1. Introduction
Hepatitis E virus (HEV) is a small enveloped or quasi-enveloped virus with an icosahedral capsid. Its genome is a single-stranded positive sense RNA with a ~7.2 bp size, expressing 3 open reading frames (ORFs): ORF1, ORF2, and ORF3 [
1,
2]. It is recognized as a biological hazard that must be controlled and is an etiological agent in viral hepatitis [
3]. According to the World Health Organization (WHO), HEV is a public health concern worldwide. It causes an estimated 20 million of infections per year, leading to 3,3 million of symptomatic cases and 44,000 deaths, accounting for 3.3% of the mortality due to viral hepatitis. Vulnerable populations for hepatitis E infection include immunosuppressed people and pregnant women [
1,
4]. HEV is mainly transmitted via fecal oral routes, blood transfusion, the environment through contaminated crops and exposure to wastewater from toilets or via floods in raining season [
5]. Eight HEV genotypes divided into several subtypes (HEV-1 to HEV-8) have been described, of which four main HEV genotypes are known to infect humans [
6]. Genotypes 1 and 2 infecting only humans are transmitted through fecal–oral routes and are responsible for large HEV outbreaks in resource limited countries. In contrast, genotypes 3 and 4 circulating primarily among mammalian animals are responsible for zoonotic transmission [
6,
7,
8,
9]. In industrialized countries and in areas with good sanitation and water supply, only sporadic cases occur caused by genotype 3 acquired through zoonotic HEV infection by ingestion of contaminated foodstuffs and particularly by eating undercooked contaminated meat (pig flesh, raw liver, sausages, etc.) [
5,
10]. However, in developing countries, most of studies reported poor sanitation and bad water supply as the main factor risk associated with HEV infection [
1,
3,
11].
HEV is a zoonotic pathogen which has a large host range, being found in humans, as well as in a wide range of domestic and wild mammals (pig, wild boar, cow, deer, rabbit, camel) [
12]. The domestic pig (
Sus scrofa domesticus) is an important reservoir host of HEV, and is a source of contamination for the consumer after consuming of raw or undercooked pork products [
13,
14]. The liver is the target organ for HEV and is where it replicates, but it was also detected in several other tissues [
15]. Infected pigs are usually without any apparent clinical symptoms, although in some cases, mild to moderate, acute, self-limiting hepatitis can occur [
15,
16]. Consequently, HEV-infected pigs enter the slaughterhouse as healthy animals; hence, their tissues and meat can pose a risk to human consumers. Pork liver and its derivatives are more frequently found to be positive for HEV-RNA and therefore are the most obvious sources of foodborne HEV [
15,
17,
18,
19]. Many studies reported detection of HEV in 1.3%, 1.9%, 11% and 6.5% of pig livers sold in retail stores in England, Japan, the USA and The Netherlands respectively [
20,
21,
22,
23]. On the other hand, several reports describe human cases of Hepatitis E associated with foodborne transmission involving products containing undercooked raw pork liver [
22,
24,
25,
26]. Furthermore, the virus has also been found in muscle tissue, which could be a source of human infection [
27].
Indeed, in African countries, few studies focused on HEV as a zoonotic and a foodborne pathogen. On the other hand, HEV problematic in foodstuffs mainly in domestic fauna is poorly covered in West Africa particularly in Senegal, in which we note the lack of data related to the national prevalence of HEV. In view of all the above-mentioned aspects, it is becoming necessary to elucidate the possible sources of zoonotic HEV contamination in Senegal in order to understand the risks associated with the consumption of pork products. The study aimed to assess the presence of HEV in pork meat and liver sold in retail in Saint-Louis.
2. Materials and Methods
2.1. Sampling
This one-site study is part of an HEV molecular surveillance program in Senegal using a One Health approach. It represents the phase II of a pilot study conducted by Diouara et
al., and focusing on the HEV seroprevalence and associated risks factors in pregnant women in Senegal [
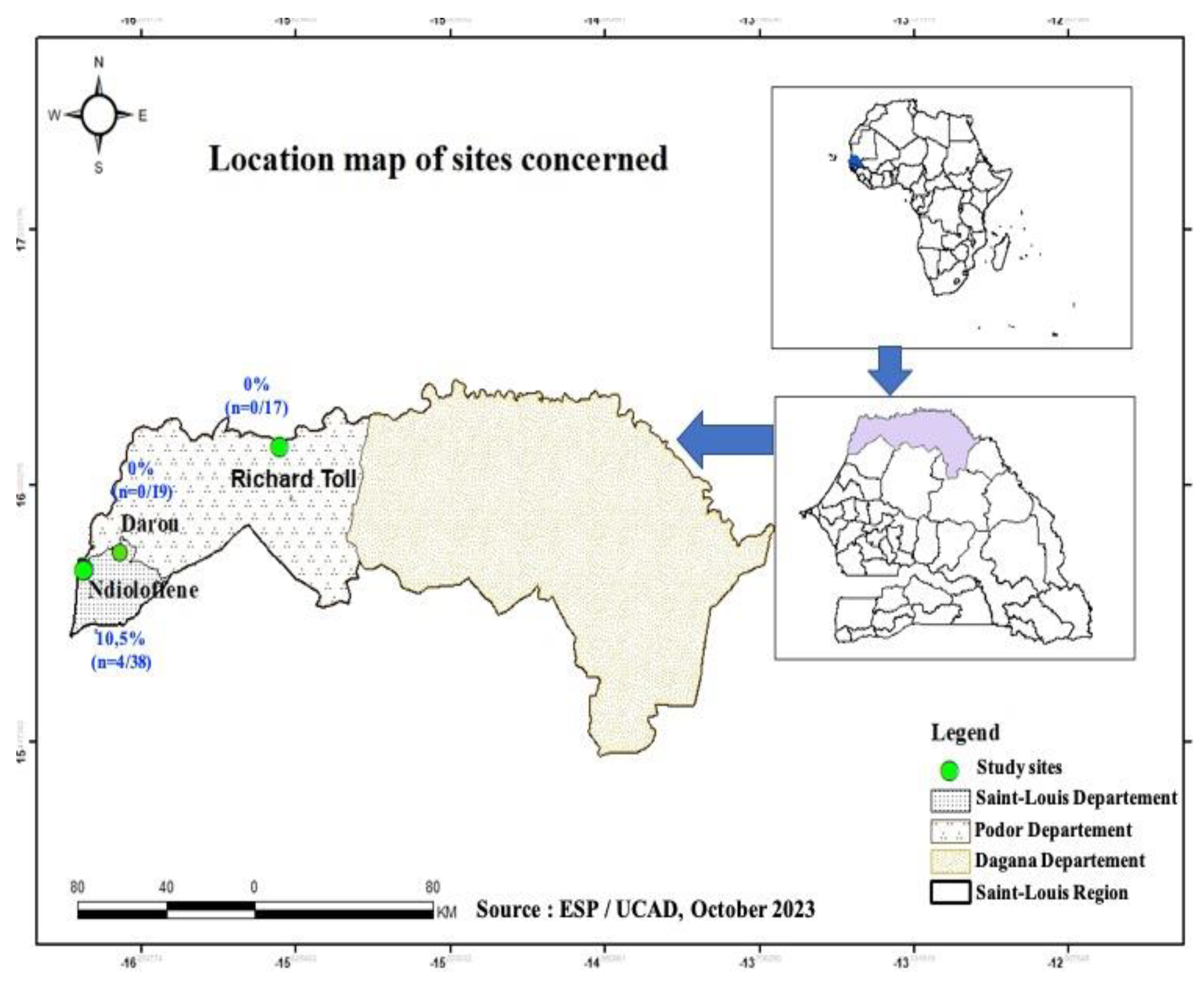
1]. In this present study, samples of pork meat and liver were collected between 20 and 25 August 2022 in three locations, Ndioloffène, Darou and Richard-Toll all in Saint-Louis region (
Figure 1). Pork is often sold there in houses or bars. The latter are well known to consumers, so these sites were chosen on the basis of their reputation, their capacity to receive consumers daily, and pork availability during our sampling period.
Samples were obtained by purchasing approximately 250 g of pork meat and/or liver, with a minimum of 3 and a maximum of 5 samples per vendor to ensure good repre-sentativeness and according to the recommendations for sampling food matrices [
28]. Samples were then placed in individual sterile zipped coded bags and transported to the laboratory in isothermal transport bags maintained between 4 – 8° C to preserve the integrity of products in accordance with validated procedures [
1,
29]. Once in the laboratory, the samples were stored in the GRBA-BE biobank at -20°C until their use.
2.2. Extraction of Total Nucleic Acid (TNA)
We minced 2 g of each sample using sterile bistoury blades under the Class II Biosafety Cabinet. This mass was suspended in sterile physiological water (9% NaCl) and then vigorously vortexed until a homogeneous paste was obtained. This was followed by centrifugation at 3000 rpm for 10 minutes to remove tissue debris. The supernatant containing any viral particles was then used for nucleic acids extraction. TNA were extracted with the Purelink™ Viral DNA/RNA Mini kit (Invitrogen, Life Technologies™) according to the manufacturer's instructions. Elution volume were 50 μL and TNA samples were stored at -80°C until further analysis.
2.3. RT-PCR
For HEV detection, we targeted the ORF2 genomic region with the specific primers pair HE040 (5'-CCCTTRTCCTGCTGAGCRTTCTC-3' [R=A or G]) and HE044 (5'-CAAGGHTGGCGYTCKGTTGAGAC-3' [H=A, T or C; Y=T or C; and K=G or T]) as described by
Mizuo et al., [
30]. One-step RT-PCR was performed using the One-Taq™ One-Step RT-PCR kit (New England Biolabs) with the following thermocycling program: 48°C for 25 min for the reverse transcription step; an initial denaturation step of 2 min at 94°C followed by 40 cycles of 94°C for 15 s; 55°C for 30 s and 68°C for 1 min, and a final extension step of 5 min at 68°C. The amplicons obtained were then visualized by electrophoresis on a 1% agarose gel with an expected size of 506 bp. The manipulation was validated when the positive control showed a band at the expected size and when the negative control showed no band.
2.4. Sequencing and phylogenetic analysis
Oxford Nanopore Technologies (ONT) sequencing libraries were prepared from ORF2 genomic region amplicon using the Rapid barcoding sequencing (SQK-RBK110.96) kit according to the manufacturer's protocol; sequencing was undertaken on the MinIon MK1C device with R9.4.1(FLO-MIN 106) flowcells for 5 h. A minimal Qscore of 7 were considered. At the end, the raw fastQ files were recovered for sequence analysis.
For the phylogenetic analysis, partial sequences of HEV ORF-2 capsid (started to position 5895 and ends at position 6382 relative to NC_001434 reference sequence) were subject to multiple sequence alignment with MUSCLE and gap positions removed by using Gblocks program on SEAVIEW software v5.0.4 [
31]. A set of HEV reference sequences downloaded from GenBank was used for this purpose. Phylogenetic trees were inferred by the Maximum Likelihood method with PhyML v3.1 [
32]. The tree was generated under the best-fit nucleotide substitution model GTR + G + I determined by MODELTEST in MEGA X [
33]. The Subtree-Pruning-Regrafting heuristic search was applied for optimal tree topology. Branch supports determined by the approximate likelihood ratio test method (aLRT) SH-like option [
34]. Phylogenetic tree was read and edited with Figtree [
35].
2.5. Statistical analysis
Data obtained were analyzed using Excel and R version 4.2.3 (2023-03-15). Positivity rates of HEV-RNA were calculated and 95% confidence intervals were obtained using proportionality tests. In order to measure possible associations, Fisher's exact test was performed, and a difference was considered statistically significant when the p-value was less than 0.05.
3. Results
In this study, samples were collected in 3 locations in Saint-Louis, namely: Ndioloffène, Darou and Richard-Toll. The
Figure 1 shows the sampling sites.
A total of 74 samples were collected at Saint-Louis in the following distribution: 65 pork meat samples and 09 pork liver samples. The table 1 shows the distribution of samples by collection site and by sample type. All pig liver samples (n=9) collected in this study came from Ndioloffène. As for the pork meat samples, n=29, n=17 and n=19 were collected at Ndioloffène, Richard Toll and Darou respectively (
Table 1,
Figure 1).
The RT-PCR results showed an overall HEV-RNA prevalence of 5.4% (n=4); 95% CI [1.7%-14%] in pork products. According to the sample type, pig meat and liver samples show 3.1%; 95% CI [0.53%-11.6%] and 22.2%; 95% CI [3.9%-59.8%] of HEV-RNA positivity respectively. In our study, while higher HEV-RNA positivity rate was found in pork liver samples, no statistically significant association was found with the types of sample (meat or liver) analysed (p-value = 0.0699) (
Table 2). It should be noted that all HEV-RNA positive samples were collected at Ndioloffène i.e., a positivity rate of 10.5% (n=4); 95% CI [3.4%-25.7%] when considering the only site. No HEV positivity was found in samples collected at Darou and Richard-Toll (
Table 3). Statistical analysis reveals no significant difference between the collection site and prevalence of HEV in pork products screened (p-value=0.1698).
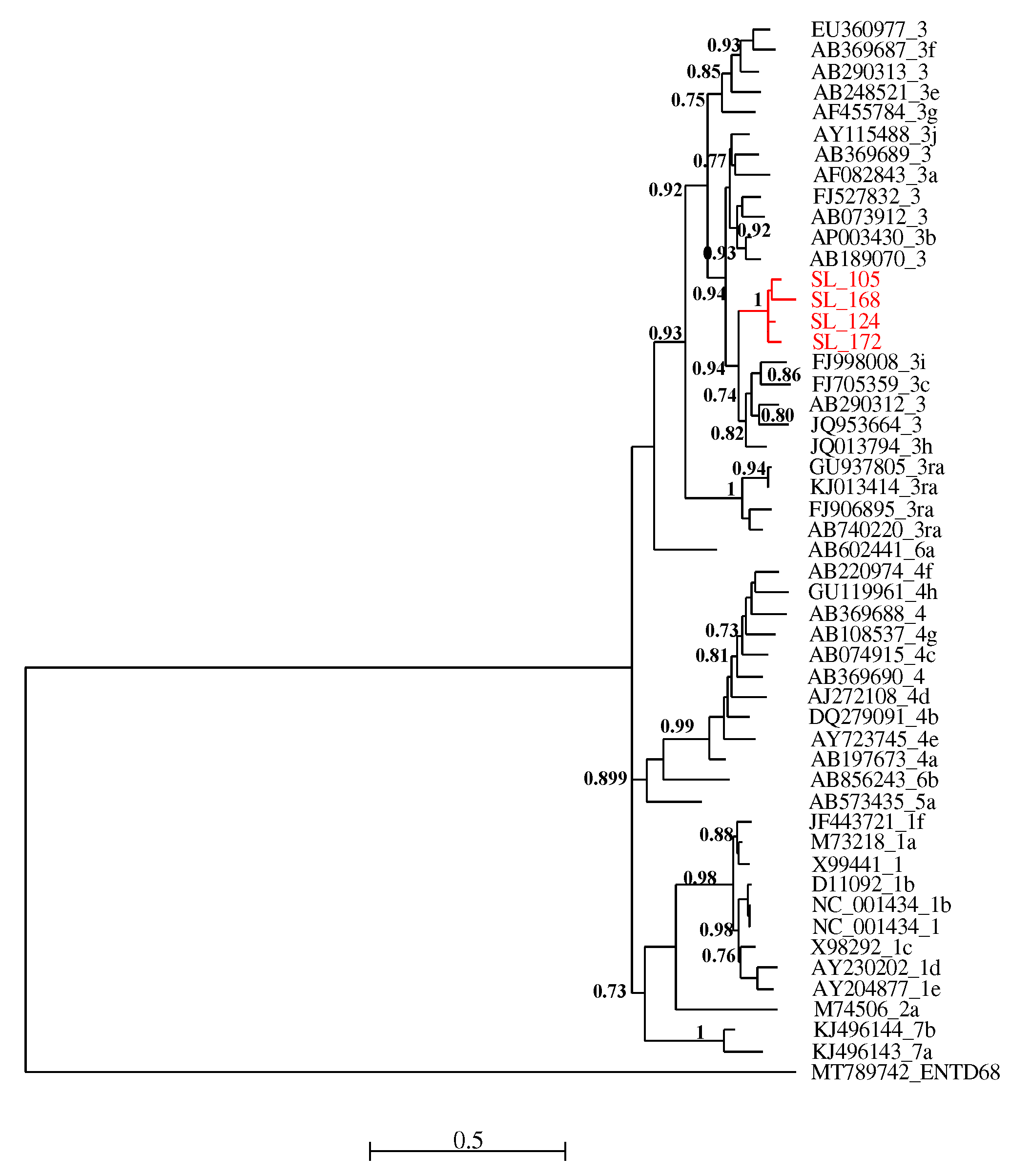
All four RT-PCR-positive samples were successfully sequenced and analyzed with HEV reference sequences. Phylogenetic relationships showed that these new viral strains belonged to genotype 3 and clustered together with maximum aLRT value = 1.
Figure 2.
Maximum likelihood (ML) tree [based on the genomic region 5895 – 6382 relative to NC_001434 reference sequence] showing the relationships between newly HEV generated partial sequences (ORF-2) isolates from pigs in Senegal (red font) and reference sequences (black font). The tree was constructed using 47 isolates (with tags: Accessions number_genotypes) under the GTR nucleotide substitution model with 4 Gamma categories using PhyML [
32] on seaview software v5.0.4 [
31]. SH-like branches support > 0,70 are indicated.
Figure 2.
Maximum likelihood (ML) tree [based on the genomic region 5895 – 6382 relative to NC_001434 reference sequence] showing the relationships between newly HEV generated partial sequences (ORF-2) isolates from pigs in Senegal (red font) and reference sequences (black font). The tree was constructed using 47 isolates (with tags: Accessions number_genotypes) under the GTR nucleotide substitution model with 4 Gamma categories using PhyML [
32] on seaview software v5.0.4 [
31]. SH-like branches support > 0,70 are indicated.
4. Discussion
This study aimed to assess the presence of HEV in pork meat and pork liver samples sold in retail in Saint-Louis, Senegal. This is the first report stating the detection of HEV RNA in meat products from domestic fauna intended for human consumption in Senegal. The results reveal the presence of HEV-RNA in 5.4%; 95% CI [1.7%-14%] of pork products (meat and liver samples) collected in the Saint-Louis region. This rate is high compared to what was reported by Modiyinji et
al. in their well conducted review paper. Authors found a specific prevalence of 3.5% in pigs while the overall prevalence of HEV-RNA observed in animals (n=6983, including 13 species) was 1.8% in Africa [
36]. According to sample types, the highest prevalence was obtained from pig liver samples (22.2%) vs 3.1% in pig meat samples. This observation is not surprising since the liver is known to be the target organ and the site of HEV replication [
15].
Although no statistically significant association was found between HEV-RNA positivity and type of sample, several studies have shown a higher prevalence of HEV in pig liver samples comparatively to other food matrix [
18,
22,
23,
37,
38,
39,
40]. This confirms the existence of a zoonotic reservoir of HEV in the pig population. According to data from dedicated literature, many human HEV cases in industrialized countries are related to the consumption of so-called "high-risk" products, i.e. pork products consumed raw or not well cooked and containing a high proportion of pork liver [
38]. Consumers should be concerned that pig liver can be contaminated with HEV and carry a risk of infection if it is not well cooked, and this emphasizes that pig liver and pork products must be controlled before entering in the chain food [
14]. This would limit the spread of HEV as a foodborne pathogen. In this study, the overall prevalence of HEV RNA in pork meat samples collected (3.1%) is high when compared to several other studies screening the same matrix [
22,
39,
40]. However, Di Bartolo et
al. reported a similar rate that was found in the present study, i.e., 3% of HEV-RNA positive samples from Czech Republic out of 112 pork meat sampled at slaughterhouses from Czech Republic, Italy, and Spain [
41]. However, the authors suggested that the presence of the HEV genome was probably due to cross-contamination during slaughtering rather than real virus replication in muscle. The presence of HEV-RNA in pork products probably reflects endogenous HEV particles in infected liver and/or viremic blood [
42]. This suggests that there is less risk of contamination when eating pork meat.
In view of these results, our study reveals a possible circulation of HEV in domestic fauna in Saint-Louis. In addition, our previous study conducted among pregnant women showed an overall high HEV seroprevalence of 7.8% in Senegal, with the specific HEV-IgG and HEV-IgM rates of 10.5% and 0.5% respectively in Saint-Louis [
1]. Therefore, our findings support the need for HEV surveillance using a One Health approach, taking into account the environment, human and animals’ compartments. Considering HEV as a model of zoonotic pathogen affecting these different compartments, more in-depth and integrated investigations will be relevant to mitigate the associated risks of emergence.
Phylogenetic analysis of the isolated sequences pointed out an exclusive presence of genotype 3. Similar results have been reported in Cameroon [
43], in Ghana [
44] and in Burkina Faso [
45]. Domestic pigs (
Sus scrofa domesticus) represent the most important reservoir of the zoonotic genotypes HEV-3 and HEV-4 [
46]; and several studies have also reported the global distribution of genotype 3 [
47]. In many cases, the identification of these genotypes was also made on ORF2 sequences [
43], but in shorter sizes. In Senegal, as in neighboring countries, these are the first data describing the presence of HEV in food matrices, particularly meat products. HEV genetic diversity data previously known was limited to genotype 2b found in humans [
48].
Given that samples were collected according to their availability in Saint-Louis, the main limitation of this study remains the sample size, especially the pork liver samples . In addition, we were limited by the number of collection sites due to the fact that there are no public pork slaughterhouses in Saint-Louis and that pork is slaughtered and sold in detail in clandestine houses or bars without any regulation or control. These places are known through a very limited network and are little known by the population as a whole, which is the reason why it is rare to find pork sold in retail in Saint-Louis. This could be explained by the fact that the majority of the Senegalese are Muslims, a context in which pig farming is poorly regarded, especially as the consumption of pork in Senegal represents 15% of white meat and is mainly consumed by non-Muslims (representing less than 4% of the population) and expatriates [
49,
50].
Another limitation of this study was the ML tree was drawn based on the 485-nt region which was not sufficient for phylogeny reconstruction using 51 representative isolates. Thus, the characterization of the complete genomes of the isolated strains could contribute substantially to documenting the HEV genotypes circulating in Senegal. Furthermore, this pioneering study on HEV carried out on food products would have made it possible to isolate the first HEV-3 sequences of swine origin in Senegal.
5. Conclusion
These preliminary results suggest the potential circulation of HEV in domestic fauna with a contamination rate of 5.4% for pork meat and liver sold in Saint-Louis, Senegal. Therefore, consumption and handling of raw or undercooked contaminated pork meat or pork liver can lead to sporadic cases of HEV infection. On the other hand, the high level of contamination (22.2%) in the pork liver samples highlight a major risk associated with their consumption. Hence, there is a need to inform local communities of the potential circulation of HEV in domestic wildlife. Taking into account these new data, strategies to raise awareness and prevent HEV infection and its spread across the country will have to be implemented. Our results stress the need for a surveillance system that extends this investigation to national scale while increasing the number of sampling sites and analyzed products. This will ensure continuous monitoring and control of the quality of pork products from farm to fork. A routine HEV surveillance program should be implemented in domestic fauna and among the general population to better known its genetic diversity and circulation in Senegal.
Author Contributions
Conceptualization, designing and funding acquisition, A.A.M.D; supervision, A.A.M.D. and M.M.; sampling: S.D.T., S.S., S.C., A.A.M.D. and A.K.; formal analysis, S.D.T., S.S. and S.C.; validation, A.A.M.D., writing—original draft preparation, S.D.T. and A.A.M.D.; revision of the manuscript: A.A.M.D., S.D.T., A.K., S.S., S.C., F.T., C.M.N., M.D., M.N.M., M.M., S.L., H.D.N., C.T.K., A.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the International Development Research Centre (IDRC) and the Ministry of Higher Education, Research and Innovation (MESRI), which contributed to the funding of this project through FIRST, as part of the SGCI2 funding program.
Institutional Review Board Statement
This study protocol has received the approval of the Comité National d'Éthique pour la Recherche en Santé (CNERS) (N° 00000193MSAS/CNERS/SP) prior to its execution.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
We are grateful of the top management of the Ecole Supérieure Polytechnique (ESP) for their support to the GRBA-BE team.
Conflicts of Interest
The authors declare no conflict of interest.
References
- A. A. M. Diouara et al., « Hepatitis E Virus Seroprevalence and Associated Risk Factors in Pregnant Women Attending Antenatal Consultations in Senegal », Viruses, vol. 14, no 8, p. 1742, août 2022. [CrossRef]
- Y. Debing, D. Moradpour, J. Neyts, et J. Gouttenoire, « Update on hepatitis E virology: Implications for clinical practice », J. Hepatol., vol. 65, no 1, p. 200-212, juill. 2016. [CrossRef]
- J. Wang, N. Li, H. Zhang, F. Li, S. Fanning, et T. Jiang, « Detection of Hepatitis E Virus in the Pig Livers and Retail Pork Samples Collected in Selected Cities in China », Foodborne Pathog. Dis., vol. 18, no 2, p. 97-103, févr. 2021. [CrossRef]
- WHO, « Hepatitis E ». Consulté le: 11 août 2023. [En ligne]. Disponible sur: https://www.who.int/news-room/fact-sheets/detail/hepatitis-e.
- G. R. Takuissu et al., « Hepatitis E Virus in Water Environments: A Systematic Review and Meta-analysis », Food Environ. Virol., vol. 14, no 3, p. 223-235, sept. 2022. [CrossRef]
- D. B. Smith et al., « Update: proposed reference sequences for subtypes of hepatitis E virus (species Orthohepevirus A) », J. Gen. Virol., vol. 101, no 7, p. 692-698, juill. 2020. [CrossRef]
- M. A. Fatawou, M. G. Chavely, M. Y. M. Henri, K. N. Daniel, E. Z. M. Claire, et N. Richard, « First Detection and Characterization of Hepatitis E Virus in Sewage Samples in Cameroon », Food Environ. Virol., vol. 15, no 3, p. 255-261, sept. 2023. [CrossRef]
- I. Nimgaonkar, Q. Ding, R. E. Schwartz, et A. Ploss, « Hepatitis E virus: advances and challenges », Nat. Rev. Gastroenterol. Hepatol., vol. 15, no 2, Art. no 2, févr. 2018. [CrossRef]
- W. H. Van der Poel, « Food and environmental routes of Hepatitis E virus transmission », Curr. Opin. Virol., vol. 4, p. 91-96, févr. 2014. [CrossRef]
- N. Pavio, P. Kooh, V. Cadavez, U. Gonzales-Barron, et A. Thébault, « Risk factors for sporadic hepatitis E infection: a systematic review and meta-analysis », Microb. Risk Anal., vol. 17, p. 100129, avr. 2021. [CrossRef]
- A. Shrestha et al., « Prevalence and risk of hepatitis E virus infection in the HIV population of Nepal », Virol. J., vol. 14, no 1, p. 228, nov. 2017. [CrossRef]
- M. A. Purdy et al., « ICTV Virus Taxonomy Profile: Hepeviridae 2022 », J. Gen. Virol., vol. 103, no 9, sept. 2022. [CrossRef]
- J. E. Forero et al., « Phylogenetic analysis of Hepatitis E virus strains isolated from slaughter-age pigs in Colombia », Infect. Genet. Evol., vol. 49, p. 138-145, avr. 2017. [CrossRef]
- N. Thippornchai et al., « Survey of hepatitis E virus in pork products and pig stools in Nakhon Pathom Province, Thailand », Vet. Med. Sci., vol. 8, no 5, p. 1975-1981, mai 2022. [CrossRef]
- L. Milojević et al., « Screening and Molecular Characterization of Hepatitis E Virus in Slaughter Pigs in Serbia », Food Environ. Virol., vol. 11, no 4, p. 410-419, déc. 2019. [CrossRef]
- X. J. Meng, « Hepatitis E virus: Animal reservoirs and zoonotic risk », Vet. Microbiol., vol. 140, no 3, p. 256-265, janv. 2010. [CrossRef]
- M. Andraud et al., « Direct contact and environmental contaminations are responsible for HEV transmission in pigs », Vet. Res., vol. 44, no 1, p. 102, oct. 2013. [CrossRef]
- P. Colson et al., « Pig liver sausage as a source of hepatitis E virus transmission to humans », J. Infect. Dis., vol. 202, no 6, p. 825-834, sept. 2010. [CrossRef]
- A. M. I. Montone et al., « Occurrence of HEV-RNA in Italian Regional Pork and Wild Boar Food Products », Food Environ. Virol., vol. 11, no 4, p. 420-426, déc. 2019. [CrossRef]
- M. Bouwknegt, F. Lodder-Verschoor, W. H. M. Van Der Poel, S. A. Rutjes, et A. M. De Roda Husman, « Hepatitis E Virus RNA in Commercial Porcine Livers in The Netherlands », J. Food Prot., vol. 70, no 12, p. 2889-2895, déc. 2007. [CrossRef]
- A. R. Feagins, T. Opriessnig, D. K. Guenette, P. G. Halbur, et X.-J. Meng, « Detection and characterization of infectious Hepatitis E virus from commercial pig livers sold in local grocery stores in the USA », J. Gen. Virol., vol. 88, no 3, p. 912-917, 2007. [CrossRef]
- C. Feurer et al., « High load of hepatitis E viral RNA in pork livers but absence in pork muscle at French slaughterhouses », Int. J. Food Microbiol., vol. 264, p. 25-30, janv. 2018. [CrossRef]
- Y. Yazaki et al., « Sporadic acute or fulminant hepatitis E in Hokkaido, Japan, may be food-borne, as suggested by the presence of hepatitis E virus in pig liver as food », J. Gen. Virol., vol. 84, no 9, p. 2351-2357, 2003. [CrossRef]
- T.-C. Li et al., « Hepatitis E Virus Transmission from Wild Boar Meat », Emerg. Infect. Dis., vol. 11, no 12, p. 1958-1960, déc. 2005. [CrossRef]
- C. Renou, A.-M. R. Afonso, et N. Pavio, « Foodborne Transmission of Hepatitis E Virus from Raw Pork Liver Sausage, France », Emerg. Infect. Dis., vol. 20, no 11, p. 1945-1947, nov. 2014. [CrossRef]
- M. Riveiro-Barciela, B. Mínguez, R. Gironés, F. Rodriguez-Frías, J. Quer, et M. Buti, « Phylogenetic demonstration of hepatitis E infection transmitted by pork meat ingestion », J. Clin. Gastroenterol., vol. 49, no 2, p. 165-168, févr. 2015. [CrossRef]
- A. Rivero-Juarez et al., « Familial Hepatitis E Outbreak Linked to Wild Boar Meat Consumption », Zoonoses Public Health, vol. 64, no 7, p. 561-565, 2017. [CrossRef]
- ISO 17604:2015, « Microbiology of the food chain — Carcass sampling for microbiological analysis ». Consulté le: 11 décembre 2023. [En ligne]. Disponible sur: https://www.iso.org/obp/ui#iso:std:iso:17604:ed-2:v1:en.
- A. A. M. Diouara et al., « Antiretroviral treatment outcome in HIV-1-infected patients routinely followed up in capital cities and remote areas of Senegal, Mali and Guinea-Conakry », J. Int. AIDS Soc., vol. 17, no 1, p. 19315, 2014. [CrossRef]
- H. Mizuo et al., « Polyphyletic Strains of Hepatitis E Virus Are Responsible for Sporadic Cases of Acute Hepatitis in Japan », J. Clin. Microbiol., vol. 40, no 9, p. 3209-3218, sept. 2002. [CrossRef]
- M. Gouy, S. Guindon, et O. Gascuel, « SeaView Version 4: A Multiplatform Graphical User Interface for Sequence Alignment and Phylogenetic Tree Building », Mol. Biol. Evol., vol. 27, no 2, p. 221-224, févr. 2010. [CrossRef]
- S. Guindon, J.-F. Dufayard, V. Lefort, M. Anisimova, W. Hordijk, et O. Gascuel, « New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0 », Syst. Biol., vol. 59, no 3, p. 307-321, mars 2010. [CrossRef]
- S. Kumar, G. Stecher, M. Li, C. Knyaz, et K. Tamura, « MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms », Mol. Biol. Evol., vol. 35, no 6, p. 1547-1549, juin 2018. [CrossRef]
- M. Anisimova, M. Gil, J.-F. Dufayard, C. Dessimoz, et O. Gascuel, « Survey of Branch Support Methods Demonstrates Accuracy, Power, and Robustness of Fast Likelihood-based Approximation Schemes », Syst. Biol., vol. 60, no 5, p. 685-699, oct. 2011. [CrossRef]
- Rambaut, « FigTree v. 1.4.4. », ResearchGate. Consulté le: 11 décembre 2023. [En ligne]. Disponible sur: : http://tree. bio. ed. ac. uk/software/figtree/.
- A. F. Modiyinji et al., « Epidemiology of hepatitis E virus infection in animals in Africa: a systematic review and meta-analysis », BMC Vet. Res., vol. 17, no 1, p. 50, janv. 2021. [CrossRef]
- N. Pavio, T. Merbah, et A. Thébault, « Frequent Hepatitis E Virus Contamination in Food Containing Raw Pork Liver, France », Emerg. Infect. Dis., vol. 20, no 11, p. 1925-1927, nov. 2014. [CrossRef]
- M. Salines, M. Andraud, et N. Rose, « From the epidemiology of hepatitis E virus (HEV) within the swine reservoir to public health risk mitigation strategies: a comprehensive review », Vet. Res., vol. 48, no 1, p. 31, mai 2017. [CrossRef]
- E. Carella et al., « Molecular and serological investigation of Hepatitis E virus in pigs slaughtered in Northwestern Italy », BMC Vet. Res., vol. 19, no 1, p. 21, janv. 2023. [CrossRef]
- I. L. A. Boxman, C. C. C. Jansen, G. Hägele, A. Zwartkruis-Nahuis, A. S. L. Tijsma, et H. Vennema, « Monitoring of pork liver and meat products on the Dutch market for the presence of HEV RNA », Int. J. Food Microbiol., vol. 296, p. 58-64, mai 2019. [CrossRef]
- Di Bartolo et al., « Hepatitis E Virus in Pork Production Chain in Czech Republic, Italy, and Spain, 2010 - PMC ». Consulté le: 29 septembre 2023. [En ligne]. Disponible sur: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3414029/.
- M. Bouwknegt et al., « The course of hepatitis E virus infection in pigs after contact-infection and intravenous inoculation », BMC Vet. Res., vol. 5, no 1, p. 7, févr. 2009. [CrossRef]
- A. F. Modiyinji, G. M. A. M. Sanding, M. A. Atsama, C. G. Monamele, M. Nola, et R. Njouom, « Serological and molecular investigation of hepatitis E virus in pigs reservoirs from Cameroon reveals elevated seroprevalence and presence of genotype 3 », PLOS ONE, vol. 15, no 2, p. e0229073, févr. 2020. [CrossRef]
- P. El-Duah et al., « Detection and genomic characterization of hepatitis E virus genotype 3 from pigs in Ghana, Africa », One Health Outlook, vol. 2, no 1, p. 10, juill. 2020. [CrossRef]
- K. A. Traoré et al., « Hepatitis E Virus Exposure is Increased in Pork Butchers from Burkina Faso », Am. J. Trop. Med. Hyg., vol. 93, no 6, p. 1356-1359, déc. 2015. [CrossRef]
- C. Spahr, T. Knauf-Witzens, T. Vahlenkamp, R. G. Ulrich, et R. Johne, « Hepatitis E virus and related viruses in wild, domestic and zoo animals: A review », Zoonoses Public Health, vol. 65, no 1, p. 11-29, 2018. [CrossRef]
- P. P. Primadharsini, S. Nagashima, et H. Okamoto, « Genetic Variability and Evolution of Hepatitis E Virus », Viruses, vol. 11, no 5, p. 456, mai 2019. [CrossRef]
- B. D. Sadio et al., « First hepatitis E outbreak in Southeastern Senegal », Sci. Rep., vol. 12, no 1, Art. no 1, oct. 2022. [CrossRef]
- Lalèyê et al., « La filière porcine au Sénégal : commercialisation et consommation des viandes de porc et de phacochère dans les départements de Dakar, Fatick, Ziguinchor et Kolda », 2007.
- W. Ossebi et al., « Analyse zootechnique et économique des systèmes d’élevage de porcs en Casamance (Sénégal) », Rev. D’élevage Médecine Vét. Pays Trop., vol. 72, p. 13, mai 2019. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).