Submitted:
29 December 2023
Posted:
30 December 2023
You are already at the latest version
Abstract
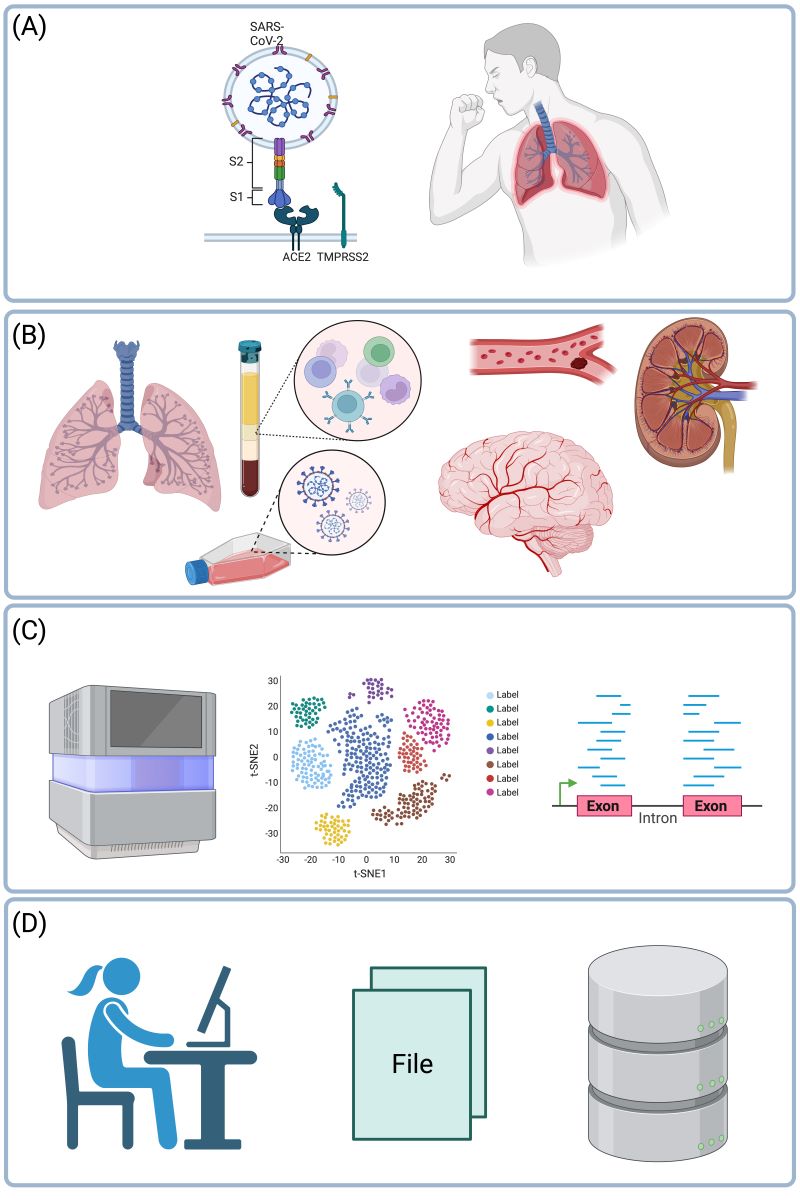
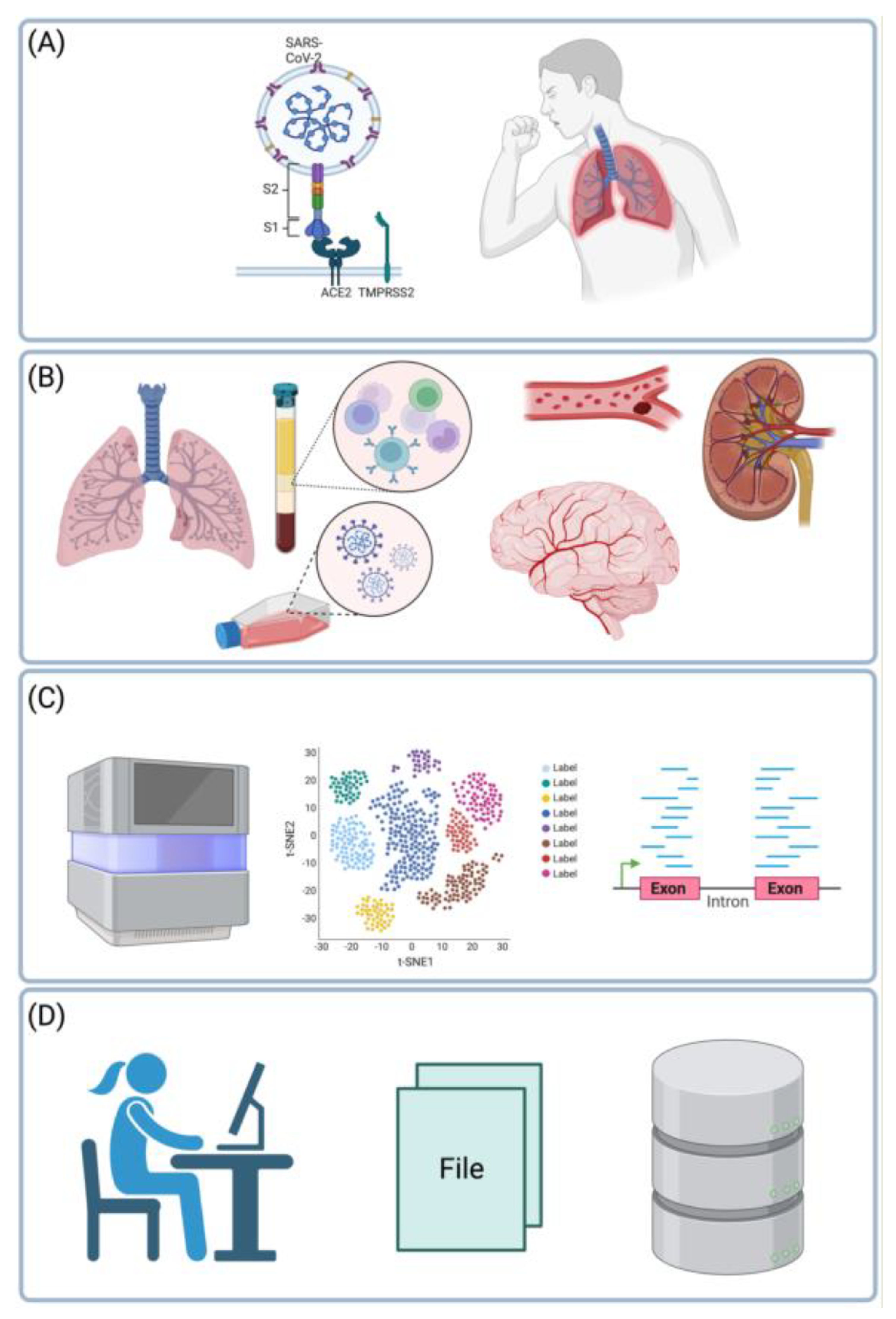
Keywords:
1. Introduction
2. Unbiased evidence from functional genomic studies of tissues and cells focusing on the etiology of COVID-19.
2.1. An in vitro study in a cell line infected with SARS-CoV-2.
2.2. Examples of in vivo transcriptional responses in bulk tissue samples exposed to SARS-CoV-2.
2.3. Examples of RNA-seq studies of individual cells in COVID-19.
2.3.1. Expression profiling of single cells in the immune system.
2.3.2. Expression profiling of the lung tissue.
2.3.3. Expression profiling of the brain, ocular epithelia, and the vasculature.
3. Meta-analyses of datasets related to the etiology of COVID-19.
4. The etiology of severe COVID-19 in the light of gene expression data.
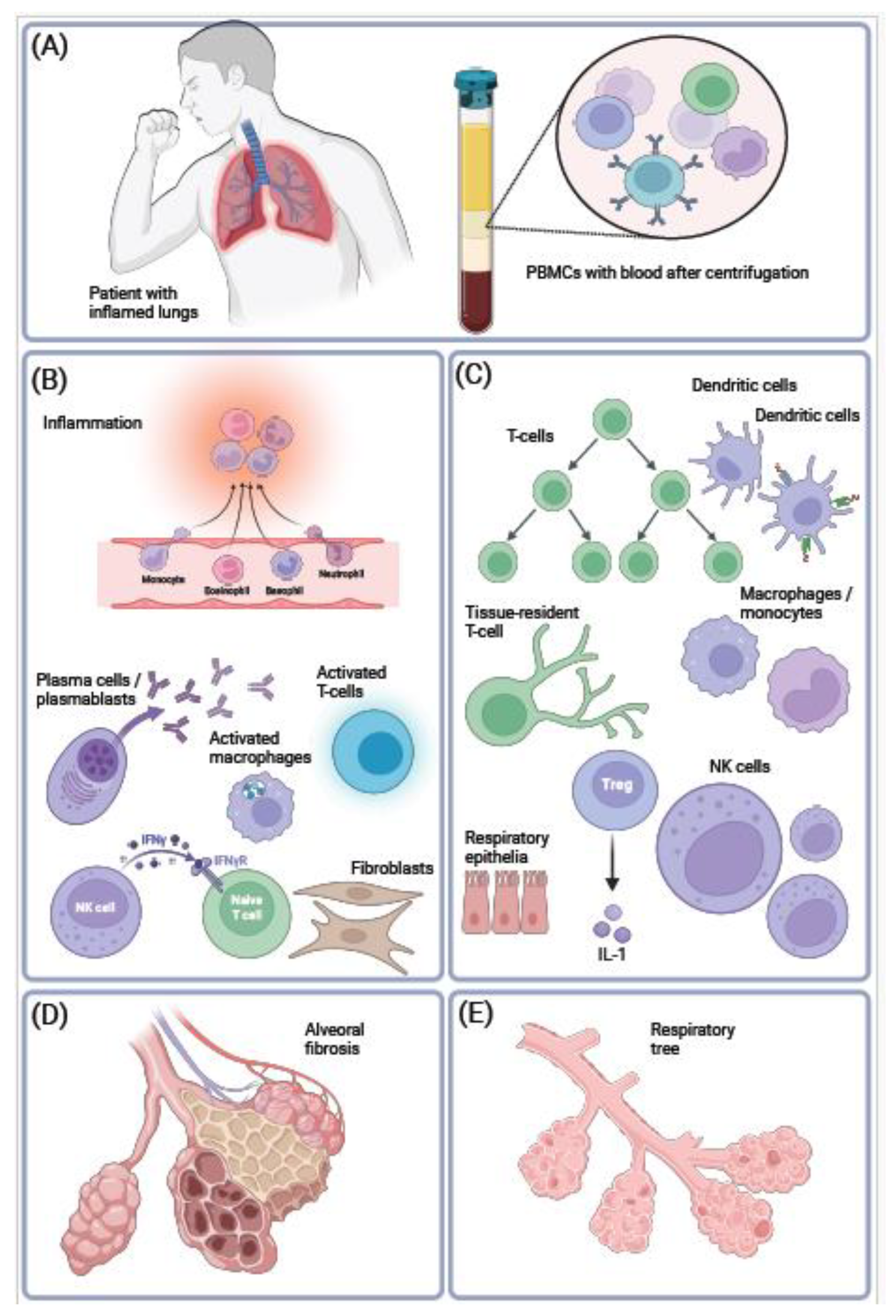
5. Conclusions.
Abbreviations.
References
- Wiersinga, W. J.; Rhodes, A.; Cheng, A. C.; Peacock, S. J.; Prescott, H. C., Pathophysiology, Transmission, Diagnosis, and Treatment of Coronavirus Disease 2019 (COVID-19): A Review. JAMA 2020, 324, (8), 782-793.
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; Niu, P.; Zhan, F.; Ma, X.; Wang, D.; Xu, W.; Wu, G.; Gao, G. F.; Tan, W., A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med 2020, 382, (8), 727-733.
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y. M.; Wang, W.; Song, Z. G.; Hu, Y.; Tao, Z. W.; Tian, J. H.; Pei, Y. Y.; Yuan, M. L.; Zhang, Y. L.; Dai, F. H.; Liu, Y.; Wang, Q. M.; Zheng, J. J.; Xu, L.; Holmes, E. C.; Zhang, Y. Z., A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, (7798), 265-269.
- Gostin, L. O.; Gronvall, G. K., The Origins of Covid-19 — Why It Matters (and Why It Doesn't). New England Journal of Medicine 2023.
- Guan, W.-j.; Ni, Z.-y.; Hu, Y.; Liang, W.-h.; Ou, C.-q.; He, J.-x.; Liu, L.; Shan, H.; Lei, C.-l.; Hui, D. S. C.; Du, B.; Li, L.-j.; Zeng, G.; Yuen, K.-Y.; Chen, R.-c.; Tang, C.-l.; Wang, T.; Chen, P.-y.; Xiang, J.; Li, S.-y.; Wang, J.-l.; Liang, Z.-j.; Peng, Y.-x.; Wei, L.; Liu, Y.; Hu, Y.-h.; Peng, P.; Wang, J.-m.; Liu, J.-y.; Chen, Z.; Li, G.; Zheng, Z.-j.; Qiu, S.-q.; Luo, J.; Ye, C.-j.; Zhu, S.-y.; Zhong, N.-s., Clinical Characteristics of Coronavirus Disease 2019 in China. New England Journal of Medicine 2020, 382, (18), 1708-1720.
- Holmes, E. C.; Goldstein, S. A.; Rasmussen, A. L.; Robertson, D. L.; Crits-Christoph, A.; Wertheim, J. O.; Anthony, S. J.; Barclay, W. S.; Boni, M. F.; Doherty, P. C.; Farrar, J.; Geoghegan, J. L.; Jiang, X.; Leibowitz, J. L.; Neil, S. J. D.; Skern, T.; Weiss, S. R.; Worobey, M.; Andersen, K. G.; Garry, R. F.; Rambaut, A., The origins of SARS-CoV-2: A critical review. Cell 2021, 184, (19), 4848-4856.
- Wu, Z.; McGoogan, J. M., Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72‚ÄØ314 Cases From the Chinese Center for Disease Control and Prevention. JAMA 2020, 323, (13), 1239-1242.
- Davis, H. E.; McCorkell, L.; Vogel, J. M.; Topol, E. J., Long COVID: major findings, mechanisms and recommendations. Nat Rev Microbiol 2023, 21, (3), 133-146.
- Sherif, Z. A.; Gomez, C. R.; Connors, T. J.; Henrich, T. J.; Reeves, W. B., Pathogenic mechanisms of post-acute sequelae of SARS-CoV-2 infection (PASC). Elife 2023, 12.
- Iba, T.; Connors, J. M.; Levy, J. H., The coagulopathy, endotheliopathy, and vasculitis of COVID-19. Inflamm Res 2020, 69, (12), 1181-1189.
- Trypsteen, W.; Van Cleemput, J.; Snippenberg, W. V.; Gerlo, S.; Vandekerckhove, L., On the whereabouts of SARS-CoV-2 in the human body: A systematic review. PLoS Pathog 2020, 16, (10), e1009037.
- Konstantinos, E.; Dimitris, V.; Koralia, P.; Periklis, G. F.; Nefeli, L.; Marios, D.; Angelos, P.; Bindu, K.; Orsalia, H.; Aikaterini, P.; Sophia, H.; Athanassios, K.; Christos, K.; Athanasios, G. T.; Laurence de, L.; Demetris, V.; Sotirios, T.; Barry, R. S.; Argyris, P.; Giovanni, B.; Ioannis, K.; Peter, J. B.; Vassilis, G. G., Pulmonary infection by SARS-CoV-2 induces senescence accompanied by an inflammatory phenotype in severe COVID-19: possible implications for viral mutagenesis. European Respiratory Journal 2022, 60, (2), 2102951.
- Ackermann, M.; Verleden, S. E.; Kuehnel, M.; Haverich, A.; Welte, T.; Laenger, F.; Vanstapel, A.; Werlein, C.; Stark, H.; Tzankov, A.; Li, W. W.; Li, V. W.; Mentzer, S. J.; Jonigk, D., Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in Covid-19. New England Journal of Medicine 2020, 383, (2), 120-128.
- Costa, T. J.; Potje, S. R.; Fraga-Silva, T. F. C.; da Silva-Neto, J. A.; Barros, P. R.; Rodrigues, D.; Machado, M. R.; Martins, R. B.; Santos-Eichler, R. A.; Benatti, M. N.; de Sá, K. S. G.; Almado, C. E. L.; Castro Í, A.; Pontelli, M. C.; Serra, L.; Carneiro, F. S.; Becari, C.; Louzada-Junior, P.; Oliveira, R. D. R.; Zamboni, D. S.; Arruda, E.; Auxiliadora-Martins, M.; Giachini, F. R. C.; Bonato, V. L. D.; Zachara, N. E.; Bomfim, G. F.; Tostes, R. C., Mitochondrial DNA and TLR9 activation contribute to SARS-CoV-2-induced endothelial cell damage. Vascul Pharmacol 2022, 142, 106946.
- Liu, F.; Han, K.; Blair, R.; Kenst, K.; Qin, Z.; Upcin, B.; Wrsdrfer, P.; Midkiff, C. C.; Mudd, J.; Belyaeva, E.; Milligan, N. S.; Rorison, T. D.; Wagner, N.; Bodem, J.; Dlken, L.; Aktas, B. H.; Vander Heide, R. S.; Yin, X. M.; Kolls, J. K.; Roy, C. J.; Rappaport, J.; Ergn, S.; Qin, X., SARS-CoV-2 Infects Endothelial Cells In Vivo and In Vitro. Front Cell Infect Microbiol 2021, 11, 701278.
- Stjepanovic, M. I.; Stojanovic, M. R.; Stankovic, S.; Cvejic, J.; Dimic-Janjic, S.; Popevic, S.; Buha, I.; Belic, S.; Djurdjevic, N.; Stjepanovic, M. M.; Jovanovic, D.; Stojkovic-Laloševic, M.; Soldatovic, I.; Bonaci-Nikolic, B.; Miskovic, R., Autoimmune and immunoserological markers of COVID-19 pneumonia: Can they help in the assessment of disease severity. Front Med (Lausanne) 2022, 9, 934270.
- Dotan, A.; Muller, S.; Kanduc, D.; David, P.; Halpert, G.; Shoenfeld, Y., The SARS-CoV-2 as an instrumental trigger of autoimmunity. Autoimmun Rev 2021, 20, (4), 102792.
- Damoiseaux, J.; Dotan, A.; Fritzler, M. J.; Bogdanos, D. P.; Meroni, P. L.; Roggenbuck, D.; Goldman, M.; Landegren, N.; Bastard, P.; Shoenfeld, Y.; Conrad, K., Autoantibodies and SARS-CoV2 infection: The spectrum from association to clinical implication: Report of the 15th Dresden Symposium on Autoantibodies. Autoimmun Rev 2022, 21, (3), 103012.
- Hikmet, F.; Mear, L.; Edvinsson, A.; Micke, P.; Uhlen, M.; Lindskog, C., The protein expression profile of ACE2 in human tissues. Mol Syst Biol 2020, 16, (7), e9610.
- Finkel Y., M. O., Nachshon A., Weingarten-Gabbay S., Morgenstern D., Yahalom-Ronen Y., Tamir H., Achdout H., Stein D., Israeli O., Beth-Din A., Melamed S., Weiss S., Israely T., Paran N., Schwartz M. & Stern-Ginossar N., The coding capacity of SARS-CoV-2. Nature 2020, 589, 125-130.
- Kim, D.; Lee, J. Y.; Yang, J. S.; Kim, J. W.; Kim, V. N.; Chang, H., The Architecture of SARS-CoV-2 Transcriptome. Cell 2020, 181, (4), 914-921 e10.
- Papa, G.; Mallery, D. L.; Albecka, A.; Welch, L. G.; Cattin-Ortola, J.; Luptak, J.; Paul, D.; McMahon, H. T.; Goodfellow, I. G.; Carter, A.; Munro, S.; James, L. C., Furin cleavage of SARS-CoV-2 Spike promotes but is not essential for infection and cell-cell fusion. PLoS Pathog 2021, 17, (1), e1009246.
- Shang, J., Wan, Y., Luo, C., Ye, G., Geng, Q., Auerbach, A., Li, F., Cell entry mechanisms of SARS-CoV-2. Proceedings of the National Academy of Sciences of the United States of America 2020, 117, (21), 11727-11734.
- Lamers, M. M. H., B. L., SARS-CoV-2 pathogenesis. Nat Rev Microbiol 2022, 20, (5), 270-284.
- Arunachalam, P. S.; Wimmers, F.; Mok, C. K. P.; Perera, R.; Scott, M.; Hagan, T.; Sigal, N.; Feng, Y.; Bristow, L.; Tak-Yin Tsang, O.; Wagh, D.; Coller, J.; Pellegrini, K. L.; Kazmin, D.; Alaaeddine, G.; Leung, W. S.; Chan, J. M. C.; Chik, T. S. H.; Choi, C. Y. C.; Huerta, C.; Paine McCullough, M.; Lv, H.; Anderson, E.; Edupuganti, S.; Upadhyay, A. A.; Bosinger, S. E.; Maecker, H. T.; Khatri, P.; Rouphael, N.; Peiris, M.; Pulendran, B., Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science 2020, 369, (6508), 1210-1220.
- Bernardes, J. P.; Mishra, N.; Tran, F.; Bahmer, T.; Best, L.; Blase, J. I.; Bordoni, D.; Franzenburg, J.; Geisen, U.; Josephs-Spaulding, J.; Köhler, P.; Künstner, A.; Rosati, E.; Aschenbrenner, A. C.; Bacher, P.; Baran, N.; Boysen, T.; Brandt, B.; Bruse, N.; Dörr, J.; Dräger, A.; Elke, G.; Ellinghaus, D.; Fischer, J.; Forster, M.; Franke, A.; Franzenburg, S.; Frey, N.; Friedrichs, A.; Fuß, J.; Glück, A.; Hamm, J.; Hinrichsen, F.; Hoeppner, M. P.; Imm, S.; Junker, R.; Kaiser, S.; Kan, Y. H.; Knoll, R.; Lange, C.; Laue, G.; Lier, C.; Lindner, M.; Marinos, G.; Markewitz, R.; Nattermann, J.; Noth, R.; Pickkers, P.; Rabe, K. F.; Renz, A.; Röcken, C.; Rupp, J.; Schaffarzyk, A.; Scheffold, A.; Schulte-Schrepping, J.; Schunk, D.; Skowasch, D.; Ulas, T.; Wandinger, K. P.; Wittig, M.; Zimmermann, J.; Busch, H.; Hoyer, B. F.; Kaleta, C.; Heyckendorf, J.; Kox, M.; Rybniker, J.; Schreiber, S.; Schultze, J. L.; Rosenstiel, P., Longitudinal Multi-omics Analyses Identify Responses of Megakaryocytes, Erythroid Cells, and Plasmablasts as Hallmarks of Severe COVID-19. Immunity 2020, 53, (6), 1296-1314 e9.
- Wilk, A. J.; Rustagi, A.; Zhao, N. Q.; Roque, J.; Martínez-Colón, G. J.; McKechnie, J. L.; Ivison, G. T.; Ranganath, T.; Vergara, R.; Hollis, T.; Simpson, L. J.; Grant, P.; Subramanian, A.; Rogers, A. J.; Blish, C. A., A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med 2020, 26, (7), 1070-1076.
- Iwamura, C.; Hirahara, K.; Kiuchi, M.; Ikehara, S.; Azuma, K.; Shimada, T.; Kuriyama, S.; Ohki, S.; Yamamoto, E.; Inaba, Y.; Shiko, Y.; Aoki, A.; Kokubo, K.; Hirasawa, R.; Hishiya, T.; Tsuji, K.; Nagaoka, T.; Ishikawa, S.; Kojima, A.; Mito, H.; Hase, R.; Kasahara, Y.; Kuriyama, N.; Tsukamoto, T.; Nakamura, S.; Urushibara, T.; Kaneda, S.; Sakao, S.; Tobiume, M.; Suzuki, Y.; Tsujiwaki, M.; Kubo, T.; Hasegawa, T.; Nakase, H.; Nishida, O.; Takahashi, K.; Baba, K.; Iizumi, Y.; Okazaki, T.; Kimura, M. Y.; Yoshino, I.; Igari, H.; Nakajima, H.; Suzuki, T.; Hanaoka, H.; Nakada, T.-A.; Ikehara, Y.; Yokote, K.; Nakayama, T., Elevated Myl9 reflects the Myl9-containing microthrombi in SARS-CoV-2-induced lung exudative vasculitis and predicts COVID-19 severity. Proceedings of the National Academy of Sciences of the United States of America 2022, 119, (33), e2203437119-e2203437119.
- Blanco-Melo, D.; Nilsson-Payant, B. E.; Liu, W.-C.; Uhl, S.; Hoagland, D.; Muller, R.; Jordan, T. X.; Oishi, K.; Panis, M.; Sachs, D.; Wang, T. T.; Schwartz, R. E.; Lim, J. K.; Albrecht, R. A.; tenOever, B. R., Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, (5), 1036-1045.e9.
- Melms, J. C.; Biermann, J.; Huang, H.; Wang, Y.; Nair, A.; Tagore, S.; Katsyv, I.; Rendeiro, A. F.; Amin, A. D.; Schapiro, D.; Frangieh, C. J.; Luoma, A. M.; Filliol, A.; Fang, Y.; Ravichandran, H.; Clausi, M. G.; Alba, G. A.; Rogava, M.; Chen, S. W.; Ho, P.; Montoro, D. T.; Kornberg, A. E.; Han, A. S.; Bakhoum, M. F.; Anandasabapathy, N.; Suárez-Fariñas, M.; Bakhoum, S. F.; Bram, Y.; Borczuk, A.; Guo, X. V.; Lefkowitch, J. H.; Marboe, C.; Lagana, S. M.; Del Portillo, A.; Tsai, E. J.; Zorn, E.; Markowitz, G. S.; Schwabe, R. F.; Schwartz, R. E.; Elemento, O.; Saqi, A.; Hibshoosh, H.; Que, J.; Izar, B., A molecular single-cell lung atlas of lethal COVID-19. Nature 2021, 595, (7865), 114-119.
- Daamen, A. R.; Bachali, P.; Owen, K. A.; Kingsmore, K. M.; Hubbard, E. L.; Labonte, A. C.; Robl, R.; Shrotri, S.; Grammer, A. C.; Lipsky, P. E., Comprehensive transcriptomic analysis of COVID-19 blood, lung, and airway. Scientific reports 2021, 11, (1), 7052-7052.
- Assou, S.; Ahmed, E.; Morichon, L.; Nasri, A.; Foisset, F.; Bourdais, C.; Gros, N.; Tieo, S.; Petit, A.; Vachier, I.; Muriaux, D.; Bourdin, A.; De Vos, J., The Transcriptome Landscape of the In Vitro Human Airway Epithelium Response to SARS-CoV-2. Int J Mol Sci 2023, 24, (15).
- Heming, M.; Li, X.; Ruber, S.; Mausberg, A. K.; Brsch, A. L.; Hartlehnert, M.; Singhal, A.; Lu, I. N.; Fleischer, M.; Szepanowski, F.; Witzke, O.; Brenner, T.; Dittmer, U.; Yosef, N.; Kleinschnitz, C.; Wiendl, H.; Stettner, M.; Meyer Zu Hrste, G., Neurological Manifestations of COVID-19 Feature T Cell Exhaustion and Dedifferentiated Monocytes in Cerebrospinal Fluid. Immunity 2021, 54, (1), 164-175 e6.
- Jackson, R. M.; Hatton, C. F.; Spegarova, J. S.; Georgiou, M.; Collin, J.; Stephenson, E.; Verdon, B.; Haq, I. J.; Hussain, R.; Coxhead, J. M.; Mudhar, H.-S.; Wagner, B.; Hasoon, M.; Davey, T.; Rooney, P.; Khan, C. M. A.; Ward, C.; Brodlie, M.; Haniffa, M.; Hambleton, S.; Armstrong, L.; Figueiredo, F.; Queen, R.; Duncan, C. J. A.; Lako, M., Conjunctival epithelial cells resist productive SARS-CoV-2 infection. Stem Cell Reports 2022, 17, (7), 1699-1713.
- Zhu, L.; Yang, P.; Zhao, Y.; Zhuang, Z.; Wang, Z.; Song, R.; Zhang, J.; Liu, C.; Gao, Q.; Xu, Q.; Wei, X.; Sun, H. X.; Ye, B.; Wu, Y.; Zhang, N.; Lei, G.; Yu, L.; Yan, J.; Diao, G.; Meng, F.; Bai, C.; Mao, P.; Yu, Y.; Wang, M.; Yuan, Y.; Deng, Q.; Li, Z.; Huang, Y.; Hu, G.; Liu, Y.; Wang, X.; Xu, Z.; Liu, P.; Bi, Y.; Shi, Y.; Zhang, S.; Chen, Z.; Wang, J.; Xu, X.; Wu, G.; Wang, F. S.; Gao, G. F.; Liu, L.; Liu, W. J., Single-Cell Sequencing of Peripheral Mononuclear Cells Reveals Distinct Immune Response Landscapes of COVID-19 and Influenza Patients. Immunity 2020, 53, (3), 685-696.e3.
- Zhang, J. Y.; Wang, X. M.; Xing, X.; Xu, Z.; Zhang, C.; Song, J. W.; Fan, X.; Xia, P.; Fu, J. L.; Wang, S. Y.; Xu, R. N.; Dai, X. P.; Shi, L.; Huang, L.; Jiang, T. J.; Shi, M.; Zhang, Y.; Zumla, A.; Maeurer, M.; Bai, F.; Wang, F. S., Single-cell landscape of immunological responses in patients with COVID-19. Nat Immunol 2020, 21, (9), 1107-1118.
- Silvin, A.; Chapuis, N.; Dunsmore, G.; Goubet, A. G.; Dubuisson, A.; Derosa, L.; Almire, C.; Hénon, C.; Kosmider, O.; Droin, N.; Rameau, P.; Catelain, C.; Alfaro, A.; Dussiau, C.; Friedrich, C.; Sourdeau, E.; Marin, N.; Szwebel, T. A.; Cantin, D.; Mouthon, L.; Borderie, D.; Deloger, M.; Bredel, D.; Mouraud, S.; Drubay, D.; Andrieu, M.; Lhonneur, A. S.; Saada, V.; Stoclin, A.; Willekens, C.; Pommeret, F.; Griscelli, F.; Ng, L. G.; Zhang, Z.; Bost, P.; Amit, I.; Barlesi, F.; Marabelle, A.; Pène, F.; Gachot, B.; André, F.; Zitvogel, L.; Ginhoux, F.; Fontenay, M.; Solary, E., Elevated Calprotectin and Abnormal Myeloid Cell Subsets Discriminate Severe from Mild COVID-19. Cell 2020, 182, (6), 1401-1418.e18.
- Schulte-Schrepping, J.; Reusch, N.; Paclik, D.; Baßler, K.; Schlickeiser, S.; Zhang, B.; Krämer, B.; Krammer, T.; Brumhard, S.; Bonaguro, L.; De Domenico, E.; Wendisch, D.; Grasshoff, M.; Kapellos, T. S.; Beckstette, M.; Pecht, T.; Saglam, A.; Dietrich, O.; Mei, H. E.; Schulz, A. R.; Conrad, C.; Kunkel, D.; Vafadarnejad, E.; Xu, C. J.; Horne, A.; Herbert, M.; Drews, A.; Thibeault, C.; Pfeiffer, M.; Hippenstiel, S.; Hocke, A.; Müller-Redetzky, H.; Heim, K. M.; Machleidt, F.; Uhrig, A.; Bosquillon de Jarcy, L.; Jürgens, L.; Stegemann, M.; Glösenkamp, C. R.; Volk, H. D.; Goffinet, C.; Landthaler, M.; Wyler, E.; Georg, P.; Schneider, M.; Dang-Heine, C.; Neuwinger, N.; Kappert, K.; Tauber, R.; Corman, V.; Raabe, J.; Kaiser, K. M.; Vinh, M. T.; Rieke, G.; Meisel, C.; Ulas, T.; Becker, M.; Geffers, R.; Witzenrath, M.; Drosten, C.; Suttorp, N.; von Kalle, C.; Kurth, F.; Händler, K.; Schultze, J. L.; Aschenbrenner, A. C.; Li, Y.; Nattermann, J.; Sawitzki, B.; Saliba, A. E.; Sander, L. E., Severe COVID-19 Is Marked by a Dysregulated Myeloid Cell Compartment. Cell 2020, 182, (6), 1419-1440 e23.
- Meckiff, B. J.; Ramirez-Sustegui, C.; Fajardo, V.; Chee, S. J.; Kusnadi, A.; Simon, H.; Eschweiler, S.; Grifoni, A.; Pelosi, E.; Weiskopf, D.; Sette, A.; Ay, F.; Seumois, G.; Ottensmeier, C. H.; Vijayanand, P., Imbalance of Regulatory and Cytotoxic SARS-CoV-2-Reactive CD4(+) T Cells in COVID-19. Cell 2020, 183, (5), 1340-1353 e16.
- Witkowski, M.; Tizian, C.; Ferreira-Gomes, M.; Niemeyer, D.; Jones, T. C.; Heinrich, F.; Frischbutter, S.; Angermair, S.; Hohnstein, T.; Mattiola, I.; Nawrath, P.; McEwen, S.; Zocche, S.; Viviano, E.; Heinz, G. A.; Maurer, M.; Kölsch, U.; Chua, R. L.; Aschman, T.; Meisel, C.; Radke, J.; Sawitzki, B.; Roehmel, J.; Allers, K.; Moos, V.; Schneider, T.; Hanitsch, L.; Mall, M. A.; Conrad, C.; Radbruch, H.; Duerr, C. U.; Trapani, J. A.; Marcenaro, E.; Kallinich, T.; Corman, V. M.; Kurth, F.; Sander, L. E.; Drosten, C.; Treskatsch, S.; Durek, P.; Kruglov, A.; Radbruch, A.; Mashreghi, M. F.; Diefenbach, A., Untimely TGF-beta responses in COVID-19 limit antiviral functions of NK cells. Nature 2021, 600, (7888), 295-301.
- Ramaswamy, A.; Brodsky, N. N.; Sumida, T. S.; Comi, M.; Asashima, H.; Hoehn, K. B.; Li, N.; Liu, Y.; Shah, A.; Ravindra, N. G.; Bishai, J.; Khan, A.; Lau, W.; Sellers, B.; Bansal, N.; Guerrerio, P.; Unterman, A.; Habet, V.; Rice, A. J.; Catanzaro, J.; Chandnani, H.; Lopez, M.; Kaminski, N.; Dela Cruz, C. S.; Tsang, J. S.; Wang, Z.; Yan, X.; Kleinstein, S. H.; van Dijk, D.; Pierce, R. W.; Hafler, D. A.; Lucas, C. L., Immune dysregulation and autoreactivity correlate with disease severity in SARS-CoV-2-associated multisystem inflammatory syndrome in children. Immunity 2021, 54, (5), 1083-1095 e7.
- You, M.; Chen, L.; Zhang, D.; Zhao, P.; Chen, Z.; Qin, E. Q.; Gao, Y.; Davis, M. M.; Yang, P., Single-cell epigenomic landscape of peripheral immune cells reveals establishment of trained immunity in individuals convalescing from COVID-19. Nat Cell Biol 2021, 23, (6), 620-630.
- Aznaourova, M.; Schmerer, N.; Janga, H.; Zhang, Z.; Pauck, K.; Bushe, J.; Volkers, S. M.; Wendisch, D.; Georg, P.; Ntini, E.; Aillaud, M.; Gendisch, M.; Mack, E.; Skevaki, C.; Keller, C.; Bauer, C.; Bertrams, W.; Marsico, A.; Nist, A.; Stiewe, T.; Gruber, A. D.; Ruppert, C.; Li, Y.; Garn, H.; Sander, L. E.; Schmeck, B.; Schulte, L. N., Single-cell RNA sequencing uncovers the nuclear decoy lincRNA PIRAT as a regulator of systemic monocyte immunity during COVID-19. Proceedings of the National Academy of Sciences of the United States of America 2022, 119, (36), e2120680119.
- Bost, P.; Giladi, A.; Liu, Y.; Bendjelal, Y.; Xu, G.; David, E.; Blecher-Gonen, R.; Cohen, M.; Medaglia, C.; Li, H.; Deczkowska, A.; Zhang, S.; Schwikowski, B.; Zhang, Z.; Amit, I., Host-Viral Infection Maps Reveal Signatures of Severe COVID-19 Patients. Cell 2020, 181, (7), 1475-1488 e12.
- Chua, R. L.; Lukassen, S.; Trump, S.; Hennig, B. P.; Wendisch, D.; Pott, F.; Debnath, O.; Thurmann, L.; Kurth, F.; Volker, M. T.; Kazmierski, J.; Timmermann, B.; Twardziok, S.; Schneider, S.; Machleidt, F.; Muller-Redetzky, H.; Maier, M.; Krannich, A.; Schmidt, S.; Balzer, F.; Liebig, J.; Loske, J.; Suttorp, N.; Eils, J.; Ishaque, N.; Liebert, U. G.; von Kalle, C.; Hocke, A.; Witzenrath, M.; Goffinet, C.; Drosten, C.; Laudi, S.; Lehmann, I.; Conrad, C.; Sander, L. E.; Eils, R., COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat Biotechnol 2020, 38, (8), 970-979.
- Yang, A. C.; Kern, F.; Losada, P. M.; Agam, M. R.; Maat, C. A.; Schmartz, G. P.; Fehlmann, T.; Stein, J. A.; Schaum, N.; Lee, D. P.; Calcuttawala, K.; Vest, R. T.; Berdnik, D.; Lu, N.; Hahn, O.; Gate, D.; McNerney, M. W.; Channappa, D.; Cobos, I.; Ludwig, N.; Schulz-Schaeffer, W. J.; Keller, A.; Wyss-Coray, T., Dysregulation of brain and choroid plexus cell types in severe COVID-19. Nature 2021, 595, (7868), 565-571.
- Delorey, T. M.; Ziegler, C. G. K.; Heimberg, G.; Normand, R.; Yang, Y.; Segerstolpe, Ö.; Abbondanza, D.; Fleming, S. J.; Subramanian, A.; Montoro, D. T.; Jagadeesh, K. A.; Dey, K. K.; Sen, P.; Slyper, M.; Pita-Ju√°rez, Y. H.; Phillips, D.; Biermann, J.; Bloom-Ackermann, Z.; Barkas, N.; Ganna, A.; Gomez, J.; Melms, J. C.; Katsyv, I.; Normandin, E.; Naderi, P.; Popov, Y. V.; Raju, S. S.; Niezen, S.; Tsai, L. T. Y.; Siddle, K. J.; Sud, M.; Tran, V. M.; Vellarikkal, S. K.; Wang, Y.; Amir-Zilberstein, L.; Atri, D. S.; Beechem, J.; Brook, O. R.; Chen, J.; Divakar, P.; Dorceus, P.; Engreitz, J. M.; Essene, A.; Fitzgerald, D. M.; Fropf, R.; Gazal, S.; Gould, J.; Grzyb, J.; Harvey, T.; Hecht, J.; Hether, T.; Jan√©-Valbuena, J.; Leney-Greene, M.; Ma, H.; McCabe, C.; McLoughlin, D. E.; Miller, E. M.; Muus, C.; Niemi, M.; Padera, R.; Pan, L.; Pant, D.; Pe‚Äôer, C.; Pfiffner-Borges, J.; Pinto, C. J.; Plaisted, J.; Reeves, J.; Ross, M.; Rudy, M.; Rueckert, E. H.; Siciliano, M.; Sturm, A.; Todres, E.; Waghray, A.; Warren, S.; Zhang, S.; Zollinger, D. R.; Cosimi, L.; Gupta, R. M.; Hacohen, N.; Hibshoosh, H.; Hide, W.; Price, A. L.; Rajagopal, J.; Tata, P. R.; Riedel, S.; Szabo, G.; Tickle, T. L.; Ellinor, P. T.; Hung, D.; Sabeti, P. C.; Novak, R.; Rogers, R.; Ingber, D. E.; Jiang, Z. G.; Juric, D.; Babadi, M.; Farhi, S. L.; Izar, B.; Stone, J. R.; Vlachos, I. S.; Solomon, I. H.; Ashenberg, O.; Porter, C. B. M.; Li, B.; Shalek, A. K.; Villani, A.-C.; Rozenblatt-Rosen, O.; Regev, A., COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature 2021, 595, (7865), 107-113.
- de Rooij, L.; Becker, L. M.; Teuwen, L. A.; Boeckx, B.; Jansen, S.; Feys, S.; Verleden, S.; Liesenborghs, L.; Stalder, A. K.; Libbrecht, S.; Van Buyten, T.; Philips, G.; Subramanian, A.; Dumas, S. J.; Meta, E.; Borri, M.; Sokol, L.; Dendooven, A.; Truong, A. K.; Gunst, J.; Van Mol, P.; Haslbauer, J. D.; Rohlenova, K.; Menter, T.; Boudewijns, R.; Geldhof, V.; Vinckier, S.; Amersfoort, J.; Wuyts, W.; Van Raemdonck, D.; Jacobs, W.; Ceulemans, L. J.; Weynand, B.; Thienpont, B.; Lammens, M.; Kuehnel, M.; Eelen, G.; Dewerchin, M.; Schoonjans, L.; Jonigk, D.; van Dorpe, J.; Tzankov, A.; Wauters, E.; Mazzone, M.; Neyts, J.; Wauters, J.; Lambrechts, D.; Carmeliet, P., The pulmonary vasculature in lethal COVID-19 and idiopathic pulmonary fibrosis at single-cell resolution. Cardiovasc Res 2023, 119, (2), 520-535.
- Ziegler, C. G. K.; Allon, S. J.; Nyquist, S. K.; Mbano, I. M.; Miao, V. N.; Tzouanas, C. N.; Cao, Y.; Yousif, A. S.; Bals, J.; Hauser, B. M.; Feldman, J.; Muus, C.; Wadsworth, M. H., 2nd; Kazer, S. W.; Hughes, T. K.; Doran, B.; Gatter, G. J.; Vukovic, M.; Taliaferro, F.; Mead, B. E.; Guo, Z.; Wang, J. P.; Gras, D.; Plaisant, M.; Ansari, M.; Angelidis, I.; Adler, H.; Sucre, J. M. S.; Taylor, C. J.; Lin, B.; Waghray, A.; Mitsialis, V.; Dwyer, D. F.; Buchheit, K. M.; Boyce, J. A.; Barrett, N. A.; Laidlaw, T. M.; Carroll, S. L.; Colonna, L.; Tkachev, V.; Peterson, C. W.; Yu, A.; Zheng, H. B.; Gideon, H. P.; Winchell, C. G.; Lin, P. L.; Bingle, C. D.; Snapper, S. B.; Kropski, J. A.; Theis, F. J.; Schiller, H. B.; Zaragosi, L. E.; Barbry, P.; Leslie, A.; Kiem, H. P.; Flynn, J. L.; Fortune, S. M.; Berger, B.; Finberg, R. W.; Kean, L. S.; Garber, M.; Schmidt, A. G.; Lingwood, D.; Shalek, A. K.; Ordovas-Montanes, J., SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell 2020, 181, (5), 1016-1035 e19.
- Smith, J. C.; Sausville, E. L.; Girish, V.; Yuan, M. L.; Vasudevan, A.; John, K. M.; Sheltzer, J. M., Cigarette Smoke Exposure and Inflammatory Signaling Increase the Expression of the SARS-CoV-2 Receptor ACE2 in the Respiratory Tract. Dev Cell 2020, 53, (5), 514-529.e3.
- Aguiar, J. A.; Tremblay, B. J.; Mansfield, M. J.; Woody, O.; Lobb, B.; Banerjee, A.; Chandiramohan, A.; Tiessen, N.; Cao, Q.; Dvorkin-Gheva, A.; Revill, S.; Miller, M. S.; Carlsten, C.; Organ, L.; Joseph, C.; John, A.; Hanson, P.; Austin, R. C.; McManus, B. M.; Jenkins, G.; Mossman, K.; Ask, K.; Doxey, A. C.; Hirota, J. A., Gene expression and in situ protein profiling of candidate SARS-CoV-2 receptors in human airway epithelial cells and lung tissue. Eur Respir J 2020, 56, (3).
- Muus, C.; Luecken, M. D.; Eraslan, G.; Sikkema, L.; Waghray, A.; Heimberg, G.; Kobayashi, Y.; Vaishnav, E. D.; Subramanian, A.; Smillie, C.; Jagadeesh, K. A.; Duong, E. T.; Fiskin, E.; Torlai Triglia, E.; Ansari, M.; Cai, P.; Lin, B.; Buchanan, J.; Chen, S.; Shu, J.; Haber, A. L.; Chung, H.; Montoro, D. T.; Adams, T.; Aliee, H.; Allon, S. J.; Andrusivova, Z.; Angelidis, I.; Ashenberg, O.; Bassler, K.; Bécavin, C.; Benhar, I.; Bergenstråhle, J.; Bergenstråhle, L.; Bolt, L.; Braun, E.; Bui, L. T.; Callori, S.; Chaffin, M.; Chichelnitskiy, E.; Chiou, J.; Conlon, T. M.; Cuoco, M. S.; Cuomo, A. S. E.; Deprez, M.; Duclos, G.; Fine, D.; Fischer, D. S.; Ghazanfar, S.; Gillich, A.; Giotti, B.; Gould, J.; Guo, M.; Gutierrez, A. J.; Habermann, A. C.; Harvey, T.; He, P.; Hou, X.; Hu, L.; Hu, Y.; Jaiswal, A.; Ji, L.; Jiang, P.; Kapellos, T. S.; Kuo, C. S.; Larsson, L.; Leney-Greene, M. A.; Lim, K.; Litviňuková, M.; Ludwig, L. S.; Lukassen, S.; Luo, W.; Maatz, H.; Madissoon, E.; Mamanova, L.; Manakongtreecheep, K.; Leroy, S.; Mayr, C. H.; Mbano, I. M.; McAdams, A. M.; Nabhan, A. N.; Nyquist, S. K.; Penland, L.; Poirion, O. B.; Poli, S.; Qi, C.; Queen, R.; Reichart, D.; Rosas, I.; Schupp, J. C.; Shea, C. V.; Shi, X.; Sinha, R.; Sit, R. V.; Slowikowski, K.; Slyper, M.; Smith, N. P.; Sountoulidis, A.; Strunz, M.; Sullivan, T. B.; Sun, D.; Talavera-López, C.; Tan, P.; Tantivit, J.; Travaglini, K. J.; Tucker, N. R.; Vernon, K. A.; Wadsworth, M. H.; Waldman, J.; Wang, X.; Xu, K.; Yan, W.; Zhao, W.; Ziegler, C. G. K., Single-cell meta-analysis of SARS-CoV-2 entry genes across tissues and demographics. Nat Med 2021, 27, (3), 546-559.
- Menon, R.; Otto, E. A.; Sealfon, R.; Nair, V.; Wong, A. K.; Theesfeld, C. L.; Chen, X.; Wang, Y.; Boppana, A. S.; Luo, J.; Yang, Y.; Kasson, P. M.; Schaub, J. A.; Berthier, C. C.; Eddy, S.; Lienczewski, C. C.; Godfrey, B.; Dagenais, S. L.; Sohaney, R.; Hartman, J.; Fermin, D.; Subramanian, L.; Looker, H. C.; Harder, J. L.; Mariani, L. H.; Hodgin, J. B.; Sexton, J. Z.; Wobus, C. E.; Naik, A. S.; Nelson, R. G.; Troyanskaya, O. G.; Kretzler, M., SARS-CoV-2 receptor networks in diabetic and COVID-19-associated kidney disease. Kidney Int 2020, 98, (6), 1502-1518.
- Zhang, F.; Mears, J. R.; Shakib, L.; Beynor, J. I.; Shanaj, S.; Korsunsky, I.; Nathan, A.; Donlin, L. T.; Raychaudhuri, S., IFN-γ and TNF-α drive a CXCL10+ CCL2+ macrophage phenotype expanded in severe COVID-19 lungs and inflammatory diseases with tissue inflammation. Genome Med 2021, 13, (1), 64.
- Chen, F.; Zhang, Y.; Sucgang, R.; Ramani, S.; Corry, D.; Kheradmand, F.; Creighton, C. J., Meta-analysis of host transcriptional responses to SARS-CoV-2 infection reveals their manifestation in human tumors. Scientific reports 2021, 11, (1), 2459.
- Garg, M.; Li, X.; Moreno, P.; Papatheodorou, I.; Shu, Y.; Brazma, A.; Miao, Z., Meta-analysis of COVID-19 single-cell studies confirms eight key immune responses. Scientific reports 2021, 11, (1), 20833.
- Müller, J. A.; Groß, R.; Conzelmann, C.; Krüger, J.; Merle, U.; Steinhart, J.; Weil, T.; Koepke, L.; Bozzo, C. P.; Read, C.; Fois, G.; Eiseler, T.; Gehrmann, J.; van Vuuren, J.; Wessbecher, I. M.; Frick, M.; Costa, I. G.; Breunig, M.; Grüner, B.; Peters, L.; Schuster, M.; Liebau, S.; Seufferlein, T.; Stenger, S.; Stenzinger, A.; MacDonald, P. E.; Kirchhoff, F.; Sparrer, K. M. J.; Walther, P.; Lickert, H.; Barth, T. F. E.; Wagner, M.; Münch, J.; Heller, S.; Kleger, A., SARS-CoV-2 infects and replicates in cells of the human endocrine and exocrine pancreas. Nature Metabolism 2021, 3, (2), 149-165.
- Jovic, D.; Liang, X.; Zeng, H.; Lin, L.; Xu, F.; Luo, Y., Single-cell RNA sequencing technologies and applications: A brief overview. Clin Transl Med 2022, 12, (3), e694.
- Chua, R. L.; Lukassen, S.; Trump, S.; Hennig, B. P.; Wendisch, D.; Pott, F.; Debnath, O.; Thürmann, L.; Kurth, F.; Völker, M. T.; Kazmierski, J.; Timmermann, B.; Twardziok, S.; Schneider, S.; Machleidt, F.; Müller-Redetzky, H.; Maier, M.; Krannich, A.; Schmidt, S.; Balzer, F.; Liebig, J.; Loske, J.; Suttorp, N.; Eils, J.; Ishaque, N.; Liebert, U. G.; von Kalle, C.; Hocke, A.; Witzenrath, M.; Goffinet, C.; Drosten, C.; Laudi, S.; Lehmann, I.; Conrad, C.; Sander, L.-E.; Eils, R., COVID-19 severity correlates with airway epithelium–immune cell interactions identified by single-cell analysis. Nature Biotechnology 2020, 38, (8), 970-979.
- Zhu, L.; Yang, P.; Zhao, Y.; Zhuang, Z.; Wang, Z.; Song, R.; Zhang, J.; Liu, C.; Gao, Q.; Xu, Q.; Wei, X.; Sun, H. X.; Ye, B.; Wu, Y.; Zhang, N.; Lei, G.; Yu, L.; Yan, J.; Diao, G.; Meng, F.; Bai, C.; Mao, P.; Yu, Y.; Wang, M.; Yuan, Y.; Deng, Q.; Li, Z.; Huang, Y.; Hu, G.; Liu, Y.; Wang, X.; Xu, Z.; Liu, P.; Bi, Y.; Shi, Y.; Zhang, S.; Chen, Z.; Wang, J.; Xu, X.; Wu, G.; Wang, F. S.; Gao, G. F.; Liu, L.; Liu, W. J., Single-Cell Sequencing of Peripheral Mononuclear Cells Reveals Distinct Immune Response Landscapes of COVID-19 and Influenza Patients. Immunity 2020, 53, (3), 685-696 e3.
- Ogura, H.; Gohda, J.; Lu, X.; Yamamoto, M.; Takesue, Y.; Son, A.; Doi, S.; Matsushita, K.; Isobe, F.; Fukuda, Y.; Huang, T.-P.; Ueno, T.; Mambo, N.; Murakami, H.; Kawaguchi, Y.; Inoue, J.-i.; Shirai, K.; Yamasaki, S.; Hirata, J.-I.; Ishido, S., Dysfunctional Sars-CoV-2-M protein-specific cytotoxic T lymphocytes in patients recovering from severe COVID-19. Nature communications 2022, 13, (1), 7063.
- Silvin, A.; Chapuis, N.; Dunsmore, G.; Goubet, A. G.; Dubuisson, A.; Derosa, L.; Almire, C.; Hénon, C.; Kosmider, O.; Droin, N.; Rameau, P.; Catelain, C.; Alfaro, A.; Dussiau, C.; Friedrich, C.; Sourdeau, E.; Marin, N.; Szwebel, T. A.; Cantin, D.; Mouthon, L.; Borderie, D.; Deloger, M.; Bredel, D.; Mouraud, S.; Drubay, D.; Andrieu, M.; Lhonneur, A. S.; Saada, V.; Stoclin, A.; Willekens, C.; Pommeret, F.; Griscelli, F.; Ng, L. G.; Zhang, Z.; Bost, P.; Amit, I.; Barlesi, F.; Marabelle, A.; Pène, F.; Gachot, B.; André, F.; Zitvogel, L.; Ginhoux, F.; Fontenay, M.; Solary, E., Elevated Calprotectin and Abnormal Myeloid Cell Subsets Discriminate Severe from Mild COVID-19. Cell 2020, 182, (6), 1401-1418 e18.
- Schulte-Schrepping, J.; Reusch, N.; Paclik, D.; Baßler, K.; Schlickeiser, S.; Zhang, B.; Krämer, B.; Krammer, T.; Brumhard, S.; Bonaguro, L.; De Domenico, E.; Wendisch, D.; Grasshoff, M.; Kapellos, T. S.; Beckstette, M.; Pecht, T.; Saglam, A.; Dietrich, O.; Mei, H. E.; Schulz, A. R.; Conrad, C.; Kunkel, D.; Vafadarnejad, E.; Xu, C. J.; Horne, A.; Herbert, M.; Drews, A.; Thibeault, C.; Pfeiffer, M.; Hippenstiel, S.; Hocke, A.; Mller-Redetzky, H.; Heim, K. M.; Machleidt, F.; Uhrig, A.; Bosquillon de Jarcy, L.; Jrgens, L.; Stegemann, M.; Glsenkamp, C. R.; Volk, H. D.; Goffinet, C.; Landthaler, M.; Wyler, E.; Georg, P.; Schneider, M.; Dang-Heine, C.; Neuwinger, N.; Kappert, K.; Tauber, R.; Corman, V.; Raabe, J.; Kaiser, K. M.; Vinh, M. T.; Rieke, G.; Meisel, C.; Ulas, T.; Becker, M.; Geffers, R.; Witzenrath, M.; Drosten, C.; Suttorp, N.; von Kalle, C.; Kurth, F.; Hndler, K.; Schultze, J. L.; Aschenbrenner, A. C.; Li, Y.; Nattermann, J.; Sawitzki, B.; Saliba, A. E.; Sander, L. E., Severe COVID-19 Is Marked by a Dysregulated Myeloid Cell Compartment. Cell 2020, 182, (6), 1419-1440 e23.
- Sinha, S.; Rosin, N. L.; Arora, R.; Labit, E.; Jaffer, A.; Cao, L.; Farias, R.; Nguyen, A. P.; de Almeida, L. G. N.; Dufour, A.; Bromley, A.; McDonald, B.; Gillrie, M. R.; Fritzler, M. J.; Yipp, B. G.; Biernaskie, J., Dexamethasone modulates immature neutrophils and interferon programming in severe COVID-19. Nature Medicine 2022, 28, (1), 201-211.
- Su, Y.; Yuan, D.; Chen, D. G.; Ng, R. H.; Wang, K.; Choi, J.; Li, S.; Hong, S.; Zhang, R.; Xie, J.; Kornilov, S. A.; Scherler, K.; Pavlovitch-Bedzyk, A. J.; Dong, S.; Lausted, C.; Lee, I.; Fallen, S.; Dai, C. L.; Baloni, P.; Smith, B.; Duvvuri, V. R.; Anderson, K. G.; Li, J.; Yang, F.; Duncombe, C. J.; McCulloch, D. J.; Rostomily, C.; Troisch, P.; Zhou, J.; Mackay, S.; DeGottardi, Q.; May, D. H.; Taniguchi, R.; Gittelman, R. M.; Klinger, M.; Snyder, T. M.; Roper, R.; Wojciechowska, G.; Murray, K.; Edmark, R.; Evans, S.; Jones, L.; Zhou, Y.; Rowen, L.; Liu, R.; Chour, W.; Algren, H. A.; Berrington, W. R.; Wallick, J. A.; Cochran, R. A.; Micikas, M. E.; Wrin, T.; Petropoulos, C. J.; Cole, H. R.; Fischer, T. D.; Wei, W.; Hoon, D. S. B.; Price, N. D.; Subramanian, N.; Hill, J. A.; Hadlock, J.; Magis, A. T.; Ribas, A.; Lanier, L. L.; Boyd, S. D.; Bluestone, J. A.; Chu, H.; Hood, L.; Gottardo, R.; Greenberg, P. D.; Davis, M. M.; Goldman, J. D.; Heath, J. R., Multiple early factors anticipate post-acute COVID-19 sequelae. Cell 2022, 185, (5), 881-895 e20.
- Varga, Z.; Flammer, A. J.; Steiger, P.; Haberecker, M.; Andermatt, R.; Zinkernagel, A. S.; Mehra, M. R.; Schuepbach, R. A.; Ruschitzka, F.; Moch, H., Endothelial cell infection and endotheliitis in COVID-19. Lancet 2020, 395, (10234), 1417-1418.
- Wen, W.; Su, W.; Tang, H.; Le, W.; Zhang, X.; Zheng, Y.; Liu, X.; Xie, L.; Li, J.; Ye, J.; Dong, L.; Cui, X.; Miao, Y.; Wang, D.; Dong, J.; Xiao, C.; Chen, W.; Wang, H., Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov 2020, 6, 31.
- Lee, J. S.; Park, S.; Jeong, H. W.; Ahn, J. Y.; Choi, S. J.; Lee, H.; Choi, B.; Nam, S. K.; Sa, M.; Kwon, J. S.; Jeong, S. J.; Lee, H. K.; Park, S. H.; Park, S. H.; Choi, J. Y.; Kim, S. H.; Jung, I.; Shin, E. C., Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci Immunol 2020, 5, (49).
- Liao, M.; Liu, Y.; Yuan, J.; Wen, Y.; Xu, G.; Zhao, J.; Cheng, L.; Li, J.; Wang, X.; Wang, F.; Liu, L.; Amit, I.; Zhang, S.; Zhang, Z., Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med 2020, 26, (6), 842-844.
- Chua, R. L.; Lukassen, S.; Trump, S.; Hennig, B. P.; Wendisch, D.; Pott, F.; Debnath, O.; Thürmann, L.; Kurth, F.; Völker, M. T.; Kazmierski, J.; Timmermann, B.; Twardziok, S.; Schneider, S.; Machleidt, F.; Müller-Redetzky, H.; Maier, M.; Krannich, A.; Schmidt, S.; Balzer, F.; Liebig, J.; Loske, J.; Suttorp, N.; Eils, J.; Ishaque, N.; Liebert, U. G.; von Kalle, C.; Hocke, A.; Witzenrath, M.; Goffinet, C.; Drosten, C.; Laudi, S.; Lehmann, I.; Conrad, C.; Sander, L. E.; Eils, R., COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat Biotechnol 2020, 38, (8), 970-979.
- Marschner, I. C., Estimating age-specific COVID-19 fatality risk and time to death by comparing population diagnosis and death patterns: Australian data. BMC Med Res Methodol 2021, 21, (1), 126.
- Wang, T.; Zhang, X.; Liu, Z.; Yao, T.; Zheng, D.; Gan, J.; Yu, S.; Li, L.; Chen, P.; Sun, J., Single-cell RNA sequencing reveals the sustained immune cell dysfunction in the pathogenesis of sepsis secondary to bacterial pneumonia. Genomics 2021, 113, (3), 1219-1233.
- Maucourant, C.; Filipovic, I.; Ponzetta, A.; Aleman, S.; Cornillet, M.; Hertwig, L.; Strunz, B.; Lentini, A.; Reinius, B.; Brownlie, D.; Cuapio, A.; Ask, E. H.; Hull, R. M.; Haroun-Izquierdo, A.; Schaffer, M.; Klingström, J.; Folkesson, E.; Buggert, M.; Sandberg, J. K.; Eriksson, L. I.; Rooyackers, O.; Ljunggren, H. G.; Malmberg, K. J.; Michaëlsson, J.; Marquardt, N.; Hammer, Q.; Strålin, K.; Björkström, N. K., Natural killer cell immunotypes related to COVID-19 disease severity. Sci Immunol 2020, 5, (50).
- Zhang, W.; Xu, X.; Fu, Z.; Chen, J.; Chen, S.; Tan, Y., PathogenTrack and Yeskit: tools for identifying intracellular pathogens from single-cell RNA-sequencing datasets as illustrated by application to COVID-19. Front Med 2022, 16, (2), 251-262.
| 1 | According to the Elsevier's abstract and citation database (Scopus) database. Retrieved on 31 July 2023. |
| 2 | According to the Scopus database. Retrieved on 31 July 2023. |
| 3 | At the China National GeneBank DataBase (db.cngb.org/cnsa). |
| 4 | This is an accession identifier (ID) in the Chinese National Genomics Data Center (NGDC). |
| 5 | In the European Genome-Phenome Archive. |
| 6 | This is an accession ID in the Chinese National Genomics Data Center (NGDC). |
| 7 | This is an accession ID in the Sequence Read Archive (SRA). |
| 8 | These are libraries for single-cell T cell-receptor (TCR) sequencing (scTCR-seq), and TCR-FACS-index-ATAC sequencing (Ti-ATAC-seq). |
| Keyword | Species | Total | NGS | Microarray | ChIP-seq |
|---|---|---|---|---|---|
| SARS-CoV-2 | All | 808 | 642 | 22 | 52 |
| H. sapiens | 596 | 472 | 16 | 42 | |
| C. sabaeus | 14 | 10 | n/a | 1 | |
| M. musculus | 16 | 14 | n/a | 2 | |
| SARS-CoV-2 | 29 | 20 | n/a | n/a | |
| COVID-19 | All | 536 | 407 | 22 | 33 |
| H. sapiens | 536 | 407 | 22 | 33 | |
| C. sabaeus | 6 | 5 | n/a | 1 | |
| M. musculus | 10 | 9 | 1 | 1 | |
| SARS-CoV-2 | 19 | 14 | n/a | n/a |
| Year of publication and reference. | Specification of the biological model and sample type. | Main conclusions of the study. | (1) GEO ID. (2) PMID. (3) Dataset size. (4) Number of samples. |
(1) IF. (2) Citations 1. |
Platform. |
|---|---|---|---|---|---|
| 2020 [29]. | Human cell lines were infected in vitro with several common viruses. Specifically, H1N1 influenza A virus (IAV), human respiratory syncytial virus (RSV), human parainfluenza virus 3 (HPIV3), and SARS-CoV-2 were used to infect human lung cancer cell lines. | Transcriptional responses did not appear to stop the replication of SARS-CoV-2. Instead, they suggested a protracted inflammatory response. | (1) GSE147507. (2) PMID 32416070. (3) 2.7 Mb. (4) 110. |
(1) IF = 67. (2) 2771 citations. |
Illumina NextSeq 500 (H. sapiens). |
| 2020 [25]. | Immune cells isolated from life COVID-19 patients were profiled with a gene expression platform. | A comprehensive atlas of immune modulations during COVID-19 was presented. The authors identified three significant immunotypes related to disease severity. | (1) GSE152418. (2) PMID 32788292. (3) 1.6 Mb. (4) 34. |
(1) IF = 48. (2) 629 citations. |
Illumina NovaSeq 6000 (H. sapiens). |
| 2020 [26]. | PBMCs from life COVID-19 patients were profiled in a longitudinal approach. (PBMCs from thirteen patients were isolated, and there were five time points profiled for each patient.) In total, scRNA-seq data were obtained for 358,930 cells, and there were 10,900 cells per sample on average. | COVID-19 was associated with a set of dynamic changes in expression patterns in PBMCs. In particular, severe disease induced both hypoxic and pro-inflammatory signalling. There was also an impaired activation of the interferon pathway. | (1) GSE161777. (2) PMID 33296687. (3) 0.5 Mb. (4) 101. |
(1) IF = 43. (2) 189 citations. |
Illumina MiSeq andIllumina NovaSeq 6000 (H. sapiens). |
| 2020 [27]. | PBMCs from 7 hospitalized patients with COVID-19, four of whom had ARDS, as well as six health controls. | Strong downregulation of human leukocyte antigen (HLA) of class I and class II, and interferon-driven inflammatory reactions in monocytes are characteristics for infection with SARS-CoV-2. | (1) GSE150728. (2) PMID 32514174. (3) 372 Mb. (4) 13. |
(1) IF = 83. (2) 896 citations. |
Illumina NovaSeq 6000 (H. sapiens). |
| 2021 [30]. | Lung cells from dead COVID-19 patients were profiled and compared with normal cells. | Lungs from individuals with COVID-19 were very inflamed, and T cell responses were impaired. There were also fewer epithelial cells in diseased lungs. Instead, there was an increase in numbers of macrophages / monocytes, neuronal cells, and fibroblasts. | (1) GSE171524. (2) PMID 33915568. (3) 1601.4 Mb. (4) 27. |
(1) IF = 70. (2) 213 citations. |
Illumina NovaSeq 6000 (M. musculus). |
| 2021 [31]. | Blood, lungs, and airways of dead COVID-19 patients were profiled. | Results of the expression screen underlined the importance of the changes in expression of immunological genes. Note that not only structural cell-types like epithelia, but also infiltrating immune cells were the source of the signal. | (1) GSE147507. (2) PMID 33782412. (3) 1800.9 Mb. (4) 110. |
(1) IF = 5. (2) 75 citations. |
Illumina NextSeq 500 (H. sapiens). |
| 2021 [33]. | CSF was collected from 7 males and one female with neuro-COVID (aged from 53 to 82). | This study pointed at the expansion of dedifferentiated monocytes, as well as at exhausted CD4+ T cells in the CSF of patients with neuro-COVID. | (1) GSE26495. (2) PMID 33382973. (3) 83 Mb. (4) 16. |
(1) IF = 43. (2) 84 citations. |
Illumina NextSeq 500, or Illumina NovaSeq 6000 (H. sapiens). |
| 2021 [32]. | Cell culture of human pancreatic islets. | As a result of the infection of cultured pancreatic islets with the SARS-CoV-2 virus, the number of insulin-secretory granules in β-cells was reduced, and glucose dependent insulin secretion was impaired. Infection was associated with morphological, transcriptional and functional changes in β-cells of the islets. As a result, glucose-stimulated insulin secretion was impaired. | (1) GSE159717. (2) PMID 33536639. (3) 3.9 Mb. (4) 4. |
(1) IF = 21. (2) 318. |
HiSeq 4000 instrument (Illumina). |
| 2022 [28]. | The study investigated tissue samples isolated from COVID-19 patients. For example, blood samples were taken for transcriptome profiling from 21 patients. In total, 57,049 single-cell transcriptomes of PBMCs were sequenced. | Endothelial injury and pathological thrombotic events were frequently a part of clinical manifestations of COVID-19. These pathological changes were positively correlated with increased activity of myeloid cells. | (1) GSE208337. (2) PMID 35895716. (3) 573.4 Mb. (4) 105. |
(1) IF = 13. (2) 7 citations. |
Illumina NovaSeq 6000 (H. sapiens ). |
| 2022 [34]. | Conjunctival epithelium was isolated from eyes of dead patients and cultured as organotypic clusters. 15,821 cell transcriptomes from 3 infected, 3 uninfected cultures were processed. | There was evidence of the entry of the coronavirus into ocular epithelial cells. At the same time, there was no evidence of productive viral replication in this cell type. | (1) GSE191232. (2) PMID 35750043. (3) 78 Mb. (4) 7. |
(1) IF = 7. (2) 2 citations. |
| Year of publication and reference. | Specification of the biological model and sample type. | Main conclusions of the study. | (1) GEO ID. (2) PMID. (3) Dataset size. (4) Number of samples. |
(1) IF. (2) Citations 2. |
Platform. |
|---|---|---|---|---|---|
| 2020 [60]. | Five COVID-19 patients and three healthy people donated their blood for isolation of PBMCs. | An atlas of single-cell gene expression was generated in both COVID-19 and influenza patients. Three signaling pathways were turned on in COVID-19: apoptosis, Signal transducer and activator of transcription 1 (STAT1), and interferon regulatory factor 3 (IRF3). Plasma cells were increased in COVID-19. | (1) CNP0001102 3. (2) PMID 32783921.(3) n/a.(4) n/a. |
(1) IF = 43. (2) 206 citations. |
DNBelab C4 library and DIPSEQ T1 sequencer. |
| 2020 [36]. | PBMCs from five healthy donors and 13 patients with COVID-19 (this included moderate, severe and convalescent cases). | COVID-19 induced acute inflammatory response with a strong induction of the interferon-alpha pathway. There were also many cytotoxic effector T cells. However, there was also evidence for disorganized interferon response and immune exhaustion in severe cases of COVID-19. | (1) HRA000150 4. (2) PMID 32788748. (3) n/a. (4) 64. |
(1) IF = 31. (2) 356 citations. |
Illumina NovaSeq 6000 (H. sapiens), scRNA-seq. |
| 2020 [44]. | This is a single-cell analysis of broncho-alveolar immune cells from patients with COVID-19. Samples from 9 patients with COVID-19 and 3 healthy controls were sequenced. | A new protocol and a computational method were developed (Viral-Track) to detect infected cells. The method showed that SARS-CoV-2 had a detrimental effect on the host immune system. | (1) GSE145926. (2) PMID 32479746. (3) 225.1 Mb. (4) 21. |
(1) IF = 67. (2) 285 citations. |
Beijing Genomics Institute MGISEQ-2000 platform, scRNA-seq. |
| 2020 [37]. | PBMCs from 10 patients (either mild, severe, or control). | Subtle changes in percentages of subtypes of neutrophils or monocytes correlate with severity of COVID-19. | (1) E-MTAB-9221. (2) 32810439 PMID.(4) 10. |
(1) IF = 67. (2) 496 citations. |
10X Chromium droplet-based platform. Illumina NovaSeq 6000 (H. sapiens), scRNA-seq. |
| 2020 [63]. | PBMCs from 18 COVID-19 patients (8 mild and 10 severe) were collected between day 3 and day 20 after symptoms were first diagnosed. A total of 48,266 single-cell expression profiles of PBMCs were analyzed along with 50,783 control PBMCs from publicly available datasets. | There are major alterations in the myeloid compartment during a SARS-CoV-2 infection. Inflammatory monocytes are increased in mild cases. Dysfunctional monocytes are increased in severe cases, and there is rapid myelopoiesis in severe cases, resulting in the production of immature neutrophils. | (1) GSE145926. (2) PMID 32479746. (3) 225.1 Mb. (4) 21. |
(1) IF = 67. (2) 805 citations. |
10X Chromium droplet-based platform. Illumina NovaSeq 6000 (H. sapiens), scRNA-seq. |
| 2020 [59]. | Nasopharyngeal and bronchial samples from 19 COVID-19 patients with either moderate or critical disease. (There were also five healthy controls.) There were transcriptional profiles of 160,528 cells obtained from a total of 36 samples. The study identified 22 different cell types and states within the epithelial cell and immune cell populations. | In critical cases, there were stronger inflammatory interactions between epithelial and immune cells. Even more lung injury, respiratory failure, and inflammatory tissue damage was observed in critical cases. | (1) EGAS00001004481 5. (2) 32591762 PMID. (4) 36. |
(1) IF = 59. (2) 635 citations. |
Illumina NovaSeq 6000 (H. sapiens), scRNA-seq. |
| 2020 [39]. | PBMCs from hospitalized and non-hospitalized COVID-19 patients, and healthy non-exposed subjects, as well as PBMCs from subjects before and/or after receiving flu vaccination. | scRNA-seq conducted for CD4+ T cells from 40 COVID-19 patients showed that hospitalization resulted in increased fraction of cytotoxic follicular helper cells and cytotoxic T helper cells, as well as a reduction in regulatory T cells. | (1) GSE152522. (2) PMID 33096020. (3) 518 Mb. (4) 78. |
(1) IF = 67. (2) 300 citations. |
Illumina NovaSeq6000(H. sapiens), scRNA-seq. |
| 2021 [40]. | 52 samples of PBMCs were obtained from 13 patients with severe COVID-19. There were also 5 samples from 5 healthy donors. After cell sorting, there were 80,325 NKs obtained from the 68 samples of peripheral blood. From these NKs single-cell transcriptomes were generated. UMAP representation divided the transcriptomes into several types, for example effector I and II, terminally differentiated, transitional, and proliferating. | Severe COVID-19 is associated with early high serum levels of TGF-beta. High levels of this cytokine early in the course of infection are associated with the inhibition of interferon-driven activation of NKs, and impaired immunological response. | (1) GSE184329. (2) PMID 34695836. (3) 174.8 Mb. (4) 13. |
(1) IF = 65. (2) 104 citations. |
Illumina NextSeq500 (H. sapiens), scRNA-seq. |
| 2021 [47]. | Single-cell atlases of 24 lung, 19 heart, 16 liver, and 16 kidney tissue samples from COVID-19 autopsies were generated. | COVID-19 is characterized by failed tissue regeneration, and pathological re-modeling of diseased tissues. In the lungs, there were more fibroblasts and less epithelial cells in COVID-19 samples. The RNA of SARS-CoV-2 was enriched in phagocytic and ECs. In the heart, there was a reduction in the number of cardiomyocytes and pericytes. There was also more vascular ECs in the heart. | (1) GSE163530. (2) PMID 33915569. (3) 40.4 Mb. (4) 1194. |
(1) IF = 65. (2) 320 citations. |
NextSeq 550, Illumina NovaSeq 6000, NextSeq 550, Nanostring GeoMx 2020 Broad COVID Platform, scRNA-seq. |
| 2021 [46]. | Brain and choroid plexus samples isolated from dead patients with severe COVID-19 were profiled at a single cell level. (There were 65,309 high-quality nuclei.) | Considerable amount of evidence was presented suggesting that long-term inflammation is a part of both COVID-19 and neurodegenerative diseases. | (1) GSE159812. (2) PMID 34153974. (3) 944 Mb. (4) 30. |
(1) IF = 70. (2) 270 citations. |
Illumina NovaSeq 6000 (M. musculus), scRNA-seq. |
| 2021 [41]. | The study profiled COVID-19, MIS-C, as well as healthy pediatric and adult individuals using scRNA-seq. The results of scRNA-seq were correlated with disease severity, flow cytometry, antigen receptor repertoire analysis, and serum proteomics. | The study defined gene expression signatures associated with MIS-C, which could find application in clinical diagnostics to predict inflammatory complications of COVID-19. In particular, MIS-C tissues had increased expression levels of S100A-family alarmins, a signature of decreased antigen presentation, and increased cytotoxicity in NK and CD8+ T cells. | (1) GSE166489. (2) PMID 33891889. (3) 826.1 Mb. (4) 54. |
(1) IF = 43. (2) 118 citations. |
Illumina NovaSeq 6000 (H. sapiens), scRNA-seq. |
| 2021 [42]. | PBMCs were isolated from 5 healthy donors, 7 individuals who recovered from moderate disease, and 3 donors who recovered from severe COVID-19. The resulting 97.315 epigenomes of PBMCs were divided by UMAP representation into monocytes, effector, memory, naive, plasma cells, or NKs. | This study catalogued patterns of global re-modeling of the chromatin accessibility landscape in convalescing COVID-19 patients. These patterns suggested establishment of immunity against SARS-CoV-2 through immunological memory. | (1) HRA000562 6, PRJNA7180097. (2) PMID 34108657. (3) n/a. (4) n/a. |
(1) IF = 28. (2) 43 citations. |
Libraries8 were processed on HiSeq X Ten platform, Illumina. |
| 2021 [32]. | To understand the role of neutrophils during COVID-19 and to elucidate the effects of dexamethasone. | A single-cell atlas of COVID-19 neutrophil states and molecular mechanisms of action of dexamethasone was generated. See http://biernaskielab.ca/COVID_neutrophil. | (1) GSE157789. (2) 34782790. (3) 360 Mb. (4) 31. |
(1) IF = 53. (2) 90 citations. |
Sequencing was performed using Illumina NovaSeq S2 and SP 100. |
| 2022 [43]. | PBMCs from seven children with MIS-C (plus six healthy controls). | Immune responses by SARS-CoV-2 are regulated by long non-coding RNAs (lncRNAs). For example, lncRNA PIRAT forms a negative feedback loop with the PU.1 transcription factor, which promotes transcription of alarmins — that is proteins promoting inflammation in response to SARS-CoV-2. Inflammation in COVID-19 is promoted by down-regulation of PIRAT, and up-regulation of lung cancer associated transcript 1 (LUCAT1) — that is a lncRNA promoting inflammation. | (1) GSE142503. (2) PMID 35998224. (3) 16.7 Mb. (4) 15. |
(1) IF = 10. (2) 6 citations. |
Illumina NovaSeq 6000 (H. sapiens), scRNA-seq. |
| 2022 [38]. | 209 COVID-19 patients (plus 100 patients with post-acute COVID-19) and 457 healthy controls were investigated for 2 to 3 months: from initial diagnosis to convalescence. The patients were investigated at the time of clinical diagnosis, acute disease, as well as at the time of convalescence. | Diabetes, viremia, and auto-immune conditions were risk factors associated with the diagnosis of PASC. | (1) E-MTAB-10129. (2) PMID 32810438. (3) 5.2 Mb. (4) 309. |
(1) IF = 67. (2) 802 citations. |
Illumina NovaSeq 6000. |
| 2023 [48]. | The study focused on single-nucleus RNA-seq performed on samples of frozen lungs from 7 deceased COVID-19 patients, 6 pairs of lungs from patients with idiopathic pulmonary fibrosis (IPF), and 12 individuals from a control group. There were 38,794 cell nuclei from a vascular fraction, which could be fractioned into 14 different subtypes of endothelial type. There were also 38,794 non-vascular nuclei. The goal was to focus on ECs and on a comparison between IPF and COVID-19. | There was an enrichment of genes involved in cellular stress, and a signature of diminished immunomodulation, and impaired vessel integrity. There was also a set of receptor-ligand interactions that were specifically enriched or depleted in either COVID-19 or IPF. | (1) GSE159585. (2) PMID 35998078. (3) 488 Mb. (4) 53. |
(1) IF = 13. (2) 7 citations. |
Illumina HiSeq 4000 / NovaSeq 6000 (H. sapiens), scRNA-seq |
| Year of publication and reference. | Goal. | Conclusions. | Datasets. | Computational methods. | (1) IF. (2) Citations.(3) PMID. |
|---|---|---|---|---|---|
| 2020 [49]. | A comprehensive meta-analysis of scRNA-seq datasets to identify cell subsets expressing ACE2 and, therefore, targeted by SARS-CoV-2. | ACE2 and TMPRSS2 promote cellular entry of SARS-CoV-2. Type I interferons, and to a lesser extent type II interferons, upregulate ACE2. Cells vulnerable to infection were identified in the lungs. | GSE148829, GSE135069, GSE19190, GSE22147. | Re-analysis with Drop-Seq Computational Protocol v2.0, Seurat. Meta-analysis with UMAP, PCA, GSEA, etc. | (1) IF = 65. (2) 1574 citations. (3) PMID 32413319. |
| 2020 [50]. | To investigate the impact of smoking on COVID-19 utilizing scRNA-seq gene expression data from lung and airway epithelial samples from human, mouse, or rat. | For ACE2 levels of protein and mRNA were highly correlated (r = 0.82, P-value < 0.0001) across 53 cell lines. Smokers had higher levels of gene expression of ACE2 in the lungs. ACE2 expression was uncorrelated with age or sex. However, inflammation in the lungs was linked to the induction of expression of ACE2. ACE2 was also stimulated in expression by interferon-signalling. | GSE132040, GSE53960, GSE53960, GSE34378, GSE53960, GSE44555, GSE132040, GSE6591, GSE80680, GSE1643, GSE18344, GSE13933, GSE22047, GSE64614, GSE76925, GSE79209, GSE121611, GSE122960, GSE134174, GSE75715, GSE39059, GSE135188, GSE57148, GSE103174, GSE2052, GSE47460, GSE43696, GSE16538, GSE3100, GSE11056, GSE86623, GSE41789, GSE32138, GSE32138, GSE47963, GSE100504, GSE51392, and GSE19392 plus some samples from TCGA, the GTEx portal, the Human Cell Atlas, the Human Protein Atlas, and the Single-Cell Expression Atlas. | The meta-analysis was performed using Python, Excel, and Graphpad Prism. Regressions were performed using Python using ordinary least squares and the statsmodels package. Analysis of single-cell expression data was performed using Python, Scanpy, and Multicore-TSNE packages. Variable genes were prioritized utilizing the Seurat function in Scanpy. The variable genes were then analyzed using PCA. | (1) IF = 14. (2) 275 citations. (3) PMID 32425701. |
| 2020 [51]. | To determine expression patterns for ACE2 or other receptors for SARS-CoV-2 in the respiratory mucosa using scRNA-seq data. | The levels of mRNA and protein of ACE2 are very low in the upper airways and in the lungs. However, there is a mechanism dynamically regulating ACE2 expression in response to SARS-CoV-2. | GSE19190, GSE11906, GSE4302, GSE67472, GSE37147, GSE108134, GSE135893, the FANTOM5 dataset, and few proteomics datasets including the Human Proteome Map. | The Cell Ranger pipeline, UMAP, Zenbu genome browser, R packages: pheatmap, Seurat, ggplot2. | (1) IF = 25. (2) 107 citations. (3) PMID 32675206. |
| 2020 [53]. | scRNA-seq data were used to investigate how kidney diseases or medications may alter ACE2 expression in kidneys. | ACE2 expression in proximal tubular epithelial cells of the kidney facilitated infection with SARS-CoV-2. | Nephrocell data were archived at http://nephrocell.miktmc.org. COVID-19 kidney data were archived at https://hb.flatironinstitute.org/covid-kidney. | scRNA-seq data were analyzed according to protocols of the Kidney Precision Medicine Project (see https://www.kpmp.org/for-researchers#protocols). | (1) IF = 8. (2) 52 citations. (3) PMID 32675206. |
| 2021 [54]. | scRNA-seq data were used to identify cellular phenotypes shared across disparate inflammatory diseases. | Similarities in gene expression and signalling between COVID-19 and other inflammatory diseases are uncovered. | GSE134809, GSE122960, GSE145926, GSE155249, GSE47189, GSE147507, GSE168710, phs001457.v1.p1, phs001529.v1.p1, phs001457.v1.p1, SCP259. | A meta-analysis and integration pipeline was constructed, which models and removes the effects of technology, tissue of origin, and donor. The meta-analysis built a reference library for immune cells in normal body and in different diseases. | (1) IF = 12. (2) 83 citations. (3) PMID 33879239. |
| 2021 [52]. | Muus et al. performed a meta-analysis of receptor genes for SARS-CoV-2 by looking at gene expression in a meta-analysis of 31 lung scRNA-seq studies. | An atlas of cell type-specific associations of age, sex, and smoking with expression levels of ACE2 (and other co-receptors for SARS-CoV-2). | SCP865, SCP895, SCP891, SCP903, SCP871, SCP870, SCP874, SCP878, SCP887, SCP899, SCP898, SCP902, SCP894, SCP869, SCP872, SCP866, SCP1240, SCP1241, SCP868, SCP877, SCP867, SCP897, SCP886, SCP879, SCP860, SCP881, SCP875, SCP876, SCP900, SCP892, SCP890, SCP889, SCP880, SCP882, SCP882, GSE102592, GSE103918, GSE104600, GSE107747, GSE108571, GSE109037, GSE110973, GSE111014, GSE111360, GSE112570, GSE112845, GSE113036, GSE114530, GSE114724, GSE114802, GSE115149, GSE115189, GSE117211, GSE117403, GSE117824, GSE118127, GSE119212, GSE119506, GSE119507, GSE119561, GSE119594, GSE120446, GSE121267, GSE121600, GSE122342, GSE122703, GSE122960, GSE123926, GSE124263, GSE124334, GSE124472, GSE124494, GSE124898, GSE125680, GSE127472, GSE128066, GSE128169, GSE128518, GSE128889, GSE129845, GSE130073, GSE130117, GSE130151, GSE130238, GSE130318, GSE130430, GSE130888, GSE131685, GSE132802, GSE133704, GSE134809, GSE135618, GSE135929, GSE136103, GSE136314, GSE136394, GSE139249, GSE139324, GSE98201. | Data generated by Chromium instrument 10X were integrated using the Cell Ranger pipeline. Datasets were post-processed using Python harmony-pytorch (for batch correction and leiden clustering) of Scanpy. | (1) IF = 87. (2) 183 citations. (3) PMID 33654293. |
| 2021 [55]. | Chen et al. used cell-line and bulk-tissue RNA-seq to detect overlap between host transcriptional responses observed in cancer and those observed in COVID-19 in response to SARS-CoV-2. | There were many similarities in host-disease interactions between cancer and COVID-19. In particular, immune cell infiltration and inflammation. | GSE147507, GSE148729, GSE36969, GSE59185, GSE68820, GSE119856, GSE115770, GSE147507, GSE146507, GSE156063. | All data were public and derived either from GEO or from TCGA. Pathway analysis was performed using GO, wikiPathways, SigTerms software. Standard statistical methods were used, for example two-sided p-values, log-transformed gene expression values, FDR correction for multiple testing, and heat-maps. | (1) IF = 4.3. (2) 12 citations. (3) PMID 33510359. |
| 2021 [56]. | Garg et al. set up a multi-dataset that included samples from 9 scRNA-seq studies. | The multi-dataset consisted of 159 samples, deriving from 7 medical procedures focused on PBMCs and 2 focused on BALF. 8 out of 20 evaluated hypotheses were confirmed. | PRJCA002413, PRJCA002564, GSE149689, PRJCA002579, GSE150728, GSE145926, GSE147143, and EGAS00001004481. | The SCANPY protocol was used for the meta-analysis. The following steps were included: normalization of the datasets, log-transformation of the data, selection of genes variable in expression levels, and PCA. Harmony was used to integrate data from different samples. UMAP was then used to cluster, visualize, and annotate gene expression data by cell type. | (1) IF = 4.3. (2) 12 citations. (3) PMID 34675242. |
| Cell or tissue type | DEG genes | DEG pathways | Reference |
|---|---|---|---|
| PBMCs. | ISG15 ubiquitin like modifier (ISG15) ↑ Interferon induced protein 44 like (IFI44L) ↑ MX dynamin like GTPase 1 (MX1) ↑ XIAP associated factor 1 (XAF1) ↑ |
Pro-inflammatory cytokines ↑ Cytokine receptors ↑ Interferon-responsive TFs ↑ Response to type 1 interferon signaling ↑ Defense response to virus signaling ↑ Endoplasm and protein-unfolding ↑ Regulation of chromosome organization ↑ DNA conformation change ↑ |
[60]. |
| Thrombospondin 1 (THBS1) ↑ |
Neutrophil degranulation ↑ Plate activation, signaling and aggregation ↑ Semaphorin interactions ↑ Crosslinking of collagen fibrils ↑ Interleuking-1 family signaling ↑ Platelet degranulation ↑ |
[28]. | |
| lncRNA LUCAT1 ↑ CXCL2 ↑ IL-6 ↑ lncRNA PIRAT ↓ |
Hematopoietic cell lineage ↑ Cytokine-cytokine receptor interaction ↑ Chemokine signaling pathway ↑ |
[43]. | |
| Immunoglobulin heavy constant alpha 1 (IGHA1) ↑ Immunoglobulin heavy constant gamma 1 (IGHG1) ↑ Lactotransferrin (LTF) ↑ Interferon regulatory factor 1 (IRF1) ↑ Signal transducer and activator of transcription 3 (STAT3) ↑ Hypoxia inducible factor 1 subunit alpha (HIF1A) ↑ RAR related orphan receptor C (RORC) ↓ |
IL-1β and vasodilatory signaling ↑ IFN-related transcripts ↑ myeloid-cell-mediated immunity ↑ neutrophil degranulation ↑ erythroid cell differentiation ↑ cell differentiation pathway ↑ hypoxic signaling ↑ inflammation and IFN signaling ↑ ribosomal structural proteins ↓ |
[26]. | |
| HLA-DPB1 and HLA-DMA in monocytes ↓ |
Type I interferon-driven inflammatory signature in monocytes ↑ |
[27]. | |
| Interferon Induced Transmembrane Protein 2 (IFITM2) ↑ Interferon-induced transmembrane protein 3 (IFITM3) ↑ Interferon-induced protein 20 (ISG20) ↑ Interferon-induced protein 15 (ISG15) ↑ |
Interferon gamma response ↑ |
[72]. | |
| Interferon-ɑ response upregulation ↑ |
HLA-class II downregulation in CD14+ monocytes ↓ |
[56]. | |
| ECs. | Heat shock protein 90 alpha family class A member 1 (HSP90AA1) ↑ Heat shock protein family A (Hsp70) member 1A (HSPA1A) ↑ TIMP metallopeptidase inhibitor 1 (TIMP1) ↑ Fibrillin 1 (FBN1) ↑ Matrix metallopeptidase 16 (MMP16) ↑ Collagen type XV alpha 1 chain (COL15A1) ↑ Indoleamine 2,3-dioxygenase 1 (IDO1) ↑ Intercellular adhesion molecule 1 (ICAM1) ↓ Interferon regulatory factor 1 (IRF1) ↓ Cadherin 5 (CDH5) ↓ Integrin subunit beta 1 (ITGB1) ↓ Member of RAS oncogene family (RAP1B) ↓ Cell division cycle 42 (CDC42) ↓ Occludin (OCLN) ↓ Vinculin (VCL) ↓ Sphingosine-1-phosphate receptor 1 (S1PR1) ↓ Protein C receptor (PROCR) ↓ Thrombomodulin (THBD) ↓ |
Genes involved in cellular stress ↑ Heat shock proteins ↑ Genes involved in antigen presentation ↑ Hypoxia signalling ↑ Extracellular matrix (ECM) interactions ↑ ECM production/remodeling ↑ Immune system regulation ↓ Vessel maintenance/integrity ↓ Inflammation ↓ Angiogenesis ↓ Cell–cell adhesion ↓ Chemokines/cytokines ↓ TNF and JAK/STAT signalling ↓ |
[48]. |
| Lungs. | ACE2 ↑ |
Interferon response ↑ |
[49,50,51]. |
| ACE2 ↑ TMPRSS2 ↑ CTSL ↑ |
Interferon response ↑ |
[52]. | |
| Lung cancer cell lines. | Interferon Gamma Receptor 1 (IFNGR1) ↑ Interferon Gamma Receptor 2 (IFNGR2) ↑ Colony Stimulating Factor 2 (CSF2) ↑ Colony Stimulating Factor 3 (CSF3) ↑ C-X-C Motif Chemokine Ligand 1 (CXCL1) ↑ C-X-C Motif Chemokine Ligand 2 (CXCL2) ↑ Interleukin 1 Alpha (IL1A) ↑ Interleukin 1 Beta (IL1B) ↑ Interleukin 6 (IL6) ↑ Tumor Necrosis Factor Superfamily Member 14 (TNFSF14) ↑ |
Type II interferon signaling ↑ Immune response ↑ Response to stress ↑ Cytokine-mediated signaling ↑ Inflammatory response ↑ Cytokine activity ↑ Extracellular space ↑ Growth factor receptor binding ↑ Response to virus ↑ |
[55]. |
| Kidney. | ACE2 ↑ |
Interferon response ↑ |
[53]. |
| Effects on DEGs. | References. |
|---|---|
| Increased interferon response, interferon signaling, interferon-responsive TFs. | [26,27,49,50,51,52,53,55,60,72]. |
| Increased immune or inflammatory responses. | [27,55,60]. |
| Increased expression of cytokines, chemokines, or receptors. | [43,48,55,60]. |
| Increased interleukin-1 family signaling. | [28,55]. |
| Increased hypoxic signalling. | [26,48]. |
| Diminished immune system regulation, angiogenesis, and vessel integrity. | [48,56]. |
| Tissue type. | Reference. | Cell type increasing in frequency in severe COVID-19. | Less common cell type. |
|---|---|---|---|
| PBMCs. | [56]. | Plasma cells. Increased B-cell clonal expansion. |
Regulatory T cells. |
| [60]. | Plasmablasts. | Lymphocytes. | |
| [72]. | Plasmablasts. | n/a | |
| [28]. | Myeloid cells. | n/a | |
| [27]. | Developing neutrophils.CD14+ monocytes.Plasmablasts. | CD16+ monocytes.Plasmacytoid dendritic cells, conventional dendritic cells, and NK cells. | |
| [39]. | Cytotoxic follicular helper cells, and cytotoxic T helper cells. | Regulatory T cells. | |
| [73]. | Highly cytotoxic NK cells containing high levels of cytotoxic proteins such as perforin. | Unarmed NK cells. | |
| [70]. | Activated macrophages. | n/a | |
| BALF. | [44]. | Recruited macrophages, monocytes, or neutrophils. | Alveolar macrophages. |
| [70]. | Neutrophils. | Basal epithelial cells. | |
| CSF. | [33]. | De-differentiated monocytes, and CD4+ T cells. | n/a |
| Lungs. | [30]. | Fibroblasts, myeloid and neuronal cells. | Antigen presenting cells, epithelia. |
| Tissue type. | Positive cell types. | Type of evidence. | Computational method. | Reference. |
|---|---|---|---|---|
| If detected, transcription of the viral genome in swabs or brush specimens was used to confirm the diagnosis of COVID-19. | n/a | The entire SARS-CoV-2 genome sequence was annotated as one viral ‘gene’. The viral gene was appended to the hg19 annotation gtf file. All reads aligning to the SARS-CoV-2 genome per sample were aggregated and divided by the total number of reads in that sample. | Transcripts were aligned to a customized reference genome in which the SARS-CoV-2 genome (Refseq-ID: NC_045512) was added as an additional chromosome to the human reference genome hg19. Viral load was calculated on the raw data matrices output by CellRanger. | [59]. |
| BALF from severe and mild COVID-19 patients. | Epithelial cells and macrophages. | Viral mRNAs were identified among scRNA-seq reads that did not map to the human genome. | Viral-Track was an R-based pipeline that utilized the STAR algorithm to align reads to the SARS-CoV-2 genomes. | [44]. |
| BALF from two severe COVID-19 patients [69]. | Epithelial cells, lymphocytes, macrophages, neutrophils. | Yeskit integrates host gene expression profiles with virus detection. | R and Python-based packages utilizing the STAR algorithm. Yeskit was an R package for data integration, clustering, identification of DEGs, functional annotation, and visualization. | [74]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).




