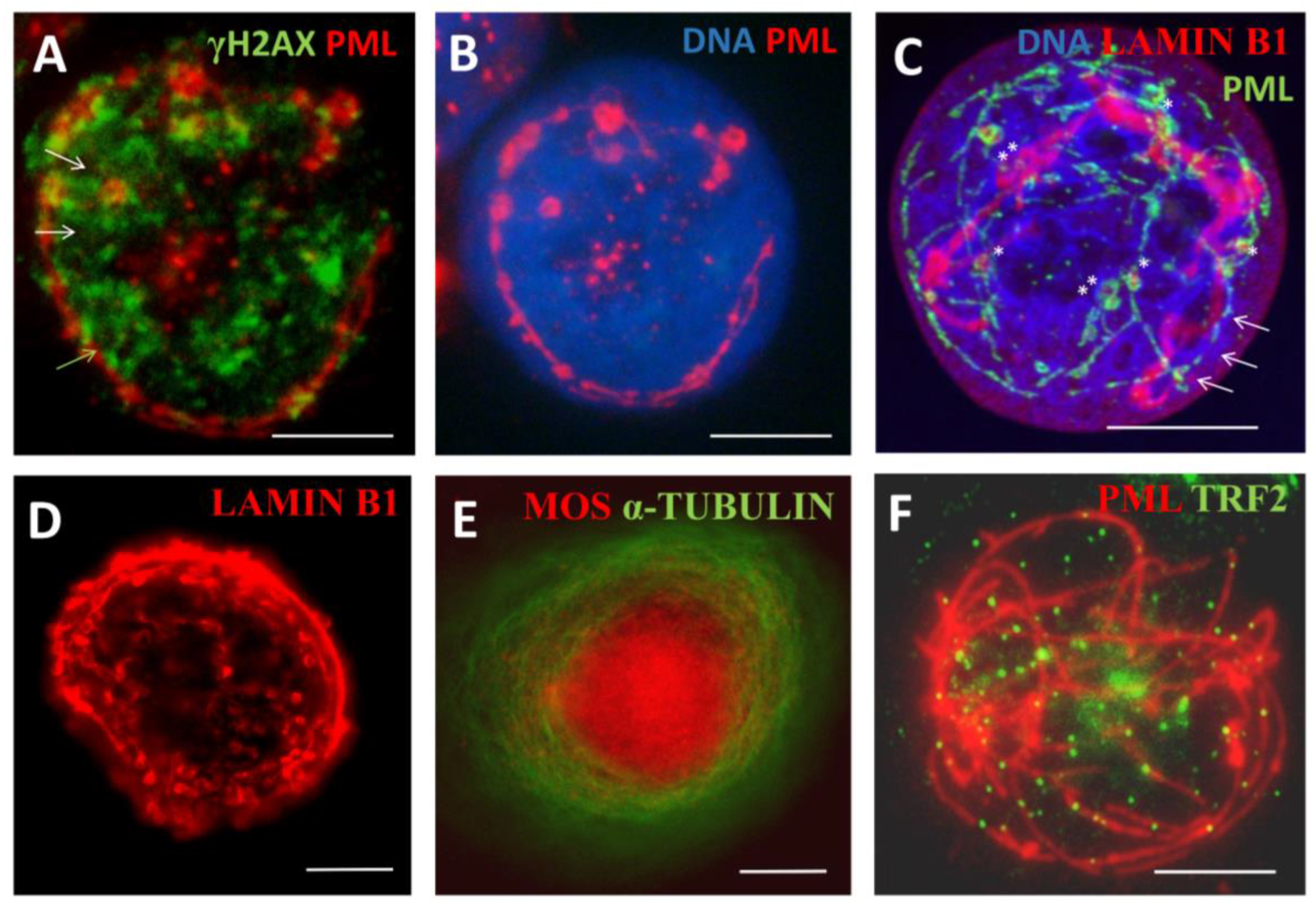
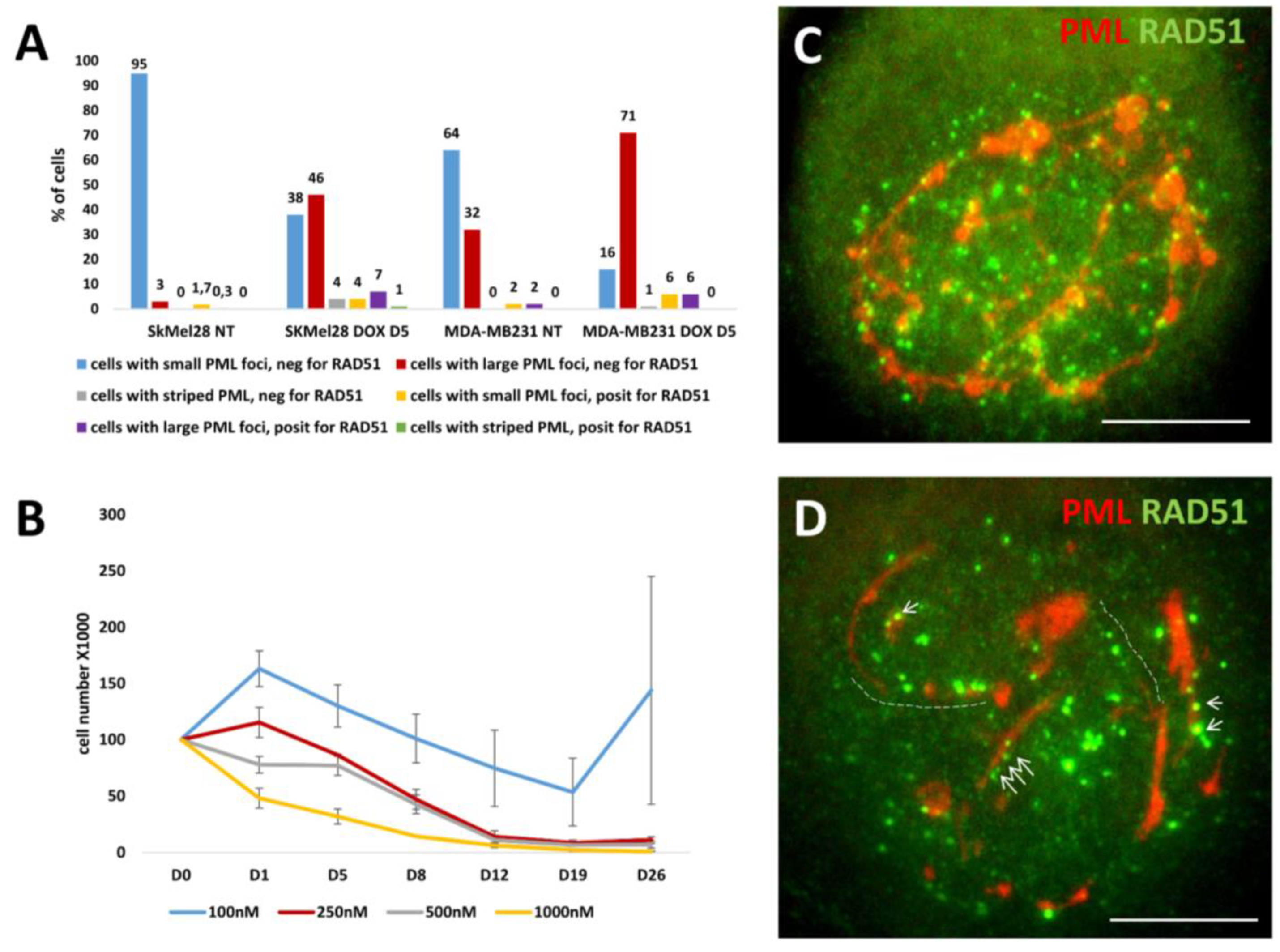
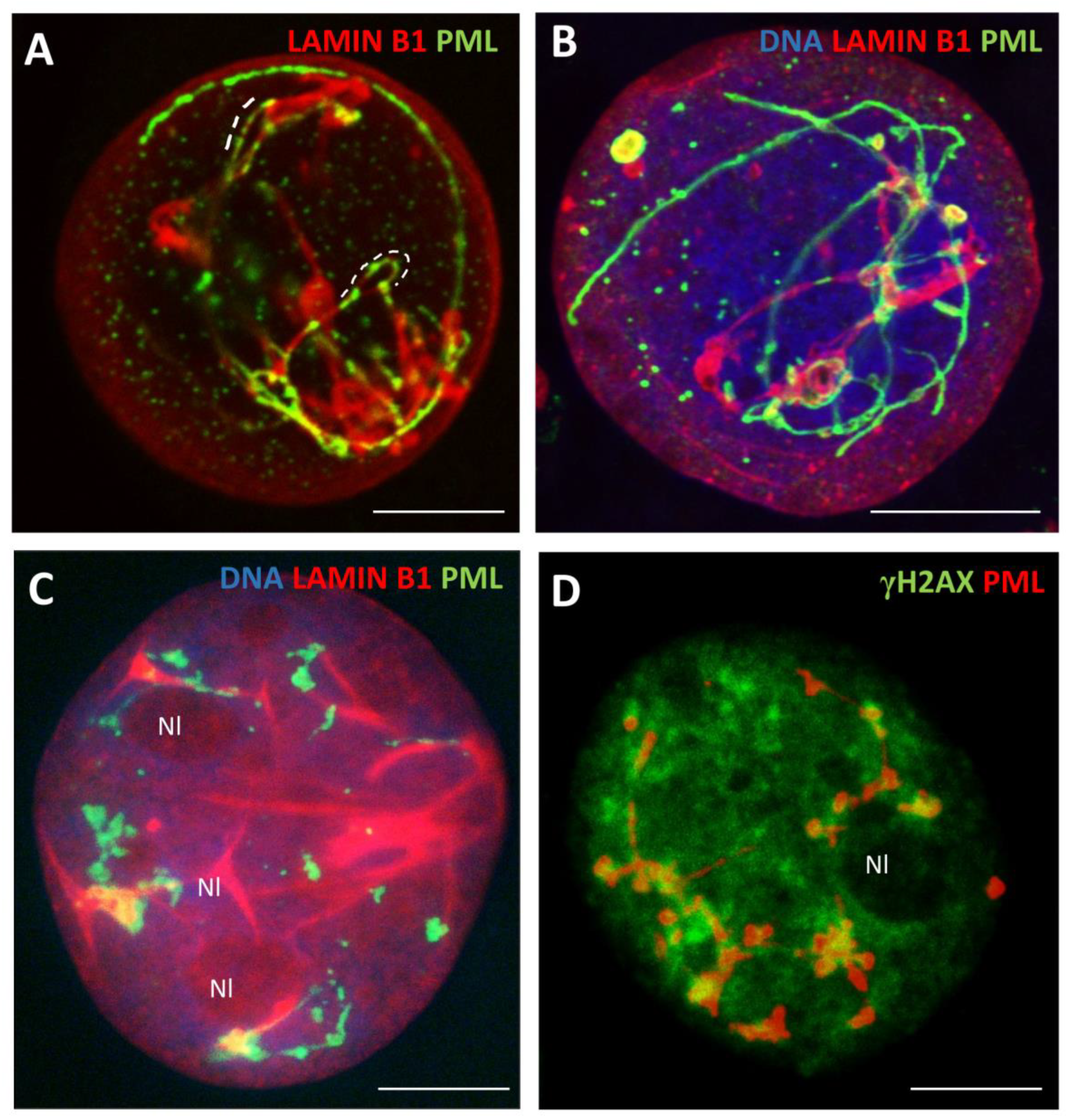
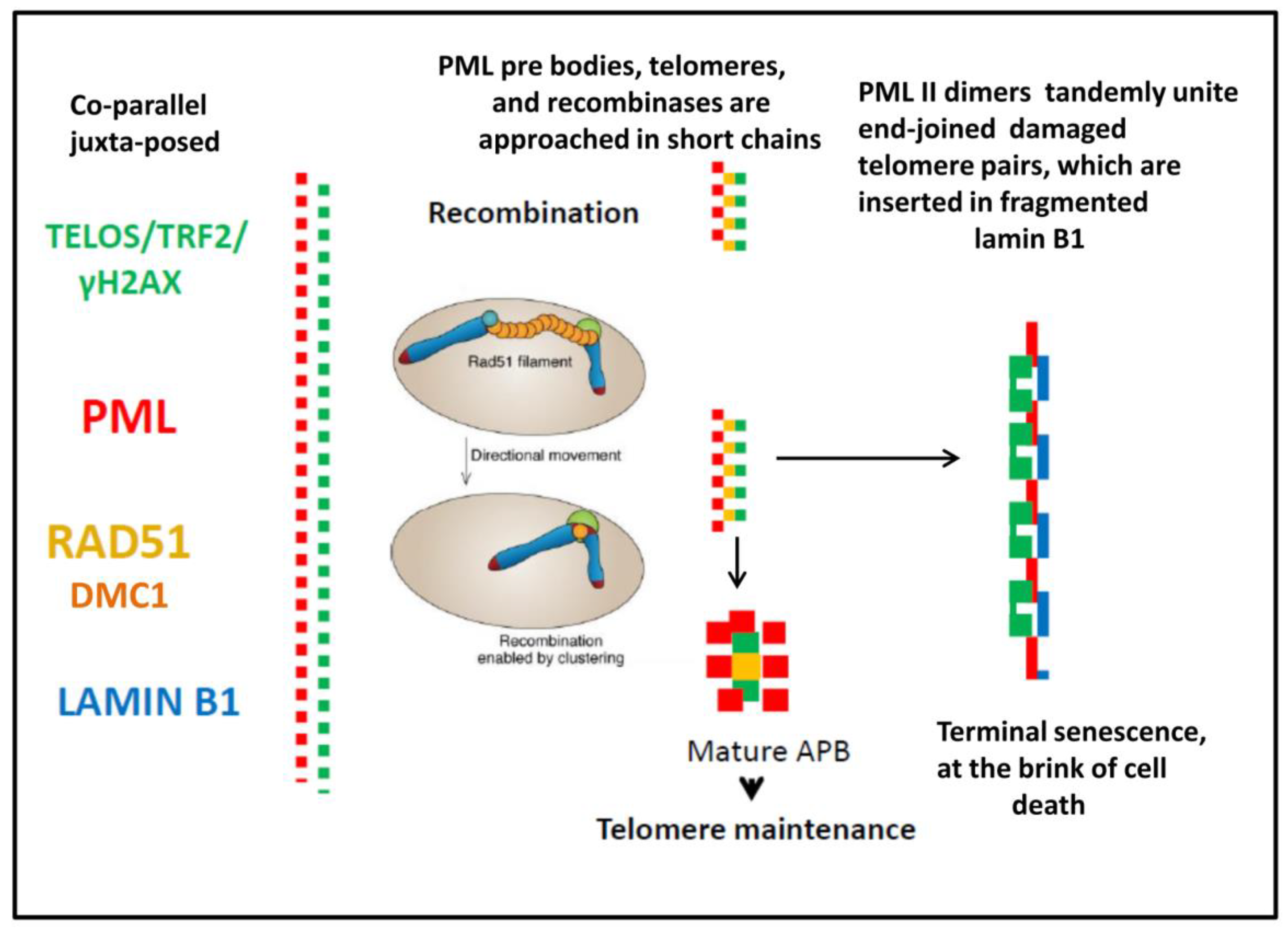
3.1. The topological relationship of the components of ALT and lamin B1 in the time course post-DOX action. DNA repair and misrepair
3.1.2. Phase III (maturation of recombinogenic APBs)
3.1.3. Phase IV: the formation of PML II isoform fibrillar structures
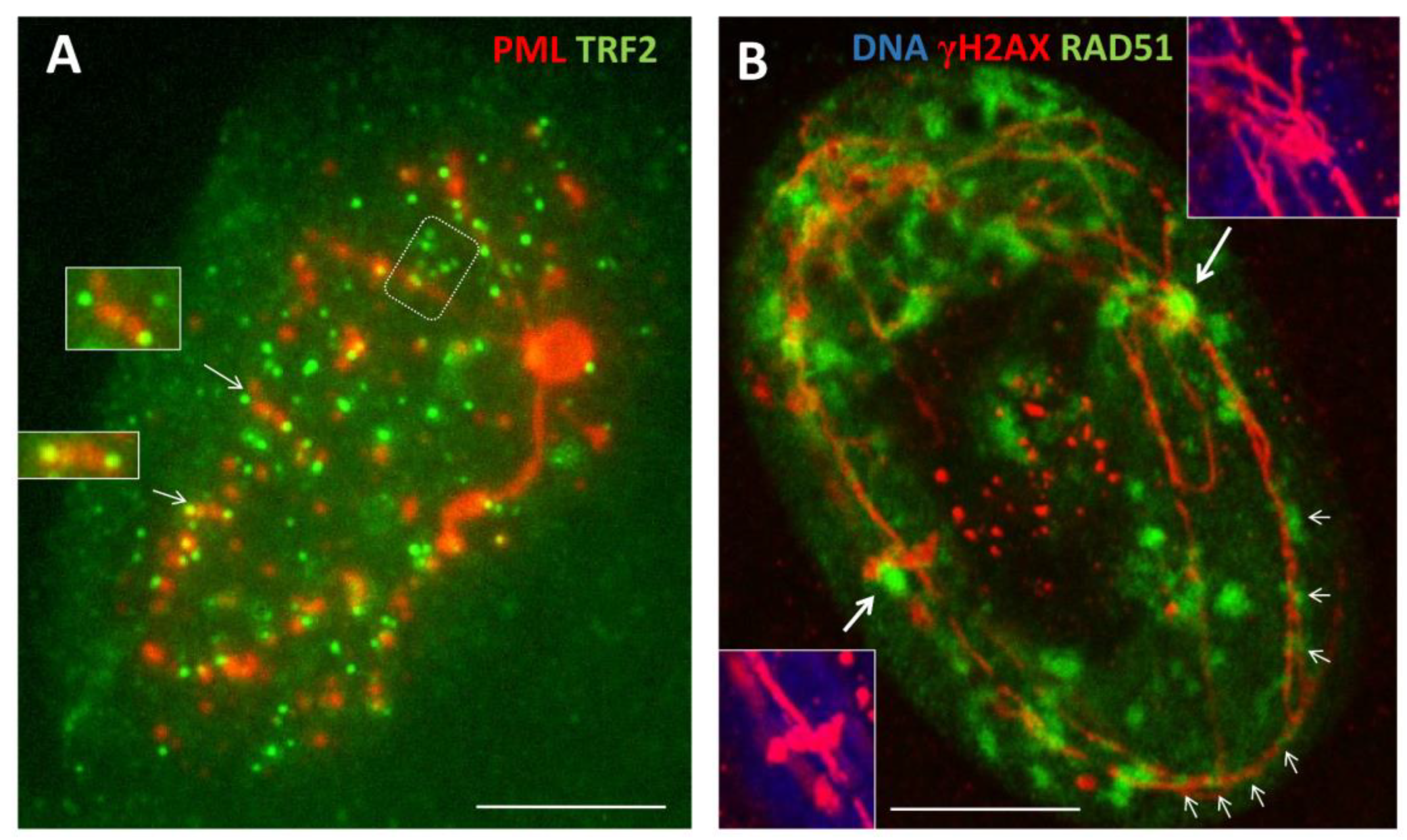
3.1.4. Thready PML and DNA recombination repair in DOX-treated cells
3.1.5. Phase V, at the brink of catastrophe
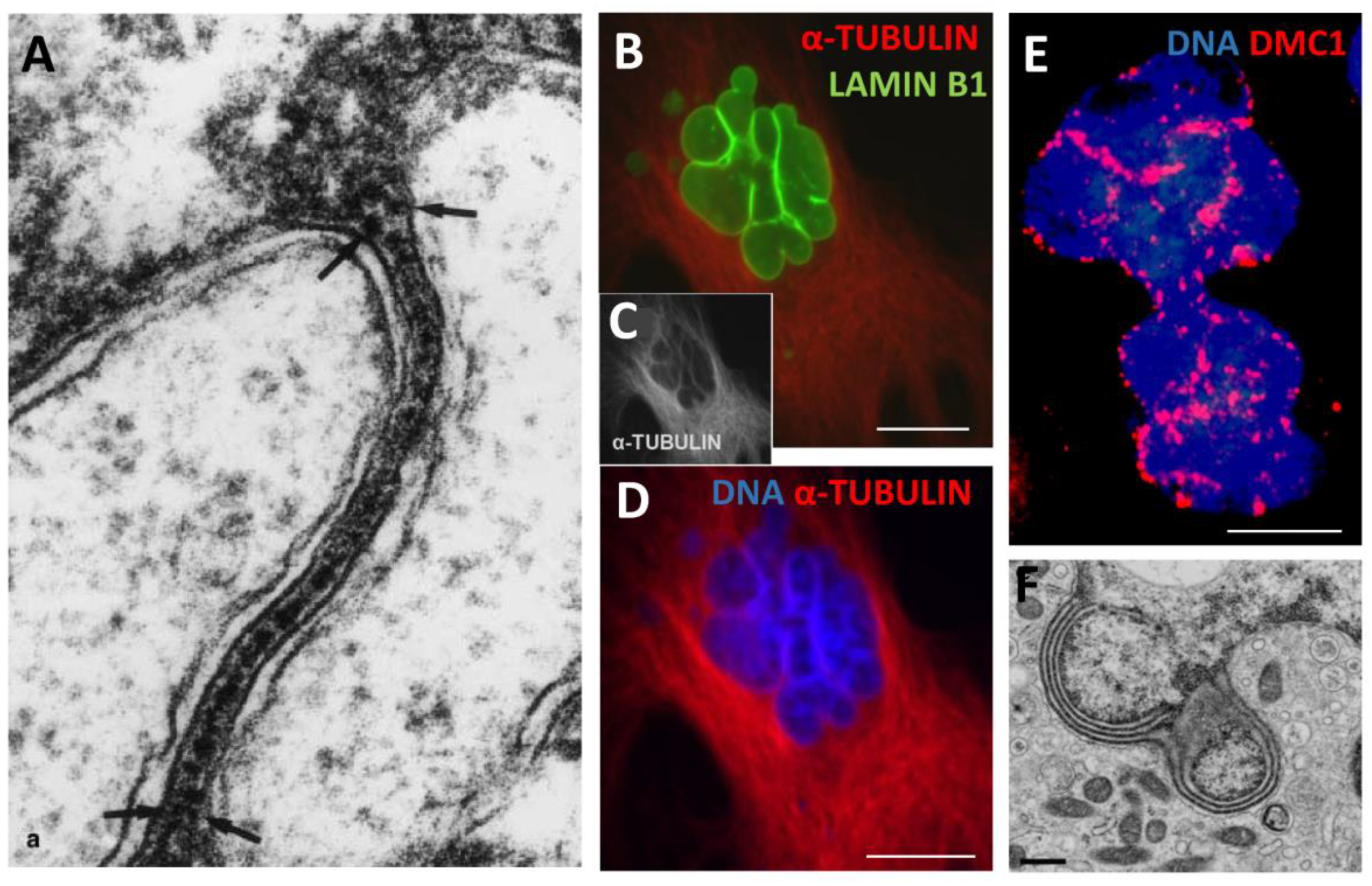
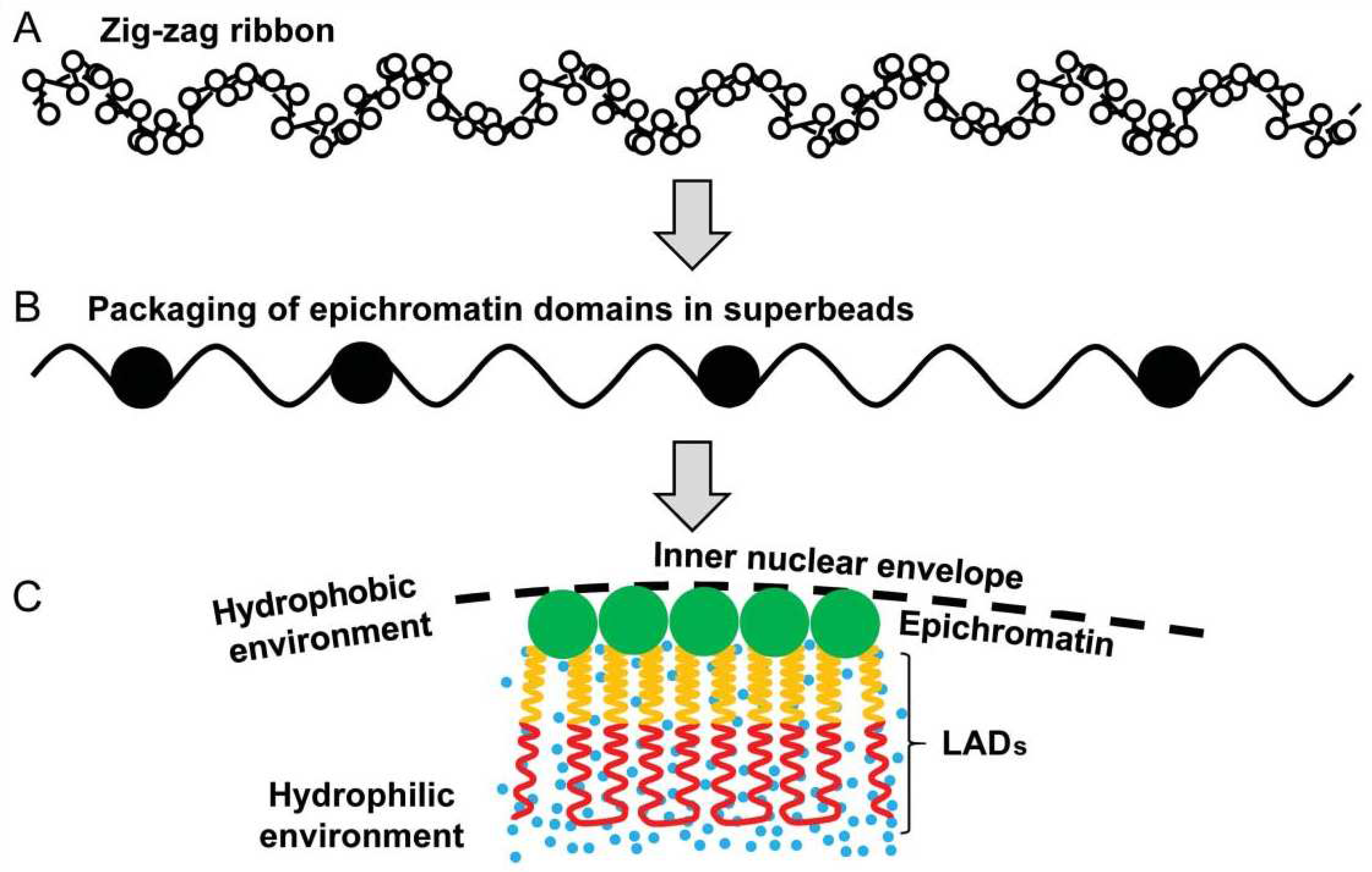
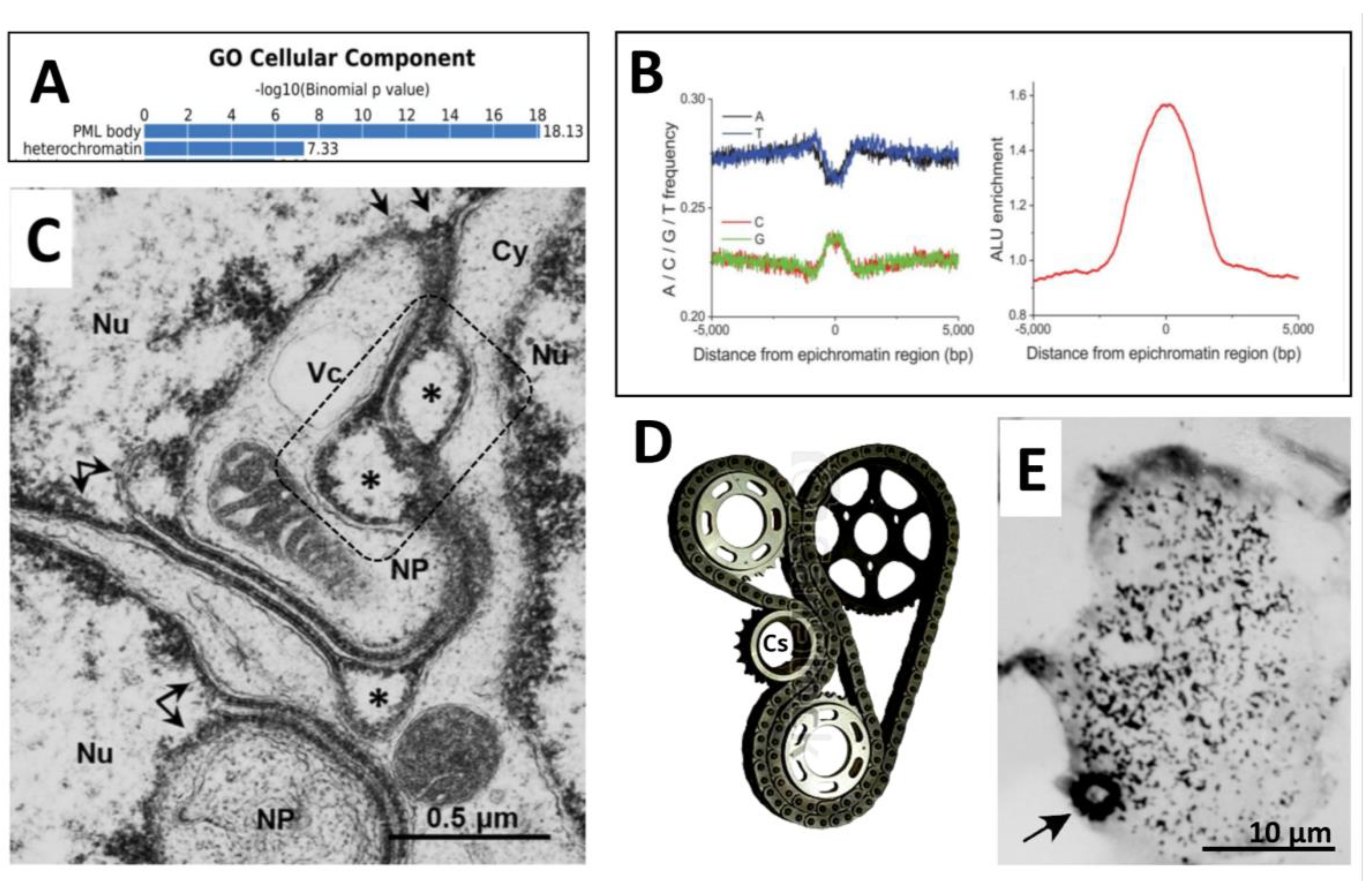
3.2.1. The epichromatin and ELCS rotating around cell nuclei
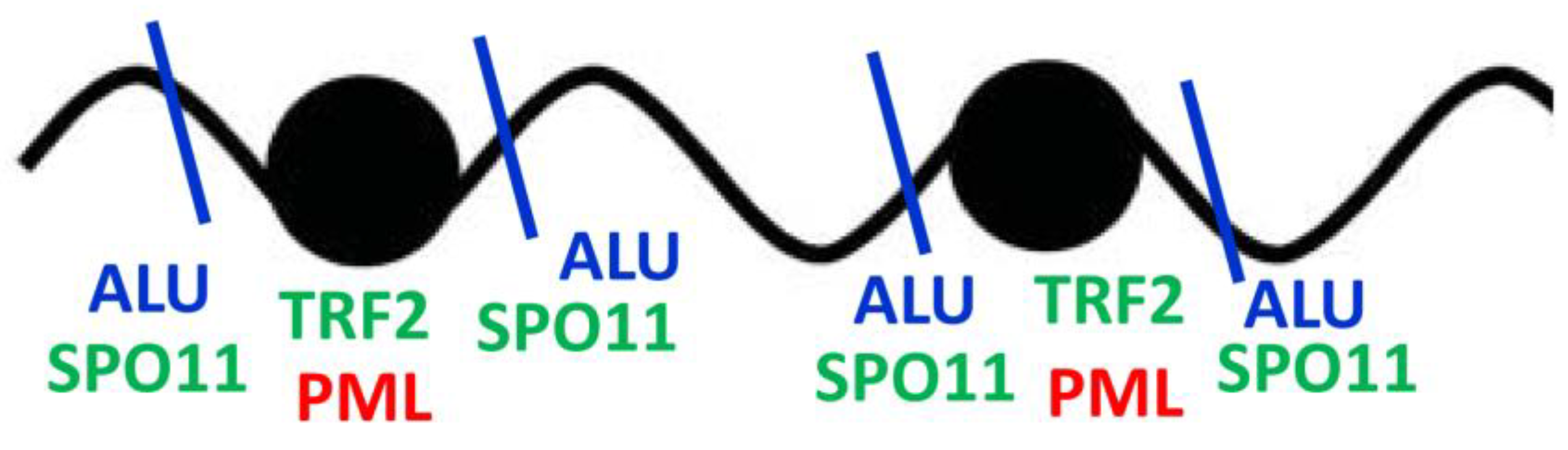
3.2.2. Chip-Seq studies: Epichromatin meets PML and ALU
3.2.3. ELCS are directly composed of telomere repeats
3.2.4. The traffic of ELCS structures from the analysis of EM pictures
4. Discussion
4.1. The telomere repair and misrepair in accelerated cellular senescence (ACS)
4.2. Telomeres, ALU flipons, and non-linear information transfer
4.3. ELCS nuclear traffic and chromothripsis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Roninson, I.B.; Broude, E.V.; Chang, B.D. If Not Apoptosis, Then What? Treatment-Induced Senescence and Mitotic Catastrophe in Tumor Cells. Drug Resist. Updat. 2001, 4, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Victorelli, S.; Passos, J.F. Telomeres and Cell Senescence - Size Matters Not. EBioMedicine 2017, 21, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Shay, J.W. Telomeres and Aging. Curr. Opin. Cell Biol. 2018, 52, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Freund, A.; Laberge, R.-M.; Demaria, M.; Campisi, J. Lamin B1 Loss Is a Senescence-Associated Biomarker. Mol. Biol. Cell 2012, 23, 2066–2075. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, A.; Pawlikowski, J.; Manoharan, I.; van Tuyn, J.; Nelson, D.M.; Rai, T.S.; Shah, P.P.; Hewitt, G.; Korolchuk, V.I.; Passos, J.F.; et al. Lysosome-Mediated Processing of Chromatin in Senescence. J. Cell Biol. 2013, 202, 129–143. [Google Scholar] [CrossRef] [PubMed]
- Campisi, J. Aging, Cellular Senescence, and Cancer. Annu. Rev. Physiol. 2013, 75, 685–705. [Google Scholar] [CrossRef] [PubMed]
- De Cecco, M.; Criscione, S.W.; Peckham, E.J.; Hillenmeyer, S.; Hamm, E.A.; Manivannan, J.; Peterson, A.L.; Kreiling, J.A.; Neretti, N.; Sedivy, J.M. Genomes of Replicatively Senescent Cells Undergo Global Epigenetic Changes Leading to Gene Silencing and Activation of Transposable Elements. Aging Cell 2013, 12, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Puig, P.-E.; Guilly, M.-N.; Bouchot, A.; Droin, N.; Cathelin, D.; Bouyer, F.; Favier, L.; Ghiringhelli, F.; Kroemer, G.; Solary, E.; et al. Tumor Cells Can Escape DNA-Damaging Cisplatin through DNA Endoreduplication and Reversible Polyploidy. Cell Biol. Int. 2008, 32, 1031–1043. [Google Scholar] [CrossRef] [PubMed]
- Lezaja, A.; Altmeyer, M. Dealing with DNA Lesions: When One Cell Cycle Is Not Enough. Curr. Opin. Cell Biol. 2021, 70, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Salmina, K.; Bojko, A.; Inashkina, I.; Staniak, K.; Dudkowska, M.; Podlesniy, P.; Rumnieks, F.; Vainshelbaum, N.M.; Pjanova, D.; Sikora, E.; et al. “Mitotic Slippage” and Extranuclear DNA in Cancer Chemoresistance: A Focus on Telomeres. Int. J. Mol. Sci. 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- Mosteiro, L.; Pantoja, C.; Alcazar, N.; Marión, R.M.; Chondronasiou, D.; Rovira, M.; Fernandez-Marcos, P.J.; Muñoz-Martin, M.; Blanco-Aparicio, C.; Pastor, J.; et al. Tissue Damage and Senescence Provide Critical Signals for Cellular Reprogramming in Vivo. Science 2016, 354. [Google Scholar] [CrossRef] [PubMed]
- Milanovic, M.; Fan, D.N.Y.; Belenki, D.; Däbritz, J.H.M.; Zhao, Z.; Yu, Y.; Dörr, J.R.; Dimitrova, L.; Lenze, D.; Monteiro Barbosa, I.A.; et al. Senescence-Associated Reprogramming Promotes Cancer Stemness. Nature 2018, 553, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Jackson, T.R.; Salmina, K.; Huna, A.; Inashkina, I.; Jankevics, E.; Riekstina, U.; Kalnina, Z.; Ivanov, A.; Townsend, P.A.; Cragg, M.S.; et al. DNA Damage Causes TP53-Dependent Coupling of Self-Renewal and Senescence Pathways in Embryonal Carcinoma Cells. Cell Cycle 2013, 12, 430–441. [Google Scholar] [CrossRef] [PubMed]
- Huna, A.; Salmina, K.; Erenpreisa, J.; Vazquez-Martin, A.; Krigerts, J.; Inashkina, I.; Gerashchenko, B.I.; Townsend, P.A.; Cragg, M.S.; Jackson, T.R. Role of Stress-Activated OCT4A in the Cell Fate Decisions of Embryonal Carcinoma Cells Treated with Etoposide. Cell Cycle 2015, 14, 2969–2984. [Google Scholar] [CrossRef] [PubMed]
- Erenpreisa, J.; Salmina, K.; Anatskaya, O.; Cragg, M.S. Paradoxes of Cancer: Survival at the Brink. Semin. Cancer Biol. 2022, 81, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Jafri, M.A.; Ansari, S.A.; Alqahtani, M.H.; Shay, J.W. Roles of Telomeres and Telomerase in Cancer, and Advances in Telomerase-Targeted Therapies. Genome Med. 2016, 8, 69. [Google Scholar] [CrossRef] [PubMed]
- Cho, N.W.; Dilley, R.L.; Lampson, M.A.; Greenberg, R.A. Interchromosomal Homology Searches Drive Directional ALT Telomere Movement and Synapsis. Cell 2014, 159, 108–121. [Google Scholar] [CrossRef] [PubMed]
- Arnoult, N.; Karlseder, J. ALT Telomeres Borrow from Meiosis to Get Moving. Cell 2014, 159, 11–12. [Google Scholar] [CrossRef] [PubMed]
- Burla, R.; La Torre, M.; Saggio, I. Mammalian Telomeres and Their Partnership with Lamins. Nucleus 2016, 7, 187–202. [Google Scholar] [CrossRef] [PubMed]
- Calderón, M.d.C.; Rey, M.-D.; Cabrera, A.; Prieto, P. The Subtelomeric Region Is Important for Chromosome Recognition and Pairing during Meiosis. Sci. Rep. 2014, 4, 6488. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, M.; Prieto, P. Telomeres and Subtelomeres Dynamics in the Context of Early Chromosome Interactions During Meiosis and Their Implications in Plant Breeding. Front. Plant Sci. 2021, 12, 672489. [Google Scholar] [CrossRef] [PubMed]
- Smalley, K.S.M.; Lioni, M.; Dalla Palma, M.; Xiao, M.; Desai, B.; Egyhazi, S.; Hansson, J.; Wu, H.; King, A.J.; Van Belle, P.; et al. Increased Cyclin D1 Expression Can Mediate BRAF Inhibitor Resistance in BRAF V600E-Mutated Melanomas. Mol. Cancer Ther. 2008, 7, 2876–2883. [Google Scholar] [CrossRef]
- Jensen, K.; Shiels, C.; Freemont, P.S. PML Protein Isoforms and the RBCC/TRIM Motif. Oncogene 2001, 20, 7223–7233. [Google Scholar] [CrossRef] [PubMed]
- Nisole, S.; Maroui, M.A.; Mascle, X.H.; Aubry, M.; Chelbi-Alix, M.K. Differential Roles of PML Isoforms. Front. Oncol. 2013, 3, 125. [Google Scholar] [CrossRef] [PubMed]
- Condemine, W.; Takahashi, Y.; Zhu, J.; Puvion-Dutilleul, F.; Guegan, S.; Janin, A.; de Thé, H. Characterization of Endogenous Human Promyelocytic Leukemia Isoforms. Cancer Res. 2006, 66, 6192–6198. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wang, L.; Qian, M.; Tang, X.; Liu, Z.; Lai, Y.; Ao, Y.; Huang, Y.; Meng, Y.; Shi, L.; et al. PML2-Mediated Thread-like Nuclear Bodies Mark Late Senescence in Hutchinson-Gilford Progeria Syndrome. Aging Cell 2020, 19, e13147. [Google Scholar] [CrossRef] [PubMed]
- Scherthan, H.; Sotnik, N.; Peper, M.; Schrock, G.; Azizova, T.; Abend, M. Telomere Length in Aged Mayak PA Nuclear Workers Chronically Exposed to Internal Alpha and External Gamma Radiation. Radiat. Res. 2016, 185, 658–667. [Google Scholar] [CrossRef] [PubMed]
- Illidge, T.M.; Cragg, M.S.; Fringes, B.; Olive, P.; Erenpreisa, J.A. Polyploid Giant Cells Provide a Survival Mechanism for p53 Mutant Cells after DNA Damage. Cell Biol. Int. 2000, 24, 621–633. [Google Scholar] [CrossRef] [PubMed]
- Pjanova, D.; Vainshelbaum, N.M.; Salmina, K.; Erenpreisa, J. The Role of the Meiotic Component in Reproduction of B-RAF-Mutated Melanoma: A Review and “brainstorming” Session. In Melanoma; IntechOpen, 2021 ISBN 9781838808785.
- Erenpreisa, J.; Salmiņa, K.; Belyayev, A.; Inashkina, I.; Cragg, M.S. Survival at the Brink. In Autophagy: Cancer, Other Pathologies, Inflammation, Immunity, Infection, and Aging; Elsevier, 2017; pp. 275–294 ISBN 9780128121467.
- Verlhac, M.H.; Kubiak, J.Z.; Weber, M.; Géraud, G.; Colledge, W.H.; Evans, M.J.; Maro, B. Mos Is Required for MAP Kinase Activation and Is Involved in Microtubule Organization during Meiotic Maturation in the Mouse. Development 1996, 122, 815–822. [Google Scholar] [CrossRef]
- Salmina, K.; Huna, A.; Kalejs, M.; Pjanova, D.; Scherthan, H.; Cragg, M.S.; Erenpreisa, J. The Cancer Aneuploidy Paradox: In the Light of Evolution. Genes 2019, 10. [Google Scholar] [CrossRef]
- Condemine, W.; Takahashi, Y.; Le Bras, M.; de Thé, H. A Nucleolar Targeting Signal in PML-I Addresses PML to Nucleolar Caps in Stressed or Senescent Cells. J. Cell Sci. 2007, 120, 3219–3227. [Google Scholar] [CrossRef] [PubMed]
- Corpet, A.; Kleijwegt, C.; Roubille, S.; Juillard, F.; Jacquet, K.; Texier, P.; Lomonte, P. PML Nuclear Bodies and Chromatin Dynamics: Catch Me If You Can! Nucleic Acids Res. 2020, 48, 11890–11912. [Google Scholar] [CrossRef] [PubMed]
- Chapman, K.B.; Filipsky, F.; Peschke, N.; Gelléri, M.; Weinhardt, V.; Braun, A.; Hausmann, M.; Cremer, C. A Comprehensive Method to Study the DNA’s Association with Lamin and Chromatin Compaction in Intact Cell Nuclei at Super Resolution. Nanoscale 2023, 15, 742–756. [Google Scholar] [CrossRef] [PubMed]
- Olins, A.L.; Rhodes, G.; Welch, D.B.M.; Zwerger, M.; Olins, D.E. Lamin B Receptor: Multi-Tasking at the Nuclear Envelope. Nucleus 2010, 1, 53–70. [Google Scholar] [CrossRef] [PubMed]
- Olins, D.E.; Olins, A.L. Nuclear Envelope-Limited Chromatin Sheets (ELCS) and Heterochromatin Higher Order Structure. Chromosoma 2009, 118, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.Y.; Lee, R.T.; Vergnes, L.; Fong, L.G.; Stewart, C.L.; Reue, K.; Young, S.G.; Zhang, Q.; Shanahan, C.M.; Lammerding, J. Cell Nuclei Spin in the Absence of Lamin b1. J. Biol. Chem. 2007, 282, 20015–20026. [Google Scholar] [CrossRef] [PubMed]
- Fais, D.; Prusov, A.N.; Polyakov, V.Y. The Anchorosome, a Special Chromatin Granule for the Anchorage of the Interphase Chromosome to the Nuclear Envelope. Cell Biol. Int. Rep. 1989, 13, 747–758. [Google Scholar] [CrossRef] [PubMed]
- Olins, A.L.; Buendia, B.; Herrmann, H.; Lichter, P.; Olins, D.E. Retinoic Acid Induction of Nuclear Envelope-Limited Chromatin Sheets in HL-60. Exp. Cell Res. 1998, 245, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Davies, H.G. Electron-Microscope Observations on the Organization of Heterochromatin in Certain Cells. J. Cell Sci. 1968, 3, 129–150. [Google Scholar] [CrossRef] [PubMed]
- Ghadially, F.N. Nucleus. In Ultrastructural Pathology of the Cell and Matrix; Elsevier, 1988; pp. 1–180 ISBN 9780407015715.
- Olins, A.L.; Ishaque, N.; Chotewutmontri, S.; Langowski, J.; Olins, D.E. Retrotransposon Alu Is Enriched in the Epichromatin of HL-60 Cells. Nucleus 2014, 5, 237–246. [Google Scholar] [CrossRef]
- Xu, P.; Mahamid, J.; Dombrowski, M.; Baumeister, W.; Olins, A.L.; Olins, D.E. Interphase Epichromatin: Last Refuge for the 30-Nm Chromatin Fiber? Chromosoma 2021, 130, 91–102. [Google Scholar] [CrossRef]
- Erenpreisa, J.; Ivanov, A.; Cragg, M.; Selivanova, G.; Illidge, T. Nuclear Envelope-Limited Chromatin Sheets Are Part of Mitotic Death. Histochem. Cell Biol. 2002, 117, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Celli, G.B.; Denchi, E.L.; de Lange, T. Ku70 Stimulates Fusion of Dysfunctional Telomeres yet Protects Chromosome Ends from Homologous Recombination. Nat. Cell Biol. 2006, 8, 885–890. [Google Scholar] [CrossRef] [PubMed]
- Paddock, S.W.; Albrecht-Buehler, G. Rigidity of the Nucleus during Nuclear Rotation in 3T3 Cells. Exp. Cell Res. 1988, 175, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Franklin, R.E.; Gosling, R.G. Molecular Configuration in Sodium Thymonucleate. Nature 1953, 171, 740–741. [Google Scholar] [CrossRef] [PubMed]
- Erenpreisa, J.; Krigerts, J.; Salmina, K.; Selga, T.; Sorokins, H.; Freivalds, T. Differential Staining of Peripheral Nuclear Chromatin with Acridine Orange Implies an A-Form Epichromatin Conformation of the DNA. Nucleus 2018, 9, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Erenpreisa, J.; Salmina, K.; Krigerts, J.; Selga, T.; Freivalds, T. The Nuclear Envelope-Associated Epichromatin and Its Sheets Are Composed of DNA A-Form Packed as Nucleosomal Superbeads Which Construct Vehicles for the Gear-Wheel Nuclear Traffic. Biopolym. Cell 2019, 35, 167–167. [Google Scholar] [CrossRef]
- Erenpreisa, J.; Huna, A.; Salmina, K.; Jackson, T.R.; Cragg, M.S. Macroautophagy-Aided Elimination of Chromatin: Sorting of Waste, Sorting of Fate? Autophagy 2012, 8, 1877–1881. [Google Scholar] [CrossRef] [PubMed]
- Olins, A.L.; Langhans, M.; Monestier, M.; Schlotterer, A.; Robinson, D.G.; Viotti, C.; Zentgraf, H.; Zwerger, M.; Olins, D.E. An Epichromatin Epitope: Persistence in the Cell Cycle and Conservation in Evolution. Nucleus 2011, 2, 47–60. [Google Scholar] [CrossRef]
- Gould, T.J.; Tóth, K.; Mücke, N.; Langowski, J.; Hakusui, A.S.; Olins, A.L.; Olins, D.E. Defining the Epichromatin Epitope. Nucleus 2017, 8, 625–640. [Google Scholar] [CrossRef] [PubMed]
- Teif, V.B.; Mallm, J.-P.; Sharma, T.; Mark Welch, D.B.; Rippe, K.; Eils, R.; Langowski, J.; Olins, A.L.; Olins, D.E. Nucleosome Repositioning during Differentiation of a Human Myeloid Leukemia Cell Line. Nucleus 2017, 8, 188–204. [Google Scholar] [CrossRef] [PubMed]
- Cebrián-Silla, A.; Alfaro-Cervelló, C.; Herranz-Pérez, V.; Kaneko, N.; Park, D.H.; Sawamoto, K.; Alvarez-Buylla, A.; Lim, D.A.; García-Verdugo, J.M. Unique Organization of the Nuclear Envelope in the Post-Natal Quiescent Neural Stem Cells. Stem Cell Reports 2017, 9, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Vicari, M.R.; Bruschi, D.P.; Cabral-de-Mello, D.C.; Nogaroto, V. Telomere Organization and the Interstitial Telomeric Sites Involvement in Insects and Vertebrates Chromosome Evolution. Genet. Mol. Biol. 2022, 45, e20220071. [Google Scholar] [CrossRef] [PubMed]
- Baker, D.J.; Sedivy, J.M. Probing the Depths of Cellular Senescence. J. Cell Biol. 2013, 202, 11–13. [Google Scholar] [CrossRef] [PubMed]
- Boateng, K.A.; Bellani, M.A.; Gregoretti, I.V.; Pratto, F.; Camerini-Otero, R.D. Homologous Pairing Preceding SPO11-Mediated Double-Strand Breaks in Mice. Dev. Cell 2013, 24, 196–205. [Google Scholar] [CrossRef] [PubMed]
- Prieler, S.; Chen, D.; Huang, L.; Mayrhofer, E.; Zsótér, S.; Vesely, M.; Mbogning, J.; Klein, F. Spo11 Generates Gaps through Concerted Cuts at Sites of Topological Stress. Nature 2021, 594, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, L.; Wang, J.; Hao, Y.; Xiao, Q.; Teng, J.; Shen, S.; Zhang, Y.; Feng, Y.; Bao, S.; et al. Conversion between Duplicated Genes Generated by Polyploidization Contributes to the Divergence of Poplar and Willow. BMC Plant Biol. 2022, 22, 298. [Google Scholar] [CrossRef] [PubMed]
- Maciver, S.K.; Koutsogiannis, Z.; de Obeso Fernández Del Valle, A. “Meiotic Genes” Are Constitutively Expressed in an Asexual Amoeba and Are Not Necessarily Involved in Sexual Reproduction. Biol. Lett. 2019, 15, 20180871. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.A.; Eichler, E.E. Primate Segmental Duplications: Crucibles of Evolution, Diversity and Disease. Nat. Rev. Genet. 2006, 7, 552–564. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.; Yeou, S.; Lee, N.K. DNA Bending Force Facilitates Z-DNA Formation under Physiological Salt Conditions. J. Am. Chem. Soc. 2022, 144, 13137–13145. [Google Scholar] [CrossRef]
- Shubernetskaya, O.; Skvortsov, D.; Evfratov, S.; Rubtsova, M.; Belova, E.; Strelkova, O.; Cherepaninets, V.; Zhironkina, O.; Olovnikov, A.; Zvereva, M.; et al. Interstitial Telomeric Repeats-Associated DNA Breaks. Nucleus 2017, 8, 641–653. [Google Scholar] [CrossRef] [PubMed]
- Herbert, A. ALU Non-B-DNA Conformations, Flipons, Binary Codes and Evolution. R Soc Open Sci 2020, 7, 200222. [Google Scholar] [CrossRef] [PubMed]
- Bosco, N.; de Lange, T. A TRF1-Controlled Common Fragile Site Containing Interstitial Telomeric Sequences. Chromosoma 2012, 121, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Zaitceva, V.; Kopeina, G.S.; Zhivotovsky, B. Anastasis: Return Journey from Cell Death. Cancers 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Mardin, B.R.; Drainas, A.P.; Waszak, S.M.; Weischenfeldt, J.; Isokane, M.; Stütz, A.M.; Raeder, B.; Efthymiopoulos, T.; Buccitelli, C.; Segura-Wang, M.; et al. A Cell-Based Model System Links Chromothripsis with Hyperploidy. Mol. Syst. Biol. 2015, 11, 828. [Google Scholar] [CrossRef] [PubMed]
- Cortés-Ciriano, I.; Lee, J.J.-K.; Xi, R.; Jain, D.; Jung, Y.L.; Yang, L.; Gordenin, D.; Klimczak, L.J.; Zhang, C.-Z.; Pellman, D.S.; et al. Comprehensive Analysis of Chromothripsis in 2,658 Human Cancers Using Whole-Genome Sequencing. Nat. Genet. 2020, 52, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Bassaganyas, L.; Beà, S.; Escaramís, G.; Tornador, C.; Salaverria, I.; Zapata, L.; Drechsel, O.; Ferreira, P.G.; Rodriguez-Santiago, B.; Tubio, J.M.C.; et al. Sporadic and Reversible Chromothripsis in Chronic Lymphocytic Leukemia Revealed by Longitudinal Genomic Analysis. Leukemia 2013, 27, 2376–2379. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.J.; Liu, G.; Heng, H.H. Experimental Induction of Genome Chaos. Methods Mol. Biol. 2018, 1769, 337–352. [Google Scholar] [PubMed]
- Hu, Q.; Valle-Inclan, J.E.; Dahiya, R.; Guyer, A.; Mazzagatti, A.; Maurais, E.G.; Engel, J.L.; Cortés-Ciriano, I.; Ly, P. Non-Homologous End Joining Shapes the Genomic Rearrangement Landscape of Chromothripsis from Mitotic Errors. bioRxiv 2023. [Google Scholar] [CrossRef] [PubMed]
- Comaills, V.; Castellano-Pozo, M. Chromosomal Instability in Genome Evolution: From Cancer to Macroevolution. Biology 2023, 12. [Google Scholar] [CrossRef] [PubMed]
- Erenpreisa, J.; Vainshelbaum, N.M.; Lazovska, M.; Karklins, R.; Salmina, K.; Zayakin, P.; Rumnieks, F.; Inashkina, I.; Pjanova, D.; Erenpreiss, J. The Price of Human Evolution: Cancer-Testis Antigens, the Decline in Male Fertility and the Increase in Cancer. Int. J. Mol. Sci. 2023, 24. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, R.A. Coming Full Circle-from Endless Complexity to Simplicity and Back Again. Cell 2014, 157, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Erenpreisa, J.; Giuliani, A. Resolution of Complex Issues in Genome Regulation and Cancer Requires Non-Linear and Network-Based Thermodynamics. Int. J. Mol. Sci. 2019, 21. [Google Scholar] [CrossRef] [PubMed]
- McCLINTOCK, B. The Origin and Behavior of Mutable Loci in Maize. Proc. Natl. Acad. Sci. U. S. A. 1950, 36, 344–355. [Google Scholar] [CrossRef]
| Antibody against | Description | Specificity/Immunogen | Used concentration | Product no. and manufacturer |
|---|---|---|---|---|
| ⍺-Tubulin | Mouse monoclonal | Recognizes an epitope located at the C-terminal end of the ⍺-tubulin isoform in a variety of organisms | 1:1000 | T5168, Sigma-Aldrich, St. Louis, MO, USA |
| DMC1 | Rabbit polyclonal | Recombinant fragment corresponding to a region within amino acids 1 and 206 of DMC1 | 1:100 | PA5-21472, Thermo fisher scientific, Waltham, MA, USA |
| γ-H2AX | Rabbit polyclonal | Recognizes mammalian, yeast Drosophila melanogaster and Xenopus laevis γ-H2AX | 1:200 | 4411-PC-100, Trevigen, Gaithersburg, MD, USA |
| γ-H2AX | Mouse monoclonal | Synthetic peptide sequence surrounding phosphorylated Ser140 | 1:200 | MA1-2022, Pierce, Waltham, MA, USA |
| LAMIN B1 | Rabbit polyclonal | Peptide mapping at the C-terminus of Lamin B1 of human origin | 1:200 | ab1604, Abcam, Cambrige, UK |
| LC3 | Rabbit polyclonal | The details of the immunogen for this antibody are not available | 1:100 | ab63817, Abcam, Cambrige, UK |
| MOS (C237) | Rabbit polyclonal | Epitope mapping at the C-terminus | 1:50 | sc-86, Santa Cruz, Dallas, TX, USA |
| PML | Rabbit polyclonal | A synthetic peptide corresponding to the N-terminus of the Human PML/RNF71/TRIM19 | 1:200 | PA5-80910, Thermo fisher scientific, Waltham, MA, USA |
| PML | Mouse monoclonal | Epitope corresponding to amino acids 37-51 mapping near the N-terminal of PML of human origin | 1:200 | sc-966, Santa Cruz, Dallas, TX, USA |
| RAD51 | Mouse monoclonal | Recombinant full length protein corresponding to Human Rad51 aa 1-338 | 1:50 | ab213, Abcam, Cambrige, UK |
| SPO11 | Mouse monoclonal | raised against amino acids 97-396 mapping at the C-terminus of Spo11 of human origin | 1:50 | sc-377161, Santa Cruz, Dallas, TX, USA |
| TRF2 | Mouse monoclonal | His-tagged, fusion-protein, corresponding to full-length TRF2 (Telomeric Repeat binding Factor 2) | 1:100 | 05-521, Millipore, Temecula, CA, USA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





